Abstract
Mitigating global infectious disease requires diagnostic tools that are sensitive, specific, and rapidly field-deployable. Here, we demonstrate that the Cas13-based SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) platform can detect Zika virus (ZIKV) and dengue virus (DENV) in patient samples at concentrations down to 1 copy/μl. We develop HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases), a protocol that pairs with SHERLOCK for viral detection directly from bodily fluids, enabling instrument-free DENV detection directly from patient samples in < 2 hours. We further demonstrate that SHERLOCK can distinguish the 4 DENV serotypes as well as region-specific strains of ZIKV from the 2015–2016 pandemic. Finally, we report the rapid design and testing (<1 week) of instrument-free assays to detect clinically relevant viral single nucleotide polymorphisms.
Recent viral outbreaks have highlighted the challenges of diagnosing viral infections, particularly in areas far from clinical laboratories. Viral diagnosis was especially difficult during the 2015–2016 Zika virus (ZIKV) pandemic; low viral titers and transient infection(1, 2), combined with limitations of existing diagnostic technologies, contributed to ZIKV circulating for months before the first cases were confirmed clinically(3–5). An additional challenge for viral diagnostics is differentiating between related viruses that cause infections with similar symptoms, like ZIKV and dengue virus (DENV) (1). More generally, existing nucleic acid detection methods are very sensitive and rapidly adaptable, but most require extensive sample manipulation and expensive machinery(1, 6–8). In contrast, antigen-based rapid diagnostic tests require minimal equipment, but have lower sensitivity and specificity, and assay development can take months(9–11). An ideal diagnostic would combine the sensitivity, specificity, and flexibility of nucleic acid diagnostics with the speed and ease of use of antigen-based tests. Such a diagnostic could be rapidly developed and deployed in the face of emerging viral outbreaks, and would be suitable for disease surveillance or routine clinical use in any context.
The Cas13-based nucleic acid detection platform SHERLOCK has the potential to address the key challenges associated with viral diagnostics. SHERLOCK combines isothermal amplification using Recombinase Polymerase Amplification (RPA)(12) with highly specific Cas13-based detection (Fig. 1A) (13). Cas13, an RNA-guided ribonuclease, provides specificity through CRISPR RNA (crRNA):target pairing, and additional sensitivity due to signal amplification by Cas13’s collateral cleavage activity(14, 15).
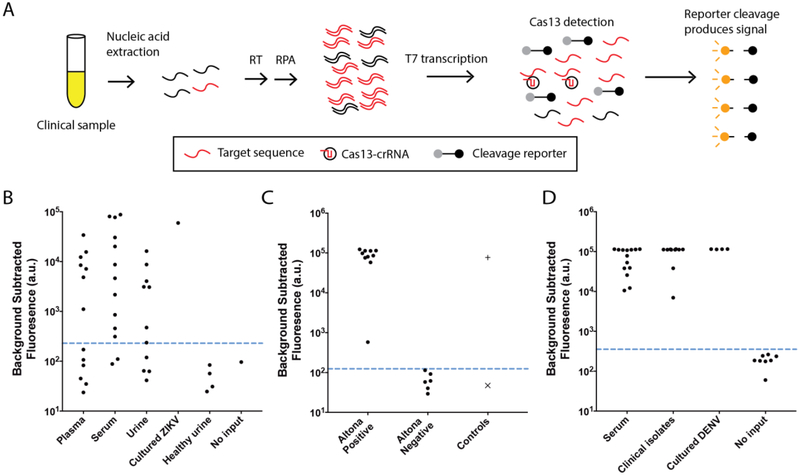
Fig. 1. ZIKV and DENV detection from patient samples and clinical isolates.
(A) Schematic of SHERLOCK. Nucleic acid is extracted from clinical samples, and the target is amplified by RPA using either RNA or DNA as input (RT-RPA or RPA, respectively). RPA products are detected in a reaction containing T7 RNA polymerase, Cas13, a target-specific crRNA, and an RNA reporter that fluoresces when cleaved. We tested SHERLOCK on (B) cDNA derived from 37 patient samples collected during the 2015–2016 ZIKV pandemic and (C) cDNA from 16 patient samples that were compared head-to-head with the Altona RealStar Zika Virus RT-PCR assay. +: ZIKV seed stock cDNA (3×102 cp/μl) and ×: no input. (D) SHERLOCK on RNA extracted from 24 DENV-positive patient samples and clinical isolates. Dashed blue line: threshold for presence or absence of ZIKV or DENV (see Methods).
For SHERLOCK to excel at viral detection in any context, it should be paired with methods enabling direct detection from patient samples with a visual readout. Here, we test the performance of SHERLOCK for ZIKV and DENV detection on patient samples and develop HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases), a method to enable rapid, sensitive detection of ZIKV and DENV directly from bodily fluids with a colorimetric readout, recently shown as part of SHERLOCKv2(16). Additionally, we design SHERLOCK assays to distinguish multiple viral species and strains and identify clinically relevant mutations.
Detection of ZIKV and DENV in patient samples provides a stringent test of the sensitivity of SHERLOCK and its tolerance of viral diversity. Our ZIKV SHERLOCK assay had single-copy (1 cp/μl) sensitivity when tested on seedstock cDNA (Fig. S1). We evaluated its performance on 40 cDNAs derived from samples collected during the 2015–2016 ZIKV pandemic, 37 from patient samples with suspected ZIKV infections and 3 from mosquito pools (Figs. 1B, S2, Table S1). For 16 samples from these patients, we benchmarked SHERLOCK by comparing its sensitivity and specificity to other nucleic acid amplification tests including the commercially available Altona Realstar Zika Virus RT-PCR assay (Figs. 1C, S3–5, Table S2, see Supplementary Text). Of the 10 samples tested positive by the Altona assay, all 10 were detected by SHERLOCK (100% sensitivity); the other 6 samples were negative by both assays (100% specificity, 100% concordance). Our ZIKV assay had no false positives when tested on healthy urine and water (Fig. 1B). We then validated the ability of SHERLOCK to detect DENV, a related but more diverse flavivirus with similar symptoms to ZIKV infection. All 24 RT-PCR-positive DENV RNA samples were confirmed DENV positive after 1 hour of detection (Figs. 1D, S6–7, Table S3). SHERLOCK sensitively and specifically detects viral nucleic acids extracted from ZIKV and DENV patient samples.
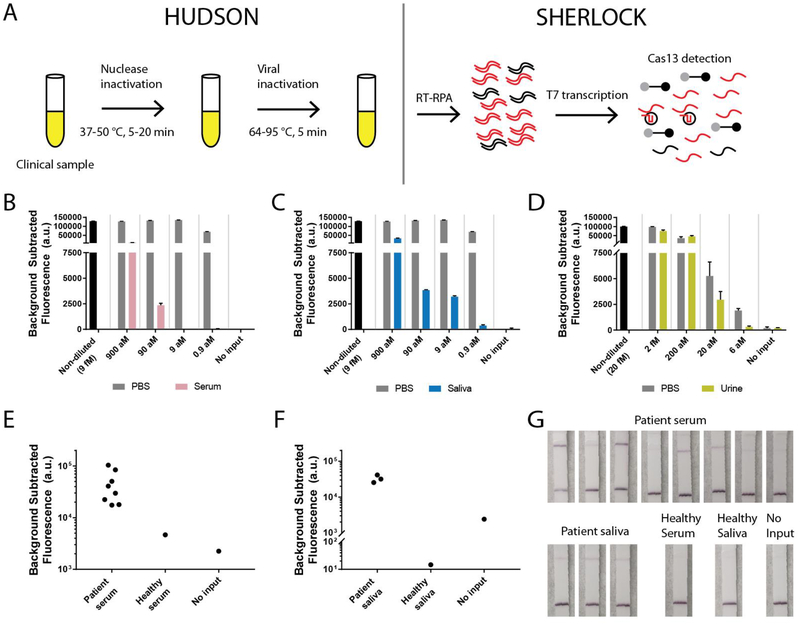
Although SHERLOCK excels at detecting extracted nucleic acids, a field-deployable, rapid diagnostic test should not require an extraction step to detect viral nucleic acid in bodily fluids. Many viruses are shed in urine or saliva, including ZIKV and DENV, and sampling is not invasive(2, 7). To detect viral nucleic acid directly from bodily fluids via SHERLOCK, we developed HUDSON, a method to lyse viral particles and inactivate the high levels of RNases found in bodily fluids using heat and chemical reduction (Figs. 2A, S8)(17). HUDSON-treated urine or saliva could be directly added to RPA reactions without dilution or purification (blood products were diluted 1:3 to avoid solidification during heating), without inhibiting subsequent amplification or detection. HUDSON and SHERLOCK enabled sensitive detection of free ZIKV nucleic acid spiked into urine, whole blood, plasma, serum, or saliva (Figs. S9–12). To mimic clinical infection, where viral nucleic acid is encapsulated in infections particles, we spiked infectious ZIKV particles into bodily fluids. HUDSON combined with SHERLOCK (Figs. S13–14) permitted sensitive detection of ZIKV RNA from infectious particles at 90 aM (45 cp/μl) in whole blood (Fig, S15) or serum (Fig. 2B), 0.9 aM (~1 cp/μl) in saliva (Fig. 2C), and 20 aM (10 cp/μl) in urine. Total turnaround time was < 2 h with fluorescent and colorimetric readout (Fig. 2D, Fig. S16). The sensitivity of HUDSON and SHERLOCK is comparable to ZIKV RNA concentrations observed in patient samples, which range from 1–1,000 cp/μl(1, 2). HUDSON, paired with the pan-DENV SHERLOCK assay, detected DENV in whole blood, serum, and saliva (Figs. S17–18). DENV was detected directly from 8 of 8 patient serum samples (Fig. 2E) and 3 of 3 patient saliva samples tested (Fig 2F), with a total turnaround time < 1 hour from saliva despite lower viral titers than in serum (7). We directly detected DENV with a colorimetric readout using lateral flow strips (Fig. 2G), showcasing a HUDSON to SHERLOCK pipeline that can detect ZIKV or DENV directly from bodily fluids with minimal equipment.
Fig. 2. Direct detection of ZIKV and DENV in bodily fluids with HUDSON and SHERLOCK.
(A) Schematic of direct viral detection using HUDSON and SHERLOCK. (B-C) Detection of ZIKV RNA in particles diluted in healthy human serum (pink, B) or healthy human saliva (blue, C). Same PBS control used in A and B as experiments were performed together. (D) Detection of ZIKV RNA in particles diluted in healthy human urine (yellow). Black: non-diluted viral stock. Grey: dilutions in PBS. Error bars indicate 1 S.D. based on 3 technical replicates. (E-F) Detection of DENV RNA directly from patient serum (E) and saliva samples (F). (G) Lateral flow detection of DENV from samples shown in (E-F). All samples were treated with TCEP/EDTA prior to heating.
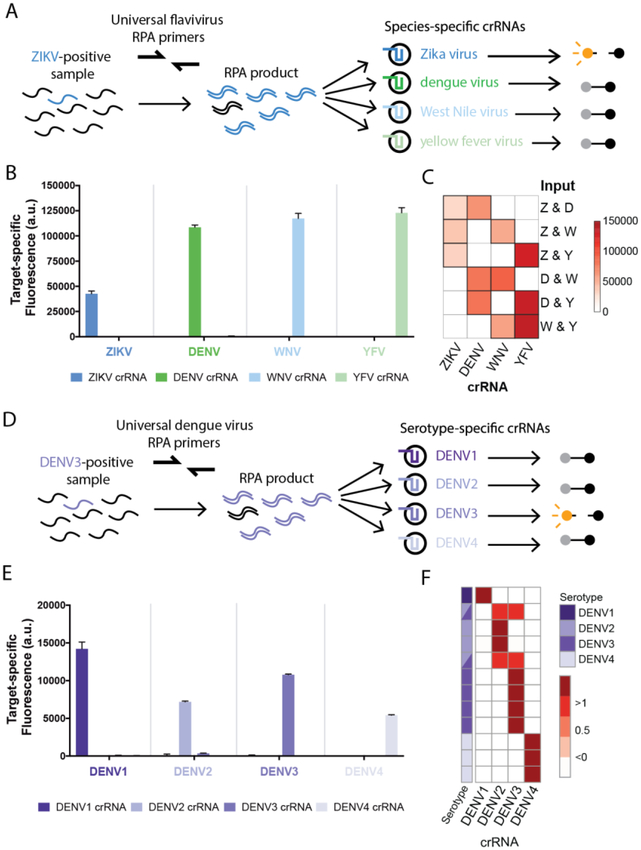
Because many genetically and antigenically similar flaviviruses co-circulate and have similar symptoms, we developed diagnostic panels to distinguish related viral species and serotypes. We identified conserved regions within ZIKV, DENV, West Nile virus (WNV), and yellow fever virus (YFV) genomes and designed a flavivirus panel with universal-flavivirus RPA primers that can amplify any of the 4 viruses, and species-specific crRNAs (Fig. 3A). This panel detected synthetic ZIKV, DENV, WNV, and YFV DNA targets with < 0.22% off-target fluorescence (Fig. 3B, Figs. S19–20, see Methods), and identified the presence of all pairwise combinations of these 4 viruses, demonstrating the ability to detect mock co-infections (Fig. 3C, Figs. S21–22). We also designed a panel using DENV-specific RPA primers and serotype-specific crRNAs (Fig. 3D) that could distinguish between DENV serotypes 1–4 with < 3.2% off-target fluorescence (Fig. 3E, Figs. S23–24). This low level of off-target fluorescence allows for 100% specificity in differentiating between serotypes, providing an alternative to current serotype identification approaches (8). The DENV panel confirmed the serotypes of 12 RT-PCR-serotyped patient samples or clinical isolates (Fig 3F, Fig. S25), and identified 2 clinical isolates with mixed infection, a commonly observed phenomenon(18). SHERLOCK can therefore be extended to differentiate between related viruses or serotypes using a single amplification reaction.
Fig. 3. Multivirus panels can differentiate viral species and serotypes.
(A-C) Panel of 4 related flaviviruses (A) used to detect individual viral targets (B) or paired viral targets (C) using species-specific crRNAs with 3 hours of detection. (D-E) Identification of DENV serotypes 1–4 using serotype-specific crRNAs (D), tested using synthetic targets with 3 hours of detection (E). (F) Identification of DENV serotypes in extracted RNA from patient samples. Each row is a sample, each column is a crRNA, target-specific fluorescent values are normalized by row. Purple: DENV serotype identified. Synthetic targets were used at 104 cp/μl. Error bars indicate 1 S.D. based on 3 technical replicates. We expect off-target crRNAs to have close to zero target-specific fluorescence (see Methods). Primer, crRNA, and target sequences are in Tables S5–7.
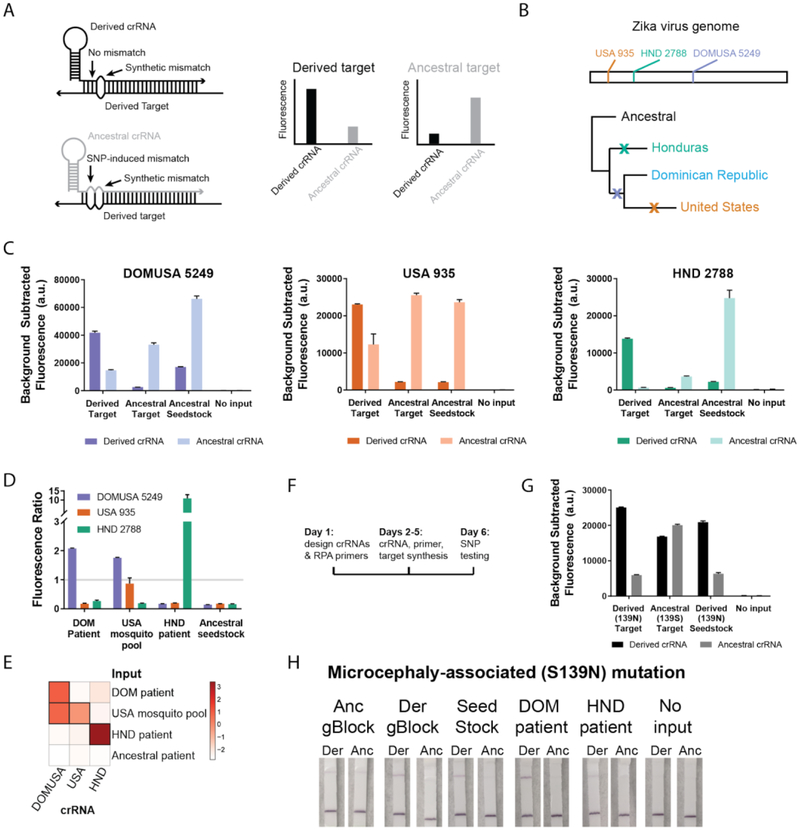
SHERLOCK is uniquely poised for field-deployable variant identification, which would allow real-time tracking of microbial threats. Genotyping of single nucleotide polymorphisms (SNPs) typically involves PCR and either fluorescent- or mass spectrometry-based detection, requiring extensive sample processing, expensive equipment, and limiting field-deployability (19). SHERLOCK can identify SNPs by placing a synthetic mismatch in the crRNA near the SNP, testing each target with an ancestral-specific and derived-specific crRNA (Fig. 4A)(13). We designed diagnostics for 3 region-specific SNPs from the 2015–2016 ZIKV pandemic (Fig. 4B) and identified these SNPs in synthetic targets, a viral seedstock, and cDNA samples from Honduras, the Dominican Republic, and the United States (Fig. 4C–E). These results demonstrate that SHERLOCK can identify SNPs in samples from the ZIKV pandemic and highlight the single-nucleotide specificity of SHERLOCK.
Fig. 4. Identification of adaptive and functional ZIKV mutations.
(A) SHERLOCK assays for SNP identification. Dark colors: derived crRNAs, light colors: ancestral crRNAs. (B-C) Three region-specific SNPs from the 2015–2016 ZIKV pandemic, including genomic locations of the SNPs and a simplified phylogenetic tree, tested using synthetic targets (104 cp/μl) and viral seedstock cDNA (3×102 cp/μl). (D-E) Identification of region-specific SNPs in ZIKV cDNA samples from the Dominican Republic (DOM), United States (USA), and Honduras (HND). The fluorescence ratio (derived crRNA fluorescence divided by ancestral crRNA fluorescence) for each SNP in each sample is shown in a bar plot (log2-transformed data in a heatmap). (F) Timeline for developing a SHERLOCK assay for a new SNP (more detail in Fig. S26). (G-H) Identification of a microcephaly-associated ZIKV mutation (PrM S139N) by fluorescent and colorimetric detection. In all panels, error bars indicate 1 S.D. based on 3 technical replicates.
Rapid identification of emerging drug resistance and other clinically relevant mutations for viruses such as ZIKV and HIV would have great utility. A ZIKV point mutation in the PrM region (S139N) recently associated with fetal microcephaly was used as a test case for the rapid development of assays for variant identification (20). Within a week of the report’s publication (Fig. 4F, Fig. S26), we developed multiple SHERLOCK assays for the S139N mutation (Fig. 4G, Fig. S27), and could identify the mutation in patient samples from the 2015–2016 ZIKV pandemic with a visual readout (Fig. 4H). To further illustrate the ease of developing SHERLOCK diagnostics for many clinically relevant mutations, we designed and tested assays for the six most commonly observed drug-resistance mutations in HIV reverse transcriptase(21) in 1 week (Fig. S28). These examples underscore that SHERLOCK could be used for monitoring clinically relevant variants in near real time.
Combining HUDSON and SHERLOCK, we have created a field-deployable viral diagnostic platform with high performance and minimal equipment or sample processing requirements. This platform is as sensitive and specific as amplification-based nucleic acid diagnostics (12, 22–26), with similar speed and equipment requirements to rapid antigen tests(9–11). Furthermore, this approach can be easily adapted to detect virtually any virus present in bodily fluids, scaled to enable multiplexed detection(16), and the reagents can be lyophilized for cold-chain independence(13). Cas13-based detection is a promising next-generation diagnostic strategy with the potential to be implemented almost anywhere in the world to enable effective, rapid diagnosis of viral infections.
Supplementary Material
Acknowledgements:
We thank S. Schaffner, A. Lin, and other Sabeti lab members for useful feedback, J. Strecker for providing SUMO protease, and the Florida Department of Health, Miami-Dade County Mosquito Control, and Boca Biolistics for support with patient and mosquito samples.
Funding: We acknowledge funding from the Howard Hughes Medical Institute (HHMI), the Broad Institute CBTS Shark Tank, NIH U19AI110818, and DARPA D18AC00006. The views, opinions, and/or findings expressed should not be interpreted as representing the official views or policies of the DOD or U.S. Government. Approved for public release; distribution is unlimited. C.M. is supported by HHMI, O.O.A. by a Paul and Daisy Soros Fellowship and NIH F30 NRSA 1F30-CA210382, S.I. and S.F.M. by NIH NIAID R01AI099210, I.B. and L.G. by NIH AI 100190, F.Z. by NIH grants (1R01-HG009761, 1R01-MH110049, 1DP1-HL141201); HHMI; the New York Stem Cell, Allen, and Vallee Foundations; the Tan-Yang Center at MIT; and J. and P. Poitras, and R. Metcalfe. F.Z. is a New York Stem Cell Foundation–Robertson Investigator. M.L.N. is supported by FAPESP (Grant#13/21719–3) and a CNPq Research Fellow.
Footnotes
Competing interests: C.M., C.A.F., P.C.S., J.S.G., O.O.A., and F.Z are co-inventors on patent applications filed by the Broad Institute relating to work in this manuscript.
Data and materials availability: All data are available in the manuscript or the supplementary material. Details on MTAs related to the sharing of clinical samples are described in the Methods section.
References
- 1.Faye O et al. , One-step RT-PCR for detection of Zika virus. J. Clin. Virol 43, 96–101 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Paz-Bailey G et al. , Persistence of Zika Virus in Body Fluids - Preliminary Report. N. Engl. J. Med (2017), doi: 10.1056/NEJMoa1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metsky HC et al. , Zika virus evolution and spread in the Americas. Nature 546, 411–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faria NR et al. , Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546, 406–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grubaugh ND et al. , Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 546, 401–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faye O et al. , Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol. J 10, 311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andries A-C et al. , Value of Routine Dengue Diagnostic Tests in Urine and Saliva Specimens. PLoS Negl. Trop. Dis 9, e0004100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waggoner JJ et al. , Comparison of the FDA-approved CDC DENV-1–4 real-time reverse transcription-PCR with a laboratory-developed assay for dengue virus detection and serotyping. J. Clin. Microbiol 51, 3418–3420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosch I et al. , Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med 9 (2017), doi: 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balmaseda A et al. , Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc. Natl. Acad. Sci. U. S. A 114, 8384–8389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priyamvada L et al. , Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. U. S. A 113, 7852–7857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piepenburg O, Williams CH, Stemple DL, Armes NA, DNA detection using recombination proteins. PLoS Biol 4, e204 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gootenberg JS et al. , Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abudayyeh OO et al. , C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.East-Seletsky A et al. , Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gootenberg JS et al. , Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science (2018), doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weickmann JL, Glitz DG, Human ribonucleases. Quantitation of pancreatic-like enzymes in serum, urine, and organ preparations. J. Biol. Chem 257, 8705–8710 (1982). [PubMed] [Google Scholar]
- 18.Requena-Castro R, Reyes-López MÁ, Rodríguez-Reyna RE, Palma-Nicolás P, Bocanegra-García V, Molecular detection of mixed infections with multiple dengue virus serotypes in suspected dengue samples in Tamaulipas, Mexico. Mem. Inst. Oswaldo Cruz 112, 520–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Misra A, SNP Genotyping: Technologies and Biomedical Applications. Annu. Rev. Biomed. Eng 9, 289–320 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Yuan L et al. , A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science, eaam7120 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Rhee S-Y et al. , Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31, 298–303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Y et al. , Coupling Sensitive Nucleic Acid Amplification with Commercial Pregnancy Test Strips. Angew. Chem. Int. Ed Engl 56, 992–996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eboigbodin KE, Brummer M, Ojalehto T, Hoser M, Rapid molecular diagnostic test for Zika virus with low demands on sample preparation and instrumentation. Diagn. Microbiol. Infect. Dis 86, 369–371 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Pardee K et al. , Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 165, 1255–1266 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Chotiwan N et al. , Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci. Transl. Med 9 (2017), doi: 10.1126/scitranslmed.aag0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Ness J, Van Ness LK, Galas DJ, Isothermal reactions for the amplification of oligonucleotides. Proceedings of the National Academy of Sciences 100, 4504–4509 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matranga CB et al. , Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol 15, 519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abd El Wahed A et al. , Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLoS One 10, e0129682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donald CL et al. , Full Genome Sequence and sfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. PLoS Negl. Trop. Dis 10, e0005048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Signor LDCC, Phylogeny of Zika Virus in Western Hemisphere, 2015. Emerg. Infect. Dis 22, 933–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambeth CR, White LJ, Johnston RE, de Silva AM, Flow cytometry-based assay for titrating dengue virus. J. Clin. Microbiol 43, 3267–3272 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.