Abstract
Amyotrophic Lateral Sclerosis (ALS) is a lethal neurodegenerative disorder that primarily affects motor neurons. Dominant mutations in the RNA binding protein Fused in Sarcoma (FUS) have been identified as causative factors of ALS. Mutation, R495X, results in a premature stop codon and induces an aggressive disease phenotype by a largely unknown process. Here, we employ CLIP-Seq, RNA-Seq and Ribo-Seq in cultured neurons expressing R495X or wild-type FUS to identify the mutation effects on the FUS targetome and on the neuronal transcriptome at the expression and translation level, simultaneously. We report that, unlike wild-type FUS that binds on precursor mRNAs (pre-mRNAs), R495X binds mature mRNAs in the cytoplasm. R495X has a moderate effect on target mRNA expression and its binding induces only modest expression changes. In contrast, we find that R495X controls the translation of genes that are associated with mitochondria function and results in significant reduction of mitochondrial size. Importantly, we show that introduction of the 4FL mutation that alters binding of R495X to RNA, partially abrogates R495X-induced effects on mRNA translation, mitochondrial size and neurotoxicity. Our findings uncover a novel RNA-mediated pathway of FUS R495X-induced neurotoxicity that affects mitochondria morphology and provide insight to previous studies associating mitochondria dysfunction to ALS.
Introduction
Amyotrophic Lateral Sclerosis (ALS) is a lethal neurological disease characterized by progressive neurodegeneration of the upper and lower motor neurons1. Patients usually progress to fatal paralysis and die from dysfunction of the respiratory system. Dominant mutations in Fused in Sarcoma (FUS) gene have been identified in familial cases of ALS (FALS)2–4. FUS encodes an RNA binding protein composed of 526 amino acids; it localizes primarily to the nucleus and consists of seven domains: QGSY-rich, Gly-rich, RNA recognition motif (RRM), Arg-Gly-Gly1 (RGG1), Zinc finger (ZnF), RGG2 and nuclear localization signal (NLS)4. The majority of the disease-causing mutations localize at the C-terminal region, disrupt the NLS and result in abnormal FUS cytoplasmic localization. FUS mislocalization has been hypothesized to disrupt its nuclear function and promote gain of function in the cytoplasm of affected neurons5–7. R495X, a FUS mutation in which the Arginine at position 495 changes into a premature stop codon, results in complete loss of the NLS and shows a severe disease phenotype8,9. Despite the severity of the phenotype, the mechanistic effects and the implications in neurons are still unknown.
Previously, we uncovered the genome-wide RNA binding profile of FUS in neurons and reported that it preferentially binds evolutionarily conserved introns of other RNA-binding proteins, altering their expression10. Similarly, other groups have shown that FUS controls the mRNA splicing of its target genes6,11–15. However, these studies identified only minor phenotypic effects, even after FUS knockdown/knockout experiments, raising the possibility that the disease phenotype is the result of a gain instead of loss of function. Overexpression of wild-type FUS in mice, Drosophila melanogaster and rats results in neurotoxicity16–18, while disease mutant FUS causes motor neuron degeneration via toxic gain of function in mice19. Furthermore, the gene expression profile that results from overexpression of the disease-associated FUS mutant in HEK293T cells is closer to that of wild-type FUS overexpression than knockdown20 indicating that cytoplasmic accumulation results in a gain of function. This is further supported by the inability of wild-type FUS to rescue mutant FUS phenotype in C. elegans21.
Recently, several reports demonstrated that FUS can form liquid droplets that are separated from solution in vitro and in vivo. The formation of these droplets is a reversible process that is dependent on the FUS low complexity domain22,23, concentration and incubation temperature. FUS mutations have been shown to accelerate the liquid-to-solid phase transition converting reversible to irreversible hydrogels24,25 and impairing ribonucleoprotein granule formation. However, it is still unclear how this relates to the disease phenotype and how FUS gain of function leads to neurodegeneration.
To examine the aggressive disease phenotype of FUS R495X and identify the involved molecular pathways, we probed its target RNA repertoire and the gene expression and translation in neurons. We find that R495X has an altered gene binding profile which leads to neurotoxicity, perturbs translation of transcripts associated with mitochondrial function and causes reduction of the mitochondria size. By altering the binding between RNA targets and R495X, we reveal a partial abrogation of the effects of the mutation, supporting RNA dysregulation as an important player in FUS R495X mediated neurotoxicity.
Results
FUS R495X mutant mislocalizes to the cytoplasm
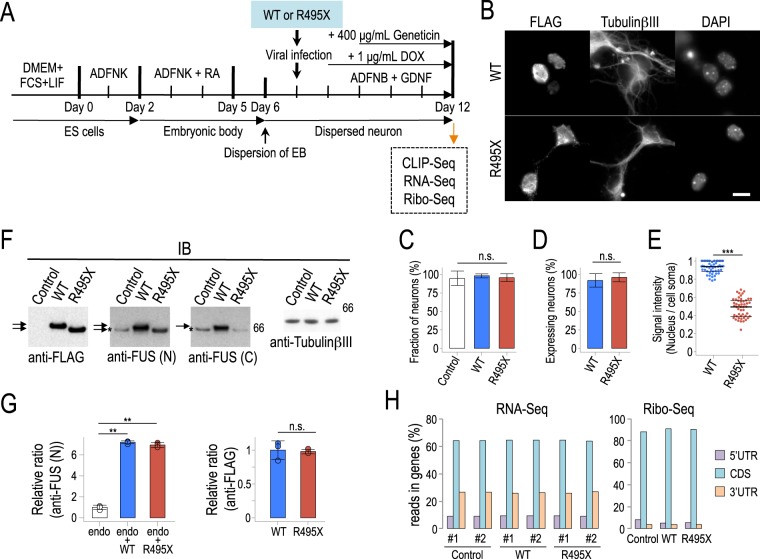
To investigate the consequences of the R495X mutant in neurons, we set up an inducible expression system based on lentiviral transduction of FLAG-tagged human R495X (NFLAG-hFUSR495X, R495X) or wild-type FUS (NFLAG-hFUSWT, WT) in neurons that we differentiated from mouse Embryonic Stem (ES) cells10. After doxycycline (DOX) induction and Geneticin selection we obtain a highly enriched population of neurons expressing the transgenes (Fig. 1A). We analyzed the localization of exogenous proteins by immunostaining using anti-FLAG along with anti-TubulinβIII antibody and DAPI staining (Fig. 1B). We confirmed that the fraction of neurons was unaffected (Fig. 1C, Supplementary Fig. S1) by WT or R495X overexpression and that the exogenous proteins were expressed in the majority of neurons (Fig. 1D, Supplementary Fig. S1) highlighting the quality of our selection process. We found that while WT localized to the nucleus, R495X mislocalized to the cytoplasm (Fig. 1B), as expected due to loss of NLS, and consistent with a previous report9. Quantification of the fluorescence intensity showed that 93% of WT and only 48% of R495X was detected in the nuclei (Fig. 1E).
Figure 1.
CLIP-Seq, RNA-Seq and Ribo-Seq on human FUS WT- or R495X-expressing neurons. (A) Schematic of the experimental design for CLIP-Seq, RNA-Seq and Ribo-Seq library preparation from mouse ES cell derived neurons. (B) Immunostaining for cellular localization of NFLAG-hFUSWT (WT) and NFLAG-hFUSR495X (R495X) in neurons. Anti-FLAG (left), anti-TubulinβIII (middle) and DAPI stain (right) are shown. Scale bar, 10 μm. (C) Bar plot for fraction of neurons over all cells for Control, WT- and R495X-expressing cells. Error bars indicate standard deviation (N = 18). n.s., not significant (p = 0.253), one-way ANOVA test. (D) Bar plot for percentage of neurons expressing WT and R495X. Error bars indicate standard deviation (N = 18). n.s., not significant (p = 0.115), two-tailed Student’s t-test. (E). Beeswarm plot for ratio of nuclear localization of WT and R495X in neurons. Ratio represents fluorescence intensity in the nucleus over cell soma. N = 50. ***p = 2.83 × 10−40, two-tailed Student’s t-test. Black bars represent median, 25th and 75th percentiles. (F) Immunoblots showing expression levels using the indicated antibodies for FLAG, amino- (FUS(N)) and carboxyl-terminal FUS (FUS(C)) and TubulinβIII. Arrows and asterisk indicate exogenous and endogenous FUS proteins, respectively. Protein standard is shown on the right. (G) Quantification of endogenous (endo) and exogenous WT and R495X FUS proteins detected by anti-FUS (N) (left) and anti-FLAG (right). N = 3. **p < 0.0001 one-way ANOVA and p < 0.01 in post Tukey’s HSD test for left graph. n.s., not significant (p = 0.827) two-tailed Student’s t-test for right graph. (H) Distribution of gene mapping reads in the 5′UTR, coding sequence (CDS) and 3′UTR for RNA-Seq and Ribo-Seq.
Probing the FUS targetome, gene expression and translation in R495X-expressing neurons
To evaluate R495X effects on the RNA target repertoire and on gene expression and translation levels, we performed high-throughput sequencing after crosslinking and immunoprecipitation (CLIP-Seq), RNA-Seq and Ribosome profiling (Ribo-Seq) in neurons expressing WT or R495X. First, we analyzed the expression levels of the exogenous proteins by immunoblot using anti-FLAG, anti-FUS amino-terminal, and anti-FUS carboxyl-terminal antibodies (Fig. 1F). Anti-FLAG antibody showed that R495X migrated faster than WT, consistent with its shorter length. The anti-FUS amino-terminal antibody recognized all proteins, including endogenous FUS, while the anti-FUS carboxyl-terminal antibody recognized WT and endogenous FUS but not R495X. Our results indicate that WT and R495X were expressed at similar levels, ~7 times higher than endogenous FUS (Fig. 1G). Consistent with a previous report12, we observed a reduction of endogenous mouse FUS protein in both WT and R495X expressing neurons (Fig. 1F and Supplementary Fig. S6), indicating that FUS autoregulation is active in our system. After CLIP of FLAG-tagged proteins and 5′end RNA radiolabeling, we observed a specific smear in both WT and R495X autoradiographs corresponding to bound and crosslinked RNA (Supplementary Fig. S2). More than 75% of CLIP sequenced reads mapped to the mouse genome for both WT and R495X, demonstrating the high CLIP specificity and showing that both exogenous proteins bound to RNA in neurons. Comparing FUS WT targetome to a control FUS CLIP library expressed at endogenous levels10 we found significant correlation (R = 0.6, Pearson’s correlation, p < 10-16) indicating that overexpression of the human FUS homolog only modestly affected gene binding. Similarly, for RNA-Seq, approximately 60% and 35% of the sequenced reads mapped in the coding sequence and UTRs, respectively, while the corresponding percentages for Ribo-Seq were 90% and 6% (Fig. 1H). This is consistent with the expectation of higher ribosome occupancy in the coding region and illustrates the protocol specificity.
R495X binds mature mRNAs in contrast to WT that binds precursor mRNAs
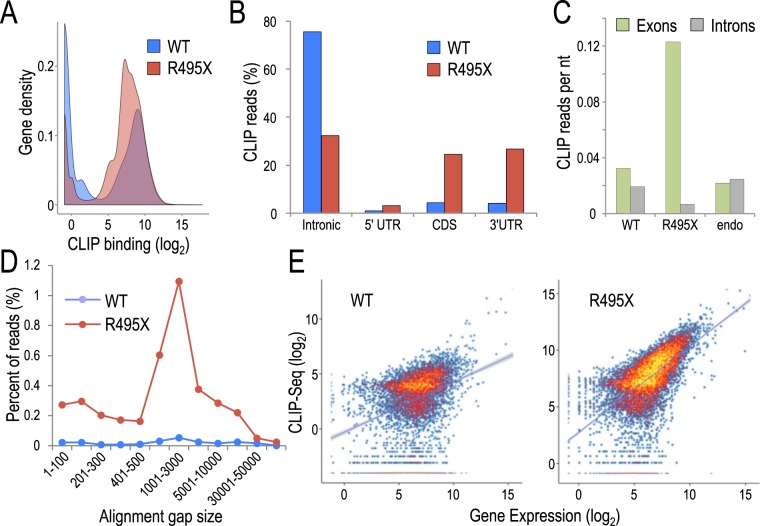
Next we wished to identify potential differences in the gene targeting profile of WT and R495X. We found that R495X bound to substantially more genes than WT, likely due to its cytoplasmic localization (Fig. 2A). Analysis of the genic distribution of CLIP reads showed that more than ~75% of WT reads derived from introns, whereas only ~10% from exons (Fig. 2B). In contrast, only ~32% of R495X CLIP reads mapped in introns and ~54% in exons. To evaluate the binding preference for these regions, excluding stochastic differences attributed to their length, we normalized the number of reads in exons and introns by the aggregate length of these elements. We found that R495X bound substantially more on exonic regions, in contrast to WT and endogenous FUS10 that showed no binding preference (Fig. 2C).
Figure 2.
Identification of RNA binding property of R495X compared to WT. (A) Gene density plot of CLIP binding for WT (blue) and R495X (red). (B) Distribution of CLIP-Seq reads in introns, 5′UTR, coding sequence (CDS) and 3′UTR for WT (blue) and R495X (red). (C) Distribution of CLIP-Seq reads in exons (green) and introns (grey) normalized by the total length of these elements for WT, R495X and endogenous FUS (endo). (D). Alignment gap size distribution of CLIP-Seq reads for WT (blue) and R495X (red) reads. Only alignment gaps overlapping annotated introns are used. (E) Scatter plot for FUS binding versus gene expression levels for WT (left) and R495X (right). Color indicates gene density with yellow and blue indicating higher and lower values respectively.
Our findings indicate that, contrary to WT that binds on pre-mRNAs, R495X mostly binds on mature mRNAs and this is attributable to R495X mutation and not FUS overexpression. To further test this, we reasoned that R495X binding on mature mRNAs would be reflected by increased binding on exon-exon junctions. We quantified exon-exon junction binding by measuring the sequencing reads that align on the genome with alignment gap sizes of 1,000 to 3,000 nt, corresponding to spliced intronic sequences26. Comparing the two conditions, we found that R495X bound on exon-exon junctions at substantially higher levels than WT (Fig. 2D) further supporting that R495X mostly binds on mature mRNAs.
We hypothesized that R495X binding is mostly directed by protein localization in the cytoplasm and less by specific acquired binding preferences. We compared CLIP binding with gene expression levels and observed a substantially higher positive correlation (R = 0.61, Pearson’s correlation) for R495X than WT (R = 0.34, Pearson’s correlation) (Fig. 2E) indicating that R495X mostly binds on mRNAs in an expression-directed way. We also interrogated the CLIP data for overrepresented sequences using a 6mer enrichment approach27. Similar to previous reports6,10–12,15 we found modestly overrepresented sequences that only explained a small fraction of the CLIP reads (Supplementary Tables S1 and S2). Collectively, our results show a distinct RNA binding profile for the mutant, binding on mature mRNAs in the cytoplasm, guided mostly by protein localization and less by sequence-specificity.
WT and R495X have modest effect on gene expression
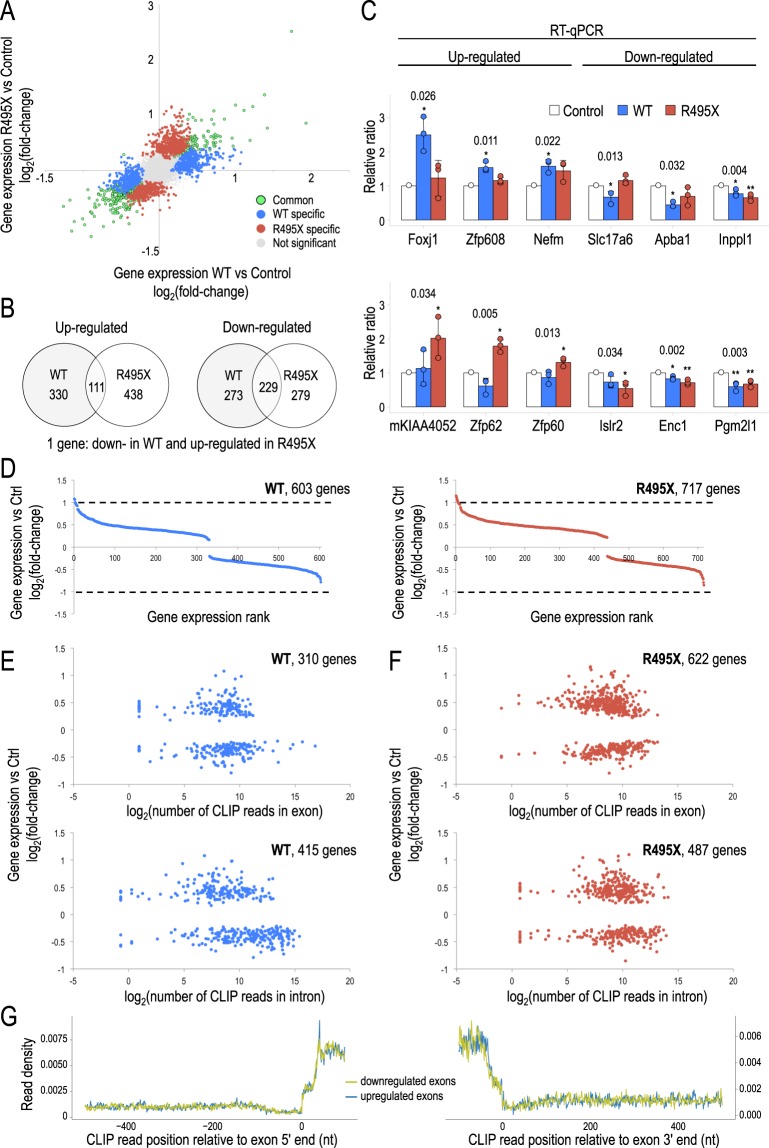
To identify gene expression changes specifically induced by R495X and to deconvolute the mutation from the overexpression effect, we first probed gene expression changes in WT and R495X compared to a control library, prepared from non-transduced neurons, and focused on unique changes for each condition (Fig. 3A). Overall, at a significance level of 0.05, we identified 603 (up: 330 in Supplementary Table S3, down: 273 in Supplementary Table S4) and 717 (up: 438 in Supplementary Table S5, down: 279 in Supplementary Table S6) differentially expressed genes for WT and R495X compared to Control, respectively (Fig. 3B). We identified 340 (up: 111, down: 229) genes differentially expressed consistently in both conditions and only 1 gene in non-congruent direction between WT and R495X (Supplementary Table S7). We confirmed the observed changes by RT-qPCR for three, randomly selected, up- and down-regulated genes specific to WT or R495X (Fig. 3C). All tested genes showed significant expression changes in concordance with the RNA-Seq results. To gain insight into the potential function of the differentially expressed genes we performed gene ontology (GO) enrichment analysis for up- and down-regulated genes in WT (Supplementary Table S8) and R495X (Supplementary Table S9). WT was enriched for neuronal function terms such as ionotropic glutamate receptor signaling pathway whereas R495X for cellular processes such as regulation of cellular component organization.
Figure 3.
Differential gene expression analysis for WT- and R495X-expressing neurons. (A) Scatter plot of WT versus control and R495X versus control differential gene expression. Differential gene expression is quantified as log2(fold-change). (B) Venn diagram showing the number of up- (left) and down- (right) regulated genes in WT- and R495X-expressing neurons. (C) Bar plots of expression measured by RT-qPCR for up- (left) and down- (right) regulated genes that are differentially expressed in WT- (top) or R495X-expressing neurons (bottom). Values indicate P-values of one-way ANOVA test. *p < 0.05, **p < 0.01, post Tukey’s HSD test. Error bar indicates standard deviation (N = 3). (D) Gene differential expression for ranked list of differentially expressed genes for WT- (left) and R495X-expressing neurons (right). Dashed lines indicate 2-fold difference. (E,F) Scatter plot of gene differential expression versus FUS binding levels for WT (E) and R495X (F). FUS binding is quantified in exons (top) and introns (bottom). (G) R495X CLIP binding density upstream (left) and downstream (right) of up- and down-regulated exons. Position 0 corresponds to the 5′ end (left) and 3′ end (right) of exons.
Interestingly, we found that only 2 and 7 differentially expressed genes in WT and R495X, respectively, had expression changes higher than 2-fold compared to control. In both conditions, more than 89% (WT: 536/603, R495X: 670/717) of differentially expressed genes had differences smaller than 1.5-fold. To examine the effect of R495X on gene alternative splicing we analyzed the differential expression of exons and found only 110 exons on 99 genes with significant changes specific to R495X. Interestingly, the majority of them (81%) had less than 2-fold difference (Supplementary Table S10). Similarly, for WT we identified only 129 differentially expressed exons for 80 genes, 62% of which with less than 2-fold change (Supplementary Table S11). GO analysis for genes with differentially expressed exons in R495X-expressing neurons (Supplementary Table S12) revealed few enriched categories including second-messenger-mediated signaling, while more terms including cell-cell adhesion were enriched in WT-expressing neurons (Supplementary Table S13). Our results indicate that expression and splicing effects attributable either to overexpression or mutation are modest (Fig. 3D).
To further support this finding we tested whether the observed changes could be explained by R495X binding. To account for binding differences in exonic and intronic regions we quantified binding levels independently for the two classes. For WT, 310 and 415 of the 603 differentially expressed genes were bound in exons and introns, respectively (Fig. 3E, Supplementary Table S14). For R495X, the corresponding numbers were 622 and 487 of the 717 differentially expressed genes (Fig. 3F, Supplementary Table S15). In both conditions, comparing the differential expression with CLIP binding levels, we found no correlation (R2 < 0.2, Pearson’s correlation). We also found no correlation (R2 < 0.1, Pearson’s correlation) when we plotted exon differential expression against FUS binding indicating that, in general, R495X binding did not mediate substantial splicing changes (Supplementary Fig. S3). To account for potential intronic, instead of exonic, FUS binding that affects upstream or downstream exon selection, we measured R495X binding around up- and down-regulated exons (Fig. 3G). We found that the binding density was identical for up- and down-regulated exons and there was no preferential binding at any position around the exons. Again, we found a prominent density increase within the exons highlighting the mature mRNA binding preference of R495X. Collectively our results suggest that the majority of the observed gene expression and splicing changes are probably due to secondary effects and not directly related to FUS binding.
R495X disturbs the translation efficiency of mitochondrial associated genes
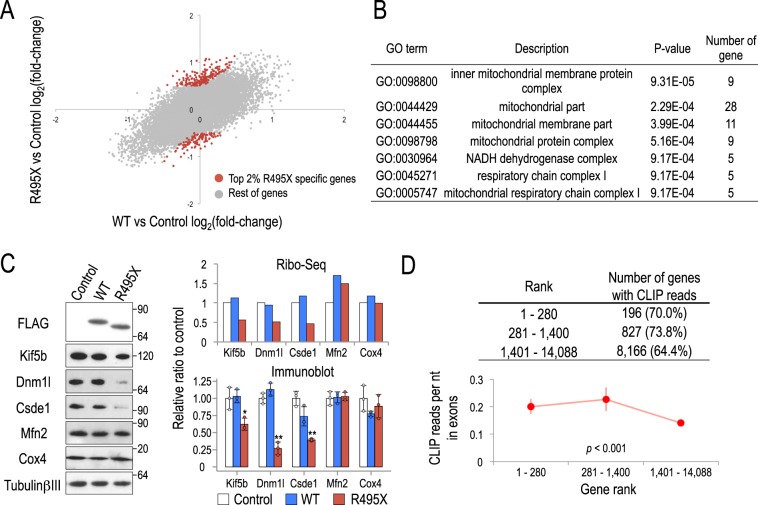
To examine R495X effects on mRNA translation we analyzed differential translation between WT and control and R495X and control libraries. Similar to RNA-Seq, the control Ribo-Seq library was prepared from non-transduced neurons that express endogenous FUS. To select R495X-specific differentially translated genes we first identified genes with no difference between WT and control (−0.5 < log2(fold-change) <0.5), selected those with high differential translation between R495X and control (log2(fold-change) <−0.5 or >0.5) and ranked them by p-value. Out of the top 2% (280) differentially translated genes (Fig. 4A and Supplementary Table S16) only 33 (~12%) displayed significant expression differences indicating that differential translation cannot be attributed to changes in mRNA abundance. Interestingly, GO analysis revealed that the top differentially translated genes were associated with mitochondria function (Fig. 4B and Supplementary Table S17) raising the possibility that R495X-induced protein changes affect mitochondria.
Figure 4.
Differential gene translation analysis for WT- and R495X-expressing neurons. (A) Scatter plot of WT versus control and R495X versus control differential gene translation. The top 2% (280) differentially translated genes that are specific to R495X are shown in red. (B) Gene ontology enriched terms for the top 2% (280) differentially translated genes specific to R495X compared to the remaining genes. (C) Immunoblots of differentially translated genes in R495X-expressing neurons (left). Bar plots for gene translational levels quantified from Ribo-Seq (top right) and protein band intensities quantified from immunoblots (bottom right). Protein standard is shown on the right. Band intensities were normalized by TubulinβIII and the average value for control was set to 1. P-values in one-way ANOVA are, 0.01, < 0.0001, 0.001, 0.725 and 0.257 for Kif5b, Dnm1l, Csde1, Mfn2 and Cox4, respectively. *p < 0.05, **p < 0.01 to control in post Tukey’s HSD test. Error bar indicates standard deviation (N = 3). (D) Number of genes that are bound by R495X for gene sets of decreasing differential translation in R495X (top) and R495X binding density in gene exons for the same gene sets (bottom). Error bars indicate standard error. P-value, one-way ANOVA test.
To further test this, we used immunoblot to measure the protein levels of the mitochondria associated genes Kif5b, Dnm1l and Csde1 along with Mfn2 and Cox4 that are also mitochondria related but their translation was unaffected in R495X. We found that the protein levels of Kif5b (p < 0.05), Dnm1l (p < 0.01) and Csde1 (p < 0.01) were significantly reduced in R495X-expressing neurons compared to control and WT, consistent with the ribosome profiling results (Fig. 4C). As expected, Mfn2 and Cox4 showed no significant difference (p > 0.05) in any combination. Interestingly, we also found that R495X targeted a marginally higher percent and had a significantly higher binding density on the top ranked differentially translated genes than the lower ranked (Fig. 4D). Importantly, we also observed a similar protein level reduction of Csde1 and Dnm1l in neurons expressing substantially less R495X, indicating that this phenotype is not a result of abnormally high R495X expression (Supplementary Fig. S6). Collectively, our results indicate that R495X binds and changes the translation profile of a set of target genes, particularly related to mitochondria function.
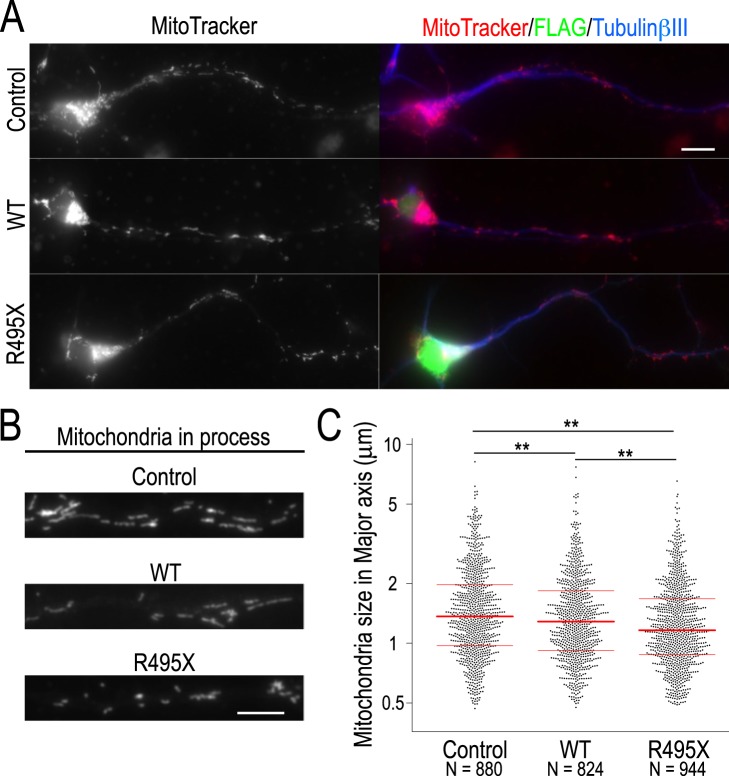
R495X reduces mitochondria size
We observed that Dnm1l translation, a known regulator of mitochondrial fission28, was significantly reduced in R495X-expressing neurons (Fig. 4C) raising the possibility that mitochondria morphology might be altered. To test this, we visualized mitochondria with MitoTracker staining and subjected them to immunostaining with anti-FLAG and anti-TubulinβIII antibodies. We found that, in all conditions, mitochondria were localized mainly in the cell soma but were also distributed in the neuronal processes (Fig. 5A). Measuring mitochondria size by quantifying their major axis length, we found that mitochondria that were localized in the processes of R495X-expressing neurons were smaller than those in control and WT-expressing neurons (Fig. 5B). The average length of the major mitochondrial axis was 1.63 ± 0.03 μm, 1.50 ± 0.03 μm and 1.38 ± 0.02 μm for control and WT- and R495X-expressing neurons respectively. We found the difference to be statistically significant for all combinations (Fig. 5C, p < 0.01). We observed a consistent result in neurons expressing lower levels of R495X, while no significant shortening of mitochondria was observed in neurons expressing lower levels of WT (Supplementary Fig. S7), suggesting that FUS overexpression results in mitochondria shortening and R495X significantly increases the effect.
Figure 5.
Mitochondria size for control, WT- and R495X-expressing neurons. (A) Images of mitochondria stained with MitoTracker (left) and merged images stained with anti-FLAG antibody (green), anti-TubulinβIII (blue) and MitoTracker (red) (right) for control (top), WT- (middle) and R495X-expressing neurons (bottom). Scale bar, 10 μm. (B) Magnified images of MitoTracker stained neuronal processes for control (top), WT- (middle) and R495X-expressing neurons (bottom). Scale bar, 5 μm. (C) Beeswarm plot for mitochondria major-axis length in control, WT- and R495X-expressing neurons. The number of analyzed mitochondria is indicated at the bottom. Red lines indicate the median, 25th and 75th percentiles. p < 0.0001, one-way ANOVA. **p < 0.01, post Tukey’s HSD test.
4FL mutation in the RNA recognition motif of R495X alters its binding
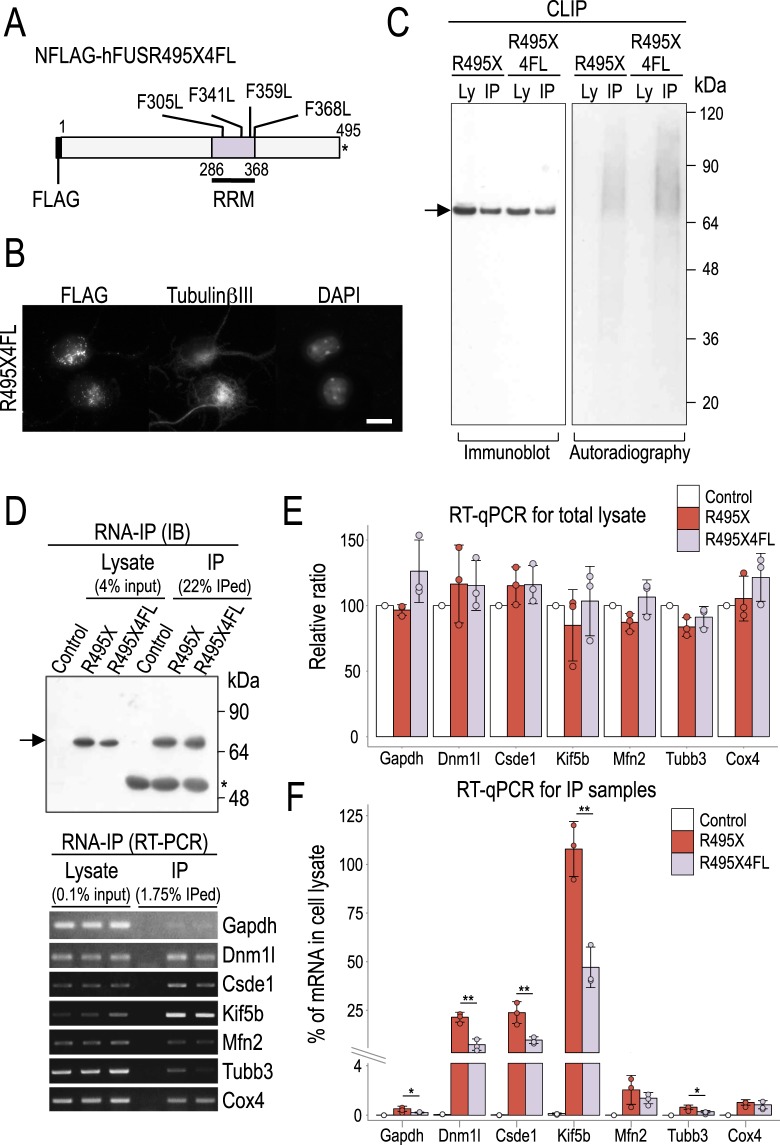
To test the specificity of the observed effects with respect to the RNA binding of R495X we substituted the Phenylalanines at positions 305, 341, 359 and 368, within the FUS RRM, with Leucine (4FL). A previous report had shown that the introduction of 4FL in WT FUS reduces its RNA binding ability29. To test this, we prepared a construct coding for the amino-terminally FLAG tagged R495X4FL mutant (NFLAG-hR495X4FL, R495X4FL, Fig. 6A) and expressed it in neurons. Immunostaining revealed that R495X4FL localization was similar to R495X (Fig. 6B). Using CLIP to compare the RNA binding of R495X4FL to R495X, we found that, unexpectedly, both proteins showed indistinguishable results highlighted by a smear of bound RNA in the autoradiography, indicating that R495X4FL could efficiently bind RNA (Fig. 6C).
Figure 6.
The RNA binding profile of R495X4FL mutant. (A) Schematic of the NFLAG-hFUSR495X4FL (R495X4FL) mutant structure. The 4 Phenylalanines at the indicated positions of the FUS RRM were substituted to Leucines. Asterisk indicates stop codon. (B) Immunostaining images for anti-FLAG (left), anti-TubulinβIII (middle) and DAPI stain (right) show the cellular localization of R495X4FL in neurons. Scale bar, 10 μm. (C) Cell lysate (Ly) and CLIP samples (IP) with a rabbit anti-FLAG polyclonal antibody from neurons expressing R495X or R495X4FL were analyzed by immunoblot with a mouse monoclonal anti-FLAG antibody (left panel) and autoradiography (right panel). Arrow indicates FLAG tagged proteins. The numbers at the right indicate protein standards. (D) Immunoblot of cell lysate and IP samples using anti-FLAG antibody (top). Arrow indicates FLAG tagged proteins. The numbers at the right indicate protein standards. Asterisk indicates IgG heavy chain. RT-PCR using RNA samples from cell lysate and IP samples for the genes indicated at the right (bottom). (E) Gene expression levels measured by qPCR from total RNA from cell lysate. Control expression was set to 100% and the relative gene expression was measured. Error bar indicates standard deviation (N = 3). No statistically significant difference was observed for any gene. P-values in one-way ANOVA are, 0.077, 0.574, 0.250, 0.579, 0.079, 0.051 and 0.243 for Gapdh, Dnm1l, Csde1, Kif5b, Mfn2, Tubb3 and Cox4, respectively. (F) Gene expression levels measured by qPCR for RNA from IP samples. mRNA abundance is measured as percent of input. Lower part corresponds to [0, 4]%, while upper part to [4, 125]%. P-values in one-way ANOVA are, 0.006, < 0.0001, 0.0004, < 0.0001, 0.035, 0.001 and 0.004 for Gapdh, Dnm1l, Csde1, Kif5b, Mfn2, Tubb3 and Cox4, respectively and *p < 0.05 and **p < 0.01 between R495X and R495X4FL in post Tukey’s HSD test. Error bar indicates standard deviation (N = 3).
Previous reports have shown that FUS binds RNA through the Gly-rich, RGG1, ZnF and RGG230,31 in addition to the RRM. Therefore, we hypothesized that while the binding ability of R495X4FL might be largely unaffected, its binding specificity might be disturbed. To test this, we performed RNA-immunoprecipitation (RNA-IP) for 5 mitochondria associated genes along with Gapdh and Tubb3. Both R495X4FL and R495X were specifically precipitated and RT-PCR products showed a specific band for each tested gene in the Lysate and IP samples (Fig. 6D). We used qPCR to quantify the relative abundance of PCR products setting the mRNA abundance of the control total cell lysate to 100%. We observed no significant difference between gene expression levels under any condition (p > 0.05, Fig. 6E), indicating that neither R495X4FL nor R495X affected the mRNA abundance of the tested genes. Then, we measured the mRNA IP abundance as a percent of mRNA abundance in total cell lysate. We observed that R495X4FL bound with significantly (p < 0.05) lower affinity than R495X on 5 out of 7 tested genes (Dnm1l, Csde1, Kif5b, Gapdh and Tubb3) and with marginally lower affinity on the other two (Mfn2 and Cox4) (Fig. 6F) indicating that 4FL altered the binding features of R495X.
4FL mutation partially rescues R495X-induced mitochondria shortening and neurotoxicity
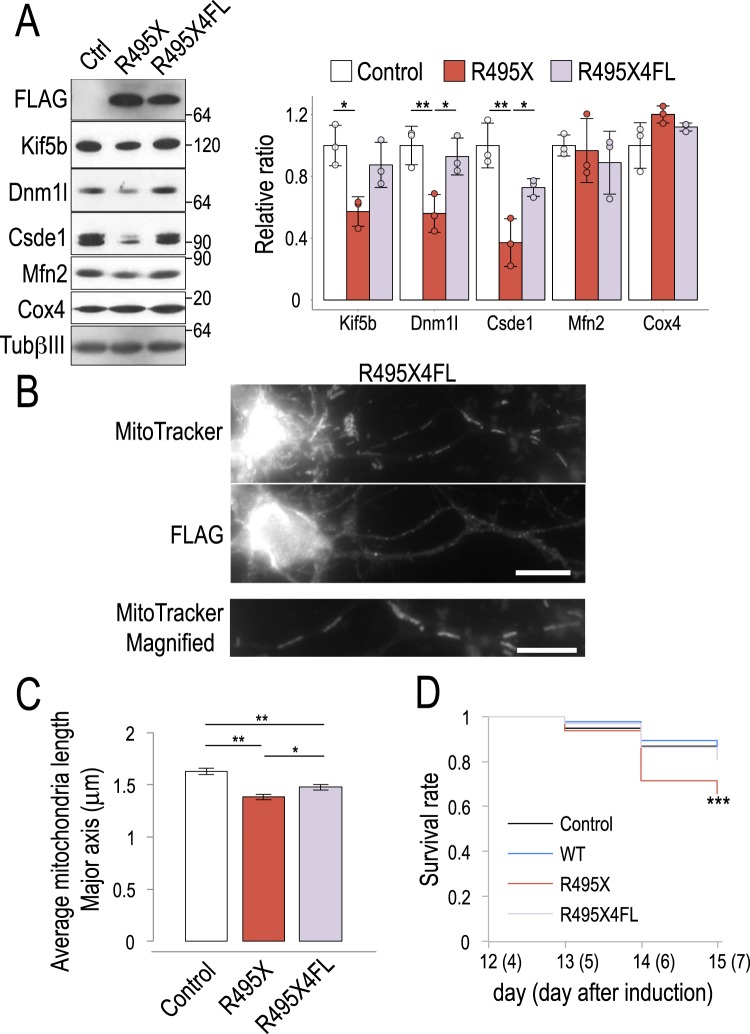
To test whether the altered binding specificity of R495X4FL also changed the protein output of the affected genes, we prepared cell lysates from control, R495X- and R495X4FL-expressing neurons and analyzed them by immunoblot. We found that Dnm1l, Csde1 and Kif5b had no significant protein level changes in R495X4FL compared to control which indicates that 4FL reverted the observed protein effects induced by R495X (Fig. 7A). To test whether 4FL could also rescue the R495X-induced mitochondria size reduction in neuronal processes, we stained neurons expressing R495X4FL with MitoTracker and employed immunostaining with anti-FLAG antibody (Fig. 7B). Interestingly, we found very few short mitochondria compared to R495X-expressing neurons (Fig. 5B). Quantification of the mitochondria size revealed that, in R495X4FL-expressing neurons, mitochondria were significantly longer than R495X with an average major axis length of 1.48 ± 0.03 μm (Fig. 7C). The observed length was close to WT FUS but still smaller than the control, indicating that FUS abnormal binding, due to overexpression- or mutation-induced mislocalization, affects mitochondria size. Interestingly, similar to WT, R495X4FL at lower expression level also did not result in significant reduction of mitochondria size (Supplementary Fig. S7) further highlighting the importance of the RRM in the phenotype. To test whether cells would compensate for the reduced mitochondria size by increasing their number, we measured mitochondria in neuronal processes but found no significant difference (Supplementary Fig. S4).
Figure 7.
R495X4FL partially rescues R495X induced mitochondria shortening and neurotoxicity. (A) Immunoblots for control, R495X- and R495X4FL-expressing neurons (left) and bar plot for normalized protein band intensities (right). Average band intensity for control was set to 1. Protein standard is shown on the right. Error bars indicate standard deviation (N = 3). P-values in one-way ANOVA are, 0.015, 0.009, 0.003, 0.731 and 0.093 for Kif5b, Dnm1l, Csde1, Mfn2 and Cox4, respectively and *p < 0.05 and **p < 0.01 in post Tukey’s HSD test. (B) Staining images using MitoTracker (top) and anti-FLAG antibody (middle) with magnified images of MitoTracker stained neuronal processes (bottom) of an R495X4FL-expressing neuron. Scale bar, 10 μm (middle) and 5 μm (bottom). (C) Average mitochondria major-axis length for control, R495X- and R495X4FL-expressing neurons. 880 mitochondria from 10 cells expressing R495X4FL were analyzed. Data for control and R495X are copied from Fig. 5C. Error bars indicate standard error. P-value in one-way ANOVA is < 0.0001 and *p < 0.05 and **p < 0.01 in post Tukey’s HSD test. (D) Cell survival curve for control, WT-, R495X- and R495X4FL-expressing neurons. The number of surviving cells at days 13–15 are expressed as the ratio over those surviving at day 12 (y-axis). The numbers in parenthesis indicate the day after induction. ***p < 0.003, log-rank test.
Then, we wished to test the toxicity of R495X to neurons. We analyzed the survival rate for Control, WT-, R495X- and R495X4FL-expressing cells and found that R495X induced cell death at significantly higher level compared to control and WT (p < 0.003) indicating that the mutation has adverse consequences on neurons. Strikingly we found that R495X4FL showed reduced evidence of cell death indicating that abolishing the RRM-directed targeting specificity of FUS can rescue the observed neurotoxicity (Fig. 7D). Collectively, our results show that 4FL partially rescues mitochondria size and neurotoxicity highlighting the key role of the RRM and the interaction of FUS with target RNA.
Discussion
FUS mutation R495X results in severe ALS phenotype. We used CLIP-Seq, RNA-Seq and Ribo-Seq to comprehensively evaluate the mutation outcome, in neurons. We set to identify its impact on the FUS targetome and on gene expression and translation simultaneously. We revealed key differences on RNA binding and showed that R495X binds on mature mRNAs in the cytoplasm, in contrast to WT that binds on precursor mRNAs. Our finding is corroborated by a previous report that showed that FUS R521G mutant also mislocalizes to the cytoplasm and has weak binding in gene introns6. To evaluate whether the abnormal binding of R495X affects mRNA expression we performed RNA-Seq in control, WT- and R495X-expressing neurons and showed that R495X has modest effects on target gene expression. Furthermore we found that there is no correlation between R495X binding and differential expression raising the possibility that any observed changes are mostly indirect effects. This is supported by reports that implicate FUS to pre-mRNA processing but not gene expression regulation6,10–15 and other studies that found minor expression changes between FUS mutants and WT7,32–34. Consistent with our results, reports using mice embryonic brain expressing NLS-deleted FUS (FUS ΔNLS) or HEK293 cells expressing R495X identified only few genes with more than 1.5-fold expression differences32,33. Other reports using spinal cord from R521C transgenic mice7 and P525L expressed motor neurons from gene edited iPS cells34 showed cell adhesion terms enriched in differentially expressed genes, similar to our results for WT but not R495X.
Interestingly, by measuring gene translation changes in control, WT- and R495X-expressing neurons we identified several genes with translation level changes specific to R495X. We found that these genes were highly targeted by R495X and the majority of them had no expression changes, indicating that R495X binding affects gene translation directly. Moreover, we found that the differentially translated genes are associated with mitochondrial functions and in R495X-expressing neurons mitochondria are significantly shorter than in control and WT-expressing neurons. Consistent with our results, it has been shown that genes targeted by R521G and R521H FUS mutants in HEK293 cells, are enriched for mitochondria related terms6. Mitochondria dysfunction has been linked to ALS pathology and several studies have demonstrated that mitochondria morphology is disturbed in neurons of ALS patients and ALS model animals35–38. Also, ALS associated FUS mutants such as R521L, R521G, R521H, R521C, P525L and R495Qfs527X have been shown to induce shortening and fragmentation of mitochondria39–44 and studies involving human WT FUS transgenic mice showed motor neuron loss16 coupled with abnormalities in mitochondrial vacuolation and a loss of synaptic vesicles45. Nevertheless, more studies in disease models maintaining physiological FUS expression levels would be needed to directly show association of mitochondria shortening to ALS pathology. While precise molecular mechanisms are still lacking, we found that genes with high translational efficiency were enriched in mitochondria-related GO terms (Supplementary Table S18), raising the possibility that R495X affects translational efficiency.
To assess the role of the R495X RNA binding activity on mitochondria shortening, we introduced 4FL mutation to perturb the FUS RRM and disrupt the interaction with RNA. We found that while R495X4FL could still bind RNA, its binding specificity was significantly diminished and it bound to mitochondria associated genes significantly less than R495X. Also, compared to control, the protein levels of R495X-targeted genes were significantly reduced in R495X but not when 4FL was introduced. This indicates that R495X can disturb the translation of non-physiologically targeted mRNAs and affect mitochondria morphology and potentially function. Finally, we find that the introduction of 4FL partially rescues mitochondria shortening and abrogates the observed neurotoxicity induced by R495X. This is in agreement with past studies indicating that 4FL suppresses toxicity in yeast46 and neurodegeneration in flies29. Notably, we find that, in contrast to WT and R495X4FL, even very low R495X expression levels are sufficient to trigger mitochondria size reduction. However, our study does not distinguish the transcriptional, translational and targetome effects at varying FUS expression levels and further studies will be required to specify the exact changes. Our results reveal key aspects of R495X effect on mitochondria shortening and neurotoxicity and importantly highlight the involvement of the FUS RRM interaction with RNA in the process.
Materials and Methods
Maintenance and differentiation of mouse embryonic stem (ES) cells
Mouse ES cells (E14Tg2A.4, RRID:MMRRC_015890-UCD) were maintained and differentiated into neurons as previously described10. Briefly, cells were maintained in gelatin-coated dish with ES cell medium supplied with 10% FCS (Hyclone, SH30071.03). For differentiation, cells were cultured with ADFNK medium for two days, then medium was replaced with ADFNK supplied with 5 μM Retinoic Acid and cultured for three days. After the culture, medium was exchanged with neuron medium (ADFNB supplied with 5 ng/mL GDNF). Formed embryonic bodies were dispersed into single neurons by Trypsinization and seeded onto poly-L-lysine (Sigma) and Laminin (Thermo) coated plates with fresh neuron medium.
Preparation of plasmids and viral supernatant
Human FUS WT or R495X with amino-terminal FLAG tag were amplified by PCR with primers; forward, 5′- CCCGGGACTAGTCACCATGGACTACAAGGACGACGATGACAAAATGGCCTCAA-3′, and reverse for WT, 5′- GACGCGGCCGCCTACAGATCCTCTTCTGAGATGAGTTTTTGTTAATA-3′, or for R495X, 5′- TACGCGGCCGCTCAGAAGCCTCCACGGTCCC-3′. 1st cDNA was prepared from total RNA isolated from HEK293T cells with SuperScriptIII (Thermo) and used as template for amplifications. The PCR products were digested with SpeI/NotI and cloned into pEN_Tmcs vector (https://www.addgene.org/25751/). The obtained plasmids were recombinated into pSLIK-Neo (https://www.addgene.org/25735/) vector with LR ClonaseII (Thermo). To obtain lentivirus supernatant, pSLIK vectors were transfected into HEK293T cells with pMD2G (https://www.addgene.org/12259/) and psPAX2 (https://www.addgene.org/12260/) vectors using Lipofectamine2000 (Thermo) and the produced lentivirus was precipitated from conditioned media by a centrifugation at 100,000 × g for 90 min. The viral pellet was resuspended with 1/100 volume of input of neuron medium and stored at −80 °C until use.
Library preparation
The dispersed neurons were infected with lentivirus encoding NFLAG-hFUSWT or NFLAG-hFUSR495X at day 7. One day later, cells were treated with neuron medium together with 1 μg/mL doxycycline (Chemicon) and two days later with 400 μg/mL Geneticin (Thermo). The cells were incubated until day 12.
CLIP-Seq
For CLIP-Seq library preparation, cells (1 × 107 cells in a 10 cm plate) at day 12 were irradiated with 400 mJ/cm2 UV, and then collected by a centrifugation, 5,000 × g for 5 min and used for library preparation as previously described47. For immunoprecipitation, 40 μL of a rabbit polyclonal anti-DYKDDDDK Tag antibody (CellSignaling, #2368) per sample was used. Libraries were sequenced with Illumina HiSeq, 50 nt, single-end. For WT and R495X we sequenced ~33 and ~25 million reads, respectively.
RNA-Seq
Cells at day12 were treated with 1 mL Trizol (Thermo) to isolate total RNA following the manufacturer’s protocol. The Ribosomal RNA was depleted by Ribo-Zero Gold kit (Illumina), and then submitted to Hokkaido System Science Co., Ltd to prepare libraries and sequenced with Illumina HiSeq, 101 nt, paired-end. We sequenced 42 to 46 million reads per sample.
Ribo-Seq
Ribo-Seq libraries were prepared with cells (1 × 107 in a 10 cm plate) at day 12 as previously described48 with some modifications. Briefly, cells were washed with PBS and collected with lysis buffer (20 mM Tris pH7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1% TritonX-100 and 25 U/mL Turbo DNase I (Thermo, AM2238)) and incubated on ice for 10 min. The cell lysate was passed through 27 G needle 10 times, and supernatant was collected by a centrifugation, 10,000 × g for 10 min. Obtained cell lysate was incubated with 1,000 U of RNaseI (Thermo, AM2294) for 45 min at room temperature. The reaction was stopped by addition of 14 μL of SUPERase.In (Thermo, AM2694). Monosomes were collected by an ultracentrifugation with 1 M Sucrose cushion, 200,000 × g for 4 hrs in TLA-100.3 (Beckman). The ribosome protected mRNA fragments (RPFs) were extracted from monosome pellet with Trizol. Ribosomal RNA was depleted with Ribo-Zero Gold kit (Illumina) and RPFs were fractionated with 15% denaturing PAGE to obtain a range from 25 to 35 nt. RPFs were extracted from gel, treated with alkaline phosphatase (Roche) and ligated to the 3′ linker conjugated with biotin47. After the 3′ linker ligation, T4 polynucleotide kinase (NEB) was directly added to the ligation reaction and incubated at 37 °C for 20 min followed by incubation at 65 °C for 20 min with an addition of EDTA at final 10 mM. Samples were subjected to library preparation as described for CLIP-Seq. Libraries were sequenced with Illumina HiSeq, 50 nt, single-end. We sequenced 27 to 29 million reads per sample.
CLIP-Seq and Ribo-Seq data preprocessing
The adaptor sequences were trimmed from the reads using the cutadapt software. The adaptor AGGGAGGACGATGCGG was trimmed from the 5′-end of reads and GTGTCAGTCACTTCCAGCGGTCGTATGCCGTCTTCTGCTTG was trimmed from the 3′-end allowing for 0.25 error rate. Only reads longer than 15-nt after adaptor trimming were retained. The reads were aligned to the mouse genome (mm9) using the STAR aligner (v2.4.1c)49 and the following parameters –outFilterMultimapScoreRange 0 –alignIntronMax 50000 –outFilterIntronMotifs RemoveNoncanonicalUnannotated –outFilterMatchNmin 15 –outFilterMatchNminOverLread 0.9 –sjdbOverhang 100. Reads mapping to areas annotated by RepeatMasker as ribosomal RNAs or tRNAs were excluded from further analysis. Aligned reads were loaded into an SQLite3 database for further processing with CLIPSeqTools27 and were annotated with information whether they overlap with elements from RepeatMasker (downloaded from UCSC) and genes (from UCSC gene model annotation file). RNA-Seq data preprocessing. The adaptor sequences were trimmed from the reads using the cutadapt software in paired-end mode. The adaptor AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC was trimmed from the 3′-end of mate 1 reads and AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTA was trimmed from the 3′-end of mate 2 reads allowing for 0.25 error rate. Paired-end alignment was performed using STAR with the same parameters as for CLIP and Ribo-Seq. Downstream preprocessing steps were performed as for CLIP and Ribo-Seq. Gene read count normalization. Read count normalization was performed from the raw counts using the DESeq250 package in R.
Gene ontology enrichment analysis
For identification of GO terms enriched in differentially expressed or translated genes, we compared differentially changed genes (up- and down-regulated) against all other expressed genes using GOrilla (http://cbl-gorilla.cs.technion.ac.il). For identification of GO terms enriched in highly translated genes, we first calculated gene translation efficiency by normalizing Ribo-Seq with RNA-Seq for control samples as in51. We then compared the top 20% to all other genes (13,542 genes).
Immunoblot
Immunoblot was performed as previously described10 using the following antibodies; mouse monoclonal anti-FLAG (Sigma, M2, 1/10,000), anti-Kif5b (Chemicon, MAB1614, 1/5,000), anti-Dnm1l (BD, 611112, 1/10,000) antibodies and Tuj1 (1/10,000), and rabbit polyclonal anti-FUS N-terminal (Bethyl, A300-302A, 1/10,000), anti-FUS C-terminal (raised against 510–523 a.a. of human FUS, 1/10,000), anti-Csde1 (Bethyl, A303–160A-T, 1/2,000), anti-Cox4 (Cell Signaling, #4844, 1/10,000) and anti-Mfn2 (Cell Signaling, #9482, 1/2,000) antibodies. Protein band quantification was performed with ImageJ (NIH). Uncropped images are shown in Supplementary Fig. S5.
Immunofluorescence and image analysis
5 × 104 cells of neurons on 8-well chamber (Thermo, #155409) were fixed with 3.7% paraformaldehyde in PBS for 10 min, then treated with 0.1% triton X-100 and 1% BSA in PBS for 10 min followed by three times wash with PBS. The cells were blocked with 3% BSA in PBS for 10 min. Primary antibody was diluted in 1% BSA in PBS and incubated with the fixed cells for O/N at 4 °C. After three times wash with PBS, cells were incubated with secondary antibody for 1 hr at room temperature. Then, the cells were washed three times with PBS and mounted with ProLong Gold with DAPI (Thermo, P36931). The used primary antibodies were mouse monoclonal anti-FLAG (Sigma, M2, 1/10,000) and rabbit polyclonal anti-TubulinβIII (Cell Signaling, #5568, 1/5,000). The secondary antibodies used were donkey anti-mouse IgG Alexa Fluor 488, goat anti-rabbit IgG Alexa Fluor 546 and goat anti-rabbit IgG Alexa Fluor 647 antibodies (Thermo). To stain mitochondria, live cells on plate were incubated with MitoTracker Red CMXRos (Thermo, M7512, 10 nM) for 45 min and then fixed. Images were obtained under a fluorescence microscope. To quantify the major mitochondrial axis length, the obtained images were processed with macro, Mitochondrial Morphology52, in ImageJ. To quantify the signal intensities in subcellular fractions, we first isolated 20 μm x 20 μm squares from the cell soma region. The signal that co-localized with DAPI staining was defined as nuclear, while the signal that co-localized with anti-TubulinβIII antibody was defined as cytoplasmic. The intensity was quantified with ImageJ and represented as the ratio of nuclear to cell soma. We counted the fraction of cells stained with anti-TubulinβIII and DAPI over those stained with DAPI alone to quantify the neuronal content. Similarly, we also counted the fraction of cells stained with anti-FLAG in the same areas described above to quantify the fraction of cells expressing exogenous proteins.
RT-qPCR
Total RNA was isolated from neurons expressing WT, R495X, and control cells using Trizol, and then treated with DNaseI (Promega, M6101) for 30 min at 37 °C, extracted with Acid Phenol Chloroform (Ambion, AM9722) and recovered by ethanol precipitation. 250 ng of total RNA was subject for reverse transcription using SuperScriptIII (Thermo). 0.5 μL of 1st cDNA prepared above was used as template for qPCR using THUNDERBIRD SYBR qPCR Mix (TOYOBO) with specific primer sets (forward, reverse); Foxj1 (5′-GAACTGCCTCCCAAGACAGG-3′, 5′-CCTTGGGCTTGAGGGAACAT-3′), Zfp608 (5′-CTCCGGTGGTCACCTCTATG-3′, 5′-GACCGAGTCCGAACCAAAAG-3′), Nefm (5′-CAAGTGGGAAATGGCTCGTC-3′, 5′-TGTCGGTGTGTGTACAGAGG-3′), Slc17a6 (5′-AGGATATATCGCATCGCGGC-3′, 5′-CCATAATGCACTCTGGCTGC-3′), Apba1 (5′-TACAGTGACCTGCTCAACACC-3′, 5′-CTTCACTCGGGACTGGTTCTT-3′), Inppl1 (5′-AGTTCCTGACCTTCTTGTCCC-3′, 5′-TGCTACTCCTTCCTGCTCAGA-3′), mKIAA4052 (5′-GTAGCGTCCTCTCTGCACTCT-3′, 5′-GGACCGAGAGGAGATTTCACTG-3′), Zfp62 (5′- GTGAAGAAACTGCTGTACAAAGA-3′, 5′-GAGCAGGGTGTACGTTTCCA-3′), Zfp60 (5′-CAGTGGTGGGACGTTGCATT-3′, 5′-GCCATTACTAACACTGACGCC-3′), Islr2 (5′-ACAGATGCCAGAAGCTCCCT-3′, 5′-TGACCACTCCTAGCAAAGCC-3′), Enc1 (5′-GGAACAGAGACGCCCTTCGT-3′, 5′-TTCAGGCCACCACTGAACAT-3′), Pgm2l1 (5′-TTCTGTGTGGAACCTCCGTG-3′, 5′-TGGCTGGCTGCTATCATATCC-3′), Tubb3 (5′- GCGCCTTTGGACACCTATTC-3′, 5′-CTCCACTCATGGTGGCAGAC-3′), Csde1 (5′- TGGCCTACAACATCACACCC-3′, 5′- TATCTGGTCCCCTTGGCTGA-3′), Mfn2 (5′- TGGTCTCCATGGTTACTGGC-3′, 5′- GCGTATTCCACAAACTGGCG-3′), Dnm1l (5′- AACACGATTGAAGGAACCGC-3′, 5′- CTGGGCTCTTCTAGACGCTT-3′), Cox4 (5′- CAGGGGCACCAATGAATGGA-3′, 5′- ATAGTCCCACTTGGCGGAGA-3′), Kif5b (5′- GGAGGCAAGCAGTCGTAAAC-3′, 5′- AAACAGGGCCGCAGTTGTAA-3′) and Gapdh (5′- CAATGTGTCCGTCGTGGATCT-3′, 5′- GTCCTCAGTGTAGCCCAAGAT-3′). qPCR reaction was carried out using Mx3005P qPCR System (Agilent Technologies). The results were analyzed as previously described10.
RNA-immunoprecipitation
Cells expressing R495X, R495X4FL or control were lysed with 100 μL of RSB200 (20 mM Tris pH 7.4, 200 mM NaCl and 2.5 mM MgCl2) supplied with 0.5% NP40 on ice for 20 min. Cell lysate was cleared by a centrifugation at 12,000 × g for 3 min. 4 μL of cell lysate was used for immunoblot and 20 μL of lysate was mixed with 100 μL of water and 380 μL of Trizol-LS (Thermo) for total RNA isolation. 70 μL of cell lysate was precleared with 5 μL of Protein G Dynabeads (Thermo) in 170 μL buffer in total for 30 min by rotation at 4 °C. For IP, 10 μL of Protein G Dynabeads was washed twice with RSB200 supplied with 0.5% NP40 and incubated with 1 μL of anti-FLAG antibody (M2, Sigma, F1804) for 1 hr by rotation at 4 °C. The beads were washed three times with buffer, mixed with precleared cell lysate and rotated for 3 hrs at 4 °C. After incubation, beads were washed five times with buffer and saved 8 μL of 35 μL suspension in buffer for immunoblot. The rest of the immunoprecipitated sample was resuspended in 500 μL of Trizol (Thermo). Total and IP RNA was isolated with Trizol. 250 ng of total RNA and RNA from IP were used for reverse transcription with SuperScriptIII. We confirmed that no non-specific RNA binding to beads was observed with our method using a normal mouse IgG (Supplementary Fig. S8).
Cell survival curve
Neurons grown on 96-well plate (5 × 104/well) were infected with lentivirus at day 0. Medium was replaced every day until day 7 after induction with 100 μL of fresh medium supplied with DOX (1 μg/mL final). The cell positions were recorded from day 4 to day 7 using a microscope. Stacked images from day 4 to 7 were aligned by the StackReg ImageJ (NIH) plug-in. We chased cells by TrackMate in ImageJ and analyzed the obtained results by the “survival” package in R, in which we tested statistical significance by log-rank test. We analyzed 2324, 3207, 1995 and 1903 cells for Control, WT-, R495X- and R495X4FL-expressing neurons, respectively.
Statistical analysis
For all of statistical analyses, One-Way ANOVA and Tukey HSD Test was applied (http://vassarstats.net/anova1u.html) unless otherwise noted. Statistical significance was concluded for p-values < 0.05.
Electronic supplementary material
Acknowledgements
The authors thank Dr. Zissimos Mourelatos for his mentoring and insightful discussions and suggestions. The authors also thank all former and current members of the Mourelatos lab for their helpful suggestions. This work was supported by The Mitsubishi Foundation, JSPS KAKENHI Grant (15K07918) to T.N. and a Brody family fellowship to M.M.
Author Contributions
T.N. and M.M. directed experiments. T.N. performed all wet experiments. M.M. performed all bioinformatics analyses. T.N. and M.M. analyzed data, and wrote the manuscript.
Data Availability
The datasets generated in the current study are available in the Sequence Read Archive (SRA) under project ID: GSE106386
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tadashi Nakaya, Email: Tadashi.Nakaya@pharm.hokudai.ac.jp.
Manolis Maragkakis, Email: emarag@pennmedicine.upenn.edu.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33964-0.
References
- 1.Ibrahim F, Nakaya T, Mourelatos Z. RNA dysregulation in diseases of motor neurons. Annu Rev Pathol. 2012;7:323–352. doi: 10.1146/annurev-pathol-011110-130307. [DOI] [PubMed] [Google Scholar]
- 2.Kwiatkowski TJ, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 3.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orozco D, et al. Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO Rep. 2012;13:759–764. doi: 10.1038/embor.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoell JI, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu H, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waibel S, Neumann M, Rabe M, Meyer T, Ludolph AC. Novel missense and truncating mutations in FUS/TLS in familial ALS. Neurology. 2010;75:815–817. doi: 10.1212/WNL.0b013e3181f07e26. [DOI] [PubMed] [Google Scholar]
- 9.Bosco DA, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakaya T, Alexiou P, Maragkakis M, Chang A, Mourelatos Z. FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns. RNA. 2013;19:498–509. doi: 10.1261/rna.037804.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishigaki S, et al. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Liu S, Liu G, Ozturk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9:e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda A, et al. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 2015;29:1045–1057. doi: 10.1101/gad.255737.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell JC, et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013;125:273–288. doi: 10.1007/s00401-012-1043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, et al. Expression of human FUS protein in Drosophila leads to progressive neurodegeneration. Protein Cell. 2011;2:477–486. doi: 10.1007/s13238-011-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C, et al. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet. 2011;7:e1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, et al. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun. 2016;7:10465. doi: 10.1038/ncomms10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Blitterswijk M, et al. Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq. PLoS One. 2013;8:e60788. doi: 10.1371/journal.pone.0060788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami T, et al. ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism. Hum Mol Genet. 2012;21:1–9. doi: 10.1093/hmg/ddr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han TW, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami T, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel A, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 26.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 27.Maragkakis M, Alexiou P, Nakaya T, Mourelatos Z. CLIPSeqTools–a novel bioinformatics CLIP-seq analysis suite. RNA. 2016;22:1–9. doi: 10.1261/rna.052167.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daigle JG, et al. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerga A, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen CD, et al. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J Mol Biol. 2011;407:273–283. doi: 10.1016/j.jmb.2010.12.041. [DOI] [PubMed] [Google Scholar]
- 32.Convertini P, et al. Genome wide array analysis indicates that an amyotrophic lateral sclerosis mutation of FUS causes an early increase of CAMK2N2 in vitro. Biochim Biophys Acta. 2013;1832:1129–1135. doi: 10.1016/j.bbadis.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scekic-Zahirovic J, et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 2016;35:1077–1097. doi: 10.15252/embj.201592559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Santis, R. et al. FUS Mutant Human Motoneurons Display Altered Transcriptome and microRNA Pathways with Implications for ALS Pathogenesis. Stem Cell Reports (2017). [DOI] [PMC free article] [PubMed]
- 35.Ahtoniemi T, Jaronen M, Keksa-Goldsteine V, Goldsteins G, Koistinaho J. Mutant SOD1 from spinal cord of G93A rats is destabilized and binds to inner mitochondrial membrane. Neurobiol Dis. 2008;32:479–485. doi: 10.1016/j.nbd.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki S, Iwata M. Ultrastructural study of synapses in the anterior horn neurons of patients with amyotrophic lateral sclerosis. Neurosci Lett. 1996;204:53–56. doi: 10.1016/0304-3940(96)12314-4. [DOI] [PubMed] [Google Scholar]
- 37.Magrane J, Cortez C, Gan WB, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum Mol Genet. 2014;23:1413–1424. doi: 10.1093/hmg/ddt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tradewell ML, et al. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- 40.Huang C, et al. Entorhinal cortical neurons are the primary targets of FUS mislocalization and ubiquitin aggregation in FUS transgenic rats. Hum Mol Genet. 2012;21:4602–4614. doi: 10.1093/hmg/dds299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng J, et al. FUS Interacts with HSP60 to Promote Mitochondrial Damage. PLoS Genet. 2015;11:e1005357. doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, et al. PINK1 and Parkin are genetic modifiers for FUS-induced neurodegeneration. Hum Mol Genet. 2016;25:5059–5068. doi: 10.1093/hmg/ddw310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoica R, et al. ALS/FTD-associated FUS activates GSK-3beta to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep. 2016;17:1326–1342. doi: 10.15252/embr.201541726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naumann M, et al. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun. 2018;9:335. doi: 10.1038/s41467-017-02299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.So E, et al. Mitochondrial abnormalities and disruption of the neuromuscular junction precede the clinical phenotype and motor neuron loss in hFUSWT transgenic mice. Hum Mol Genet. 2018;27:463–474. doi: 10.1093/hmg/ddx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Z, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vourekas A, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol15, 550, doi:s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed]
- 51.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in the current study are available in the Sequence Read Archive (SRA) under project ID: GSE106386