Abstract
OBJECTIVE
We evaluated whether the increasing number of genetic loci for coronary artery disease (CAD) identified in the general population could be used to predict the risk of major CAD events (MCE) among participants with type 2 diabetes at high cardiovascular risk.
RESEARCH DESIGN AND METHODS
A weighted genetic risk score (GRS) derived from 204 variants representative of all the 160 CAD loci identified in the general population as of December 2017 was calculated in 5,360 and 1,931 white participants in the Action to Control Cardiovascular Risk in Diabetes (ACCORD) and Outcome Reduction With Initial Glargine Intervention (ORIGIN) studies, respectively. The association between GRS and MCE (combining fatal CAD events, nonfatal myocardial infarction, and unstable angina) was assessed by Cox proportional hazards regression.
RESULTS
The GRS was associated with MCE risk in both ACCORD and ORIGIN (hazard ratio [HR] per SD 1.27, 95% CI 1.18–1.37, P = 4 × 10−10, and HR per SD 1.35, 95% CI 1.16–1.58, P = 2 × 10−4, respectively). This association was independent from interventions tested in the trials and persisted, though attenuated, after adjustment for classic cardiovascular risk predictors. Adding the GRS to clinical predictors improved incident MCE risk classification (relative integrated discrimination improvement +8%, P = 7 × 10−4). The performance of this GRS was superior to that of GRS based on the smaller number of CAD loci available in previous years.
CONCLUSIONS
When combined into a GRS, CAD loci identified in the general population are associated with CAD also in type 2 diabetes. This GRS provides a significant improvement in the ability to correctly predict future MCE, which may increase further with the discovery of new CAD loci.
Introduction
The risk of coronary artery disease (CAD) is between two and five times higher in patients with type 2 diabetes than in the general population, making CAD the most common long-term complication of type 2 diabetes (1,2). Given the millions of people affected by diabetes in the U.S. and worldwide, the great burden of increased morbidity and mortality associated with CAD as well as the costs, financial and otherwise, of preventive programs (3,4), there is a crucial need for new predictive tools to identify patients with diabetes who are at especially high risk for this complication so that intensive prevention strategies can be targeted at them.
Several clinical characteristics such as age, male sex, hypertension, dyslipidemia, and smoking are known risk factors for CAD in subjects both with and without diabetes (5–7). Genetic factors are also known to contribute to CAD, and 160 different loci associated with increased CAD risk have been identified to date in the general population through large genome-wide association studies (GWAS) involving hundreds of thousands of individuals (8–18). Whether these loci are associated with CAD and/or predict incident major CAD events (MCE) in people who are, on average, at high cardiovascular risk, such as patients with diabetes, is unknown at this time. Also unclear is whether these genetic variants may have clinical utility in identifying individuals with diabetes at especially high MCE risk who may benefit from special preventive interventions. A previous report suggested minimal improvements in CAD prediction by the addition of five CAD-associated single nucleotide polymorphisms (SNPs) to a clinical prediction model (19), but whether the outcome may be different with the current larger pool of CAD loci deserves investigation, especially in consideration of the possible interactions between the diabetic milieu and genetic variants on CAD risk (20,21).
In this study, we examined the association between the currently available genetic markers and CAD in people with type 2 diabetes. We also evaluated the extent to which these genetic markers improve the prediction of incident MCE when added to conventional cardiovascular risk factors and assessed whether a genetic risk score (GRS) derived from these SNPs could identify subsets of patients with type 2 diabetes with different responses to intensive glucose, blood pressure, and lipid control in the Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial.
Research Design and Methods
Study Subjects
The ACCORD trial was aimed at evaluating whether intensive treatments targeting hyperglycemia, elevated blood pressure, and dyslipidemia could reduce the risk of major adverse cardiovascular events among patients with high-risk type 2 diabetes (22). A total of 10,251 participants with type 2 diabetes were recruited from seven clinical networks in the U.S. and Canada between 2001 and 2005 and randomized in a 1:1 ratio to receive intensive glucose-lowering treatment, aiming for an HbA1c level below 6%, or to standard treatment, aiming for an HbA1c level between 7.0% and 7.9%. Half of the participants were additionally randomized to a subtrial of intensive versus standard antihypertensive therapy (ACCORD-BP) (23) and the other half to a subtrial of fenofibrate + simvastatin versus simvastatin alone (ACCORD-Lipid) (24). All study participants provided written informed consent according to local regulations. The present cohort study was limited to 5,360 self-reported white participants who provided consent to genetic studies.
The Outcome Reduction With Initial Glargine Intervention (ORIGIN) trial (25), conducted worldwide between 2003 and 2011, investigated the effect of titrated basal insulin versus standard care and of n-3 fatty acid supplements versus placebo on major cardiovascular events occurrence among 12,537 participants with impaired fasting glucose, impaired glucose tolerance or type 2 diabetes, and high cardiovascular risk. After a median follow-up time of 6.2 years, the study reported lack of significant cardiovascular effect of these two treatments. The current study was limited to 1,931 self-reported white participants who provided consent for genetic studies.
Outcomes
Baseline history of CAD was defined as the presence of positive history for myocardial infarction, coronary revascularization procedures, angina, and/or documented ischemic changes on a graded exercise tolerance test or cardiac imaging. Incident MCE—one of the prespecified secondary outcomes of the ACCORD trial (22)—were defined as fatal CAD events, nonfatal myocardial infarction, or unstable angina occurring during the trial. In the ORIGIN trial, MCE were also defined as fatal and nonfatal myocardial infarction or unstable angina occurring during the study.
Genotyping and Quality Control
Two-hundred and four independent SNPs were selected for their genome-wide significant association with CAD in GWAS and exome-wide association studies published as of December 2017, representing 160 known CAD loci (8–18) (Supplementary Table 1). The ADTRP-C6orf105 locus was not included in the analysis since it was detected only in Han Chinese (26) and did not harbor variants associated with CAD at GWAS significant levels in any of the more recent studies (8–18). When multiple SNPs had been reported to be associated with CAD at a given locus (defined as a 1-Mb window on either side), the SNP with the highest significant association with CAD reported in literature (sentinel SNP) was selected first, followed by any other SNP at that locus (independent SNP) that was not in linkage disequilibrium (R2 < 0.2) with the sentinel SNP. Linkage disequilibrium between SNPs was assessed with the LDlink tool (https://analysistools.nci.nih.gov/LDlink/?tab=home) based on genotyped data originating from phase 3 (version 5) of the 1000 Genomes Project among subjects with European ancestry.
Since these CAD loci were discovered in populations in which the majority of participants were of European ancestry (8–18), the current study was limited to self-reported white participants with genetic data.
For ACCORD, genotypes for the 204 SNPs were derived from the study’s GWAS data set, the generation of which has been described in detail elsewhere (27). Sixty-seven of the 204 SNPs considered in the study were genotyped and the remaining 137 imputed at high quality (median info score 0.98, interquartile range 0.95–0.99). For ORIGIN, genotypes were derived from a GWAS data set generated by means of the HumanCoreExome BeadChip 12 v1.0 and v1.1 (Illumina), which evaluated 551,839 markers and allowed the imputation of 30 million SNPs using Impute v2.3.1. Sixty-four of the 204 SNPs were genotyped and 138 were imputed at high quality (median info score 0.96, interquartile range 0.88–0.99). Two of the 204 SNPs (rs112470402 and rs507666) were not available in the genetic data set (neither were proxies).
Statistical Analysis
Single-SNP Analyses
The association between each SNP and history of CAD was tested in ACCORD by logistic regression using an additive genetic model adjusted for age, sex, clinical network, and principal components of population structure. To account for multiple comparisons, study-wide significance was defined according to a false discovery rate <0.05. Post hoc power calculations (Quanto v1.2.4) showed that depending on the effect allele frequency, single SNP analyses were powered (1 – β = 80%) to detect a study-wide significant association (α = 2 × 10−4) with history of CAD with an odds ratio [OR] ≥1.21, ≥1.36, and ≥2.48 for SNPs with effect allele frequency of 50%, 10%, and 1%, respectively. The corresponding figures to detect effects at the nominal significance level (α = 0.05) were OR ≥1.13, ≥1.22, and ≥1.76. The number of SNPs showing the same effect direction as that reported in the literature was computed and compared with the binomial distribution of the effect directions expected on the basis of 204 trials conducted under the null hypothesis of equal chances of the effect going in either direction.
GRS Analysis
A weighted GRS was derived from the genotypes by adding the numbers of risk alleles at each variant after weighing them for the effect magnitudes reported for each variant in the literature. In ACCORD a 204-SNP–derived GRS was used, while in ORIGIN a 202-SNP–derived GRS was used (since 2 SNPs were not available in ORIGIN genetic data set). To aid interpretation of the GRS, the GRS were standardized to a mean of 0 and an SD of 1.
The association of the GRS with history of CAD was tested by logistic regression analysis adjusting for age, sex, clinical network, and principal components of population structure. The association between GRS and MCE during follow-up was evaluated by means of Cox proportional hazards regression. Four different regression models of increasing complexity were tested: model 1 including GRS, age, sex, and ACCORD study covariates (assignment to randomized treatment arms, clinical network, genotyping platform, and principal components of population structure); model 2 with further adjustment for history of CAD at baseline; model 3 with further adjustment for the 10-year American College of Cardiology (ACC)/American Heart Association (AHA) atherosclerotic cardiovascular disease (ASCVD) risk estimator (6) (which includes total and HDL cholesterol, smoking habits, systolic blood pressure, hypertension treatment, and history of diabetes); model 4 with further adjustments for duration of diabetes, HbA1c levels at baseline, and family history of cardiovascular disease. The assumption of proportionality of the hazards was formally tested by testing the significance of time × predictor interaction terms added to the models. The assumption was respected in all cases.
Kaplan-Meier curves were generated to illustrate the association between GRS tertile and incidence of MCE. Influences of clinical characteristics at baseline (age, sex, history of CAD, family history of CAD, median duration of diabetes and of HbA1c levels at baseline) or of different treatments during the trial on the relationship between GRS and incident MCE were tested by means of Cox proportional hazards models including the clinical characteristic or intervention, the GRS, and a “GRS × clinical characteristic/intervention” interaction term.
Power calculations showed that the GRS was sufficiently powered (1 – β = 80%) to detect a significant association (α = 0.05) with incident MCE with a hazard ratio (HR) per SD increment as small as 1.13. Analyses of “GRS × treatment” interaction had 80% power (α = 0.05) to detect coefficients of interaction with glycemic, blood pressure, and fibrate interventions ≥0.24, ≥0.36, and ≥0.32, respectively (which would be in the range of “gene × treatment” interactions estimates detected in previous study, e.g., 0.27 for gene × glycemic intervention on nonfatal myocardial infarction or 0.33 for gene × fenofibrate interaction on major cardiovascular events [27,28]).
Replication in ORIGIN Study
Validation of the association between GRS and MCE was sought by means of Cox proportional hazards regression in the ORIGIN study. The same models tested in ACCORD were evaluated in ORIGIN: model 1, including GRS, age, sex, and ORIGIN study covariates (assignment to randomized treatment arms and principal components of population structure); model 2, with further adjustment for history of CAD at baseline; and model 3, with further adjustment for the 10-year ACC/AHA ASCVD risk estimator. Model 4 was not tested since information on family history of cardiovascular disease was not available in ORIGIN. Power analyses showed that the GRS was powered (1 – β = 80%) to detect a significant association (α = 0.05) with incident MCE event with an HR per SD increment as small as 1.26.
Model Performance for Predicting Risk of Incident MCE
Model performance in terms of MCE prediction was compared in ACCORD across three models: model 1, a genetic model including the GRS alone; model 2, a clinical model based on conventional clinical predictors of CAD and including history of CAD, age, sex, the ACC/AHA ASCVD risk estimator, and the ACCORD study covariates; and model 3, a clinical + genetic model in which the GRS was added to the clinical model. Receiver operating characteristic (ROC) curves, plotting sensitivity against 1 – specificity, were constructed for all possible cutoffs for the three models. The area under the ROC curve (AUC), or C-statistic, was estimated as a measure of the probability that a randomly selected individual who experienced an event had a higher predicted risk than an event-free person. As alternative measures of risk discrimination, we also calculated the relative integrated discrimination improvement (rIDI) (29), which represents the increase in discrimination slopes provided by the new model relative to the old model, and the category-free net reclassification index (NRI) (29), which examines whether the predicted probabilities of individuals with and without events move in the right directions (upward and downward, respectively) from the old to the new model. The analyses testing the improvement in predictive performance were computed using logistic regression models with methods developed by Kennedy and Pencina (30).
All the analyses were performed using the SAS software (version 9.4, SAS Institute Inc., Cary, NC), and all the tests were two-sided. Graphs were edited with GraphPad Prism version 7.2.
Results
Genetic Associations With Baseline Characteristics and Prevalent CAD
Baseline characteristics of the ACCORD participants included in this study are shown in Table 1. The distribution of the GRS (weighted and rescaled to a range from 0 to 408) showed that participants carried an average of 187.4 ± 8.1 risk alleles. GRS distributions were similar in the two glycemic treatment arms (P = 0.63). A higher GRS was associated with younger age, lower levels of plasma HDL cholesterol, and higher glomerular filtration rate (this difference was not significant after adjustment for age, P = 0.2). The GRS was also strongly associated with a positive family history of cardiovascular disease and a positive CAD history at study entry. Specifically, each GRS SD increment was associated with a 40% increase in the odds of a positive CAD history (OR 1.40, 95% CI 1.32–1.49, P = 3 × 10−27), with participants with a GRS in the third and second tertile having 101% (OR 2.01, 95% CI 1.73–2.37) and 35% (95% CI 1.16–1.57) higher odds of CAD, respectively, as compared with participants with GRS in the lowest tertile (Supplementary Fig. 1). In individual loci analysis, 32 SNPs were nominally associated with CAD, although none of them reached study-wide significance (Supplementary Table 2). Overall, 74% of the SNPs included in the study (151/204, P value for deviation from 50% = 2 × 10−12) showed a trend for an association with prevalent CAD that went in the same direction as in the general population.
Table 1.
Characteristics of self-reported white participants with genetic data available included in the ACCORD trial
| Characteristic | All participants | GRS tertile |
|||
|---|---|---|---|---|---|
| Low risk | Intermediate risk | High risk | P | ||
|
N |
5,360 |
1,787 |
1,786 |
1,787 |
|
| Age (years) |
62.8 ± 6.5 |
63.2 ± 6.4 |
62.8 ± 6.5 |
62.3 ± 6.5 |
0.0003 |
| Female |
1,888 (35.2) |
626 (35.0) |
636 (35.6) |
626 (35.0) |
0.9 |
| Ever smoker |
3,348 (62.5) |
1,097 (61.4) |
1,138 (63.7) |
1,113 (62.3) |
0.3 |
| BMI (kg/m2) |
33.0 ± 5.2 |
33.1 ± 5.1 |
32.9 ± 5.2 |
32.9 ± 5.2 |
0.5 |
| Duration of diabetes (years) |
10.7 ± 7.5 |
10.7 ± 7.4 |
10.9 ± 7.6 |
10.5 ± 7.3 |
0.3 |
| Baseline HbA1c (%) |
8.2 ± 1.0 |
8.24 ± 0.95 |
8.19 ± 0.96 |
8.21 ± 0.95 |
0.4 |
| Fasting plasma glucose (mg/dL) |
178.5 ± 50.9 |
179.2 ± 52.0 |
178.3 ± 49.8 |
178.0 ± 50.9 |
0.8 |
| HDL cholesterol (mg/dL) |
40.3 ± 10.3 |
40.7 ± 10.4 |
40.5 ± 10.5 |
39.6 ± 9.9 |
0.005 |
| Total cholesterol (mg/dL) |
183.0 ± 40.0 |
182.8 ± 39.5 |
182.3 ± 40.1 |
183.9 ± 40.4 |
0.5 |
| LDL cholesterol (mg/dL) |
102.9 ± 32.3 |
102.4 ± 31.9 |
102.2 ± 32.2 |
104.2 ± 32.7 |
0.11 |
| Triglycerides (mg/dL) |
203.7 ± 122.3 |
203.2 ± 120.6 |
201.5 ± 121.0 |
206.4 ± 125.2 |
0.5 |
| Statin treatment |
3,425 (64.1) |
1,070 (60.0) |
1,147 (64.4) |
1,208 (67.9) |
<0.0001 |
| Systolic BP (mmHg) |
135.2 ± 16.5 |
135.4 ± 16.6 |
135.0 ± 16.4 |
135.2 ± 16.5 |
0.8 |
| Diastolic BP (mmHg) |
74.1 ± 10.2 |
74.3 ± 10.3 |
74.0 ± 10.2 |
74.0 ± 10.2 |
0.6 |
| BP medication |
4,419 (82.4) |
1,454 (81.4) |
1,477 (82.7) |
1,488 (83.3) |
0.3 |
| Creatinine (mg/dL) |
0.90 ± 0.22 |
0.91 ± 0.22 |
0.91 ± 0.22 |
0.90 ± 0.22 |
0.3 |
| Glomerular filtration rate (mL/min/1.7 m2) |
88.1 ± 21.6 |
87.4 ± 21.1 |
87.8 ± 21.6 |
89.2 ± 22.1 |
0.04 |
| Family history of CVD |
2,629 (50.6) |
826 (47.8) |
858 (49.5) |
945 (54.7) |
0.0001 |
| History of CAD |
1,643 (30.7) |
429 (24.0) |
531 (29.7) |
683 (38.2) |
<0.0001 |
| Rescaled weighted GRS |
187.4 ± 8.1 |
178.6 ± 4.1 |
187.4 ± 2.0 |
196.3 ± 4.4 |
N/A |
| Standardized GRS | 0.0 ± 1.0 | –1.1 ± 0.5 | 0.0 ± 0.2 | 1.1 ± 0.5 | N/A |
Data are mean ± SD or n (%) unless otherwise noted. To convert the values for glucose to millimoles per liter, multiply by 0.05551. To convert the values for cholesterol to millimoles per liter, multiply by 0.02586. To convert the values for triglycerides to millimoles per liter, multiply by 0.01129. To convert the values for creatinine to micromoles per liter, multiply by 88.4. P values for comparison across tertiles were calculated based on ANOVA or χ2 test. BP, blood pressure; CVD, cardiovascular disease; N/A, not applicable.
GRS and Risk of Incident Coronary Events
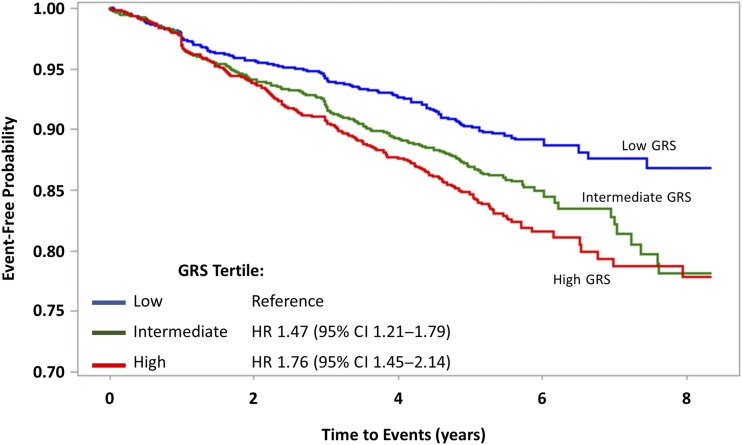
A total of 675 participants enrolled in the ACCORD study suffered one or more MCE over a median follow-up of 4.7 years. The GRS was significantly associated with the risk of experiencing one of these MCE (HR per SD increment 1.27, 95% CI 1.18–1.37, P = 4 × 10−10). As reported in Table 2 (and illustrated graphically by means of Kaplan-Meier curves in Fig. 1), participants with a GRS in the third tertile of the distribution had a 76% higher risk of experiencing an MCE event as compared with those having a GRS in the lowest tertile (HR 1.76, 95% CI 1.45–2.14). This association was attenuated, but still significant, after further adjustments for clinical predictors (Table 2). Similar results were also obtained when alternative cardiovascular outcomes of ACCORD (e.g., revascularization procedures) were considered (Supplementary Table 3). Exploratory analyses among nonwhite participants showed significant association of the GRS with MCE among 469 participants of Asian origin but not among self-reported African American or Hispanic participants (Supplementary Table 4).
Table 2.
GRS effect on MCE over follow-up in the ACCORD and ORIGIN studies
| Model 1 (N = 5,360) |
Model 2 (N = 5,360) |
Model 3 (N = 5,322) |
Model 4 (N = 5,104) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||
| ACCORD |
Per risk allele | 1.03 (1.02–1.04) | 4 × 10−10 | 1.02 (1.01–1.03) | 7 × 10−6 | 1.02 (1.01–1.03) | 3 × 10−5 | 1.02 (1.01–1.03) | 8 × 10−5 |
| Per SD | 1.27 (1.18–1.37) | 4 × 10−10 | 1.19 (1.10–1.28) | 7 × 10−6 | 1.18 (1.09–1.27) | 3 × 10−5 | 1.17 (1.08–1.27) | 8 × 10−5 | |
| By tertile of GRS | |||||||||
| Low risk | Ref. | Ref. | Ref. | Ref. | |||||
| Medium risk | 1.47 (1.21–1.79) | 2 × 10−4 | 1.39 (1.14–1.69) | 1 × 10−3 | 1.39 (1.14–1.69) | 1 × 10−3 | 1.38 (1.12–1.69) | 2 × 10−3 | |
| High risk |
1.76 (1.45–2.14) |
1 × 10−8 |
1.53 (1.26–1.86) |
2 × 10−5 |
1.49 (1.23–1.82) |
6 × 10−5 |
1.47 (1.20–1.80) |
2 × 10−4 |
|
| Model 1 (N = 1,931) |
Model 2 (N = 1,931) |
Model 3 (N = 1,931) |
Model 4 (N/A) |
||||||
| ORIGIN | Per risk allele | 1.04 (1.02–1.06) | 2 × 10−4 | 1.04 (1.02–1.06) | 3 × 10−4 | 1.04 (1.02–1.06) | 5 × 10−4 | N/A | |
| Per SD | 1.35 (1.16–1.58) | 2 × 10−4 | 1.33 (1.14–1.56) | 3 × 10−4 | 1.32 (1.13–1.55) | 5 × 10−4 | |||
| By tertile of GRS | |||||||||
| Low risk | Ref. | Ref. | Ref. | ||||||
| Medium risk | 1.55 (1.03–2.35) | 4 × 10−2 | 1.51 (0.99–2.28) | 5 × 10−2 | 1.51 (1.00–2.29) | 5 × 10−2 | N/A | ||
| High risk | 1.90 (1.27–2.83) | 2 × 10−3 | 1.84 (1.23–2.74) | 3 × 10−3 | 1.80 (1.21–2.69) | 4 × 10−3 | |||
Model 1, adjusted for study variables (assignment to study treatment arms and principal components of population structure) plus age and sex; model 2, model 1 covariates plus history of CAD; model 3, model 2 covariates plus ACC/AHA ASCVD score; model 4, model 3 covariates plus HbA1c, duration of diabetes, and family history of cardiovascular disease. N/A, not applicable.
Figure 1.
Kaplan-Meier curves for MCE according to genetic risk category.
Replication in the ORIGIN Study
The analysis of 1,931 participants in the ORIGIN trial, 163 of whom had one or more MCE during follow-up, confirmed the results in ACCORD. Each SD increment in GRS was associated with a 35% increase in MCE risk (95% CI 16–58), which was similar to the 27% increase in risk seen in ACCORD (P for difference between the two HRs = 0.5). Participants with a GRS in the third tertile of the distribution had 90% higher risk of experiencing an MCE event as compared with those having a GRS in the lowest tertile (HR 1.90, 95% CI 1.27–2.83), as shown in Table 2. Adjustment for clinical predictors modestly attenuated this association. Similar results were obtained with an expanded outcome including coronary revascularization procedures (276 total events) (Supplementary Table 5).
GRS × Environment Interactions
No significant differences were found in the effect of the GRS when the ACCORD study population was stratified according to age, sex, family history of cardiovascular disease, history of CAD, or duration of diabetes (Supplementary Fig. 2). A nominally significant larger effect of the GRS was observed among participants with baseline HbA1c levels above the median as compared with those with values below the median (P for interaction = 0.02). Such difference was not observed when participants were stratified based on their average on-trial HbA1c levels (Supplementary Table 6). Similarly, none of the ACCORD randomized interventions (i.e., intensive vs. standard glycemic control, fenofibrate + simvastatin vs. placebo + simvastatin, and intensive vs. standard blood pressure control) showed a significant interaction with the GRS on MCE (all P for interaction > 0.1). The same analyses, illustrated from a different perspective in Supplementary Fig. 3, showed that the treatments compared in the trial had similar effects on MCE across GRS tertiles. A sensitivity analysis excluding 219 participants with follow-up (time to event or to censoring) ≤1 year, in order to allow for sufficient time for the intervention to be modified by genetic background, provided similar results (all P for interaction > 0.4).
MCE Prediction Model Performance, Discrimination, and Calibration
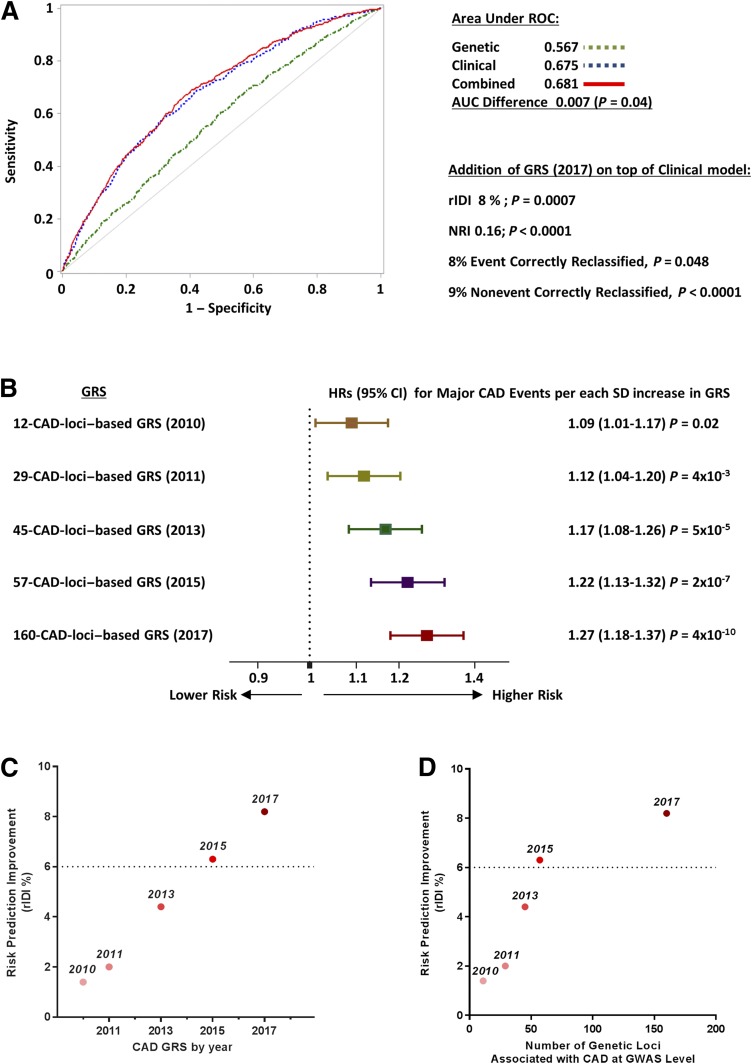
The AUC for a genetic prediction model of incident MCE including the GRS alone was 0.57 as compared with 0.67 for a clinical prediction model including history of CAD, 10-year ASCVD risk score, age, sex, and other ACCORD study covariates. Addition of GRS to the clinical model marginally improved the AUC (AUC difference +0.007, P = 0.04) (Fig. 2A). Despite the minimal increase in AUC, the clinical + genetic prediction model showed a substantial increase in the rIDI (rIDI +8%, P = 0.0007) along with an NRI of 0.16 (P < 0.0001), with 8% of events and 9% of nonevents correctly reclassified by the new model. Goodness-of-fit test (Hosmer and Lemeshow) showed adequate calibration for both clinical and clinical + genetic models (P = 0.4 and P = 0.2, respectively). Results were similar if the GRS was added to an expanded clinical prediction model that also included HbA1c levels, duration of diabetes, and family history of cardiovascular disease (AUC increase 0.007, P = 0.03; rIDI +7%, P = 0.002; NRI +13%, P = 0.002).
Figure 2.
A: ROC curves for the predictive performance of MCE using GRS (green), clinical predictors (blue), or the combination of them (red) in the ACCORD trial (5,322 subjects included in the analysis; 667 events). Clinical predictors include history of CAD, ASCVD risk score, age, sex, and ACCORD study covariates. B: Association with MCE of different GRS based on the increasing number of CAD SNPs. GRS are ordered from top to bottom in chronological order (years are reported in brackets), with an increasing number of CAD loci for each GRS. C and D: Improvement in discrimination for MCE with GRS from 2010 to 2017 (plotted against year of discovery in C and against the number of CAD loci included in each GRS in D). The y-axis shows the percent increase in rIDI when each GRS was added to the model including clinical predictors.
Comparison of the Performances of Different GRS With Increasing Number of CAD Loci
Finally, we tested whether the inclusion of the newly identified CAD loci and variants available in December 2017 and included in the 204-SNP–based GRS (representing 160 CAD loci) improved incident MCE prediction as compared with the GRS based on the smaller number of CAD variants available in 2010 (13-SNP–based GRS, representing 12 CAD loci), 2011 (30-SNP–based GRS, representing 29 CAD loci), 2013 (53-SNP–based GRS, representing 45 CAD loci), or 2015 (70-SNP–based GRS, representing 57 CAD loci). The variants included in each of these GRS are listed in Supplementary Table 1. As shown in Fig. 2B, the increasing number of CAD loci and variants from 2010 to 2017 was paralleled by an increasingly stronger association between GRS and MCE. Whereas the 12-CAD-loci GRS from 2010 was associated with a 9% increase (95% CI 1–17) in the odds of MCE per SD, the corresponding value for the 160-CAD-loci GRS from 2017 was 27% (95% CI 18–37) (P for differences between HRs = 0.002). Consistent with this, there was a linear improvement from 2010 to 2017 in the GRS discrimination of participants experiencing MCE from those with no events during follow-up (Fig. 2C and Supplementary Fig. 4). However, when the improvement in discrimination was plotted against the number of CAD loci included in each GRS (Fig. 2D), a tendency toward a plateau in this relationship was observed, indicating that the improvement in discrimination observed during the past 2 years required the addition of a much larger number of new CAD loci than in previous years.
Conclusions
Our understanding of the genetic architecture of CAD has been constantly improving during the past decade in parallel with the identification of an increasing number of genetic loci associated with this multifactorial disease. In the general population, the number of genetic loci harboring common variants (i.e., having minor allele frequency ≥1%) that are robustly associated with CAD has increased from 12 in 2010 to 160 in 2017 (8–18). In this study, we found that a GRS capturing the information provided by all the CAD loci identified to date in the general population was strongly associated with the prevalence and incidence of CAD also among subjects with type 2 diabetes. This association was independent of classical clinical cardiovascular risk factors, family history of cardiovascular disease, previous history of CAD, and duration of diabetes and did not influence the effectiveness of interventions aimed at decreasing cardiovascular morbidity such as intensive glycemic and blood pressure control and lipid-lowering treatment with fenofibrate. Interestingly, while clinical risk factors clearly outperformed the GRS as predictors of incident MCE, addition of the GRS to a clinical prediction model provided a modest but significant improvement in the discrimination of participants who developed events from those who did not. Such improvement depended on the number of loci captured by the GRS, with the most updated list of CAD loci clearly outperforming GRS based on the smaller number of CAD loci available in previous years.
These results have several important implications. First, they confirm the overall transferability of CAD loci from the general to the population with type 2 diabetes. Few studies have previously investigated this concept. In 2011, Qi et al. (19) reported that a GRS based on five CAD loci from the general population was significantly associated with prevalent or incident CAD among subjects with type 2 diabetes. Similarly, in the Diabetes Heart Study, a GRS based on 28 CAD loci was associated with history of cardiovascular disease and with levels of coronary artery calcification (31). More recently, a GRS based on 153 SNPs associated with CAD at genome-wide significant levels or surviving a 5% false discovery rate in the general population (8) was found to be significantly associated with incident cardiovascular events (including stroke) in the Look AHEAD trial (32). Taken together, these findings from the ACCORD and ORIGIN studies indicate that the genetic loci that influence CAD risk in the general population generally do so also in the presence of diabetes. In other words, the high cardiovascular risk conferred by the exposure to the diabetic milieu does not appear to override the genetic factors influencing CAD risk in the general population. Thus, any future therapeutic advancements that may stem from the study of the mechanistic links between these 160 loci and atherosclerosis are likely to also benefit patients with type 2 diabetes. This is in addition to the benefits that may arise from the study of CAD loci that are specific to type 2 diabetes such as GLUL (20).
Second, our findings indicate that the information provided by the CAD loci included in the GRS can be used to improve the identification of individuals with diabetes at high risk of MCE events. While this improvement may appear modest (+0.01 increase in AUC, 8% increase in rIDI, and 8% of events and 9% of nonevents correctly reclassified by the continuous NRI), it is substantial in the context of the thresholds commonly used to add new biomarkers to established prediction tools (33,34). For instance, during the development of the widely used ACC/AHA pooled cohort equations to assess ASCVD risk (6), new biomarkers were deemed to be worth their addition to the equation based on an rIDI improvement of 6% or more—a criterion that would have been met by the GRS described in the current study. It is also interesting to note that, as observed in the general population (35–37), the improvement in risk discrimination provided by the GRS has been increasing over the years with the addition of new CAD loci, despite the increasingly smaller effects of the newly identified loci. We can therefore reasonably expect further prediction improvements deriving from the identification of additional CAD loci through the larger studies that are currently under way (38). While our data suggest that the improvement in discrimination provided by each newly discovered locus is progressively declining, the greater power of the current studies may compensate for this by increasing the rate of discovery of new CAD loci. Better prediction models will help maximize cost-effectiveness, by directing preventive interventions to high-risk subjects, and may also enhance the power of clinical trials, by allowing the selection of individuals with a high likelihood to experience an event during the study (39). At the clinical level, disclosure and discussion of information about genetic risk with patients could lead to better shared decisions on treatment and higher adherence to them (40).
While the GRS described in this article may help identify patients at high risk of MCE, it does not seem to be useful for personalizing their treatment, at least with regard to the interventions investigated in ACCORD. In this subcohort of the trial (including only white participants who provided genetic consent, i.e., ∼80% of all self-reported white participants), intensive glycemic control and fenofibrate treatment were associated with 15% and 19% reductions in MCE risk, respectively (HR 0.85, 95% CI 0.73–0.97, P = 0.04, and HR 0.82, 95% CI 0.67–1.00, P = 0.05). These effects were similar across GRS classes, suggesting that the genetic heterogeneity captured by the CAD-GRS does not generally overlap with the heterogeneity observed in the response to these treatments. Consistent with this tenet is the fact that the two loci influencing the cardiovascular response to intensive glycemic control in ACCORD that were recently identified by our group (27) are not among the 160 CAD loci discovered to date. Nonetheless, this does not exclude the possibility that variants at some of these loci, in particular those involved in the pathways directly targeted by treatments, may individually influence treatment outcomes. An example of this is the common gain-of-function mutation in the LPL gene coding for lipoprotein lipase (an established CAD locus), which we have recently found to influence the cardiovascular response to fenofibrate (28), a PPAR-α agonist with known effects on LPL expression and lipoprotein lipase activity (41).
This study, conducted in the rigorous clinical trial setting of ACCORD and ORIGIN, with precise definitions and classifications of CAD and MCE, is one of the largest efforts to examine the association between a GRS derived from the general population and coronary events in patients with type 2 diabetes. However, the results of this study should be considered in the context of its potential limitations. One concerns the exclusive focus on non-Hispanic whites and on genetic loci discovered in this ethnic group due to the relatively small number of nonwhites in ACCORD. As the genetic architecture of CAD and linkage disequilibrium patterns may differ across races, one cannot extrapolate these findings to other racial or ethnic groups. A second limitation concerns the possibility of a false negative result regarding the GRS × treatment interaction due to the fact that gene × treatment interactions on CAD outcomes may be easier to detect in primary prevention studies, as suggested by previous reports concerning statins (42,43). Since more than one-third of ACCORD participants were in secondary prevention and all others had multiple cardiovascular risk factors, one cannot exclude an influence of the GRS on treatment responses among subjects with lower cardiovascular risk. For these reasons, our results appear to be generalizable to self-reported white subjects with type 2 diabetes at high cardiovascular risk, as confirmed also by the replication of these findings in the ORIGIN study. Finally, despite the relatively large sample size, we had limited power to analyze the effects of individual SNPs. Thus, even if the association of the GRS with CAD and MCE was highly significant, we could draw limited conclusions about the transferability of individual CAD loci from the general population to type 2 diabetes.
Conclusion
In conclusion, a GRS combining information from the 160 CAD loci identified in the general population as of December 2017 was strongly associated with CAD history and MCE incidence in participants with type 2 diabetes from the ACCORD and ORIGIN studies. This GRS provided a significant improvement in the performance of a prediction model based on clinical risk factors. Given the trend in GRS performance observed from 2010 to 2017 as well as the exponential increase in the size of genetic studies, we can reasonably expect that the discovery of additional CAD loci in coming years will further improve our ability to identify patients with type 2 diabetes at especially high cardiovascular risk and will provide novel insights into the genetic heterogeneity of CAD among subjects with and without diabetes.
Supplementary Material
Article Information
Acknowledgments. The authors thank the investigators, staff, and participants of the ACCORD study for their support and contributions and for giving us access to this rich data set.
Funding and Duality of Interest. The ACCORD genome-wide association analysis was supported by National Institutes of Health (NIH) National Heart, Lung, and Blood Institute grants R01-HL110400 (to A.D.) and R01-HL110380 (to J.B.B.) as well as National Institute of Diabetes and Digestive and Kidney Diseases grant P30-DK36836 (through the Advanced Genomics and Genetics Core of the Diabetes Research Center at the Joslin Diabetes Center). The project described was also supported by the NIH National Center for Advancing Translational Sciences through grant award number UL1TR001111. J.B.B. was also supported by the NIH National Center for Advancing Translational Sciences through grant award number UL1TR001111. R.J.S. was supported by a Health Senior Scholar award from Alberta Innovates – Health Solutions. H.C.G. is supported by the McMaster-Sanofi Population Health Institute Chair in Diabetes Research and Care. The ACCORD study (ClinicalTrials.gov identifier NCT00000620) was supported by National Heart, Lung, and Blood Institute contracts N01-HC-95178, N01-HC-95179, N01-HC-95180, N01-HC-95181, N01-HC-95182, N01-HC-95183, and N01-HC-95184 as well as IAA #Y1-HC-9035 and IAA #Y1-HC-1010. Other components of the NIH, including the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute on Aging, and the National Eye Institute, contributed funding. The Centers for Disease Control and Prevention funded substudies within ACCORD on cost-effectiveness and health-related quality of life. General Clinical Research Centers and Clinical and Translational Science Awards provided support at many sites. The ORIGIN study (ClinicalTrials.gov identifier NCT00069784) was funded by Sanofi. No other potential conflicts of interest relevant to this article were reported.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funders.
Author Contributions. M.L.M., H.G., H.S.S., and A.D. contributed to study conception and design; acquisition, analysis, and interpretation of data; and drafting and final approval of the manuscript. M.P., J.S., C.M., T.H., P.B., A.A.M.-R., D.M.R., and S.M.M. contributed to acquisition of data and revising and final approval of the manuscript. R.J.S., P.K., J.B.B., M.J.W., H.C.G., J.C.M., and G.P. contributed to study conception and design, interpretation of data, and revising and final approval of the manuscript. A.D. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data Availability. The ACCORD database is available upon request from the National Heart, Lung, and Blood Institute Biologic Specimen and Data Repository (https://biolincc.nhlbi.nih.gov/studies/accord/). The ACCORD GWAS data have been deposited in the database of Genotypes and Phenotypes (dbGAP, Study Accession phs001411.v1.p1).
Prior Presentation. Parts of this study were presented in abstract form at the 77th Scientific Sessions of the American Diabetes Association, 9–13 June 2017, San Diego, CA.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc18-0709/-/DC1.
References
- 1.Kannel WB, McGee DL. Diabetes and cardiovascular risk factors: the Framingham study. Circulation 1979;59:8–13 [DOI] [PubMed] [Google Scholar]
- 2.Sarwar N, Gao P, Seshasai SR, et al.; Emerging Risk Factors Collaboration . Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies [published correction appears in Lancet 2010;376:958]. Lancet 2010;375:2215–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho NH, Shaw JE, Karuranga S, et al. IDF Diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract 2018;138:271–281 [DOI] [PubMed] [Google Scholar]
- 4.Booth GL, Bishara P, Lipscombe LL, et al. Universal drug coverage and socioeconomic disparities in major diabetes outcomes. Diabetes Care 2012;35:2257–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation 1998;97:1837–1847 [DOI] [PubMed] [Google Scholar]
- 6.Goff DC Jr, Lloyd-Jones DM, Bennett G, et al.; American College of Cardiology/American Heart Association Task Force on Practice Guidelines . 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines [published correction appears in J Am Coll Cardiol 2014;63:3026]. J Am Coll Cardiol 2014;63:2935–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American Diabetes Association Cardiovascular disease and risk management. Sec. 9. In Standards of Medical Care in Diabetes—2017. Diabetes Care 2017;40(Suppl. 1):S75–S87 [DOI] [PubMed] [Google Scholar]
- 8.CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stitziel NO; Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators . Variants in ANGPTL4 and the risk of coronary artery disease. N Engl J Med 2016;375:2306. [DOI] [PubMed] [Google Scholar]
- 10.Verweij N, Eppinga RN, Hagemeijer Y, van der Harst P. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci Rep 2017;7:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deloukas P, Kanoni S, Willenborg C, et al.; CARDIoGRAMplusC4D Consortium; DIAGRAM Consortium; CARDIOGENICS Consortium; MuTHER Consortium; Wellcome Trust Case Control Consortium . Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schunkert H, König IR, Kathiresan S, et al.; CARDIoGRAM Consortium . Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339–344 [DOI] [PubMed] [Google Scholar]
- 14.Webb TR, Erdmann J, Stirrups KE, et al.; Wellcome Trust Case Control Consortium; MORGAM Investigators; Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators . Systematic evaluation of pleiotropy identifies 6 further loci associated with coronary artery disease. J Am Coll Cardiol 2017;69:823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howson JMM, Zhao W, Barnes DR, et al.; CARDIoGRAMplusC4D; EPIC-CVD . Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet 2017;49:1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson CP, Goel A, Butterworth AS, et al.; EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group . Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385–1391 [DOI] [PubMed] [Google Scholar]
- 17.Klarin D, Zhu QM, Emdin CA, et al.; CARDIoGRAMplusC4D Consortium . Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet 2017;49:1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi L, Parast L, Cai T, et al. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J Am Coll Cardiol 2011;58:2675–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi L, Qi Q, Prudente S, et al. Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA 2013;310:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cahill LE, Levy AP, Chiuve SE, et al. Haptoglobin genotype is a consistent marker of coronary heart disease risk among individuals with elevated glycosylated hemoglobin. J Am Coll Cardiol 2013;61:728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buse JB, Bigger JT, Byington RP, et al.; ACCORD Study Group . Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial: design and methods. Am J Cardiol 2007;99(12A):21i–33i [DOI] [PubMed] [Google Scholar]
- 23.Cushman WC, Evans GW, Byington RP, et al.; ACCORD Study Group . Effects of intensive blood-pressure control in type 2 diabetes mellitus. N Engl J Med 2010;362:1575–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginsberg HN, Elam MB, Lovato LC, et al.; ACCORD Study Group . Effects of combination lipid therapy in type 2 diabetes mellitus. N Engl J Med 2010;362:1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosch J, Gerstein HC, Dagenais GR, et al.; ORIGIN Trial Investigators . n-3 fatty acids and cardiovascular outcomes in patients with dysglycemia. N Engl J Med 2012;367:309–318 [DOI] [PubMed] [Google Scholar]
- 26.Wang F, Xu CQ, He Q, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet 2011;43:345–349 [DOI] [PubMed] [Google Scholar]
- 27.Shah HS, Gao H, Morieri ML, et al. Genetic predictors of cardiovascular mortality during intensive glycemic control in type 2 diabetes: findings from the ACCORD clinical trial. Diabetes Care 2016;39:1915–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morieri ML, Shah H, Doria A; the Action to Control Cardiovascular Risk in Diabetes (ACCORD) Genetic Study Group . Variants in ANGPTL4 and the risk of coronary artery disease. N Engl J Med 2016;375:2304–2305 [DOI] [PubMed] [Google Scholar]
- 29.Pencina MJ, D’Agostino RB Sr, D’Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157–172; discussion 207–212 [DOI] [PubMed] [Google Scholar]
- 30.Kennedy KF, Pencina MJ. A SAS macro to compute added predictive ability of new markers predicting a dichotomous outcome [article online], 2010. NC State University. Available from http://analytics.ncsu.edu/sesug/2010/SDA07.Kennedy.pdf. Accessed 29 November 2017
- 31.Raffield LM, Cox AJ, Carr JJ, et al. Analysis of a cardiovascular disease genetic risk score in the Diabetes Heart Study. Acta Diabetol 2015;52:743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Look AHEAD Research Group Prospective association of a genetic risk score and lifestyle intervention with cardiovascular morbidity and mortality among individuals with type 2 diabetes: the Look AHEAD randomised controlled trial. Diabetologia 2015;58:1803–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Assimes TL, Salfati EL, Del Gobbo LC. Leveraging information from genetic risk scores of coronary atherosclerosis. Curr Opin Lipidol 2017;28:104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sniderman AD, Pencina M, Thanassoulis G. Limitations in the conventional assessment of the incremental value of predictors of cardiovascular risk. Curr Opin Lipidol 2015;26:210–214 [DOI] [PubMed] [Google Scholar]
- 35.Antiochos P, Marques-Vidal P, McDaid A, Waeber G, Vollenweider P. Association between parental history and genetic risk scores for coronary heart disease prediction: the population-based CoLaus study. Atherosclerosis 2016;244:59–65 [DOI] [PubMed] [Google Scholar]
- 36.de Vries PS, Kavousi M, Ligthart S, et al. Incremental predictive value of 152 single nucleotide polymorphisms in the 10-year risk prediction of incident coronary heart disease: the Rotterdam Study. Int J Epidemiol 2015;44:682–688 [DOI] [PubMed] [Google Scholar]
- 37.Abraham G, Havulinna AS, Bhalala OG, et al. Genomic prediction of coronary heart disease. Eur Heart J 2016;37:3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet 2017;18:331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan P, O’Donnell CJ. Reducing cardiovascular risk using genomic information in the era of precision medicine. Circulation 2016;133:1155–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kullo IJ, Jouni H, Austin EE, et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES clinical trial). Circulation 2016;133:1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duval C, Muller M, Kersten S. PPARα and dyslipidemia. Biochim Biophys Acta 2007;1771:961–971 [DOI] [PubMed]
- 42.Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 2015;385:2264–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natarajan P, Young R, Stitziel NO, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 2017;135:2091–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.