Abstract
Background:
Acinetobacter baumannii is a major cause of hospital care-acquired infections, and this bacterium poses a significant challenge to health care worldwide. At King Fahd Hospital of the University (KFHU), Al Khobar, Saudi Arabia, there had been a significant increase in the number of cases of A. baumannii infections.
Objective:
The objective of this study was to determine the clonal relationship between A. baumannii collected from different specimens of patients admitted to KFHU using the enterobacterial repetitive intergenic consensus–polymerase chain reaction (ERIC-PCR) fingerprinting method.
Materials and Methods:
A. baumannii strains were isolated from a total of 59 specimens from inpatients admitted to KFHU between January and September 2014. These specimens were mainly collected from wound, rectal and throat swabs and transtracheal aspiration. ERIC-PCR fingerprinting was used to determine the clonal relationship between the different isolated strains.
Results:
Using ERIC-PCR fingerprinting genotype analysis, 51 strains of A. baumannii were clustered into seven groups, while the remaining 8 were single strains. The genetic relatedness of A. baumannii isolated from admitted patients was high, indicating cross-transmission within the hospitalized patients.
Conclusion:
This study found that the increase in the incidence of A. baumannii in patients at KFHU was likely due to the spread of seven epidemic clones, thereby highlighting the need for intensifying the infection control measures to prevent nosocomial transmission of A. baumannii. These results also demonstrate that ERIC-PCR is a reliable and rapid method for studying the clonal similarity between A. baumannii isolated from different clinical specimens.
Keywords: Acinetobacter baumannii, clonal relationship, enterobacterial repetitive intergenic consensus-polymerase chain reaction, genetic typing, Saudi Arabia
INTRODUCTION
Acinetobacter baumannii is an important Gram-negative coccobacilli classified as an opportunistic bacterial pathogen that is responsible for nearly about 9% of all nosocomial infections.[1] In fact, the Infectious Diseases Society of America has classified A. baumannii as one of the six most important multidrug-resistant microorganisms in hospitals worldwide.[2,3] A. baumannii is currently believed to be responsible for causing a wide range of severe nosocomial infections, including pneumonia, skin and soft tissue infections, wound infections, urinary tract infections and ventilator- and nosocomial-associated pneumonia, especially among patients in the intensive care unit with high mortality rates.[4,5,6,7,8,9] This bacterium is unique because it exhibits resistance against multiple antibiotics. Further, A. baumannii can survive for long periods in hospital environments, enabling its transmission between patients, either through human reservoirs or through inanimate materials.[4,10,11] A. baumannii can also persist for long periods in the environmental surfaces of hospitals and can be isolated in hospitals from air, tap water faucets, bedside urinals, gloves, ventilators and angiography catheters.[9,12]
Molecular typing is becoming a paradigm for understanding the fundamental mechanisms of Acinetobacter infections in hospital settings to investigate the spreading, the clonality relationship among bacterial strains and its geographical spread. In infection control, molecular typing methods are currently an important tool to measure and identify the source of the original infection in hospitals outbreaks. Molecular typing methods have been used to investigate the nosocomial spread of A. baumannii outbreaks in hospital settings worldwide. Some examples of these techniques are plasmid typing, ribotyping, pulsed-field gel electrophoresis (PFGE) and polymerase chain reaction (PCR)-based fingerprinting.[13,14,15,16,17] As compared with other methods, PCR-based fingerprinting methods are easier to perform and cost effective.
A vast increase in the number of cases of A. baumannii outbreaks had been observed at King Fahd Hospital of the University (KFHU), Al Khobar, Saudi Arabia. These cases were derived from different patient specimens. Accordingly, this study was conducted to investigate the relationship between A. baumannii strains collected from different specimens using enterobacterial repetitive intergenic consensus (ERIC)-PCR DNA fingerprinting. The results of this study would help determine if the increase in the incidence of A. baumannii in patients was due to the spread of epidemic clones as well determine the effectiveness of ERIC-PCR fingerprinting technique for genotyping of A. baumannii.
MATERIALS AND METHODS
A total of 59 A. baumannii strains were collected from the clinical microbiology laboratory at KFHU between January and September 2014. The 59 strains of A. baumannii were isolated from inpatient specimens: wound swabs (n = 14), transtracheal aspiration (n = 11), rectal swabs (n = 6), throat swabs (n = 5), urine (n = 5), sputum (n = 4), pleural fluid (n = 4), blood (n = 4), intravenous catheter tips (n = 3), nasal swabs (n = 1), skin swabs (n = 1) and abscess specimens (n = 1) [Table 1]. Genomic DNA was prepared using a DNA template extracted using the standard boiling lysis method described elsewhere.[18] A pure, single colony was inoculated in 10 ml of Luria-Bertani broth with 0.5% NaCl (w/v) and the solution was placed in an incubator shaker at 37°C with a rotary speed of 250 rpm for 18 h. The overnight-grown bacterial culture was pelleted and resuspended in a 1.5-ml microcentrifuge tube of sterilized distilled water and boiled in a water bath at 100°C for 15 min. After boiling, the tubes were centrifuged, and 2 μl of the supernatant was used as the DNA template.
Table 1.
Distribution of Acinetobacter baumannii strain specimens (n = 59)
| Specimen | Number of specimen (%) |
|---|---|
| Urine | 5 (8.5) |
| Blood | 4 (6.8) |
| Sputum | 4 (6.8) |
| Abscess | 1 (1.7) |
| Skin swab | 1 (1.7) |
| Nasal swab | 1 (1.7) |
| Rectal swab | 6 (10.2) |
| Throat swab | 5 (8.5) |
| Wound swab | 14 (23.7) |
| Pleural fluid | 4 (6.8) |
| Transtracheal aspiration | 11 (18.6) |
| Intravenous catheter tips | 3 (5.0) |
The primers used for ERIC-PCR were ERIC-1 (5′ TGTAAGCTCCTGG GGATTCAC 3′) and ERIC-2 (5′ AAGTAAGTGACTGGGGTGAGCG 3′), as described by Coudron et al.[19] The amplification reaction volume was 25 μl, and the cycling conditions were as follows: an initial denaturation at 94°C for 5 min, followed by 35 cycles at 95°C for 1 min, 52°C for 1 min, 72°C for 5 min and a final extension at 72°C for 10 min. The amplified products were resolved through electrophoresis and analyzed on 1.5% agarose gel-stained ethidium bromide. The clonal relatedness between the strains of A. baumannii was analyzed using ERIC-PCR fingerprinting with the BioNumerics fingerprint data software package (Applied Maths, Sint-Martens-Latem, Belgium). Normalization steps were included in the analysis of DNA polymorphism patterns produced by ERIC-PCR fingerprinting to ensure an adequate gel-to-gel banding pattern comparison. The process of band scoring was used to identify bands in each lane that combined to make the fingerprint based on the stringency threshold and optimization settings. Dendrograms were generated for the ERIC-PCR gels using the Dice similarity coefficient and the unweighted pair group method using arithmetic averages, with 1% optimization and 1% position tolerance.[20] A. baumannii strains with a similarity exceeding 90% were considered to be clonally related.
The Institutional Review Board at Imam Abdulrahman Bin Faisal University provided ethical approval for this study (IRB-2017-01-203).
RESULTS
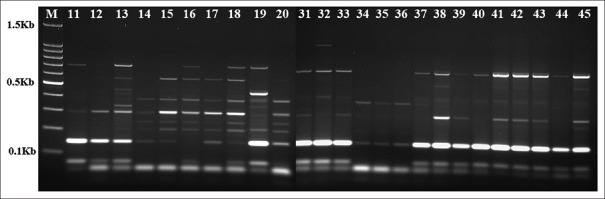
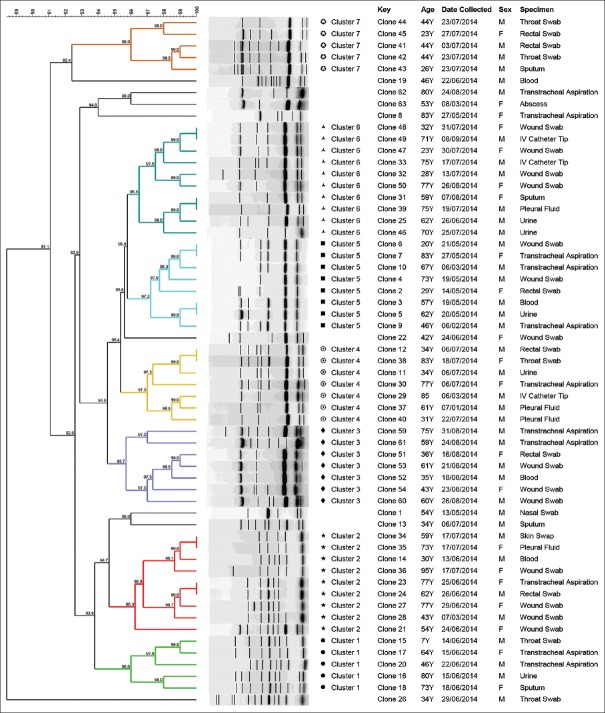
ERIC-PCR fingerprinting grouped A. baumannii strains isolated from different specimens and from the same period of isolation and location [Figure 1]. Among the 59 collected strains of A. baumannii, 51 strains were grouped in 7 clusters (Clusters 1–7) and 8 single isolates were observed at a similarity level of 97% [Figure 2]. Interestingly, within these seven clusters, Clusters 1, 3 and 7 comprised A. baumannii strains isolated from different specimens during the same months (June, July and August); Clusters 2, 4, 5 and 6 comprised A. baumannii strains isolated during the entire course of the study, except in April, in which no strains were islolated.
Figure 1.
Representative patterns of Acinetobacter baumannii obtained by enterobacterial repetitive intergenic consensus-polymerase chain reaction, with the numbers indicating the corresponding strains (Lane M is the DNA molecular weight marker)
Figure 2.
Dendrogram of enterobacterial repetitive intergenic consensus–polymerase chain reaction using BioNumerics fingerprint data software and unweighted pair group method with arithmetic averages at 97% similarity on 59 strains of Acinetobacter baumannii; the different clusters at 97% similarity are arbitrarily designated as Clusters 1–7, of which Clusters 2 and 6 are the largest group representing the most prevalent clone of A. baumannii and its variants among the isolates
DISCUSSION
A. baumannii is one of the emerging nosocomially acquired pathogens and is responsible for different types of infections, ranging from minor soft tissue infections to more severe infections such as ventilator-associated pneumonia and bacteremia.[16] Bacterial genomes containing repeat sequences such as the ERIC sequence can be used as molecular biological tools to assess the clonal variability of many bacterial isolates.[21,22] ERIC-PCR fingerprinting is one of the fastest molecular typing techniques to differentiate between A. baumannii and other strains of Gram-negative bacteria responsible for hospital-acquired infections.[9,19,23,24,25] The findings of this study are similar to that of studies reported by Ying et al. and Hammoudi et al., who were also able to cluster A. baumannii strains based on their genetic relatedness using ERIC-PCR.[9,25]
The results of this study indicate an influence of epidemiological relatedness on the clustering of A. baumannii strains, as seven clusters of A. baumannii strains with high relatedness were recovered from the same period of isolation and location [Figure 2]. This indicates that there was cross-transmission within hospitalized patients. Based on our results, it can be stated that ERIC-PCR is a reliable method to demonstrate the clonal relatedness between A. baumannii recovered from different specimens isolated from different inpatients.
Another interesting result found in this study was that Clusters 1, 3 and 7 were only, although highly, prevalent during June–August, whereas Clusters 2, 4, 5 and 6 were prevalent during the entire course of the study. This difference in prevalence patterns between clusters could be because the strains in Clusters 1, 3 and 7 are either only prevalent during those months or that there had been an outbreak of these strains. However, further analyses are required to validate these differences in prevalence patterns.
The results of this study revealed that ERIC-PCR is a reliable technique for discriminating intraspecific variations. In addition, according to the authors, ERIC-PCR is a very convenient and fast method, thereby making using it advantageous compared with other PCR- and non-PCR-based fingerprinting such as PFGE for A. baumannii. Further, the obtained ERIC patterns exhibited a high genetic similarity, ranging from 91% to 99%, and generated a larger number of DNA fingerprints among the strains of A. baumannii. Therefore, this PCR-based target repetitive element method is more suitable than rapid typing, in addition to this method being more cost effective than other DNA fingerprint methods.[9]
CONCLUSION
Using ERIC-PCR, this study demonstrated a high level of genetic similarity and relationship among the strains of A. baumannii isolated from different clinical specimens at KFHU, Al-Khobar, Saudi Arabia. This highlights the need for intensifying infection control measures to prevent nosocomial transmission of A. baumannii at KFHU. Our study also found that ERIC-PCR fingerprinting exhibits an excellent discriminatory capability of A. baumannii strains.
Financial support and sponsorship
This work was supported by Grant no. 2013203 from the Deanship of Scientific Research at Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia.
Conflict of interest
There are no conflicts of interest.
REFERENCES
- 1.Joly-Guillou ML. Clinical impact and pathogenicity of Acinetobacter. Clin Microbiol Infect. 2005;11:868–73. doi: 10.1111/j.1469-0691.2005.01227.x. [DOI] [PubMed] [Google Scholar]
- 2.Talbot GH, Bradley J, Edwards JE, Jr, Gilbert D, Scheld M, Bartlett JG, et al. Bad bugs need drugs: An update on the development pipeline from the antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin Infect Dis. 2006;42:657–68. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 3.Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: Evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. As nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–65. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villari P, Iacuzio L, Vozzella EA, Bosco U. Unusual genetic heterogeneity of Acinetobacter baumannii isolates in a university hospital in Italy. Am J Infect Control. 1999;27:247–53. doi: 10.1053/ic.1999.v27.a96961. [DOI] [PubMed] [Google Scholar]
- 6.Smolyakov R, Borer A, Riesenberg K, Schlaeffer F, Alkan M, Porath A, et al. Nosocomial multi-drug resistant Acinetobacter baumannii bloodstream infection: Risk factors and outcome with ampicillin-sulbactam treatment. J Hosp Infect. 2003;54:32–8. doi: 10.1016/s0195-6701(03)00046-x. [DOI] [PubMed] [Google Scholar]
- 7.Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacter baumannii: Laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti Infect Ther. 2013;11:395–409. doi: 10.1586/eri.13.21. [DOI] [PubMed] [Google Scholar]
- 8.McConnell MJ, Actis L, Pachón J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev. 2013;37:130–55. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 9.Ying C, Li Y, Wang Y, Zheng B, Yang C. Investigation of the molecular epidemiology of Acinetobacter baumannii isolated from patients and environmental contamination. J Antibiot (Tokyo) 2015;68:562–7. doi: 10.1038/ja.2015.30. [DOI] [PubMed] [Google Scholar]
- 10.Aygün G, Demirkiran O, Utku T, Mete B, Urkmez S, Yilmaz M, et al. Environmental contamination during a carbapenem-resistant Acinetobacter baumannii outbreak in an Intensive Care Unit. J Hosp Infect. 2002;52:259–62. doi: 10.1053/jhin.2002.1300. [DOI] [PubMed] [Google Scholar]
- 11.Russotto V, Cortegiani A, Raineri SM, Giarratano A. Bacterial contamination of inanimate surfaces and equipment in the Intensive Care Unit. J Intensive Care. 2015;3:54. doi: 10.1186/s40560-015-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkshoorn L, Aucken HM, Gerner-Smidt P, Kaufmann ME, Ursing J, Pitt TL, et al. Correlation of typing methods for Acinetobacter isolates from hospital outbreaks. J Clin Microbiol. 1993;31:702–5. doi: 10.1128/jcm.31.3.702-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gräser Y, Klare I, Halle E, Gantenberg R, Buchholz P, Jacobi HD, et al. Epidemiological study of an Acinetobacter baumannii outbreak by using polymerase chain reaction fingerprinting. J Clin Microbiol. 1993;31:2417–20. doi: 10.1128/jcm.31.9.2417-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vila J, Marcos MA, Jimenez de Anta MT. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. Baumannii complex. J Med Microbiol. 1996;44:482–9. doi: 10.1099/00222615-44-6-482. [DOI] [PubMed] [Google Scholar]
- 15.Rafei R, Kempf M, Eveillard M, Dabboussi F, Hamze M, Joly-Guillou ML, et al. Current molecular methods in epidemiological typing of Acinetobacter baumannii. Future Microbiol. 2014;9:1179–94. doi: 10.2217/fmb.14.63. [DOI] [PubMed] [Google Scholar]
- 16.Biswas I. Genetic tools for manipulating Acinetobacter baumannii genome: An overview. J Med Microbiol. 2015;64:657–69. doi: 10.1099/jmm.0.000081. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed SS, Alp E. Genotyping methods for monitoring the epidemic evolution of A. Baumannii strains. J Infect Dev Ctries. 2015;9:347–54. doi: 10.3855/jidc.6201. [DOI] [PubMed] [Google Scholar]
- 18.Elhadi N, Alsamman K. Incidence and antimicrobial susceptibility pattern of extended-spectrum-β-lactamase-producing Escherichia coli isolated from retail imported mackerel fish. Afr J Biotechnol. 2015;14:1954–60. [Google Scholar]
- 19.Coudron PE, Moland ES, Thomson KS. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol. 2000;38:1791–6. doi: 10.1128/jcm.38.5.1791-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sneath PH, Sokal RR. San Francisco, CA: W. H. Freeman; 1973. Numerical Taxonomy: The Principles and Practice of Numerical Classification. [Google Scholar]
- 21.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–31. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalla-Costa LM, Irino K, Rodrigues J, Rivera IN, Trabulsi LR. Characterisation of diarrhoeagenic Escherichia coli clones by ribotyping and ERIC-PCR. J Med Microbiol. 1998;47:227–34. doi: 10.1099/00222615-47-3-227. [DOI] [PubMed] [Google Scholar]
- 23.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–13. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez MS, Traglia GM, Lin DL, Tran T, Tolmasky ME. Plasmid-mediated antibiotic resistance and virulence in gram-negatives: The Klebsiella pneumoniae paradigm. Microbiol Spectr. 2014;2:1–5. doi: 10.1128/microbiolspec.PLAS-0016-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammoudi D, Moubareck CA, Hakime N, Houmani M, Barakat A, Najjar Z, et al. Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int J Infect Dis. 2015;36:56–61. doi: 10.1016/j.ijid.2015.05.015. [DOI] [PubMed] [Google Scholar]