Abstract
Polyketide synthases (PKSs) are microbial multienzymes for the biosynthesis of biologically potent secondary metabolites. Polyketide production is initiated by the loading of a starter unit onto an integral acyl carrier protein (ACP) and its subsequent transfer to the ketosynthase (KS). Initial substrate loading is achieved either by multidomain loading modules or by the integration of designated loading domains, such as starter unit acyltransferases (SAT), whose structural integration into PKS remains unresolved. A crystal structure of the loading/condensing region of the nonreducing PKS CTB1 demonstrates the ordered insertion of a pseudodimeric SAT into the condensing region, which is aided by the SAT-KS linker. Cryo-electron microscopy of the post-loading state trapped by mechanism-based crosslinking of ACP to KS reveals asymmetry across the CTB1 loading/condensing region, in accord with preferential 1:2 binding stoichiometry. These results are critical for re-engineering the loading step in polyketide biosynthesis and support functional relevance of asymmetric conformations of PKSs.
Polyketide synthases (PKSs) are multienzymes that underlie the biosynthesis of potent bioactive compounds such as antibiotics or toxins1. Their product diversity is generated by the sequential3 action of PKS modules, each responsible for polyketide elongation by condensation of acyl building blocks and subsequent reduction and further modification. Homodimeric PKS modules are organized into a mandatory-condensing and an optional-mod- ifying region2–5 and integrate acyl carrier proteins (ACPs) for shuttling covalently tethered intermediates. Condensing regions consist of an acyltransferase (AT) domain and a ketosynthase (KS) domain. The AT is responsible for loading acyl-CoA-derived extender units onto the phosphopantetheine (Ppant)-modified holo-ACP, whereas the KS catalyzes the elongation of a polyketide precursor by decarboxylative Claisen condensation with the ACP-bound acyl extender unit. Modifying regions contain up to three canonical types of domains, the ketoreductase (KR), the dehydratase (DH), and the enoyl-reductase (ER), which sequentially modify the β-keto position of the KS condensation product. In the presence of all three canonical modifying domains (KR, DH, and ER), a fully saturated product is generated, whereas in non- or partially reducing modules various combinations of the modifying domains can maintain the β-keto group or optionally reduce or dehydrate it to a hydroxyl, alkene, or alkane. PKS modules act either iteratively (iPKS) or sequentially with directed substrate transfer between linked modules in multimodular assembly lines (modPKS). Specialized loading modules or domains act as gatekeepers and initiate polyketide synthesis by starter unit loading6.
Iterative nonreducing (NR)-PKSs, which have reducing modifying domains, elongate successive poly-β-ketone intermediates until a final length—determined by the KS—is reached7. The nonreduced polyketide is cyclized by a product template (PT) domain to form an aromatic or fused aromatic ring system that is released from the ACP by a thioesterase (TE). In NR-PKSs, biosynthesis is initiated by starter unit acyltransferase (SAT) domains, which load the ACP with acyl starter units obtained from coenzyme A (CoA) thioesters or from ACPs of other multienzymes8,9. The ACP transfers the starter unit to the active site cysteine of the KS and then the Ppant arm of the ACP is charged with a malonyl extender unit by a malonyl-CoA-specific acyl-transferase (MAT) in all subsequent elongation cycles. NR-PKSs share a common architecture and conserved interdomain interactions as demonstrated by functional domain swaps of ACPs, PTs, and loading/condensing regions (SAT-KS-MAT) of homologous enzymes in deconstruction experiments7,10. In some NR-PKSs, ACPs occur as functionally equivalent tandem didomains10,11.
PKSs are dimeric multienzymes with structural segregation of condensing and modifying regions2,12, similar to the mammalian fatty acid synthase (mFAS)5. However, the structural integration of dedicated loading modules or specialized substrate-loading domains, as exemplified by the NR-PKS SAT-KS-MAT core region, remains unknown. The KS-MAT didomains of NR-PKSs are homologs of the corresponding condensing regions in other PKSs and mFAS, which have been structurally characterized as isolated dido-mains2,4,5,13–15. However, in NR-PKSs, soluble expression of KS-MAT is only achieved by including the SAT domain in a tridomain construct16,17, indicating that SAT, KS, and MAT form an integrated loading/condensing region architecture. SATs can be successfully isolated18 and swapped in the context of the condensing region environment19. Together with an ACP, the SAT-KS-MAT core region of NR-PKSs is catalytically competent for synthesis of the complete polyketide backbone in vitro7,10,17. Early crosslinking experiments demonstrated binding of NR-PKS ACP domains to the dimeric KS preferentially with a 1:2 stoichiometry20.
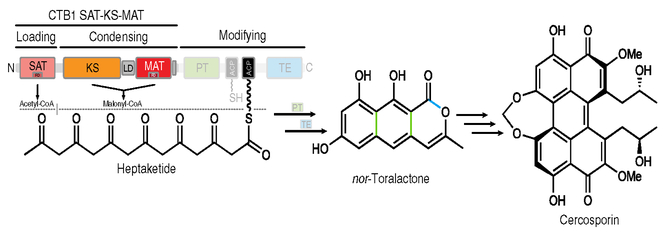
To study PKS substrate-loading architectures and the organization of NR-PKSs, we focused on the SAT-KS-MAT loading/condensing region of the NR-PKS CTB1. CTB1 produces nor−toralactone, the aromatic polyketide precursor to cercosporin in the fungus Cercospora nicotianae21–22 (Fig. 1). Cercosporin is a light activated plant toxin and protein kinase C inhibitor essential for C. nicotianae infectivity23. CTB1 contains five enzymatic domains as well as tandem ACPs (ACP1-ACP2). For initiation of CTB1 biosynthesis, acetyl-CoA is loaded via the SAT6 onto an ACP and transferred to the KS. The iterative action of the KS-MAT condensing region produces an ACP-tethered heptaketide (C14), which is subsequently cyclized by the PT and released as a pyrone by the TE24 (Fig. 1).
Fig. 1 |. Domain organization and catalytic scheme of the cercosporin PKS CTB1.
The domain organization is shown schematically with functional regions indicated. The CTB1 SAT-KS-MAT and ACP2 constructs used for structural studies are shown in full color; other domains are shown in faded colors. The iterative synthesis of nor−toralactone involves initial loading of the starter-unit acetyl-CoA by the SA T as well as six cycles of condensation by the KS using malonyl-CoA loaded by the MAT. The linear product is cyclized and released by the PT and TE, respectively, as indicated by colored bonds in nor−toralactone. Downstream processing of nor−toralactone yields cercosporin. FD, ferredoxin-like subdomain.
Here we report the crystal structure of the CTB1 SAT-KS-MAT region, which reveals a pseudodimeric arrangement of SAT domains nested in a cleft formed by the KS and MAT domains. To specifically resolve the transient interactions in the substrate-loading step, we employed mechanism-based crosslinking25 to trap a functional state of an ACP-KS interaction for cryo-electron microscopy (EM) structure determination.
Results
Crystal structure of an integrated loading architecture.
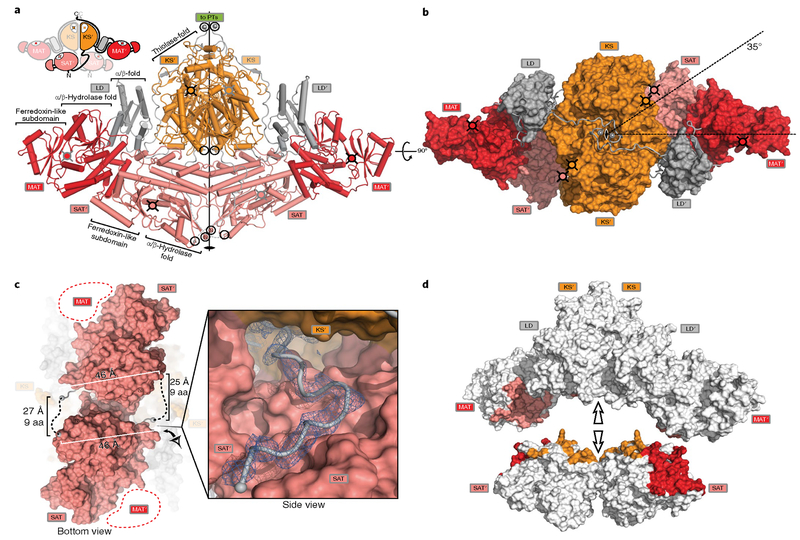
The 2.8-Å; resolution crystal structure of SAT-KS-MAT reveals a compact dimer of rhomboid shape (Fig. 2; Supplementary Table 1; Supplementary Fig. 1). The two KS domains dimerize, and each monomer of the KS laterally connects to one MAT through a small linker domain (LD). The individual domain structures (Supplementary Table 2a) and the overall arrangement of the KS-LD-MAT domains (Fig. 2a,b) resemble condensing-region structures from mFAS5,15, fully-reducing iPKS2 and several modPKS4,13,14, but differ from the architecture of the PikAIII modPKS that was resolved at intermediate resolution by cryo-EM12,26 (Supplementary Fig. 2). The SAT has structural homology with other ATs, with r.m.s. deviations of Cα atom positions between 2.5–3.2 Å for 62–75% aligned amino acids (aa) (Supplementary Table 2a) and an active site located in a cleft between the a/p-hydrolase core fold and a ferredoxin-like subdomain. The C terminus of the SAT domain is connected via a 26-aa linker to the N terminus of the KS domain (Fig. 2c). Nine aa of this linker remain disordered in the crystal structure, but the connectivity is unambiguously determined on the basis of maximal linker length and spatial restraints (Fig. 2c). In an intertwined arrangement, the SAT domain linked to the KS of the first protomer is nested by the KS-LD-MAT domains of the second protomer (Fig. 2a). The clefts formed between the MAT and KS domains, which have been suggested to guide the interactions of the upstream ACP with the KS in modPKSs27, are partially occupied by the SAT domains. The active site entrance of each SAT domain points into this cleft toward the other protomer’s KS active site, which is located at a distance of 56 Å.
Fig. 2 |. Crystal structure and interdomain interactions in CTB1 SAT-KS-M AT.
a, Cartoon representation of the SA T-KS-M A T crystal structure (front view); the intertwined domain connectivity is indicated in the inset (top left). Active sites are marked by cross-hatched filled-in circles, termini of SA T and KS aligned along the two-fold axis are indicated by open circles, and N and C termini of SA T-KS-M A T are additionally labeled. b, Top view of SAT-KS-M AT; domains are shown in surface representation and linkers as cartoon. Active sites, termini and two-fold axis are indicated as in a; the angle between the longest axes of the KS-LD-MAT and SAT (pseudo-) dimers is indicated. c, The SAT-KS linker. Bottom view of the crystal structure as in b highlighting the connectivity of the SA T-KS linker and euclidean distances. The inset provides a close-up view from the side of the C-terminal region of the SAT-KS linker, which is involved in contacts to KS’ and SAT. An Fo–Fc shaked omit difference map is shown at 2.5σ. d, Interdomain interfaces between the SAT domains and the K S-LD-MAT region of CTB1 are mapped onto a split-surface representation of the SAT-KS-MAT structure by coloring according to the interaction partner. The SA T domains have been separated from the KS-LD-MAT for representation.
The arrangement of the two SAT domains is stabilized by interfaces with surrounding domains (Fig. 2d and Supplementary Fig. 3a). The two SAT domains form a direct pseudodimer interface of 324 Å2 that is devoid of hydrogen bonds or salt bridges (Supplementary Table 2b and Supplementary Fig. 3b,c). The C-terminal end of the SAT-KS linker adds an average interface area of 493 Å2 by binding to a surface groove of the opposite SAT (Fig. 2c and Supplementary Table 2b). The largest interface with the highest predicted stability is the one formed between the SAT and MAT of opposite chains, covering an average of 957 Å2 (Fig. 2d and Supplementary Fig. 3d,e) and a network of at least two salt bridges and five hydrogen bonds. The interface of the SAT domain to the other protomer’s KS domain is considerably smaller (184 Å2; Supplementary Fig. 3f,g). Notably, the SAT contacts the KS and MAT of the other protomer without interfacing with the connecting LD.
The SAT pseudodimer is arranged at a 35° angle relative to the condensing-region KS-MAT dimer (Fig. 2b), but the two-fold symmetry axes of KS and SAT are aligned. The N and C termini of the KS and SAT domains are arranged in close proximity to the twofold axis (Fig. 2a). Although CTB1 is an iPKS, the locations of the N and C termini at opposite ends of the molecule would be compatible with extended connectivity in an assembly line. The only other known structure of a NR-PKS SAT, which was excised as an isolated domain from CazM18, reveals a C-terminal linker element that ends in a position similar to that of the SAT-KS in CTB1 despite a lack of sequence conservation in the linker (Supplementary Fig. 4a,b). By contrast, the excised loading AT domain of the avermectin modPKS (AVES1)28, which features completely different connectivity via an ACP to the downstream KS domain, has a distinct linker architecture resembling the post-MAT linker in PKS and FAS condensing regions (Supplementary Fig. 4c,d). Together, these structural comparisons, in agreement with functional domain swaps19, support a similar organization among NR-PKS loading domains. The active sites of the loading/condensing region reveal high structural conservation with their closest structural and functional neighbors (Supplementary Table 2a). The SAT has a conserved Cys-His dyad (Supplementary Fig. 5a), which resembles the catalytic Ser-His dyad in AT domains of PKSs and mFASs. Similarly, the active sites of the MAT (Supplementary Fig. 5b) and KS (Supplementary Fig. 5c) are conserved. The KS active sites are connected by a hydrophobic tunnel. The tunnel connection is relatively narrow (Supplementary Fig. 5d) compared to other PKSs and mFASs connected tunnels, which may be involved in chain-length determination.
Cryo-EM analysis of a trapped ACP-KS post-loading state.
To analyze the impact of substrate loading, we trapped CTB1 SAT-KSMAT in an initial post-loading state by mechanism-based cross-linking20,25 and analyzed it by cryo-EM (Supplementary Fig. 6). In CTB1, the SAT mediates loading of acetyl or, less efficiently, propionyl moieties onto ACP6 for subsequent transfer to the KS active site cysteine. A Ppant analog linked to a selectively reactive α-bro-mopropionyl moiety was synthesized (Supplementary Note 1) and enzymatically loaded onto isolated CTB1 ACP2 (Supplementary Fig. 6a), which is functionally equivalent to CTB1 ACP1 (Supplementary Fig. 6b) and shares a higher sequence identity (a CP2: 40% vs. ACP1: 30%) with the previously characterized PksA ACP20,29. For crosslinking, the loaded ACP2 was added to a variant of CTB1 SAT-KS-MAT in which the SAT and MAT active sites were mutated to strictly direct crosslinking to the KS (denoted as SAT°-KS-MAT°). Loading of only a single ACP2 to the dimeric SAT°-KSMAT° was kinetically favored over two-site loading, as previously observed for related NR-PKSs that contain only a single ACP20 (Supplementary Fig. 6c). Using an excess of ACP2 and an extended reaction time, > 90% crosslinking was achieved. Negative-stain EM revealed mostly dimeric particles, whereas cryo-EM grid preparation led to substantial monomerization of SAT°-KS-MAT°=ACP2 (= indicates chemical crosslink; Supplementary Figs. 6d and 7). Under optimized conditions, 60% of all particles were observed as intact dimers (Supplementary Fig. 6e,f).
Single-particle analysis of dimeric SAT°-KS-MAT°=ACP2 provided a map at 7.1 Å overall resolution and was interpreted by rigid-body and real-space refinement using an all-atom model based on the crystal structure of SAT-KS-MAT and a homology model of ACP2 based on the NMR structure of PksA ACP29. Due to the limited resolution and the absence of an authentic high-resolution structure, the analysis for ACP2 is restricted to conserved structural elements (Supplementary Table 1; for more details see Online methods). Single-particle processing including reference-free maximum likelihood classification provided a single, predominant conformation of SAT°-KS-MAT° = ACP2 (for details see methods). Although the sample used for grid preparation was >90% crosslinked to ACP2, only a single ACP position was clearly resolved in an overall asymmetric structure (Fig. 3a-c). ACP2 is bound in a cleft between the LD and the KS-KS′ dimer. The N-terminal end of helix II in ACP2, which contains the Ppant-analog-bearing DSL motif, points directly toward the KS active site entrance. For ACP interactions in modPKSs, two different interaction sites have been proposed for the transthioesterification from the upstream ACP and elongation with the canonical ACP in modPKSs27. Both proposed sites are buried in the KS-LD-AT clefts of the DEBS PKS module five, whereas the ACP in CTB1 sits on top of the KS-LD-MAT cleft and therefore diverges from both proposed models.
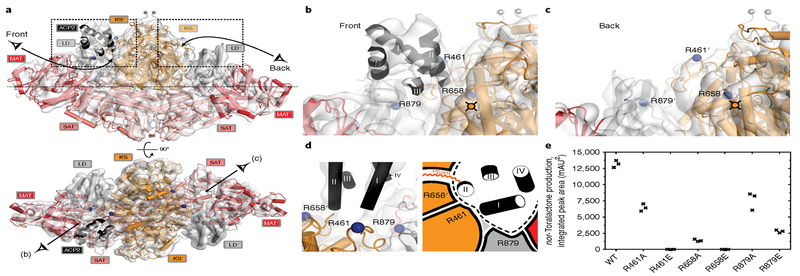
Fig. 3 |. Asymmetric cryo-EM structure of CTB1 SAT°-KS-MAT °= ACP2.
a, Reconstructed volume and model of SAT °-KS-MAT °= ACP2 in front and top views. In the asymmetric structure, ACP2 is only resolved at one of the two active sites in dimeric CTB1 SAT °-KS-MAT °. The canonical four-helix bundle of CTB1 ACP2 is shown. Termini and views of b and c are indicated. All maps are contoured at 7.1a. b,c, Close-up of the active site (indicated) regions of the two KS domains with (b) or without (c) resolved ACP2. d, Location of residues on SAT °-KS-MAT ° interacting with ACP2 as structural (left) and schematic (right) representations. In the scheme, the approximate position of the KS active site tunnel and the crosslinker-modified serine in ACP2 are indicated. The helices of the canonical ACP four helix bundle are indicated with Roman numerals. ACP boundaries are indicated as a dotted line in the schematic inset of panel d.e, Functional analysis of CTB1 SA T-KS-M A T mutants of ACP2-interacting residues. Nor−toralactone production was quantified by integrated peak areas of HPLC chromatograms milliabsorbance units2 (mAU2) at 280 nm. Each mutant was assayed in triplicate, and data from each replicate are presented.
At the second KS active site entrance, where no ACP is visualized, the surroundings of the substrate entry tunnel are more disordered (Supplementary Fig. 8). Presuming that ACP2 is still crosslinked to the second site, binding of ACP2 to one KS active site may prevent ordered and competent ACP binding to the second active site by allosteric effects. Such negative feedback is supported by activity assays of 50% crosslinked samples of the related Pks4, which contains a single ACP and exhibits only 20% activity instead of the expected 50% activity relative to a noncrosslinked sample20.
For the SAT-KS-MAT region, amino acids involved in ACP2 interactions can be identified based on the authentic higher-resolution crystal structure, whereas the limited resolution of the EM reconstruction precludes assignment of interface amino acids in ACP2. The LD core around residue Arg879 and the LD-MAT linker interact with the loop between helix I and II in ACP2. In accord with this proposed interaction, mutations of Arg879 to alanine or glutamate reduce activity in deconstruction experiments for the complete CTB1 PKS (Fig. 3d,e and Supplementary Fig. 9) but do not completely abolish production of nor-toralactone. ACP2 interactions at the KS-KS′ active site entrance (Fig. 3b,c) involve two positively charged residues: Arg461 is strictly conserved in NR-PKSs and Arg658′ is conserved as a positively charged arginine or histidine in the CTB1 clade (Supplementary Fig. 10a,b). The mutations R461A or R658A considerably reduce activity, whereas the corresponding charge-inversion mutations to glutamate completely abolish nor−toralactone production, strongly suggesting that the two arginines mediate critical interactions with negatively charged groups in ACP2 (Fig. 3e and Supplementary Fig. 9).
Discussion
Interactions of an ACP with a multienzyme KS domain have previously been resolved in the crystal structure of yeast FAS (yFAS) at 3.1 A resolution30 and in the cryo-EM reconstruction of the PikAIII modPKS at 8.6 Å and 7.3 Å resolutions12 (Supplementary Fig. 11a–e). Both structures reveal strictly symmetric ACP interactions, but only the heterododecameric yFAS fulfills the distance restraints needed for a productive ACP-KS interaction with solvent-accessible surface (SAS) distances of the ACP Ppant attachment point to the KS active site cysteine of approximately 27 Å (Supplementary Fig. 11c,f-i). The position and orientation of ACP relative to KS is conserved between yFAS and the CTB1 NR-PKS despite a completely different integration of the KS domain in a barrel-like scaffold and the presence of an unusual four-helical extension of the canonical ACP domain in yFAS. The corresponding SAS distance in SAT°-KS-MAT°=ACP2 is 28 Å, which is in agreement with a covalently trapped functional interaction (Supplementary Fig. 11c,f). The ACP-KS interaction observed in PikAIII, which is nonproductive based on SAS distances (Supplementary Fig. 11h,i), however, is considerably different. The ACP docking surface observed in CTB1, and verified by mutant activity assays in deconstruction experiments, is partially blocked in PikAIII due to the divergent location of the LD domain (Supplementary Fig. 11d,e).
The cryo-EM solution structure of SAT°-KS-MAT°=ACP2 reveals considerably greater asymmetry than the crystal structure without ACP2, strongly suggesting that the predominant asymmetric conformation derives from ACP2 binding by either an induced-fit or conformational selection mechanism31. Relative to the crystal structure, the SATs are translocated individually toward the central KS domains, not as a rigid dimer (Supplementary Fig. 12a,b). ACP2 binding causes an upward movement of the proximal LD and MAT regions by 4.4 Å and 8.4 Å, respectively, (Fig. 4 and Supplementary Video 1) resulting in an opening of the interface between the SAT and MAT domains (Fig. 2d and Supplementary Fig. 12c). Coordinated with this separation, the proximal SAT domain moves toward the KS by 6.9 A, substantially enlarging the SAT-KS interface, whereas the second SAT is pushed (by 4.5 Å) against the second pair of KS and MAT domains (Supplementary Fig. 12d). Because large fragments and domains of various NR-PKSs can be functionally interchanged in deconstruction experiments, coupling of structural changes and catalysis might be representative for many NR-PKSs7,10.
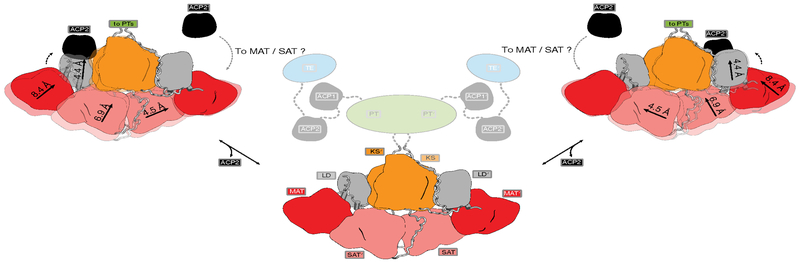
Fig. 4 |. Schematic illustration of suggested modes of conformational coupling in CTB1.
ACP2 preferentially binds to a single KS active site. ACP2 binding results in coupled conformational changes across the dimeric CTB1, disfavoring the productive ACP2-KS interaction in the other protomer and re-aligning active sites for ACP2 interaction and substrate shuttling. Such conformational coupling resembles a V-twin engine mechanism for alternating KS-ACP2 interactions. Regions of full-length CTB1, not included in the reported crystal structure, are represented as cartoon in faded colors and with linkers indicated as dotted lines.
Conformational asymmetry and coupling have previously been observed in iteratively acting, fully reducing iPKS2 and mFASs5,32,33. The asymmetric conformational states observed in these systems, as well as in SAT°-KS-MAT°=ACP2, are derived by local hinge-bending or shear motions, maintaining the individual domain structures, the key conserved interfaces, and the overall interdomain architecture. For non-iteratively acting modPKSs, however, no information is currently available on functional asymmetry. By contrast, symmetric large-scale conformational transitions that require partial refolding of conserved domains have been visualized by EM analysis of distinct functional states of the modPKS PikAIII on the basis of a divergent condensing region architecture12,26. Functional asymmetry in the interaction of ACP with dimeric enzymatic domains has previously been suggested in the form of a seesaw-type conformational coupling for a bacterial hexameric dehydratase (FabZ) involved in fatty acid biosynthesis34 that binds ACPs in a 6:3 ratio. However, a stoichiometrically ACP-crosslinked structure of the bacterial dimeric dehydratase FabA demonstrates that asymmetry is not a general feature in related dehydratases even though this structure also revealed increased disorder for the second ACP, as indicated by the crystallographic B factors25. A distinct type of persistent structural asymmetry is observed in the iterative actinorhodin type II PKS, in which asymmetry is implemented by heterodimerization of one active KS protomer with a catalytically inactive KS homolog, the chain length factor (CLF)35. Iterative elongation is catalyzed only at the KS active site, and the CLF determines maximum chain length by restricting the available volume of the substrate binding tunnel35.
Here, the crosslinked interaction of an ACP induces conformational asymmetry across the CTB1 SAT-KS-MAT loading/condensing region. Binding of an early elongation intermediate, mimicked here by the α-bromopropionyl moiety, may cause local asymmetry around the KS active sites. However, the observed overall asymmetry is more plausibly explained by binding of the ACP, which directly connects the KS active site to the surrounding domains via the LD, which is part of the ACP binding interface.
Substrate elongation in NR-PKSs occurs rapidly, and reactive intermediates remain bound in the active site tunnel of the KS, as suggested by the absence of detectable ACP-bound chain elongation intermediates in PksA17. After initial loading, however, the ACP shuttles between the KS and the MAT active site for delivery of malonyl extender units. Presuming that ACP binding is linked to overall asymmetry in the SAT-KS-MAT region, productive binding of an extender-unit-charged ACP (malonyl-ACP) to one KS active site would disfavor binding to the other KS active site in every elongation cycle. Product elongation by the dimeric KS would occur with alternating asymmetric states that are analogous to a V-twin engine, favoring substrate loading by the MAT on one side and elongation by the KS on the other side of the dimer (Fig. 4 and Supplementary Video 1). Considering that the ACP binding sites are located proximal to the attachment points for the downstream modification (PT) and release (TE) domains, ACP could plausibly contribute to a coordination of substrate shuttling and overall product formation analogous to conformational coupling2 and large scale rearrangements12,26 observed in modifying regions of other PKSs. However, further analysis will be required to dissect the relative contributions of substrate loading and ACP interaction on asymmetry in iterative and modular PKSs.
Understanding the architectures and dynamic properties of PKSs remains a central challenge in natural product research. Although all canonical types of PKS domains have been studied in detail in isolation, understanding substrate loading and transfer, as well as the functional integration of individual domains into large multienzymes or even assembly lines remains a challenge. Our analysis of CTB1 SAT-KS-MAT now reveals the organization of an integrated loading architecture in a PKS and suggests a mechanism for conformational coupling between the two protomers of the dimeric PKS promoted by substrate loading to KS. These results provide an important step toward a comprehensive understanding of conformational coupling in PKS multienzyme function and afford critical insights for the dissection and re-engineering of initial substrate loading for polyketide production.
Methods
Cloning.
Plasmids used in this study were assembled using standard protocols. CTB1 SAT°-KS-MAT° was generated by site-directed introduction of the mutations C119A and S1010A. All plasmids used in this study are summarized in Supplementary Table 3. The plasmids for expression of wild-type CTB1 SATKS-MAT (pECTB1-NKA6), PT (pECTB1-PT), and TE (pECTB1-TE) have been previously described24. Primers used for assembly of new plasmids in this study are detailed in Supplementary Table 4. All expression constructs were prepared and maintained in E. coli BL21(DE3). Cut sites for CTB1 ACP2 deconstruction were chosen based on the PksA ACP solution structure29. CTB1 ACP2 was cloned as an N-terminal Thrombin-cleavable His6−tagged construct to allow differential purification throughout the crosslinking process. The desired gene sequence was amplified by PCR from pECTB1-ACP (tandem ACP, previously described)24 using CTB1-ACP2–5 and CTB1-ACP2-stop-3, and ligated into pET-28a at NdeI and NotI sites using T4 DNA ligase. Active site mutations for selective crosslinking were introduced into pECTB1-NKA6 by Gibson assembly36 of PCR-amplified fragments using standard protocol, giving pECTB1-SKM-C119A-S1010A. Mutations to CTB1 SAT-KS-MAT for interface validation were introduced via Gibson assembly in the same manner. Assembled plasmids were screened by restriction digestion and sequences confirmed by the Johns Hopkins Synthesis and Sequencing Facility (Baltimore, MD). Sequencing revealed the point mutation T321A (compared to Uniprot Q6DQW3) in all SAT containing constructs.
Protein expression and purification for crosslinking and interface validation.
Proteins were expressed as His6−tagged constructs in E. coli BL21(DE3). One liter Terrific Broth cultures supplemented w ith 25 μg/mL kanamycin (GoldBio) were inoculated with overnight starter cultures and grown at 37°C w ith shaking at 250 r.p.m. until OD 600 reached 0.7. Cultures were cold-shocked in ice water for 1 h before induction with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG; GoldBio). Expression was carried out at 19°C with shaking at 250 r.p.m. for approximately 16h. Cells were harvested by centrifugation at 4000 × g for 15 min and flash frozen in liquid nitrogen for storage at −80°C.
Cell pellets were thawed in lysis buffer A (50 mM potassium phosphate, pH 8, 300 mM NaCl, 10% (v/v) glycerol) and lysed by sonication. Lysate was cleared by centrifugation at 27,000 ×g for 25 min. The resulting supernatant was batch bound to Co2+−TALON resin (Clontech) at 4°C, typically for 1 h. The protein-bound resin was applied to a gravity-flow column and washed and eluted with lysis buffer A containing increasing concentrations of imidazole (0–100 mM), as directed by the manufacturer. Fractions containing the protein of interest were pooled and dialyzed against the appropriate buffer (50 mM Tris pH 7.5, 5% (v/v) glycerol unless otherwise indicated) at 4°C. For dialysis of CTB1 SAT°-KS-MAT°,1 mM DTT was included to maintain the KS active site cysteine in its reduced form. If necessary, the protein was concentrated in Amicon Ultra centrifuge filters (Millipore). Protein concentration was quantified based on absorbance at 280 nm on a Cary 50 UV-Vis spectrophotometer and extinction coefficients calculated using Expasy ProtParam (Supplementary Table 3). CTB1 ACP1 concentration was determined in triplicate by Bradford assay (Bio-Rad) using bovine serum albumin as a standard.
Proteolysis of tag from CTB1 ACP2-His.
To facilitate separation of ACP2 from both crosslinker loading reactions and crosslinking reactions, the N-terminal His6−tag was removed by thrombin proteolysis using a Thrombin CleanCleave Kit (Sigma Aldrich). The thrombin agarose resin was washed with reaction buffer (50 mM Tris pH 8, 10 m M CaCl2) and conditions were optimized as recommended by the manufacturer. Complete cleavage was achieved using approximately100 μL thrombin agarose resin per 50 mg CTB1 ACP2 at 2 mg/mL CTB1 ACP2 in reaction buffer. Reactions were rotated for 3 h at 25°C. Cleavage was monitored by SDS-PAGE and confirmed by M ALDI-TOF (Bruker Auto Flex III). Cleaved CTB1 ACP2 was isolated by brief centrifugation at 500 × g to pellet the thrombin resin and dialyzed in 50 mM Tris pH 7.5, 5% (v/v) glycerol at 4 °C.
Expression and purification of CTB1 SAT-KS-MAT for crystallization.
C-terminal hexahistidine (His6) tagged CTB1 SAT-KS-MAT (pECTB1-NKA6, aa 1–1293) was overexpressed with Streptomyces chaperonins (pETcoco-2A-L1SL2 plasmid)37 in E. coli BL21(DE3) pRIL. Cells were cultured in 2 × YT medium, supplemented with 0.5% (v/v) glycerol, NPS (25 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4), kanamycin (100 μg/mL), chloramphenicol (34 μg/mL), and ampicillin (100 μg/mL). An expression culture (1.5 L) was inoculated (1:20), grown at 37 °C for 2 h, cooled to 20 °C, and induced with IPTG (0.1 mM) at an OD600 of 1.0. Cells were harvested after 12 h by centrifugation (7000 × g) and resuspended in lysis buffer B (50 mM HEPES pH 7.4, 200 mM NaCl, 2.5 mM MgCl2, 40 mM imidazole, 10% (v/v) glycerol, 5 mM β-mercaptoethanol), supplemented with protease inhibitors (200 μM phenylmethylsulfonyl fluoride, 20 μM bestatin, 4μM E64, 2 μM pepstatin A, 20 μM phenantrolin, 2 μM phosphoram idon) as well as DNase I, RNase A, and lysozyme. Cells were placed on ice and lysed by sonication. The lysate was cleared by centrifugation (100,000 × g for 30 min) and immobilized by metal-affinity chromatography on a 25 mL Ni-affinity column (GenScript) pre-equilibrated with lysis buffer B. Unbound protein was eluted with five column volumes (CV) of HisA buffer (50 mM HEPES pH 7.4, 200 mM NaCl, 2.5 mM MgCl2, 40 mM imidazole, 10% (v/v) glycerol, 5 mM β-mercaptoethanol, protease inhibitors). The sample was eluted with a linear gradient to 100% HisB buffer (50 mM HEPES pH 7.4, 500 mM imidazole, 50 mM NaCl, 10%(v/v) glycerol, 2.5 mM β-mercaptoethanol, and protease inhibitors) and directly loaded on a strong anion-exchange colum n (PL-SAX 4000 Å, 10 μm, Agilent). The protein was eluted with a linear gradient from 0% AIC-A (50 m M HEPES pH 7.4, 50 mM NaCl, 10% (v/v) glycerol, 2.5 mM β-mercaptoethanol) to 100% AIC-B (50 mM HEPES pH 7.4, 1 M NaCl, 10% (v/v) glycerol, 2.5 mM β-mercaptoethanol). The protein was concentrated with Amicon Ultra centrifuge filters (Millipore) and subjected to size-exclusion chromatography (SEC; Superdex 200 16/60, GE Healthcare) using 20 m M HEPES pH 7.4, 250 mM NaCl, 5% glycerol (v/v), 1 mM dithiothreitol. Protein-containing fractions were pooled and concentrated in Amicon Ultra centrifuge filters (Millipore) to 10–15 mg/mL and frozen in liquid nitrogen.
Synthesis of α-bromopropionyl aminopantetheine.
For full synthetic details and compound characterization see Supplementary Note 1.
Phosphorylation of α-bromopropionyl aminopantetheine.
α-bromopropionyl aminopantetheine was phosphorylated enzymatically under the following conditions: 0.25 mg/mL PanK (pantothenate kinase), 2 m M α-bromopropionyl aminopantetheine, 5 mM ATP, 20 mM KCl, 10 mM MgCl2, 50 mM Tris pH 7.5 at room tem perature for approximately 20 h. Complete phosphorylation was confirmed by UPLC-ESI-MS (Waters Acquity/Xevo-G2, negative ion mode). All protein was removed from the reaction with a 3 kDa MWCO Am icon Ultra centrifuge filter (Millipore). The flow-through was flash frozen in liquid nitrogen and lyophilized to dryness. The resulting white powder was resuspended in 5% aqueous acetonitrile. α-bromopropionyl aminopantetheine was HPLC purified on an Agilent 1100 equipped with a Kinetex C18 semi-prepcolum n (250 × 10 mm, 5 μ, Phenomenex) using the following method at 4 mL/min: hold 5% solvent A/95% solvent B for 3 min, 5–35% solvent A over 17 min, 35–95% solvent A over 5 min, followed by re-equilibration to 5% solvent A, where solvent A is acetonitrile + 0.1% trifluoroacetic acid and solvent B is 0.1% trifluoroacetic acid. Purity and identity of the isolated com pound was confirmed by UPLC-ESI-MS using a Waters Acquity/Xevo-G2 in positive ion mode as above, and the pooled fractions were lyophilized to dryness.
Loading of ACP2 with α-bromoacyl crosslinker.
α-bromopropionyl aminophosphopantetheine was adenylated and subsequently loaded onto CTB1 ACP2 in a one-pot enzymatic reaction under the following conditions: 0.5 mM α-bromopropionyl aminophosphopantetheine, 0.5 mg/mL PPAT (phosphoribosyl pyrophosphate amidotransferase), 5 μM Sfp (surfactin 4’-phosphopantetheinyl transferase), 0.5 mM CTB1 ACP2, 5 mM ATP, 10 mM MgCl2, 20 mM KCl, 50 mM Tris pH 7.5, 10% glycerol. Reactions were run at 22–24°C and m onitored by MALDI-TOF (Bruker AutoFlex III). Once loading was complete, the His-tagged Sfp and PPAT were separated from untagged CTB1 ACP2 by passing the reaction solution through an equilibrated Co2+−TALON column (Clontech) and recovering the flow-through. Additionally, any unreacted crosslinker was removed by repeated concentration and dilution with a 3 kDa M W CO Am icon Ultra centrifuge filter (Millipore). Complete removal of free crosslinker was confirmed by UPLC-ESI-MS analysis.
Crosslinking of CTB1 SAT°-KS-MAT° to ACP2.
CTB1 SAT°-KS-MAT° was combined with 20-fold excess CTB1 ACP2 bearing the α-bromoacyl crosslinker in 50 mM Tris pH 7.5, 10% glycerol (v/v) at room temperature for 1 h. Crosslinking was monitored by SDS-PAGE and found to be approximately 95% complete after 1 h. Crosslinked SAT°-KS-MAT°=ACP2 was separated from remaining free SAT°-KS-MAT° by anion exchange chromatography. Q-Sepharose Fast Flow resin (GE Healthcare) was equilibrated with 50 mM Tris pH 7.5 and 10% glycerol, and the crosslinking reaction mixture was applied to the column. SAT°-KS-MAT° and SAT°-KS-MAT°=ACP2 were separated using a gradient of potassium chloride from 0–500 mM. To remove free ACP2 from the crosslinked protein, fractions containing SAT°-KS-MAT°=ACP2 were pooled and applied to an equilibrated Co2+−TALON column. ACP2 was removed in the flow-through, and pure SAT°-KS-MAT°=ACP2 was eluted with a gradient of imidazole from 2–100 mM.
In vitro enzymatic reactions.
CTB1 SAT-KS-MAT (wild-type and mutants of proposed interface residues), PT, ACP1, ACP2, and TE were purified as described above and dialyzed into 100 mM potassium phosphate pH 7 and 10% glycerol. ACP1 and ACP2 were activated enzymatically with CoA for 1 h at 25 °C under the following conditions: 2 μM Sfp, 0.5 mM CoA, 10 mM MgCl2, 200 μM ACP2, 100 mM potassium phosphate pH 7, and 10% glycerol.
To compare the biosynthetic competency of ACP1 and ACP2, specifically the ability of the ACP to interact with KS, minimal PKS reactions containing only SAT-KS-MAT and ACP were performed. ACP1 and ACP2 concentrations were determined in triplicate by the Bradford assay using bovine serum album in as a standard. 10 pM SAT-KS-MAT was combined with 10 pM ACP1, 10 μM ACP2, or 5 μM of each ACP in assay buffer (100 mM potassium phosphate pH 7, 10% glycerol, 1 mM TCEP). Reactions were initiated by addition of 0.5 mM acetyl- S-N- acetylcysteamine (SNAC) and 2 mM malonyl-SNAC and incubated for 4 h at 25 °C. The 250-μL reactions were quenched with 10 μL hydrochloric acid and extracted thrice with ethyl acetate. Extracts were pooled, dried, and resuspended in 250 μL 20% aqueous acetonitrile for HPLC analysis.
Product profiles of each enzyme mutant were analyzed on an Agilent 1200 equipped with a Prodigy ODS3 analytical column (4.6 × 250 mm, 5 μm, Phenomenex). 100 μL injections were separated by abisolvent method at 1 mL/min: 5–85% solvent A over 40 min; 85–95% solvent A over 10 min; re-equilibrate to 5% solvent A over 10 min (where solvent A is acetonitrile +0.1% formic acid and solvent B is 0.1% formic acid). Chromatograms were recorded at 280 nm, and UV-vis spectra were recorded over a range of 200–800 nm. High-resolution mass data were collected on a Waters Acquity/Xevo-G2 in positive ion mode. Previously characterized CTB1 derailment products were identified by their UV-Vis spectrum and exact mass24.
For CTB1 interface validation, the full PKS was reconstituted with each SAT-KS-MAT mutant under the following conditions: 10 μM CTB1-SAT-KS-MAT, 10 μM PT, 10 μM ACP2, and 10 μM TE were combined in assay buffer. Reactions were initiated by addition of 0.5 mM acetyl-SNAC and 2 mM malonyl-SNAC and incubated for 4 h at 25 °C. Reactions were extracted and analyzed by HPLC as above.
Crystallization, data collection and structure determination of CTB1 SAT-KS-MAT.
CTB1 SAT-KS-MAT plate-like crystals were obtained by the sitting-drop vapor-diffusion method in 0.2 M M gCl2, 0.1 M Bis–Tris propane pH 6.5, 18% (v/v) PEG 3350 at 4 °C, a drop ratio of 1.0 μL + 0.5 μL and further optimized by seeding. Crystals grew to a final size of 0.4 × 0.04 × 0.04 mm3 within 1 week. The crystals were dehydrated and cryo-preserved by successively increasing the concentration to 0.5 M MgCl2, 0.1 M Bis-Tris propane pH 6.5, 25% PEG 3350, and 22% ethylene glycol. Diffraction data of crystals in space group C2221 (a = 108.1Å, b = 230.2 Å, c = 253.8 Å) and 56% solvent content were collected at the Swiss Light Source (SLS, Villigen, Switzerland) at beamline X06SA, a tem perature of 100 K and a wavelength of 1.000 A. Data reduction was perform edusing XDS and XSCALE38, and data were analyzed using phenix.xtriage39. The structure was solved with PHASER40 using molecular replacement and the KS and AT domains of DEBS module five4 as well as the AT domain of CurL14 as search models. A n initial model was obtained in iterative cycles of rebuilding with BUCCANEER41 and density modification using solvent flattening and histogram matching in PARROT42. Iterative cycles of m anual model building and real space refinement in Coot43 and TLS refinement in Phenix39 yielded a high-quality model with Rwork/Rfree values of 0.21/0.24 at 2.8 A resolution with excellent geometry (Ramachandran favored/outliers: 97.07%/0.00%) (Supplementary Table 1a and Supplementary Fig. 1).
Electron microscopy sample preparation and data collection.
Initial sample quality was evaluated by conventional negative-stain EM. Initial cryo-EM analysis revealed a monomer-dim er equilibrium, which has previously been observed for PKS and FAS fragments2,12,14,44,45. Buffer conditions were optimized by analyzing SAT-KS-MAT by analytical size-exclusion chromatography (SEC; 4.6/250 Superose 6 Increase, GE Healthcare) at a flow rate of 0.1 m L/m in followed by grid preparation and testing of the maximal achievable particle concentration. The results indicated a fast equilibrium and a mostly dimeric species in low-salt buffer (20 mM Tris pH 7.4, 50 mM NaCl, 2.5 mM tris(2-carboxyethyl)phosphine (TCEP); Supplementary Fig. 6e). For cryo-EM, crosslinked sample was concentrated to 15–16 mg/mL, applied to analytical SEC and collected in fractions of 50 μL. For grid preparation, 4 μL of individual fractions were applied to glow discharged (30 s) lacey carbon grids (300 mesh, copper), immediately blotted for 2 s, and plunge frozen in liquid ethane using a Vitrobot (FEI, Vitrobot III). A n incubation step was omitted because of fast adsorption of the particles to the carbon. The integrity of the crosslinked sample was analyzed by denaturing PAGE of the remaining sample after grid preparation (Supplementary Fig. 6f).
Grid quality was analyzed using a Philips CM200 FEG cryo-transm ission electron microscope, operated at an acceleration voltage of 200 kV. Optimal conditions were identified for peak fractions diluted to 270 μg/m L in a tradeoff between low particle density (approximately 70 particles per m icrograph), monomer formation and particle adsorption to carbon. Final data was collected using a Titan Krios electron microscope (FEI) operated at 300 kV, a GIF Quantum LS imaging filter (Gatan) and a K2 Summit (Gatan) operating in counting mode. Images were acquired at 0.8–4.5 μm defocus and a nominal magnification of 105,000×, corresponding to a pixel size of 1.326 A (Supplementary Table 1b). Movies were collected with a total dose of approximately 90 e-/Å2 per 18 s exposure, fractionated over 60 frames.
EM data processing and analysis.
Drift correction of dose fractionated frames46 was performed using Zorro47 and global contrast transfer function (CTF) parameters were determined using CTFFIND4.1 (ref. 48). Particles were picked from aligned dose-filtered averages using the swarm semi-automated procedure as implemented in e2boxer.py49, followed by extraction from unfiltered averages using Relion-2 (ref. 50) and local CTF parameter refinement using Gctf v1.06 (ref. 51). Single-particle analysis included several rounds of reference-free maximum a posteriori−2D classification50 to remove junk particles and monomers, revealing a clearly distinguishable shape; this could be reconstructed at low resolution (Supplementary Figs. 6d and 7a). For reconstruction of the dimeric, crosslinked SAT°-KS-MAT°=ACP, an initial volume was generated using e2initialmodel. py49 and low-pass filtered to 50 Å. Exhaustive global followed by local reference-free 3D classification and alignment without symmetry were applied, yielding highly similar classes that were combined in a consensus refinement before movie refinement and particle polishing, as implemented in Relion-2 (ref. 50). Parameters for B-factor fitting as a function of movie frames have been carefully examined, as well as the different number of frames in the particle movie stacks. At this stage all 60 frames were included in the weighted particle movie stacks, as no substantial difference between inclusion of different amounts of frames per movie could be identified after applying weighting. A nother round of 2D and 3D classification yielded three almost-identical volumes that were refined as individual classes and in all possible com binations of classes. The combination of two classes (Supplementary Fig. 7a) resulted in a slightly improved m odel at 7.3 Å resolution (25,107 particles) in comparison to all three classes at 7.5 Å (44,859 particles). For the refinement of the final map at 7.1 Å resolution, weighted particle averages of 27 movie frames were used, corresponding to a total dose of 41 e−/Å 2. All reported resolution estimates are based on the 0.143 threshold criterion of the Fourier Shell Correlation (FSC)52 between two halves of the data set refined independently in RELION-2, after accounting for correlations introduced by masking53. Throughout all refinements in Relion-2 (ref. 50), particles were grouped by k-mean clustering of their CTF parameters with a minim um of 50 particles per group. The alignment was not influenced by a preferred orientation54 (Supplementary Fig. 7b,c), which was assessed by reducing the number of particles per angular sampling group to the mean of all groups by random subset deletion in four independent refinements. Later refinements were carried out using a soft m ask and solvent-corrected FSC during the refinement53. Final maps were sharpened with a sharpening B factor of −350 Å2. Local resolution was calculated with ResMap55 and indicated higher resolution for secondary structure elements in the center of the particle (Supplementary Fig. 7d,e). Therefore, masked 3D classification and refinement after signal subtraction56 was carried out, resulting in slightly improved resolution but no substantial differences in the electron density map. Map generation from coordinates as well as filtering and file type conversion was done using EMAN2 tools49. For analysis, map transformations were applied using the CCP4 tool m aprot57. Maps were resam pled using UCSF Chimera58, and difference maps were calculated by Python Num py and M RCZ59.
Cryo-EM structure refinement and modelling.
Modelling of the cryo-EM structure was started by rigid body fitting of individual domains of the CTB1 SAT-KS-MAT crystal as well as of an ACP2 hom ology m odel in Coot43. The orientation of ACP2 could be unambiguously determined based on steric requirements of the Ppant attachment site (DSL motif), a continuously connected density between ACP2 helix II and the KS active site entrance, and additional density features which can be explained by loop regions of the homology model. The model for CTB1 ACP2 was generated using SwissModel60 based on PksA ACP29, which shares 40% sequence identity and can be functionally replaced with the CTB1 tandem ACPs in in vitro assays7,10. Initial B factors of the model were scaled relative to B factors of atomic displacement parameter (ADP) refined crystal structures in the PDB at similar resolution. Prior to m ap-based real-space refinement, the model was protonated using phenix.reduce and geometry minimization with secondary structure restraints and reference coordinate restraints was applied using phenix.geometry_minimization39. Afterwards global minimization and ADP refinement with reference structure restraint was carried out against the unsharpened cryo-EM map in phenix.real_space_refine39. Reference structure and nonbonded distance restraint weights were carefully monitored and optimized. As the local resolution around the resolved ACP2 is not sufficient to unam biguously interpret possible conformational changes of loop regions relative to the isolated PksA ACP NM R structure, the loop regions were excluded from the final model after all atom refinement, without affecting model statistics (Supplementary Table 1b). The resulting model contained the regions corresponding to the canonical ACP four helix bundle (residues: 1782–1796, 1816–1828, 1839–1842, 1846–1854) and was validated using phenix.m olprobity39 with Ram achandran plot statistics of “favored/outliers”: 96.47%/0.12%. A position for a second crosslinked ACP2 could not be undoubtedly resolved in any intermediate or final EM map. The sample has been analyzed after grid preparation and is > 90% crosslinked (Supplementary Fig. 6f).
Due to residual noise around the putative second ACP2 binding site, the presence of a second ACP2 in a mostly disordered state without a solid interface, tethered only by covalent crosslinking (Supplementary Fig. 8a,b), cannot be strictly excluded. However, we cannot exclude the possibility that effects of selective adsorption of partially or fully crosslinked SAT°-KS-MAT° = ACP2 or those of selective destabilization under grid preparation also contribute to selective imaging of a single-crosslinked state with low population in the sample used for grid preparation.
Structure analysis and visualization.
Related structures were identified using PDBeFold (20% query/10% target)61, and interfaces were analyzed using QtPISA v2.1.0 (ref. 57). Transformations and coordinate m anipulations were carried out using CCP4 tools and M OLEM AN62. Bias-removal for crystallographic Fo−Fc omit maps was achieved by applying a random perturbation to coordinates (A0.2 A) and B factors (△20% of the mean overall B factor) with MOLEMAN2 (ref. 62) before refinement. Cα−r.m.s. deviation between domains of the crystal and cryo-EM structure were calculated using LSQMAN63. Structures of PikAIII were modeled according to Dutta et al.12 using Chim era58. Solvent-accessible surface distances (SAS) between Ca atoms of ACP phosphopantetheinylation and KS active site residues were calculated using Xwalk64. Figures, movies and plots were generated using Pymol (Schrodinger, v1.8.6.0) and Python.
Sequence analysis.
Sequence alignments were generated using Clustal Omega65. A Phylogenetic tree was generated using the neighboring joining algorithm in Geneious (Biomatters Limited, v8.1.6).
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
Code availability.
Custom computer code is available from the corresponding author upon reasonable request.
Data availability.
Atomic coordinates, structure factors for the reported crystal structure, and cryo-EM map have been deposited with the Protein Data Bank under accession codes 6FIJ, 6FIK and EMDB accession code EM D-4266.
Supplementary Material
Acknowledgements
We thank P. Leadlay and L. Betancor for providing plasmid pETcoco-2A-L1SL2. X-ray diffraction data were collected at beamline PXI of the Paul Scherrer Institute, Villigen, Switzerland, and cryo-EM data were collected at the BioEM facility of the University of Basel; we acknowledge excellent support by teams of both facilities. This work was supported by the Swiss National Science Foundation (SNF) project grants 138262, 159696, SNF Requip grants 145023 and 164074, and the National Institutes of Health (ES001670). D.A.H. acknowledges a fellowship from the Werner-Siemens Foundation.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41589-018-0026-3.
Reprints and permissions information is available at www.nature.com/reprints.
Correspondence and requests for materials should be addressed to T.M.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hertweck C The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Edn. Engl 48, 4688–4716 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Herbst DA, Jakob RP, Zahringer F & Maier T Mycocerosic acid synthase exemplifies the architecture of reducing polyketide synthases. Nature 531, 533–537 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Weissman KJ The structural biology of biosynthetic megaenzymes. Nat. Chem. Biol 11, 660–670 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Tang Y, Kim CY, M athews II, Cane DE & Khosla C The 2.7-angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl Acad. Sci. USA 103, 11124–11129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maier T, Leibundgut M & Ban N The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Crawford JM, Vagstad AL, Whitworth KP, Ehrlich KC & Townsend CA Synthetic strategy of nonreducing iterative polyketide synthases and the origin of the classical “starter-unit effect”. ChemBioChem 9, 1019–1023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vagstad AL et al. Combinatorial domain swaps provide insights into the rules of fungal polyketide synthase program m ing and the rational synthesis of non-native aromatic products. Angew. Chem. Int. Edn. Engl 52, 1718–1721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foulke-Abel J & Townsend CA Demonstration of starter unit interprotein transfer from a fatty acid synthase to a multidomain, nonreducing polyketide synthase. ChemBioChem 13, 1880–1884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford JM, Dancy BC, Hill EA, Udwary DW & Townsend CA Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc. Natl Acad. Sci. USA 103, 16728–16733 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newm an AG, Vagstad AL, Storm PA & Townsend CA Systematic dom ain swaps of iterative, nonreducing polyketide synthases provide a mechanistic understanding and rationale for catalytic reprogramming. J. Am. Chem. Soc 136, 7348–7362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii I, Watanabe A, Sankawa U & Ebizuka Y Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem. Biol 8, 189–197 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Dutta S et al. Structure of a modular polyketide synthase. Nature 510, 512–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Chen AY, Kim CY, Cane DE & Khosla C Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem. Biol 14, 931–943 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whicher JR et al. Cyanobacterial polyketide synthase docking domains: a tool for engineering natural product biosynthesis. Chem. Biol 20, 1340–1351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pappenberger G et al. Structure of the hum an fatty acid synthase KS-MAT didomain as a framework for inhibitor design. J. Mol. Biol 397, 508–519 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Vagstad AL, Bumpus SB, Belecki K, Kelleher NL & Townsend CA Interrogation of global active site occupancy of a fungal iterative polyketide synthase reveals strategies for maintaining biosynthetic fidelity. J. Am. Chem. Soc 134, 6865–6877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawford JM. et al. Deconstruction of iterative multidomain polyketide synthase function. Science 320, 243–246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter JM. et al. Biochemical and structural basis for controlling chemical modularity in fungal polyketide biosynthesis. J. Am. Chem. Soc 137, 9885–9893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, Chiang YM, Somoza AD, Oakley BR & Wang CC Engineering of an “unnatural” natural product by swapping polyketide synthase domains in Aspergillus nidulans. J. Am. Chem. Soc 133, 13314–13316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruegger J et al. Probing the selectivity and protein•protein interactions of a nonreducing fungal polyketide synthase using mechanism -based crosslinkers. Chem. Biol 20, 1135–1146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choquer M. et al. The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae. Mol. Plant Microbe Interact 18, 468–476 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Chung KR, Ehrenshaft M, Wetzel DK & Daub ME Cercosporin-deficient mutants by plasmid tagging in the asexual fungus Cercospora nicotianae. Mol. Genet. Genomics 270, 103–113 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Daub ME & Ehrenshaft M The photoactivated cercospora toxin cercosporin: contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol 38, 461–490 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Newman AG, Vagstad AL, Belecki K, Scheerer JR & Townsend CA Analysis of the cercosporin polyketide synthase CTB1 reveals a new fungal thioesterase function. Chem. Commun. (Camb.) 48, 11772–11774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen C et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 505, 427–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whicher JR et al. Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature 510, 560–564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapur S et al. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation. Proc. Natl Acad. Sci. USA 109, 4110–4115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F et al. Structural and functional analysis of the loading acyltransferase from avermectin modular polyketide synthase. ACS Chem. Biol 10, 1017–1025 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Wattana-amorn P et al. Solution structure of an acyl carrier protein domain from a fungal type I polyketide synthase. Biochemistry 49, 2186–2193 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Leibundgut M, Jenni S, Frick C & Ban N Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science 316, 288–290 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Changeux JP & Edelstein S Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep 3, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brignole EJ, Smith S & Asturias FJ Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat. Struct. Mol. Biol 16, 190–197 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier T, Jenni S & Ban N Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science 311, 1258–1262 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Zhang L et al. Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic m echanism of dehydratase in fatty acid biosynthesis. Cell Res 26, 1330–1344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C & Stroud RM An antibiotic factory caught in action. Nat. Struct. Mol. Biol 11, 888–893 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Betancor L, Fernandez MJ, W eissman KJ & Leadlay PF Improved catalytic activity of a purified multienzym e from a modular polyketide synthase after coexpression with Streptomyces chaperonins in Escherichia coli. ChemBioChem 9, 2962–2966 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD et al. PHENIX: a comprehensive Python-based system form acrom olecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, (213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowtan K The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Cowtan K Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P & Cowtan K Coot: m odel-building tools for molecular graphics. Acta Crystallogr. D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Edwards AL, Matsui T, Weiss TM. & Khosla C Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J. Mol. Biol 426, 2229–2245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asturias FJ et al. Structure and molecular organization of m am Malian fatty acid synthase. Nat. Struct. Mol. Biol 12, 225–232 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Li X et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLeod RA, Kowal J, Ringler P & Stahlberg H Robust image alignment for cryogenic transmission electron microscopy. J. Struct. Biol 197, 279–293 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Kimanius D, Forsberg BO, Scheres SH & Lindahl E Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Chen S et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determ ination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urnavicius L et al. The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bai XC, Rajendra E, Yang G, Shi Y & Scheres SH Sampling the conformational space of the catalytic subunit of hum any-secretase. eLife 4, e11182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Pettersen EF et al. UCSF C him era-a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 59.McLeod RA, Diogo Righetto R, Stewart A & Stahlberg H M RCZ - A file format for cryo-TEM data with fast com pression. J. Struct. Biol 201, 252–257 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Schwede T, Kopp J, Guex N & Peitsch MC SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31, 3381–3385 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krissinel E & Henrick K Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Kleywegt GJ Validation of protein models from Ca coordinates alone. J. Mol. Biol 273, 371–376 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Kleywegt GJ Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr 52, 842–857 (1996). [DOI] [PubMed] [Google Scholar]
- 64.Kahraman A, Malmstrom, L. & Aebersold, R. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics 27, 2163–2164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates, structure factors for the reported crystal structure, and cryo-EM map have been deposited with the Protein Data Bank under accession codes 6FIJ, 6FIK and EMDB accession code EM D-4266.