Abstract
To identify differences in gene expression profiles of infected cells between thyroid carcinoma (C), thyroid adenoma (A) and normal thyroid (N) epithelial cells, differentially expressed genes were identified using three pairwise comparisons with the GEO2R online tool. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis were used to classify them at the functional level. The most significant cluster in the N vs. A pairwise comparison had four hub genes: Insulin-like growth factor 2, Von Willebrand factor (VWF), multimerin 1 (MMRN1) and complement factor D (CFD). In N vs. C, the most significant cluster had 19 genes: IGF2, early growth response 2, transcription factor 3, KIT proto-oncogene receptor tyrosine kinase, SMAD family member 9, MLLT3 super elongation complex subunit, runt related transcription factor 1, CFD, actinin α 1, SWI/SNF related matrix associated actin dependent regulator of chromatin subfamily a member 4, JunD proto-oncogene AP-1 transcription factor subunit, serum response factor (SRF), FosB proto-oncogene, AP-1 transcription factor subunit, connective tissue growth factor (CTGF), SRC proto-oncogene, non-receptor tyrosine kinase, MMRN1, SRY-box 9, early growth response 3 and ETS variant 4. In A vs. C, the most significant cluster had 14 genes: BCL2-like 1, galectin 3, MCL1 BCL2 family apoptosis regulator, DNA damage inducible transcript 3, BCL2 apoptosis regulator, CTGF, matrix metallopeptidase 7, early growth response 1, kinase insert domain receptor, TIMP metallopeptidase inhibitor 1, apolipoprotein E, VWF, cyclin D1 and placental growth factor. Histological evidence was presented to confirm the makeup of the hubs prior to logistic regression analysis to differentiate benign and malignant neoplasms. The results of the present study may aid in the search for novel potential biomarkers for the differential diagnosis, prognosis and development of drug targets of thyroid neoplasm.
Keywords: thyroid neoplasm, differentially expressed genes, gene ontology, logistic regression
Introduction
Various countries worldwide, including the United States have observed an increase in the number of thyroid cancer cases. The incidence rate and mortality risk for advanced-stage papillary thyroid cancer have increased >3% annually in the past 3 decades in the United States (1,2). As the most common type of endocrine malignancy in the human body, thyroid cancer primarily consists of papillary, follicular, medullary and anaplastic carcinoma. Thyroid cancer advances with genetic and epigenetic alterations, and the consequential disarrangement of corresponding signaling pathways. Tumorigenesis is accelerated and amplified by a complex interaction of numerous secondary molecular alterations, in tumor cells and their microenvironment (3).
Typically, physical examinations or imaging studies accidentally identify thyroid cancer as nodules in the neck region. The majority of these nodules are benign, making it important to differentiate between the benign and malignant tumors. Patients with cancer require appropriately definitive treatment, which means diagnostic surgery is redundant. Fortunately, fine-needle aspiration (FNA), followed by cytological examination, allows for a relatively exact diagnosis of the type (benign, malignant, or uncertain) of nodule with suspicious ultrasound or other clinical characteristics (4–7).
However, ~25% of FNA cytology samples yield more than two types of indeterminate cytological diagnoses. In a meta-analysis, ~20% of the 25,445 samples were diagnosed as atypia of undetermined significance/follicular lesion of undetermined significance (AUS/FLUS), follicular or oncocytic neoplasm/suspicious for a follicular or oncocytic neoplasm (FN/SFN) or suspicious for malignant cells, with a mean malignant risk of 15.9, 26.1, or 75.2%, respectively (8). Such malignant risks are not low enough to postpone or cancel surgical management, or indicate definitive cancer surgery completely. Cytology to separate benign from malignant tumors has limited efficacy because of inevitable similarities among the different subtypes of thyroid lesions, and intraobserver reproducibility is always unrepeatable (9). Consequently, the majority of these patients suffer through diagnostic surgery, which is unnecessary if a diagnosis is clear-cut.
In fact, molecular cytology diagnosis is enriched in multiplatform tests for DNA, mRNA, and microRNA, which are able to accurately classify thyroid nodules and further improve the preoperative risk-based management of benign nodules with AUS/FLUS or FN/SFN cytology (10). Over a decade ago, researchers performed a review of the top 12 recommended markers, including well-known markers, such as MET, TFF3, SERPINA1, TIMP1, FN1 and TPO, as well as relatively novel or vague ones, such as TGFA, QPCT, CRABP1 and PROS1 (11).
The present study aimed to analyze the differences in gene profiles between thyroid carcinoma (TC or C), thyroid adenoma (TA or A) and normal thyroid tissue (N) to identify differentially expressed genes (DEGs) and hub genes. DEGs are used for gene ontology analysis (GO) and hub genes may be able to facilitate clinical studies following consideration of their clinical relevance.
Materials and methods
Microarray data
As an international public database that archives and shares microarray, next-generation sequencing and other forms of high-throughput functional genomics data, the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) provided the original dataset (GSE27155) for further analysis. GEO offers the gene expression profiles of GSE27155 submitted by Giordano et al (12,13), which was based on the Affymetrix GPL96 platform (Affymetrix Human Genome U133A Array). The GSE27155 dataset contained 99 samples, including 17 TA samples, 78 TC samples and four normal thyroid epithelia.
Identification of DEGs
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE27155) is an interactive online software application that allows users to compare two or more groups of samples in a certain platform in order to identify genes that are differentially expressed across different subtypes or different diseases. The results are output as a table of probes ordered by significance. In order to ensure quality control, raw data were processed using the GEOquery package R data structures, which may be used by numerous algorithms in other R packages. Distributions of value data may be viewed graphically or exported as a statistical summary table, which is useful for determining if the data are median-centered across samples and thus suitable for cross-comparison. Student's t-test was then used to identify DEGs. P<0.01 and adjusted P<0.05 (Benjamini & Hochberg's method or false discovery rate controlling procedures) were considered to indicate a statistically significant difference, and the fold change (FC) was set at 1.2.
GO and pathway enrichment analysis of DEGs
As a canonical method to annotate genes and gene products, GO analysis, including biological process (BP), cellular component (CC) and molecular function (MF), aids in identifying biological traits for high-throughput genome or transcriptome data (14,15). Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/) is a common omnibus to systematically interpret gene functions, facilitating an intensive understanding of genomic information and higher-order functional information (16). Feeding a certain gene list to the DAVID database (https://david.ncifcrf.gov/) is an essential procedure for the further functional analysis and relevant biological annotation of high-throughput genome or transcriptome data (17). To annotate DEGs at the functional level, GO enrichment and KEGG pathway analysis were performed using the DAVID online tool. P<0.05 was considered to indicate a statistically significant difference.
Protein-protein interaction (PPI) and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org/) database is an online tool for evaluating PPI data. The latest version of STRING (version 10.0) covers 184 million interactions of 9.6 million proteins from 2031 organisms. DEGs were processed in STRING to identify the most significantly interactive associations, in which a criterion of a combined score >0.4 was set as the significant level. The data from the PPI networks from STRING was analyzed with Cytoscape (version 3.6.1) software (18), in which a tool named Molecular Complex Detection (MCODE) directly illustrated the most significant clusters in the PPI network. False degree cut-off, node score cut-off, haircut, false K-core and max depth from seed were set at 2, 0.2, true, 2 and 100, respectively.
Clinical relevance of identified genes
To determine the clinical relevance of gene expression differences, clinical specimens from the Human Protein atlas (www.proteinatlas.org) were analyzed. Finally, logistic regression was performed to determine the trait (benign or malignant) of a certain neoplasm with the most differentially expressed genes (P<0.0005, FC>1.2) among the hub genes.
Results
Identification of DEGs
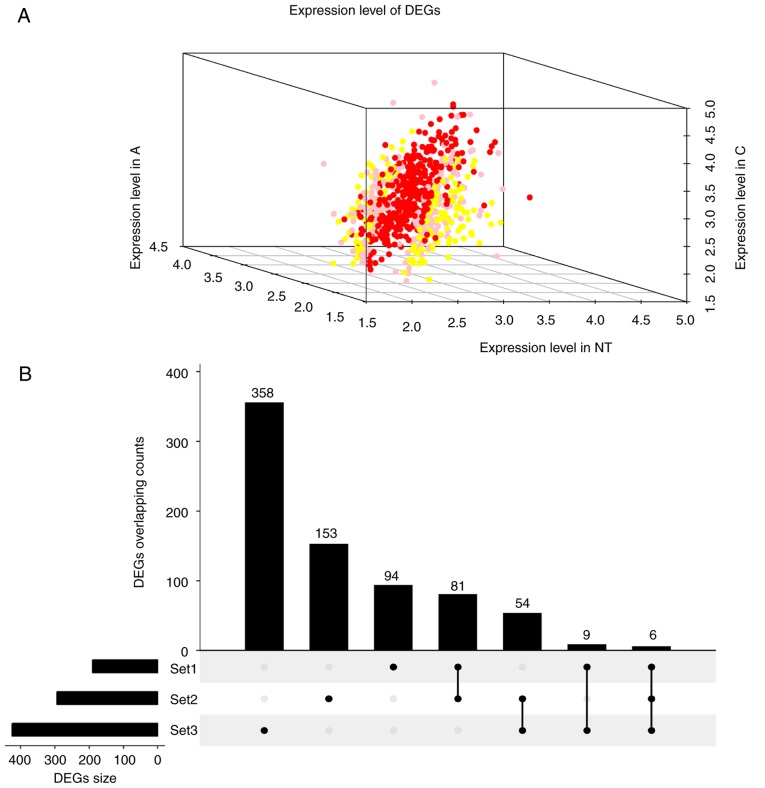
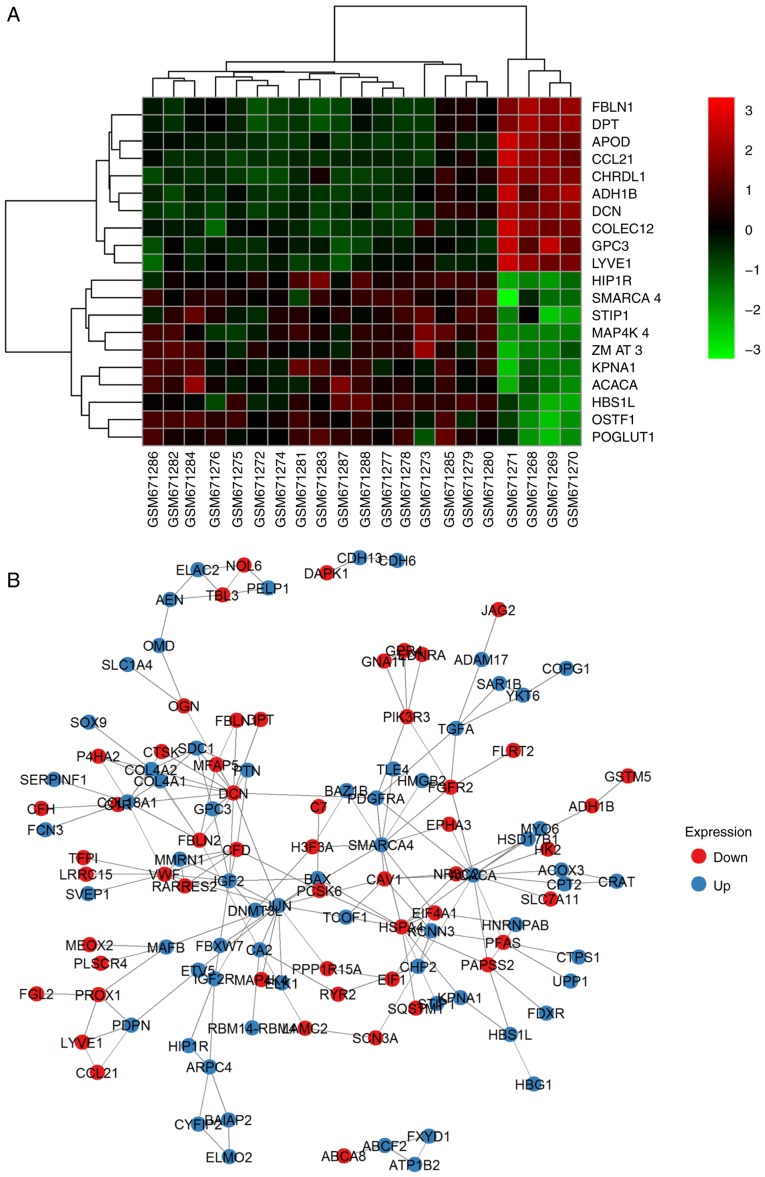
To ensure quality control on the 99 samples (17 TA samples, 78 TC samples and four normal thyroid epithelia), median-centered value data were acquired using the GEOquery R package in GEO2R, and the results were suitable for cross-comparison. Based on the GEO2R analysis of GSE27155 with the criterion of a P<0.01 cut-off, an adjusted P<0.05 and a FC>1.2, 190, 294 and 425 DEGs (Fig. 1A; DEG overlapping counts are presented in Fig. 1B) were identified in adenoma vs. normal tissue (A vs. N), carcinoma vs. normal tissue (C vs. N) and adenoma vs. carcinoma (A vs. C), respectively. The top 10 upregulated and downregulated genes are shown in Fig. 2 (representative image of A vs. N comparison). Detailed gene expression data and statistics of these DEGs are also available (data not shown). Genes without an Entrez annotation were filtered out. Four available clinical characteristics included ret/ptc translocation (N: NA: Y=57:31:11), braf t1799a mutation (N: NA: Y=40:31:28), pax8/pparg translocation (N: NA: Y=56:36:7) and kras, nras, or hras mutation (N: NA: Y=52:34:13).
Figure 1.
Expression and overlapping count of DEGs. (A) Expression of DEGs. Red foci, DEGs between C and N. Pink foci, DEGs between A and C. Yellow foci, DEGs between N and A. (B) DEGs overlapping count. Set1, DEGs counts between N and A. Set2, DEGs counts between N and T. Set3, DEGs counts between T and A. C, thyroid carcinoma; A, thyroid adenoma; N, normal thyroid epithelial cells; DEGs, differentially expressed genes.
Figure 2.
Representative DEGs and interactions of all DEGs. (A) Top 10 upregulated and downregulated genes in N vs. A. (B) Protein-protein interaction network between N and A. Red, downregulated genes. Blue, upregulated genes. A, thyroid adenoma; N, normal thyroid epithelial cells.
GO term enrichment analysis
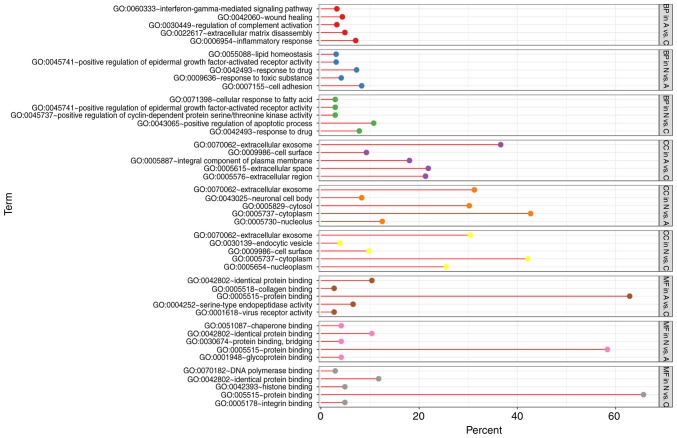
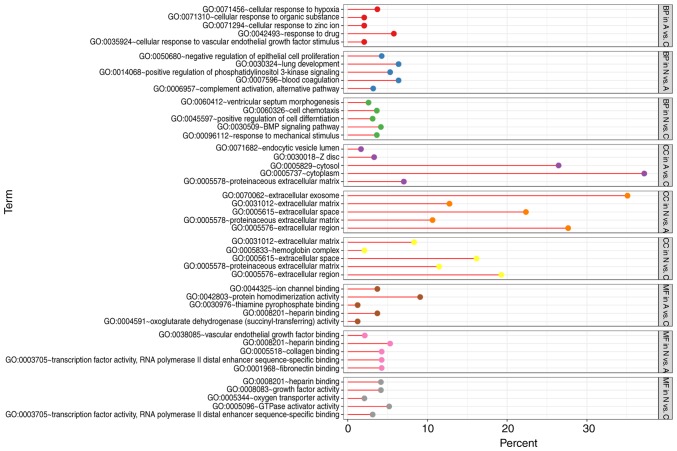
With DEGs uploaded to the online tool DAVID, the upregulated DEGs were significantly enriched in BP, including positive regulation of epidermal growth factor-activated receptor activity, response to drug, response to toxic substance, cell adhesion, and lipid homeostasis in N vs. A; positive regulation of apoptotic process, positive regulation of epidermal growth factor-activated receptor activity, response to drug, cellular response to fatty acid, and positive regulation of cyclin-dependent protein serine/threonine kinase activity in N vs. C; and positive regulation of extracellular matrix (ECM) disassembly, regulation of complement activation, wound healing, inflammatory response, and the interferon-γ-mediated signaling pathway in A vs. C. The downregulated DEGs identified using GO analysis were significantly enriched in lung development, including positive regulation of phosphatidylinositol 3-kinase signaling, complement activation, alternative pathway, blood coagulation, and negative regulation of epithelial cell proliferation in N vs. A; the BMP signaling pathway, response to mechanical stimulus, positive regulation of cell differentiation, cell chemotaxis, and ventricular septum morphogenesis in N vs. C; and cellular response to hypoxia, cellular response to organic substance, cellular response to zinc ion, cellular response to vascular endothelial growth factor stimulus, and response to drug in A vs. C (Figs. 3 and 4).
Figure 3.
GO analysis of upregulated genes. GO, gene ontology.
Figure 4.
GO analysis of downregulated genes. GO, gene ontology.
As for MF, the upregulated DEGs were involved in glycoprotein binding, protein binding/bridging, chaperone binding, identical protein binding, and protein binding in N vs. A; protein binding, DNA polymerase binding, integrin binding, and histone binding in N vs. C; and serine-type endopeptidase activity, collagen binding, and virus receptor activity in A vs. C. The downregulated DEGs were enriched in fibronectin binding, collagen binding, transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding, heparin binding, and vascular endothelial growth factor binding in N vs. A; oxygen transporter activity, transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding, heparin binding, growth factor activity, and GTPase activator activity in N vs. C; and ion channel binding, protein homodimerization activity, oxoglutarate dehydrogenase (succinyl-transferring) activity, and thiamine pyrophosphate binding in A vs. C (Figs. 3 and 4). In addition, GO CC analysis also showed that the upregulated DEGs were significantly enriched in extracellular exosome, neuronal cell body, cytoplasm, cytosol, and nucleolus in N vs. A; extracellular exosome, cytoplasm, cell surface, endocytic vesicle, and nucleoplasm in N vs. C; and extracellular exosome, extracellular space, extracellular region, integral component of plasma membrane, and cell surface in A vs. C. Downregulated DEGs were enriched in extracellular region, ECM, extracellular exosome, extracellular space, and proteinaceous ECM in N vs. A; proteinaceous ECM, extracellular region, extracellular space, and hemoglobin complex in N vs. C; and proteinaceous zecm, cytosol, Z disc, endocytic vesicle lumen, and cytoplasm in A vs. C (Figs. 3 and 4).
KEGG pathway annotation
Table I lists the most significantly enriched pathways of the upregulated and downregulated DEGs analyzed with KEGG analysis. The DEGs were annotated as an enrichment in ECM-receptor interaction, focal adhesion, protein digestion and absorption, complement and coagulation cascades, and the cAMP signaling pathway in N vs. A; and transcriptional misregulation in cancer and tyrosine metabolism in N vs. C. In addition, the DEGs were enriched in complement and coagulation cascades, proteoglycans in cancer, microRNAs in cancer, thyroid hormone synthesis, thyroid hormone signaling pathway, and the Rap1 signaling pathway in A vs. C.
Table I.
KEGG pathways enriched with DEGs of N vs. A, N vs. C, and A vs. C.
| Term | Group | Up/down regulated | Count | % | P-value | Entrez gene ID | Fold enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa04512:ECM-receptor interaction | N vs. A | Up | 5 | 5.21 | 0.004292243 | 3918, 1282, 1284, 7450, 6382 | 7.354193274 | 0.473192807 | 0.473192807 | 5.001337745 |
| hsa04510:Focal adhesion | N vs. A | Up | 6 | 6.25 | 0.020191824 | 3918, 1282, 8503, 1284, 2002, 7450 | 3.727076591 | 0.952135026 | 0.636916977 | 21.5971279 |
| hsa05222:Small cell lung cancer | N vs. A | Up | 4 | 4.17 | 0.027067112 | 3918, 1282, 8503, 1284 | 6.021786492 | 0.983237079 | 0.558561771 | 27.91339933 |
| hsa04610:Complement and coagulation cascades | N vs. A | Down | 5 | 5.32 | 7.66E-04 | 7035, 1675, 715, 3075, 730 | 11.64475902 | 0.0737502 | 0.0737502 | 0.846730468 |
| hsa04024:cAMP signaling pathway | N vs. A | Down | 5 | 5.32 | 0.031337025 | 1909, 3725, 5348, 6262, 6662 | 4.058022081 | 0.958574286 | 0.796466922 | 29.76946113 |
| hsa00982:Drug metabolism-cytochrome P450 | N vs. C | Down | 3 | 3.19 | 0.063860849 | 125, 2327, 2949 | 7.089603283 | 0.998638427 | 0.807907585 | 51.92743956 |
| hsa05203:Viral carcinogenesis | N vs. C | Up | 6 | 5.88 | 0.043011299 | 6714, 121504, 581, 87, 5902, 896 | 3.064301552 | 0.997876983 | 0.997876983 | 40.47226822 |
| hsa00350:Tyrosine metabolism | N vs. C | Down | 3 | 1.56 | 0.053641868 | 125, 316, 3081 | 7.897142857 | 0.999414543 | 0.999414543 | 47.60595747 |
| hsa05030:Cocaine addiction | N vs. C | Down | 3 | 1.56 | 0.096458334 | 3725, 2905, 2354 | 5.640816327 | 0.99999887 | 0.998937075 | 69.55265629 |
| hsa04060:Cytokine-cytokine receptor interaction | N vs. C | Down | 6 | 3.13 | 0.099426908 | 6387, 3590, 6366, 3815, 2690, 4982 | 2.403478261 | 0.999999275 | 0.991018404 | 70.7049881 |
| hsa04610:Complement and coagulation cascades | A vs. C | Up | 8 | 4.37 | 4.58E-05 | 718, 3426, 5627, 3075, 629, 5265, 1604, 5328 | 8.175096125 | 0.007482548 | 0.007482548 | 0.055516901 |
| hsa05205:Proteoglycans in cancer | A vs. C | Up | 11 | 6.01 | 4.63E-04 | 595, 2065, 8323, 4060, 7074, 2335, 1634, 960, 4233, 7474, 5328 | 3.878061224 | 0.073089834 | 0.024982191 | 0.559605893 |
| hsa04115:p53 signaling pathway | A vs. C | Up | 5 | 2.73 | 0.014250559 | 595, 2810, 64065, 637, 894 | 5.261955528 | 0.905002436 | 0.324510279 | 15.97387522 |
| hsa04918:Thyroid hormone synthesis | A vs. C | Down | 6 | 2.48 | 0.008755536 | 7849, 5578, 3708, 5172, 7173, 2778 | 4.663667042 | 0.823146922 | 0.579460968 | 10.41228645 |
| hsa04015:Rap1 signaling pathway | A vs. C | Down | 10 | 4.13 | 0.014252214 | 5909, 5578, 3397, 2321, 3791, 3815, 2778, 5228, 83660, 9223 | 2.590926134 | 0.940862276 | 0.61039768 | 16.42925112 |
| hsa04925:Aldosterone synthesis and secretion | A vs. C | Down | 6 | 2.48 | 0.015829909 | 3777, 3164, 5578, 3708, 4929, 2778 | 4.030329542 | 0.956865767 | 0.544272203 | 18.08627865 |
C, thyroid carcinoma; A, thyroid adenoma; N, normal thyroid epithelial cells; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
PPI network and the most enriched clusters
Based on the PPI network derived from the STRING database and node parameters (gene symbols, P-values and FCs) from GEO, the MCODE app in Cytoscape provided us with significant clusters in the three comparison groups. In N vs. A, four genes made up the most significant cluster (hub): Insulin-like growth factor 2 (IGF2), Von Willebrand factor (VWF), multimerin 1 (MMRN1) and complement factor D (CFD). In N vs. C, the most significant cluster had 19 genes: IGF2, early growth response 2 (EGR2), transcription factor 3 (TCF3), KIT proto-oncogene receptor tyrosine kinase (KIT), SMAD family member 9 (SMAD9), MLLT3 super elongation complex subunit (MLLT3), runt related transcription factor 1 (RUNX1), CFD, actinin α 1 (ACTN1), SWI/SNF related matrix associated actin dependent regulator of chromatin subfamily a member 4 (SMARCA4), JunD proto-oncogene, AP-1 transcription factor subunit (JUND), serum response factor (SRF), FosB proto-oncogene, AP-1 transcription factor subunit (FOSB), connective tissue growth factor (CTGF), SRC proto-oncogene, non-receptor tyrosine kinase (SRC), MMRN1, SRY-box 9 (SOX9), early growth response 3 (EGR3) and ETS variant 4 (ETV4). In A vs. C, the most significant cluster had 14 genes: BCL2-like 1 (BCL2L1), galectin 3 (LGALS3), MCL1 BCL2 family apoptosis regulator (MCL1), DNA damage inducible transcript 3 (DDIT3), BCL2 apoptosis regulator (BCL2), CTGF, matrix metallopeptidase 7 (MMP7), early growth response 1 (EGR1), kinase insert domain receptor (KDR), TIMP metallopeptidase inhibitor 1 (TIMP1), apolipoprotein E (APOE), VWF, cyclin D1 (CCND1) and placental growth factor (PGF). A representative image of A vs. N comparison is demonstrated in Fig. 2B and all of the genes are listed in Table II.
Table II.
Hub genes in N vs. A, N vs. C and A vs. C comparisons.
| Cluster | Score | Nodes | Edges | Hub genes |
|---|---|---|---|---|
| N vs. A Cluster 1 | 4 | 4 | 6 | IGF2, VWF, MMRN1, CFD |
| N vs. A Cluster 2 | 3 | 3 | 3 | PROX1, PDPN, LYVE1 |
| N vs. A Cluster 3 | 3 | 3 | 3 | EDNRA, GNA11, GPR4 |
| N vs. A Cluster 4 | 3 | 3 | 3 | BAIAP2, ELMO2, CYFIP2 |
| N vs. C Cluster 1 | 5.222 | 19 | 47 | IGF2, EGR2, TCF3, KIT, SMAD9, MLLT3, RUNX1, CFD, ACTN1, SMARCA4, JUND, SRF, FOSB, CTGF, SRC, MMRN1, SOX9, EGR3, ETV4 |
| N vs. C Cluster 2 | 5 | 5 | 10 | HBA2, HBG2, HBA1, HBB, HBD |
| N vs. C Cluster 3 | 4.5 | 5 | 9 | RERGL, OGN, DIRAS2, OMD, FMOD |
| N vs. C Cluster 4 | 3 | 5 | 6 | GRIN2C, FGFR2, DUSP4, GRIN1, CARD10 |
| A vs. C Cluster 1 | 8 | 14 | 52 | BCL2L1, LGALS3, MCL1, DDIT3, BCL2, CTGF, MMP7, EGR1, KDR, TIMP1, APOE, VWF, CCND1, PGF |
| A vs. C Cluster 2 | 7 | 11 | 35 | FOXO1, MET, CCND2, KIT, STAT1, FOS, JUN, RUNX1, MDK, FLT1, ERBB3 |
| A vs. C Cluster 3 | 5 | 5 | 10 | BDKRB2, C3, ANXA1, NMU, CCR5 |
| A vs. C Cluster 4 | 4.5 | 5 | 9 | LUM, PDE5A, PDE10A, FMOD, LRRC2 |
C, thyroid carcinoma; A, thyroid adenoma; N, normal thyroid epithelial cells.
Clinical relevance of identified genes
BCL2L1, LGALS3, MCL1, DDIT3, BCL2, MMP7, KDR, TIMP1, APOE, CCND1 and PGF demonstrated different expression levels in A vs. C. TCF3, SMAD9, ACTN1, JUND, SRF, SRC, EGR3 and ETV4 were differentially expressed in N vs. C. IGF2, MMRN1, CFD, SMARCA4 and SOX9 were significantly different in N vs. A and N vs. C. EGR2, KIT, MLLT3, RUNX1, FOSB, CTGF and EGR1 were significantly different in N vs. C and A vs. C. VWF was significantly different in N vs. A and A vs. C. Of the 19 hub genes in N vs. C, 60% (11/19), including KIT (P=6.73×10−04), SMAD9 (P=1.56×10−02), RUNX1 (P=1.86×10−02), ACTN1 (P=4.15×10−02), JUND (P=3.56×10−02), SRF (P=1.95×10−02), FOSB (P=2.94×10−02), CTGF (P=1.14×10−02), MMRN1 (P=8.32×10−09), SOX9 (P=3.32×10−02) and ETV4 (P=6.49×10−03), that performed well (R2=0.520) in the histology analysis was explored, and the results of KIT and SMAD9 expression in NT and TC are shown in Fig. 5. For hubs in the N vs. A and A vs. C comparisons, the analysis was limited as the samples were not satisfactory due to no benign neoplasms being available. Thus, more improved evidence is required to confirm the makeup of the hub genes in the future.
Figure 5.
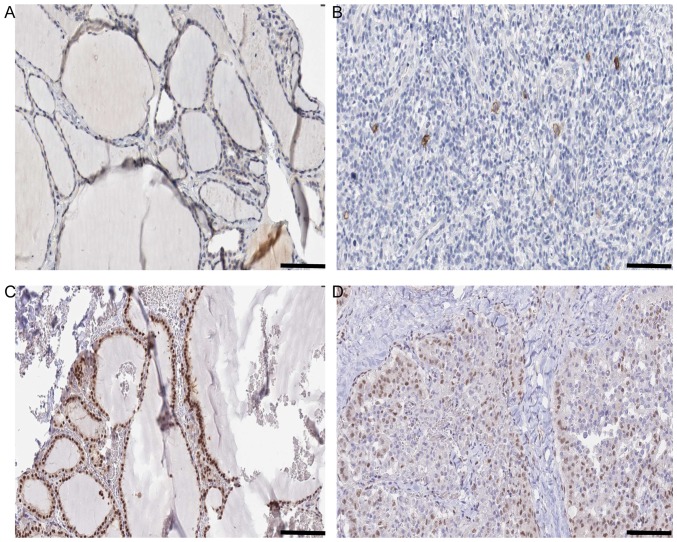
Histology information of KIT and SMAD9 in N vs. C comparison. (A) Expression of KIT in N Antibody (cat. no., HPA004471; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Sex, female; age, 22 years; Patient ID, 2146. Glandular cells Staining: Low Intensity. Weak Quantity: 75–25%. Location: Cytoplasmic/membranous. Website: http://www.proteinatlas.org/ENSG00000157404-KIT/tissue/thyroid+gland#img. (B) Expression of KIT in C Antibody (cat. no., HPA004471; Sigma-Aldrich; Merck KGaA). Sex, female; age, 77 years; Patient ID, 2479. Staining: Not detected. Intensity: Negative. Quantity: Negative. Location: None. Website: http://www.proteinatlas.org/ENSG00000157404-KIT/cancer/tissue/thyroid+cancer#img. (C) Expression of SMAD9 in N Antibody. (cat no., HPA031162; Sigma-Aldrich; Merck KGaA). Sex, female; age, 39 years; Patient ID, 1948. Staining: High. Intensity: Strong. Quantity: >75%. Location: Nuclear. Website: http://www.proteinatlas.org/ENSG00000120693-SMAD9/tissue/thyroid+gland#img. (D) Expression of SMAD9 in C Antibody. (cat. no., HPA031162; Sigma-Aldrich; Merck KGaA). Sex, male; age, 75 years; Patient ID, 3107. Staining: Medium. Intensity: Moderate. Quantity: >75%. Location: Nuclear. Website: http://www.proteinatlas.org/ENSG00000120693-SMAD9/cancer/tissue/thyroid+cancer#img. Scale bar, 100 µm.
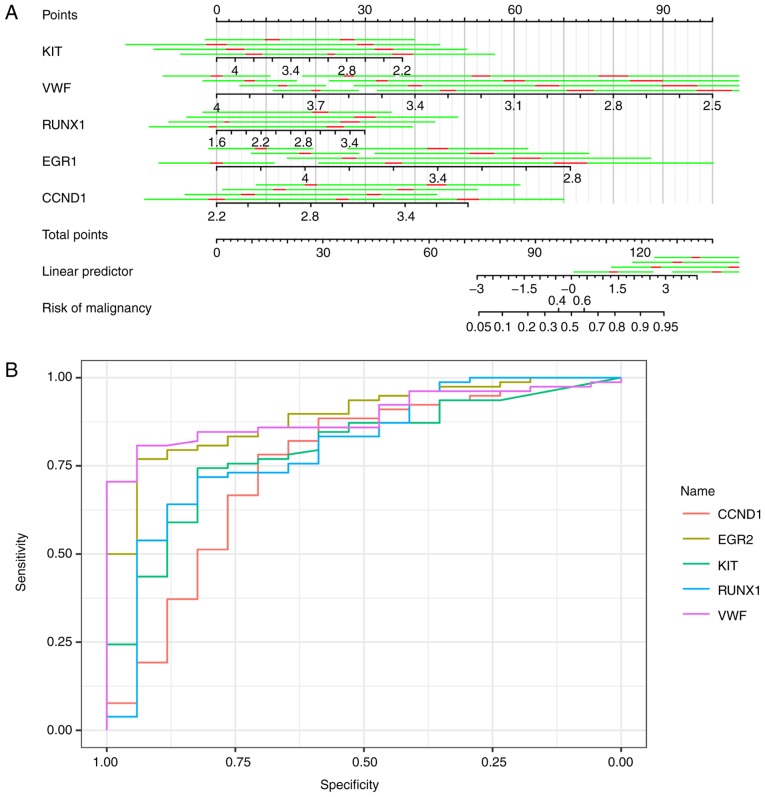
Further analysis of the hub genes in the most significant clusters in the NT vs. TA comparison indicated that VWF was a good biomarker of TA. No other gene was an evident biomarker to differentiate NT from TA as it was hard to isolate TA from TC. A similar method was used to identify EGR2, KIT, MLLT3, RUNX1, FOSB, CTGF, BCL2L1, LGALS3, MCL1, DDIT3, BCL2, CTGF, MMP7, EGR1, KDR, TIMP1, APOE, VWF, CCND1, and PGF.KIT, VWF, BCL2L1, RUNX1, EGR1, FOSB, LGALS3, KDR, EGR1, CCND1, PGF and DDIT3 were utilized in logistic regression (stepwise) to predict the trait of a certain thyroid neoplasm, as shown in Fig. 6A. Logistic regression produced estimates of KIT, VWF, RUNX1, EGR1 and CCND1 as −0.167 [standard deviation (SD), 0.07556; 95% confidence interval (CI), −0.3150276 to −0.0188324], −0.360 (SD, 0.10058; 95% CI, −0.5566868 to −0.1624132), 0.144 (SD, 0.06506; 95% CI, 0.0162424 to 0.2712776), −0.379 (SD, 0.11652; 95% CI, −0.6068892 to −0.1501308) and 0.273 (SD, 0.09865; 95% CI, 0.079686 to 0.466394), respectively. The area under the curves of KIT, VWF, RUNX1, EGR1 and CCND1 were 0.8910, 0.8906, 0.796, 0.8024 and 0.7534 respectively, with the receiving operator curve presented in Fig. 6B.
Figure 6.
Assessment of selected hub genes. (A) Nomogram to predict the malignant possibility of a thyroid neoplasm. Red, 10% CI; Green, 50% CI; R2=0.520. (B) Receiver operating characteristic curve of CCND1, EGR2, KIT, RUNX1 and VWF.
Discussion
In the present study, gene expression data of 78 TC samples, 17 TA samples and four normal thyroid epithelial tissues were extracted from the GEO database via the accession ID GSE27155. Among the 190 (with 96 upregulated in A), 294 (with 102 upregulated in C) and 425 (with 183 upregulated in C) DEGs identified between N vs. A, N vs. C and A vs. C, respectively, GO and KEGG pathway analyses were performed to gain a more improved understanding of the DEG interactions. The GO term analysis revealed that upregulated DEGs were primarily involved in ECM-receptor interaction, viral carcinogenesis, and complement and coagulation cascades, while downregulated DEGs were involved in complement and coagulation cascades, tyrosine metabolism, and thyroid hormone synthesis. To a certain extent, managing these signaling pathways may facilitate the manipulation and prediction of tumor genesis.
As a suitable marker to evaluate acute functional changes of endothelia, VWF is sensitive to changes in endothelial function (19). The association between hyperthyroidism and thromboembolism may be partially explained by the effects of thyroid hormone on receptors and transcription factors, which interferes with coagulation-involved proteins, including VWF, in numerous cell types (19). EGR2, KIT, MLLT3, RUNX1, FOSB and CTGF were recommended as useful biomarkers in the most enriched cluster in the N vs. C gene profile comparison. Over a decade ago, EGR2 was reported as one of the five genes to effectively predict malignancy with a specificity of 98.5% in the diagnosis of follicular thyroid carcinoma with logistic regression analysis (20). Together with the lack of mutations, the low or absent c-kit expression argued against an important role of c-kit in undifferentiated thyroid carcinoma cell proliferation (21), and RUNX1 was considered one of the 43 most suitable molecular markers in papillary thyroid carcinoma (22). Differential induction of Fos family genes represents the regulation of thyroid cell function by thyroid stimulating hormone (23).
A total of 14 hub genes, BCL2L1, LGALS3, MCL1, DDIT3, BCL2, CTGF, MMP7, EGR1, KDR, TIMP1, APOE, VWF, CCND1 and PGF were identified in the C vs. A comparison. The degree of upregulation of certain genes, including BCL2L1, has been associated with increased resistance to chemotherapeutic treatments, and small interfering RNA targeting BCL2L1 renders tumor cells sensitive to chemotherapeutic treatments (24,25). LGALS3 is considered as a discriminative molecular marker in FNA biopsies of benign and malignant thyroid tumors (26), and MCL1 is an important molecule in the inhibition of chemotherapy-induced apoptosis by thyroid hormone (T4) in αvβ3-expressing cancer cells (27). In the pathogenesis of FTC, a previous study indicated that DDIT3 was involved (28). Thyroid anaplastic carcinoma has been demonstrated to exhibit downregulated BCL2 expression compared with differentiated thyroid tumors, indicating that the loss of BCL2 is associated with the loss of differentiation in thyroid carcinoma. Thyroid anaplastic carcinoma appeared to be worse in prognosis compared with other subtypes of thyroid carcinoma, and the loss of BCL2 may be partially responsible (29). The In-Space FTC-133 human follicular thyroid cancer cell experiment revealed a scaffold-free formation of extraordinarily large three-dimensional aggregates of thyroid cancer cells with an altered CTGF gene under real microgravity (30). KDR is a key TKI target protein, and TIMP1 mRNA expression was demonstrated to be an independent diagnostic marker of malignant thyroid neoplasms (31,32). A study in 2011 suggested that the CCND1 gene serves an early role in thyroid tumorigenesis (33). PGF expression is associated with iodide medium and may interfere with human thyroid follicles (34). Little is known about the roles of MMP7, EGR1 and APOE in human thyroid disorders, and these genes may be false positives.
There are certain limitations of the present study. Firstly, the relatively small sample size indicates that these potential biomarkers require further assessment prior to consideration of the widespread practical application. In other words, the findings of a study using historical controls from a particular geographical region may not be applicable to newer cohorts of patients or different regions. Secondly, relying on few clinical samples as histological evidence is not robust. Lastly, an assessment of the association of potential biomarkers with other clinical characteristics, including age, sex and the stage of disease, is required. Four accessible clinical characteristics were used in the present study, including ret/ptc translocation, braf t1799a mutation, pax8/pparg translocation and kras, nras or hras mutation. However, it should be acknowledged that innate bias may exist for the lack of age, sex, and the stage of disease information.
To conclude, gene expression profiles of cancer cells in patients with TA or TC were compared with normal epithelial cells to identify DEGs, generating three pairwise comparison tables. Subsequently, GEO2R-screened DEGs were characterized using GO and pathway enrichment analysis. Further characterization of their biological functions and pathways may shed light on the general development of thyroid neoplasm at the molecular level, and identify biomarkers for differential diagnosis and drug targets, eventually leading to novel thyroid cancer therapies.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- A/TA
thyroid adenoma
- ACTN1
actinin α 1
- APOE
apolipoprotein E
- AUS/FLUS
atypia of undetermined significance/follicular lesion of undetermined significance
- BCL2
BCL2 apoptosis regulator
- BCL2L1
BCL2-like 1
- BP
biological process
- CC
cellular component
- CCND1
cyclin D1
- C/TC
thyroid carcinoma
- CFD
complement factor D
- CTGF
connective tissue growth factor
- DDIT3
DNA damage inducible transcript 3
- DEG
differentially expressed gene
- EGR1
early growth response 1
- EGR2
early growth response 2
- EGR3
early growth response 3
- ETV4
ETS variant 4
- FNA
fine-needle aspiration
- FN/SFN
follicular or oncocytic neoplasm/suspicious for a follicular or oncocytic neoplasm
- FOSB
FosB proto-oncogene, AP-1 transcription factor subunit
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- JUND
JunD proto-oncogene, AP-1 transcription factor subunit
- KDR
kinase insert domain receptor
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KIT
KIT proto-oncogene receptor tyrosine kinase
- IGF2
insulin-like growth factor 2
- LGALS3
galectin 3
- MCL1
MCL1 BCL2 family apoptosis regulator
- MCODE
Molecular Complex Detection
- MF
molecular function
- MLLT3
MLLT3 super elongation complex subunit
- MMP7
matrix metallopeptidase 7
- MMRN1
multimerin 1
- N/NT
normal tissue
- PGF
placental growth factor
- RUNX1
runt related transcription factor 1
- SMAD9
SMAD family member 9
- SMARCA4
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
- SOX9
SRY-box 9
- SRC
SRC proto-oncogene, non-receptor tyrosine kinase
- SRF
serum response factor
- STRING
Search Tool for the Retrieval of Interacting Genes
- TCF3
transcription factor 3
- TIMP1
TIMP metallopeptidase inhibitor 1
- VWF
von Willebrand factor
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81271088 and 81670926) and from the Science and Technology Commission of Shanghai Municipality, China (grant nos. 15411950303 and 14DZ2260300).
Availability of data and materials
The datasets used and/or analyzed during the current study including not shown data are available from the corresponding author on reasonable request.
Authors' contributions
MX and YM conceived the present study. MX, QW and YS designed the experiments. QW, YS and BY wrote the manuscript. MX provided platform supports. QW, HH, YZ and JL performed the experiments. QW, BY, HH, TW and CF analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in Thyroid Cancer Incidence and Mortality in the United States, 1974–2013. JAMA. 2017;317:1338–1348. doi: 10.1001/jama.2017.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renuka IV, Bala Saila G, Aparna C, Kumari R, Sumalatha K. The bethesda system for reporting thyroid cytopathology: Interpretation and guidelines in surgical treatment. Indian J Otolaryngol Head Neck Surg. 2012;64:305–311. doi: 10.1007/s12070-011-0289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baloch ZW, LiVolsi VA, Asa SL, Rosai J, Merino MJ, Randolph G, Vielh P, DeMay RM, Sidawy MK, Frable WJ. Diagnostic terminology and morphologic criteria for cytologic diagnosis of thyroid lesions: A synopsis of the National Cancer institute thyroid fine-needle aspiration state of the science conference. Diagn Cytopathol. 2008;36:425–437. doi: 10.1002/dc.20830. [DOI] [PubMed] [Google Scholar]
- 6.Gharib H. Changing trends in thyroid practice: Understanding nodular thyroid disease. Endocr Pract. 2004;10:31–39. doi: 10.4158/EP.10.1.31. [DOI] [PubMed] [Google Scholar]
- 7.Sclabas GM, Staerkel GA, Shapiro SE, Fornage BD, Sherman SI, Vassillopoulou-Sellin R, Lee JE, Evans DB. Fine-needle aspiration of the thyroid and correlation with histopathology in a contemporary series of 240 patients. Am J Surg. 2003;186:702–710. doi: 10.1016/j.amjsurg.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Nikiforov YE, Carty SE, Chiosea SI, Coyne C, Duvvuri U, Ferris RL, Gooding WE, Hodak SP, LeBeau SO, Ohori NP, et al. Highly accurate diagnosis of cancer in thyroid nodules with follicular neoplasm/suspicious for a follicular neoplasm cytology by ThyroSeq v2 next-generation sequencing assay. Cancer. 2014;120:3627–3634. doi: 10.1002/cncr.29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin HS, Komisar A, Opher E, Blaugrund SM. Follicular variant of papillary carcinoma: The diagnostic limitations of preoperative fine-needle aspiration and intraoperative frozen section evaluation. Laryngoscope. 2000;110:1431–1436. doi: 10.1097/00005537-200009000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Labourier E, Shifrin A, Busseniers AE, Lupo MA, Manganelli ML, Andruss B, Wylie D, Beaudenon-Huibregtse S. Molecular Testing for miRNA, mRNA, and DNA on Fine-Needle aspiration improves the preoperative diagnosis of thyroid nodules with indeterminate cytology. J Clin Endocrinol Metab. 2015;100:2743–2750. doi: 10.1210/jc.2015-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith OL, Melck A, Jones SJM, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol. 2006;24:5043–5051. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- 12.Giordano TJ, Au AY, Kuick R, Thomas DG, Rhodes DR, Wilhelm KG, Jr, Vinco M, Misek DE, Sanders D, Zhu Z, et al. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clin Cancer Res. 2006;12:1983–1993. doi: 10.1158/1078-0432.CCR-05-2039. [DOI] [PubMed] [Google Scholar]
- 13.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K, et al. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24:6646–6656. doi: 10.1038/sj.onc.1208822. [DOI] [PubMed] [Google Scholar]
- 14.Gene Ontology Consortium: The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burggraaf J, Lalezari S, Emeis JJ, Vischer UM, de Meyer PH, Pijl H, Cohen AF. Endothelial function in patients with hyperthyroidism before and after treatment with propranolol and thiamazol. Thyroid. 2001;11:153–160. doi: 10.1089/105072501300042820. [DOI] [PubMed] [Google Scholar]
- 20.Foukakis T, Gusnanto A, Au AY, Höög A, Lui WO, Larsson C, Wallin G, Zedenius J. A PCR-based expression signature of malignancy in follicular thyroid tumors. Endocr Relat Cancer. 2007;14:381–391. doi: 10.1677/ERC-06-0023. [DOI] [PubMed] [Google Scholar]
- 21.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 22.Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, Pezzino V, Belfiore A. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. J Clin Endocrinol Metab. 2002;87:245–254. doi: 10.1210/jcem.87.1.8142. [DOI] [PubMed] [Google Scholar]
- 23.Kambe F, Miyazaki T, Seo H. Differential induction of fos and jun family genes by thyrotropin in rat thyroid FRTL-5 cells. Thyroid. 1996;6:123–128. doi: 10.1089/thy.1996.6.123. [DOI] [PubMed] [Google Scholar]
- 24.Kim SJ, Kim JS, Park ES, Lee JS, Lin Q, Langley RR, Maya M, He J, Kim SW, Weihua Z, et al. Astrocytes upregulate survival genes in tumor cells and induce protection from chemotherapy. Neoplasia. 2011;13:286–298. doi: 10.1593/neo.11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee BS, Cha HY, Shin YS, Kim YS, Kim CH. AY4, an agonistic anti-death receptor 4 MAB, induces apoptotic cell death in anaplastic thyroid cancer cells via downregulation of Bcl-xL with reactive oxygen species generation. Endocr Relat Cancer. 2013;20:283–291. doi: 10.1530/ERC-12-0405. [DOI] [PubMed] [Google Scholar]
- 26.Karger S, Krause K, Gutknecht M, Schierle K, Graf D, Steinert F, Dralle H, Führer D. ADM3, TFF3 and LGALS3 are discriminative molecular markers in fine-needle aspiration biopsies of benign and malignant thyroid tumours. Br J Cancer. 2012;106:562–568. doi: 10.1038/bjc.2011.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin HY, Glinsky GV, Mousa SA, Davis PJ. Thyroid hormone and anti-apoptosis in tumor cells. Oncotarget. 2015;6:14735–14743. doi: 10.18632/oncotarget.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maciel RMB, Kimura ET, Cerutti JM. Pathogenesis of differentiated thyroid cancer (papillary and follicular) Arq Bras Endocrinol Metabol. 2005;49:691–700. doi: 10.1590/S0004-27302005000500009. (In Portuguese) [DOI] [PubMed] [Google Scholar]
- 29.Gupta A, Jain S, Khurana N, Kakar AK. Expression of p63 and Bcl-2 in malignant thyroid tumors and their correlation with other diagnostic immunocytochemical markers. J Clin Diagn Res. 2016;10:EC04–EC08. doi: 10.7860/JCDR/2016/13899.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietsch J, Ma X, Wehland M, Aleshcheva G, Schwarzwälder A, Segerer J, Birlem M, Horn A, Bauer J, Infanger M, et al. Spheroid formation of human thyroid cancer cells in an automated culturing system during the Shenzhou-8 Space mission. Biomaterials. 2013;34:7694–7705. doi: 10.1016/j.biomaterials.2013.06.054. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Antona C, Muñoz-Repeto I, Inglada-Pérez L, de Cubas AA, Mancikova V, Cañamero M, Maliszewska A, Gómez A, Letón R, Leandro-García LJ, et al. Influence of RET mutations on the expression of tyrosine kinases in medullary thyroid carcinoma. Endocr Relat Cancer. 2013;20:611–619. doi: 10.1530/ERC-12-0316. [DOI] [PubMed] [Google Scholar]
- 32.Kebebew E, Peng M, Reiff E, McMillan A. Diagnostic and extent of disease multigene assay for malignant thyroid neoplasms. Cancer. 2006;106:2592–2597. doi: 10.1002/cncr.21922. [DOI] [PubMed] [Google Scholar]
- 33.Bièche I, Franc B, Vidaud D, Vidaud M, Lidereau R. Analyses of MYC, ERBB2, and CCND1 genes in benign and malignant thyroid follicular cell tumors by real-time polymerase chain reaction. Thyroid. 2001;11:147–152. doi: 10.1089/105072501300042802. [DOI] [PubMed] [Google Scholar]
- 34.Yamada E, Yamazaki K, Takano K, Obara T, Sato K. Iodide inhibits vascular endothelial growth factor-A expression in cultured human thyroid follicles: A microarray search for effects of thyrotropin and iodide on angiogenesis factors. Thyroid. 2006;16:545–554. doi: 10.1089/thy.2006.16.545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study including not shown data are available from the corresponding author on reasonable request.