Abstract
Purpose of Review
Hematopoiesis is an ordered developmental process that requires dynamic regulation to warrant proper response to physiological challenges and prevent malignancies. Long noncoding RNAs are emerging as key, multi-faceted regulators of gene expression. This review explores the function of lncRNAs in the control of HSC homeostasis and hematopoietic differentiation.
Recent Findings
Multiple lncRNAs have been implicated in maintaining HSC stemness and enabling progenitors to carry out the correct programs of lineage differentiation. Specific lncRNAs have been identified that regulate the differentiation of multipotent progenitors into terminally differentiated blood cells. These lncRNAs predominantly act by assisting master regulators that drive specific differentiation programs, either by enhancing or repressing the transcription of particular genomic loci.
Summary
Long noncoding RNAs contribute to the correct differentiation and maturation of various hematopoietic lineages by assisting with the activation of transcriptional programs in a time- and cell-dependent manner.
Keywords: Hematopoiesis, long noncoding RNA, HSC, cell fate, differentiation
Introduction
In Greek myth, the Moirai were the three goddesses of fate who personified the destiny of man. They were responsible for spinning out, measuring, and then cutting the threads of life [1]. In this review article we will discuss the accumulating knowledge about the long regulatory RNA molecules which are expressed in blood cells and which akin to ‘molecular Moirai’ are involved in the determination of cell fate and lineage specification during hematopoietic development.
Hematopoiesis is an ordered biological process that has evolved to provide multicellular organisms with a lifelong supply of mature blood cells [2]. The three main blood cell types (red, white, and platelet cells) that arise through this complex developmental process mediate a diverse array of physiological functions in the body, including immune defense against pathogenic infections, transport of oxygen from lungs to peripheral tissues and prevention of blood loss after injury through clotting. Hematopoietic development starts from a pluripotent hematopoietic stem cell (HSC) that has an unlimited capacity for self-renewal and usually resides in the red bone marrow (BM). Largely quiescent, slow cycling HSCs gives rise to a small pool of multipotent progenitor (MPP) cells that display a significant loss of self-renewing ability and, thus, can only transiently support hematopoiesis. According to the prevailing model in the field, the hematopoietic pathway bifurcates downstream of the MPP into myeloid and lymphoid lineages. Common myeloid progenitor (CMP) and common lymphoid progenitor (CLP) cells, in contrast to the MPP from which they hail, have a significantly restricted lineage potential and, therefore, can sustain production of only myeloid or lymphoid cell types, respectively. Following a series of additional maturation steps, the CLP gives rise to all major subsets of lymphoid cells, including T and B lymphocytes, natural killer (NK) and innate lymphoid cells (ILCs). On the other hand, via two distinct branches in the myeloid differentiation pathway the CMP generates red blood cells and platelets (MEP), as well as several classes of innate immune cells such as granulocytes, macrophages and dendritic cells (GMP).
Careful regulation of cell fate and lineage determination decisions during hematopoiesis is required to both maintain a healthy immune system; and to enable a rapid and robust response to physiological stresses, like microbial infection or blood loss after trauma. To address this need, multiple layers of regulation that ensure the correct timing and output of blood cell differentiation have evolved. The functional goal of these regulatory circuits is to control stage- and lineage-specific transcriptional machinery that is activated in response to specific extracellular cues (e.g. growth factors and cytokines) and drives progenitor differentiation [3]. The importance of regulatory RNA in this dynamic process has started to emerge recently as a consequence of our greater understanding of the genomic complexity [4].
Advances in sequencing technology have increased our ability to interrogate the molecular mechanisms underpinning basic biological functions. One of the fruits of the genomics revolution has been a surprising realization that mammalian genomes are pervasively transcribed and contain numerous genes that encode regulatory RNAs (reviewed in [5], [4]). For the last two decades, microRNA (miRNA) has been the most studied class of regulatory RNA. These tiny (~22 nucleotides long) RNA molecules use short stretches of complementarity to recognize their target genes and attenuate their expression at the mRNA level [6]. miRNAs have been implicated in the regulation of various steps in hematopoietic development: from HSC homeostasis to lineage specification and activation of terminally differentiated cells [7–9]. By contrast, research into the longer regulatory RNA species, the so called long noncoding RNAs (lncRNAs), and their role in hematopoietic development is only now picking up steam. The slow pace in recognizing lncRNAs as important factors that control hematopoiesis is due to a variety of reasons, such as the relative difficulty in identifying lncRNA genes, the lack of a single mechanism of action and the difficulty in identifying their legitimate molecular interactions. Despite these difficulties, over the last few years our knowledge of lncRNA functions in hematopoiesis has dramatically increased, revealing a new understanding of the way this critical process is regulated.
What is a long noncoding RNA?
LncRNAs are defined as any transcribed RNA molecule that is both over 200 nucleotides in length and does not encode a protein. Although they were originally dismissed as “junk DNA”, it is now appreciated that approximately 10,000 identified lncRNA sequences in the human genome play some sort of function and are not merely being expressed due to transcriptional “noise” [10, 5]. Some estimates pin the number close to 60,000 genes [11], suggesting that lncRNAs are numerically as abundant as protein-coding genes (~20,000). Although the majority of lncRNAs are transcribed by RNA Polymerase II (RNAPII), many lncRNAs that interact with transcription factors have been found to be transcribed by RNA Polymerase III (RNAPIII). Most lncRNAs in the cell are spliced and carry 5’ cap and poly(A) structures. Interestingly, lncRNA genes are expressed at a much lower rate on average than the protein-coding genes. Most lncRNA genes are poorly conserved and expressed in a tissue-specific fashion. lncRNAs are often classified on the basis of their position in relationship to a neighboring protein-coding gene: (i) intergenic lncRNAs are located in between protein-coding gene loci; (ii) antisense lncRNAs are transcribed in the antisense direction to an overlapping protein-coding gene; (iii) intronic lncRNAs are located in the introns of protein-coding genes; and (iv) divergent lncRNAs are oriented head-to-head with a protein-coding gene and are transcribed from the same promoter. Another large category of lncRNAs is enhancer RNAs (eRNAs), which are transcribed from the DNA enhancer regions and are typically neither spliced or polyadenylated.
Because of the lack of apparent sequence-specific targeting, the dominant mechanism of action of lncRNAs is not clearly defined. Evidence in the literature suggests that it is useful to distinguish two different modes of action of lncRNAs: (i) those that utilize the lncRNA transcript to interact with other genetic elements; and (ii) those where transcription at or near the lncRNA locus itself is the driver of regulation. When viewing lncRNAs as genomic regulatory agents it is apparent that they can act in both cis and trans to their locus of origin. By contrast, the eRNA- associated loci that have been found to manipulate gene expression are always acting in cis to their genomic targets. Both these lncRNA-associated loci and lncRNAs themselves are capable of acting in this enhancer RNA role, manipulating gene expression through direct interaction with the transcriptional machinery. LncRNAs have been found to form RNA-protein, RNA-DNA and RNA-RNA interactions. Some lncRNA molecules appear to have a modular structure and, therefore, can support more than one type of biochemical interaction. The paradigmatic lncRNA interaction is the binding of a lncRNA to a chromatin modifying complex, which can either enhance or suppress transcription. Due to the difficulties in interrogating high order complexes of DNA, RNA and proteins, it is not immediately apparent if there are hard and fast rules regarding the participation of lncRNAs in these complexes. We are however able to ascertain whether or not these lncRNAs are involved in processes that manipulate gene expression through the enhancement or repression of transcriptional programs.
Long non-coding RNAs in the HSC compartment
Using expression profiling of purified HSCs, Margaret Goodell and colleagues have recently uncovered 332 lncRNA genes that are expressed in the HSC compartment [12]. A set of 159 unannotated lncRNAs from this list was found to be expressed uniquely by HSCs, but not by two kinds of terminally differentiated blood cells. The use of poly(A)-enriched RNA for sequencing (which would eliminate most eRNAs) and very strict filtering criteria to identify lncRNA genes most likely resulted in some underestimation of the number of HSC-specific lncRNAs in this study. The members of the HSC-specific lncRNA set are well expressed and display characteristics that set them apart from the protein-coding genes, such as enrichment for retrovirus-related transposable elements in their gene bodies. Furthermore, promoter regions of HSC-specific lncRNA genes were generally undermethylated and enriched in binding sites for HSC-associated transcription factors (e.g. Erg, Flil1 and Pu.1). Taken together, this evidence indicates that the newly identified HSC transcripts are indeed genuine lncRNAs. This list of HSC-enriched lncRNA genes generated by Luo et al. was largely congruent to the HSC-specific lncRNA signature that was identified by a previous study [13]. Of note, the number of lncRNAs that are apparently expressed in HSCs (about 300) seems to be roughly comparable to the number of protein-coding genes in the HSC-specific expression program [14].
To gain insight into how lncRNAs control HSC functions, Luo et al. focused on two HSC-enriched transcripts (lncHSC-1 and lncHSC-2) that both displayed strong nuclear expression. Experimental perturbation of these lncRNAs in the murine hematopoietic compartment revealed a non-overlapping regulatory role for these molecules. Transplantation of HSCs transduced with lncHSC-1 knockdown (KD) constructs resulted in an expansion of myeloid cells, concomitant with an increase in myeloid progenitors and a drop in the number of HSCs [12]. In contrast, in vivo silencing of lncHSC-2 in the HSC compartment led to an expansion of T lymphocytes along with a drop in the self-renewal capacity of HSCs. This evidence implicates these two lncRNAs as mediators of specific lineage differentiation, although their expression is restricted to the HSC compartment. The mechanisms of action of lncHSC-1 and lncHSC-2 are currently not known. The KD of these lncRNAs in HSC compartment perturbed expression of a relatively small number of genes (less than 100), with none of the observed changes affecting neighboring genes, suggesting that lncHSC-1 and lncHSC-2 might act in trans. Goodell and colleagues mapped lncHSC-2 binding sites genome-wide and found them enriched for binding sites of transcription factors associated with hematopoietic development, like E2A.
Besides lncHSC-1 and lncHSC-2, two other lncRNA genes (H19 and Xist) were previously implicated as regulators of HSC biology. The H19-Igf2 locus was initially discovered to be a parentally imprinted gene and explored in the context of epigenetic regulation of development [15]. Venkatraman et al. found that this locus was differentially regulated in long-term HSCs (LT-HSCs) compared to other progenitor cells and experimentally examined how this locus may affect HSC function [16]. They found that the differentially methylated region (DMR) of the H19-Igf2 locus controlled H19 lncRNA expression from the maternal allele, which in turn kept the paternal allele silenced. This differential expression between alleles allowed for expression of Igf2 from the paternal allele, while stable H19 expression from the maternal allele attenuated expression of its receptor Igfr1 through production of miR-675. Control of this Igf2-Igfr1 circuit is important for HSC quiescence, because sustained Igfr1 signaling drives FoxO3 inactivation and as a result stimulates cell cycle entry and proliferation. Maintenance of the Igf2-Igfr1 circuit is thus controlled by H19 expression, leading the HSC to remain in a quiescent state. When this balance is perturbed experimentally, a marked shift towards HSC differentiation and proliferation was observed [16].
Xist, one of the first lncRNA transcripts to be studied, is the major contributor to the inactivated X chromosome state in diploid XX cells. Its role in X inactivation (Xi) has been well studied, but only recently has this process been found to play a crucial role in hematopoiesis. Female mice with a pan-hematopoietic Xist deletion develop an aggressive myeloproliferative neoplasm and myelodysplastic syndrome (mixed MPN/MDS) and die prematurely [17]. This defect is the results of a progressive LT-HSC loss in the absence of Xist and is likely cell-autonomous, based on competitive repopulation studies. Xist-deficient HSCs display significant X reactivation (as evidenced by an upregulation of X-linked genes), as well as dysregulation of a large set of genetic factors, including several that are often mutated in myeloid malignancies (e.g. Tet2, Idh1, Ezh2 and Kit). Mechanistically, these findings resemble observations in some cancers with a deregulated Xist [18, 19], indicating that the role of this lncRNA in hematopoietic cells is likely to restrict the gene dosage of the X chromosome and not to target any specific set of genes for repression.
Control of myeloid development by lncRNAs
Along with the newly explored role of lncRNAs in HSC maintenance, there has been a profusion of studies identifying specific lncRNAs responsible for the differentiation of HSC progenitors into mature immune cell subtypes and their activation. Mature blood cells that derive from the CMP arise from either a granulocyte-monocyte progenitor (GMP) or a megakaryocyte-erythroid progenitor (MEP). Several studies have examined the transcriptome of defined myeloid precursors and have identified putative lncRNA transcripts involved in myeloid subset development [20–22]. Comprehensive surveys of the lncRNA landscape in mouse megakaryocyte-erythroid lineage by Mitchell Weiss and colleagues revealed about 1100 previously unannotated transcripts [21]. Strikingly, more than 80% of the mouse erythro-specific lncRNAs were not found in human erythroblasts, despite being conserved between different mouse strains. This study, along with Alvarez-Dominguez et al. took a closer look at several of these lncRNAs expressed in erythroid precursors, including Bloodlinc/alncRNA-EC7 [22]. Ex vivo knockdown experiments confirmed that expression of these lncRNAs is critical for terminal erythroid cell maturation. The first lncRNA identified to have a myeloid cell specific activity was the eosinophil granule ontogeny (EGO) gene in 2007 [23]. Wagner et al. found that this lncRNA was specifically expressed during eosinophil development and upon mature eosinophil stimulation. The mechanism of action is currently unknown, but EGO transcript levels correlate with other mRNAs essential for eosinophil development. This suggests that EGO acts as an enhancer or stabilizer of genes necessary for normal eosinophil maturation. In 2009, Zhang et al. found another lncRNA that appears to regulate granulocyte cell development [24]. Transcribed from an intergenic region of the HOXA gene locus, the lncRNA HOTAIRM1 appears to play a role in myelopoiesis. By enhancing or stabilizing expression of the HOXA1 and HOXA4 genes this lncRNA ensures that these key transcriptional regulators are expressed during normal granulocyte development.
The other two major GMP-derived lineages were also found to utilize lncRNAs as regulators of differentiation or activation. Identified in 2014 by Wang et al. the lncRNA lnc-DC was described as a significant driver of dendritic cell (DC) differentiation [25]. Unlike EGO or HOTAIRM1, lnc-DC appears to function by binding to a single protein, transcription factor STAT3, and promoting its phosphorylation. The specific mechanism has yet to be determined, but it appears that the constitutive subsequent activation of STAT3 signaling in the developing DC context promotes mature DC differentiation. In 2015, Chen et al. described the lncRNA lnc-MC, which has a unique function in macrophage development. Lnc-MC appears to act as an endogenous sponge of microRNA miR-199a-5p [26]. This microRNA targets activin A receptor type 1B (ACVR1B), a member of the TGF-β receptor superfamily that operates as a positive regulator of myeloid development. Thus, miR-199a-5p sequestration by lnc-MC elevates ACVR1B levels and promotes monocyte differentiation. Another recent discovery of a lncRNA regulator of myeloid cell fate is the lncRNA Morrbid. Kotzin et al. found that this lncRNA functions as a cis-acting negative regulator of pro-apoptotic gene Bim. Morrbid coordinates recruitment of an epigenetic silencer, the PRC2 complex, to the Bim promoter, thereby keeping Bim quiet and preventing the engagement of apoptotic programme. [27]. This anti-apoptotic effect is crucial for enabling survival of otherwise short-lived myeloid cells as evidenced by the dramatic depletion of eosinophils, neutrophils, and classical monocytes in Morrbid-deficient mice.
LncRNAs in lymphoid cell differentiation
Expression profiling of various mature and immature T cell subsets have identified several lncRNAs for which specific roles in the differentiation and maturation of T lymphocyte subsets were defined experimentally. RNA sequencing of CLP populations revealed a large array of previously undescribed lncRNAs that might contribute to B and T cell development [28]. The most well characterized of these lncRNAs have been those that play a direct role in regulating transcriptional machinery. Nuclear factor of activated T cells (NFAT) is a key transcription factor that operates downstream of the T cell receptor (TCR) and controls activation of T cells [29]. Using a lncRNA KD approach to identify noncoding RNA regulators of NFAT, Willingham et al. found that the lncRNA NRON functions as an NFAT repressor [30]. NRON is proposed to regulate nuclear trafficking of NFAT by interacting with nuclear import factors and thus prevents NFAT from inducing its activation program in T cells. Multiple lncRNAs have now been identified to play unique roles in naïve CD4+ T helper (Th) cell polarization into several effector cell types. The activity of these lncRNAs is primarily aimed at the regulation of expression of specific cytokines that drive Th cell differentiation. The lncRNAs lnc-MAF-4 and NeST have been implicated in the differentiation of the Th1 subset that promotes cell-mediated immunity [31]. These lncRNAs assist in establishing and maintaining of the Th1 program by recruiting transcriptional activators to key cytokine loci, or by antagonizing the transcriptional machinery that would drive a different developmental outcome. Not surprisingly, lncRNA regulators were also found to facilitate two other Th differentiation programs [32]. Th2 LCR and lincR-Ccr2-5’AS promote expression of the cytokines and chemokines that drive Th2 polarization, while the lncRNA Rmrp facilitates Th17 differentiation by recruiting a master regulator of the Th17 program, RORɣt, to Th17-specific loci (e.g. IL-17a) [33, 34].
Despite originating from the CLP, B cell progenitors have a markedly different lncRNA expression pattern compared to T cell progenitors. Global transcriptome analyses by Brazao et al. have revealed a unique B cell progenitor pattern of lncRNA expression [35]. Specific lncRNAs have not been characterized, but it appears that lncRNA usage in B cells is strongly controlled by the transcription factor PAX5, indicating that they might be involved in B cell differentiation. Recent work has shown that the lncRNA Xist in female cells plays a stage-specific role in early B cell development. Temporary loss of Xist appears to transiently abrogate Xi at the pro-B stage in female cells, however the mechanistic consequences of this deregulation have not been revealed [36]. Xi is restored in activated mature B cells in an YY1-dependent fashion, pointing to dynamic changes in expression of X-linked genes during B cell development.
The role of lncRNA in human leukemias
In addition to our understanding of how lncRNAs shape normal lymphoid development, more information is emerging that highlights the role of lncRNAs in malignant lymphopoiesis. Recent studies have identified a host of putative lncRNA genes that are dysregulated in various leukemias, and the proliferation of single cell transcriptomics promises more discoveries in the immediate future [20]. Analysis of several leukemias, including T cell acute lymphoblastic leukemia (T-ALL) and acute myeloid leukemia (AML), have revealed a new landscape of essential lncRNAs and their potentially novel mechanisms of action [37]. NOTCH1 activating mutations are the most frequent genetic lesions identified in T-ALL, suggesting that this signaling pathway is an essential driver of T-ALL pathogenesis [38, 39]. Through a comprehensive analysis of all noncoding transcripts dysregulated in T-ALL, Trimarchi et al. discovered the lncRNA Leukemia-induced noncoding activator RNA-1 (LUNAR1) [40]. LUNAR1 is upregulated by constitutive NOTCH1 signaling and drives T-ALL cell growth. Intriguingly, LUNAR1 achieves this effect through a cis-regulatory mechanism that promotes transcription of its neighboring gene IGF1R, which in turn promotes a proliferative cellular state. Curiously, IGF1R is also implicated in AML pathogenesis. The lncRNA IRAIN was identified as a paternally imprinted antisense transcript from the IGF1R locus [41]. IRAINs mechanism of action involves the formation of intrachromosomal DNA loops by this lncRNA at its site of transcription. This RNA:DNA interaction serves to coordinate previously distant intragenic enhancer sequences with the IGF1R locus. Analysis of AML patient samples however revealed a negative correlation between IRAIN transcription and IGF1R signaling activity. Despite our lack of clarity regarding this lncRNA in malignant states, there is still prognostic value in using the relative IRAIN transcriptional activity to stratify high and low risk AML patients. Comprehensive genomic and transcriptional analyses of AML patient samples have expanded clinically relevant lists of dysregulated lncRNAs [42, 43]. Although their precise contribution to leukemic etiology is not completely understood, there is enough data to distinguish sets of lncRNAs with positive or negative prognostic patient outcome. Recent work on AML-specific lncRNAs by Delas et al. has further refined our understanding of lncRNA-directed myeloid leukemia development [37]. Following up the identification of AML-specific lncRNAs, they utilized a library-based knockdown screen to individually study the developmental effect of these lncRNAs. This work identified a subset of 20 AML-specific lncRNAs that are required for in vivo disease development and highlighted the role that lncRNAs Pvt1 and Lilam play in AML progression but not normal myeloid cell development.
Conclusions
Despite the relative paucity of information regarding this class of noncoding RNA, it is clear that lncRNAs play an important role in regulating signaling circuits that maintain normal hematopoietic cell development and function. The lncRNAs we have highlighted here have all been found to participate in key HSC fate and differentiation pathways (Fig. 1). Due to the multifaceted nature of this class of regulators, it is important to gain a deeper understanding of their specific molecular functions. A more complete understanding of the genesis, conservation, and function of lncRNAs will reveal a plethora of information about how genomic elements are created, conserved, and functionalized along different evolutionary lineages. As lncRNAs display more lineage-specific pattern of expression than protein-coding transcripts, expression profiling of additional hematopoietic lineages is necessary for the identification and characterization of new lncRNA genes. We believe that better insight into the biology of blood cell-associated lncRNAs will create new avenues for prevention and treatment of hematopoietic malignancies and immune-mediated disorders.
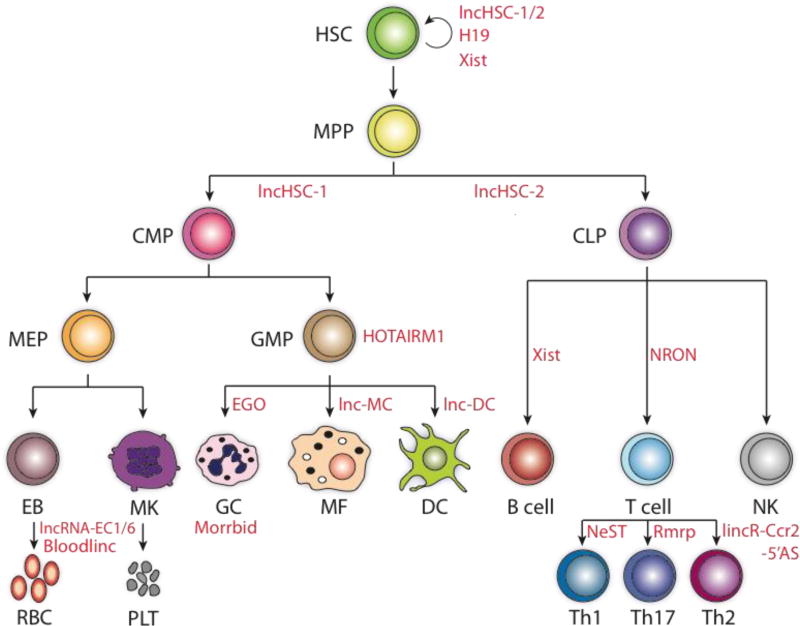
Figure 1. Control of mammalian hematopoiesis by lncRNA.
Long noncoding RNAs play key roles in maintenance of HSC homeostasis and development of terminally differentiated blood cells during hematopoiesis. LncRNA genes are listed in red. HSC, hematopoietic stem cell; MPP, multipotent progenitor cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte erythroid progenitor; GMP, granulocyte macrophage progenitor; EB, erythroblast; MK, megakaryocyte; GC, granulocyte; MF, macrophage; DC, dendritic cell; NK, natural killer; PLT, platelets; RBC, red blood cells.
Table 1.
Important lncRNAs in hematopoiesis.
| lncRNA | Cell Type | Subcellular location |
Mechanism of action | References |
|---|---|---|---|---|
| lncHSC-1/2 | HSC | Nuclear | Unknown | [12] |
| H19 | HSC | Nuclear | Allelic silencing | [16] |
| Xist | HSC/B cell | Nuclear | Allelic silencing | [18, 19, 17] |
| HOTAIRM1 | GMP | Nuclear | Enhancer/stabilizer | [24] |
| lncRNA-EC1-6 | Erythroblast | Nuclear | Unknown | [22] |
| Bloodlinc | Erythroblast | Nuclear | Enhancer | [22] |
| Morrbid | Granulocyte | Nuclear | Epigenetic silencing | [27] |
| EGO | Granulocyte | Nuclear | Enhancer/possible stablizer | [23] |
| lnc-MC | Macrophage | Cytoplasmic | microRNA sponge | [26] |
| lnc-DC | Dedritic cell | Cytoplasmic | STAT3 signaling | [25] |
| NRON | T cell | Cytoplasmic | Blocks nuclear import | [30] |
| NeST | Th1 T cell | Nuclear | Epigenetic silencing | [31] |
| Rmrp | Th17 T cell | Nuclear | Transcriptional enhancer | [34] |
| lincR-Ccr2-5’AS | Th2 T cell | Nuclear | Unknown | [33] |
Acknowledgments
This work was supported by the National Cancer Institute of the NIH under Award P30CA033572. This work was funded in part by the NIH AI125615, the Nesvig Lymphoma Research Fund at the City of Hope and the Research Career Development award (to M.P.B.) by the STOP CANCER Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Compliance with Ethical Standards
Conflict of Interest
Nathaniel Magilnick and Mark P. Boldin declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Morrison JV. Kerostasia, The Dictates of Fate, and the Will of Zeus in the Iliad. Arethusa. 1997;30:276–96. [Google Scholar]
- 2.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabezas-Wallscheid N, Eichwald V, de Graaf J, Lower M, Lehr HA, Kreft A, et al. Instruction of haematopoietic lineage choices, evolution of transcriptional landscapes and cancer stem cell hierarchies derived from an AML1-ETO mouse model. EMBO molecular medicine. 2013;5(12):1804–20. doi: 10.1002/emmm.201302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The Dimensions, Dynamics, and Relevance of the Mammalian Noncoding Transcriptome. Trends in genetics : TIG. 2017;33(7):464–78. doi: 10.1016/j.tig.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nature reviews Immunology. 2016;16(5):279–94. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 8.Montagner S, Deho L, Monticelli S. MicroRNAs in hematopoietic development. BMC immunology. 2014;15:14. doi: 10.1186/1471-2172-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nature immunology. 2008;9(8):839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 10.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Luo M, Jeong M, Sun D, Park HJ, Rodriguez BA, Xia Z, et al. Long non-coding RNAs control hematopoietic stem cell function. Cell stem cell. 2015;16(4):426–38. doi: 10.1016/j.stem.2015.02.002. Identification and characterization of lnc-HSC1/2 in hematopoietic stem cells. The first HSC-specific lncRNAs whose functions have been experimentally verified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell stem cell. 2014;15(4):507–22. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell stem cell. 2007;1(5):578–91. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 16.Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500(7462):345–9. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727–42. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaligne R, Heard E. X-chromosome inactivation in development and cancer. FEBS letters. 2014;588(15):2514–22. doi: 10.1016/j.febslet.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Su Q, Liu GQ, Gong L, Zhang W, Zhu SJ, et al. Skewed X chromosome inactivation of blood cells is associated with early development of lung cancer in females. Oncol Rep. 2006;16(4):859–64. [PubMed] [Google Scholar]
- 20.Schwarzer A, Emmrich S, Schmidt F, Beck D, Ng M, Reimer C, et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nature communications. 2017;8(1):218. doi: 10.1038/s41467-017-00212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–37. doi: 10.1182/blood-2013-12-544494. Analysis and verification of functional lncRNAs in erythropoietic lineage development and confirmed the identity of erythro-lineage specific lncRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–81. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109(12):5191–8. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–34. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–3. doi: 10.1126/science.1251456. Identification of the lncRNA lnc-DC which is important for dendritic cell differentiation. [DOI] [PubMed] [Google Scholar]
- 26.Chen MT, Lin HS, Shen C, Ma YN, Wang F, Zhao HL, et al. PU.1-Regulated Long Noncoding RNA lnc-MC Controls Human Monocyte/Macrophage Differentiation through Interaction with MicroRNA 199a-5p. Mol Cell Biol. 2015;35(18):3212–24. doi: 10.1128/MCB.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537(7619):239–43. doi: 10.1038/nature19346. Discovery and functional verification of the myeloid cell lncRNA Morrbid which acts to repress a target locus in an allele specific manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nature immunology. 2015;16(12):1282–91. doi: 10.1038/ni.3299. Distinct profiles of lncRNA landscape in B and T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macian F. NFAT proteins: key regulators of T-cell development and function. Nature reviews Immunology. 2005;5(6):472–84. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 30.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 31.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spurlock CF, 3rd, Tossberg JT, Guo Y, Collier SP, Crooke PS, 3rd, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nature communications. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nature immunology. 2013;14(11):1190–8. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528(7583):517–22. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Brazao TF, Johnson JS, Muller J, Heger A, Ponting CP, Tybulewicz VL. Long noncoding RNAs in B-cell development and activation. Blood. 2016;128(7):e10–9. doi: 10.1182/blood-2015-11-680843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syrett CM, Sindhava V, Hodawadekar S, Myles A, Liang G, Zhang Y, et al. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 2017;13(10):e1007050. doi: 10.1371/journal.pgen.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delas MJ, Sabin LR, Dolzhenko E, Knott SR, Munera Maravilla E, Jackson BT, et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. eLife. 2017;6 doi: 10.7554/eLife.25607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tosello V, Ferrando AA. The NOTCH signaling pathway: role in the pathogenesis of T-cell acute lymphoblastic leukemia and implication for therapy. Therapeutic advances in hematology. 2013;4(3):199–210. doi: 10.1177/2040620712471368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Current opinion in genetics & development. 2004;14(1):48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158(3):593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Li W, Sun Y, Yu D, Wen X, Wang H, et al. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 2014;42(15):9588–601. doi: 10.1093/nar/gku549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez-Dominguez JR, Hu W, Gromatzky AA, Lodish HF. Long noncoding RNAs during normal and malignant hematopoiesis. International journal of hematology. 2014;99(5):531–41. doi: 10.1007/s12185-014-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–84. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]