Abstract
We previously reported that overexpression of an O-type glycosyltransferase, GALNT6 (polypeptide N-acetylgalactosaminyltransferase 6) played critical roles in mammary carcinogenesis. To further investigate the biological function of GALNT6, we screened a substrate protein(s) of GALNT6 using a VVA (Vicia villosa agglutinin) lectin (specific to GalNAc-Ser/Thr) pull-down method followed by mass spectrometry analysis. Here we report GRP78 (glucose-regulated protein 78, also known as HSPA5, heat shock 70 kDa protein 5), which is highly expressed in cancer cells and indicated to play important roles in various cellular processes including ER (endoplasmic reticulum) stress and autophagy, as a novel substrate of GALNT6. We found that GALNT6-induced O-glycosylation is critical for the stability of GRP78, its subcellular localization in ER, and its anti-apoptotic function. Furthermore, we demonstrated that overexpression of GRP78 could be important for Golgi-to-ER relocation of GALNT6. Collectively, our study revealed biological significances of O-glycosylation of GRP78 protein, which might play significant roles in the survival of cancer cells, and thus provided a new insight in cancer cell death and useful information for development of anti-cancer treatment targeting the GALNT6-GRP78 pathway.
Abbreviations: GALNT6, polypeptide N-acetylgalactosaminyltransferase 6; VVA, Vicia villosa agglutinin; GalNAc, N-acetylgalactosamine; GRP78, glucose-regulated protein 78; ER, endoplasmic reticulum
Introduction
O-glycosylation is a protein modification at serine or threonine residue of protein, and enhanced O-glycosylation is often observed in malignant cells [1], [2], [3]. The mechanism to cause such aberrant O-glycosylation in cancers is not well understood, but emerging evidence has suggested that the expression of polypeptide GalNAc (N-acetylgalactosamine)-transferases (GalNAc-Ts), which control the initiation of O-glycosylation, was markedly up-regulated in cancer cells [4], [5], [6], [7]. We previously indicated that GALNT6 (polypeptide N-acetylgalactosaminyltransferase 6, GalNAc-T6) was involved in O-glycosylation of mucins and could be a promising molecular target for development of a new class of anti-cancer drugs for breast and pancreatic cancers [7], [8], [9]. Studies from other groups also indicated that GALNT6 could be a potential biomarker in breast cancer progression and metastasis [5], [6]. However, it is still not well understood how GALNT6 promotes carcinogenesis since its O-glycan substrates are not fully clarified. Therefore, identification and functional analysis of GALNT6 substrates should be important for better understanding of the oncogenic pathways medicated by GALNT6 and for obtaining the fundamental information for development of GALNT6-targeted and/or its substrate-targeted therapies.
GRP78 (glucose-regulated protein 78, also known as HSPA5, heat shock 70 kDa protein 5) is highly expressed in various types of human cancer, and was shown to be important for tumor formation and progression in vitro and in vivo [10]. GRP78 is involved in many cellular processes including the endoplasmic reticulum (ER) stress which activates the unfolded protein response (UPR) to alleviate this stress and restore ER homeostasis [11], [12]. GRP78 controls a cross-talk between apoptosis and autophagy [13], [14], and exhibits oncogenic activities by promoting cell proliferation, survival of cancer cells, angiogenesis, metastasis, and drug resistance [15], [16].
In this study, we report that GALNT6 is involved in O-glycosylation and stabilization of GRP78 protein and that O-glycosylation was essential for proper subcellular localization and anti-apoptotic function of GRP78. In addition, we demonstrated that overexpression of GRP78 could enhance Golgi-to-ER relocation of GALNT6. Our findings revealed biological significances of O-glycosylation in GRP78 protein, which seemed to be important for the survival of cancer cells, and thus provided a new insight in efficient induction of cancer cell death by targeting the GALNT6-GRP78 pathway.
Materials and Methods
Cell Culture
Human cancer cell lines MCF7, T47D, MDA-MB-435S, HeLa, and human embryonic kidney 293T cell were purchased from American Type Culture Collection (ATCC) and cultured according to the manufacturer's protocols. Three HeLa cell-derived cell lines stably expressing HA-tagged wild-type GALNT6 protein (HeLa-GALNT6-WT), HA-tagged enzyme-dead H271D-substituted GALNT6 protein (HeLa-GALNT6-H271D), and empty vector (HeLa-Mock) were established as previously described [7]. MCF7 stable cells of mock, wild-type GRP78 (GRP78-WT), and GRP78 with alanine-substitutions at O-glycosylation sites (T85A, T151A, T166A, T184A, and T203A) were newly generated. Briefly, no insert (mock), GRP78-WT, or alanine-substituted GRP78 in pCAGGS-3xFlag expression vectors were transfected into MCF7 cells by using TransIT-BrCa Transfection Reagent (Mirus). Forty-eight hours after transfection, cells were selected under incubation with culture medium containing 0.5 mg/ml of G418 (Geneticin). Two or 3 weeks later, single clones were picked up for further study. Similarly, no insert (mock) and GALNT6-WT in pCAGGS-HA expression vectors were transfected into MDA-MB-435S, and polyclone stable cells were generated by selection of positively transfected cells using 0.8 mg/ml of G418. Transfection of plasmids in the study was done by using TransIT-BrCa Transfection Reagent (Mirus), Lipofectamine 2000 (Life Technologies), or FuGENE 6 (Roche) reagents according to the manufacturer's protocols.
Identification of GALNT6’s Substrates
To detect proteins harboring the Tn antigen (GalNAc-Ser/Thr), we did western blot using biotinylated Vicia villosa agglutinin (VVA) lectin (1:1000, Vector Laboratories) and Streptavidin-HRP (1:10,000, Thermo Scientific) as described previously [7]. For screening candidate O-glycan subtracts of GLANT6, total proteins from HeLa-Mock, −GALNT6-WT, and -H271D stable cells were extracted and then performed pull-down assay using biotin-conjugated VVA-lectin (specific to GalNAc-Ser/Thr) and streptavidin-conjugated agarose (Invitrogen), followed by elution with 100 mM of GalNAc, according to manufacturer's protocol. Considering a possibility of incomplete elution of O-glycan proteins, we examined both eluted protein fraction as well as finally precipitated proteins with the Streptavidin-conjugated agarose beads. The isolated proteins were visualized by the SilverQuest Staining Kit (Invitrogen). Protein bands that were specifically observed in the HeLa-GALNT6-WT lane were excised with a clean, sharp scalpel and the extracted proteins were applied for PMF (Peptide Mass Fingerprint) analysis using MALDI-TOF MS (Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry). Expression levels of the genes that encode identified proteins were examined by semi-quantitative RT-PCR as described previously [7]. Primer sets and PCR conditions are provided in Supplementary Table 1.
Gene Cloning and Mutagenesis
Flag-tagged full-length wild type GRP78 expression vector (pCAGGS-GRP78-WT-3xFlag) was constructed according to the protocol described previously [7], [17]. Primer sets are described in Supplementary Table 2. To generate expression vectors for alanine-substituted GRP78 (T85A, T151A, T166A, T184A, and T203A) that correspond to the candidate O-glycosylation sites, we performed two-step mutagenesis PCR [7], using four primers: a primer set for GRP78 wild-type cloning and another set for mutant (harboring a mutated nucleotide in the middle of the primer) as shown in Supplementary Table 2. For construction of GRP78 fragments (GRP78–1-280, GRP78–125-500 and GRP78–281-654), we performed PCR by using GRP78 wild-type plasmid as a template DNA. The primer sets were described in Supplementary Table 2. Sequences of all constructs were confirmed by BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) and ABI3500XL (Life Technologies), and protein expression of these plasmids was also confirmed by western blot.
Western Blot
Western blot was performed as described previously [17]. Finally, protein bands were visualized by ECL or ECL prime detection reagents (GE Healthcare). The primary antibodies used in this study were: anti-human GRP78 polyclonal antibody (1:1000, Santa Cruz), anti-human GALNT6 polyclonal antibody (1:1000, Sigma-Aldrich), anti-Flag M2 monoclonal antibody (1:1000, Sigma-Aldrich), anti-HA monoclonal antibody (1:1000, Roche), anti-PARP-1 antibody (1:1000, Santa Cruz), anti-caspase 7 (1:1000, Cell signaling), anti-PERK (1:1000, Cell signaling), anti-IRE1α (1:1000, Cell signaling), anti-ATF6 (1:1000, Cell signaling), and anti-β-actin monoclonal antibody (1:10,000, Sigma-Aldrich). The secondary antibodies were goat anti-rabbit, anti-rat, and anti-mouse IgG-HRP secondary antibodies (1:10,000~ 1:30,000, Santa Cruz). Intensity of protein band was quantified by ImageJ software as previously described [8].
Immunoprecipitation (IP)
Cell extracts were prepared by adding CelLytic M reagent (Sigma-Aldrich) with 1% of Protease Inhibitor Cocktail Set III (Calbiochem) according to the manufacturer's protocols. Extracts were pre-cleared by incubation with 40 μl of rec-Protein A- or G-Sepharose 4B Conjugate (Invitrogen) and 2 μg of rabbit or mouse IgG (Santa Cruz) at 4°C for 1 h. For pulling down endogenous GRP78 or Flag-tagged GRP78 proteins, pre-cleared cell extracts were then incubated with 2 μg of anti-GRP78 (Proteintech) or anti-Flag M2 monoclonal antibody (Sigma-Aldrich) at 4°C for overnight followed by 40 μl of rec-Protein A- or G-Sepharose 4B Conjugate at 4°C for 2 h, respectively. For pulling down HA-GALNT6, pre-cleared cell extracts were incubated with 40 μl of monoclonal anti-HA-Agarose (Sigma-Aldrich) at 4°C for overnight. The beads were then spun down and washed 4 times with 1 ml of cell lysis buffer. Finally, immunoprecipitated proteins were released from the beads by boiling in sample buffer for 2 min or by adding elution buffer.
Immunocytochemistry
Immunocytochemistry was performed as previously described [17], [18]. The first antibodies used in this study included: anti-Flag M2 monoclonal antibody (1:500, Sigma-Aldrich), anti-HA monoclonal antibody (1:500, Roche), anti-GRP78 (1:25, Proteintech) and anti-PDI monoclonal antibody (1:100, Cell signaling). The secondary antibodies were: Alexa Fluor 488 Anti-Mouse or -Rabbit IgG antibodies (1:500–1000, Life Technologies) and Alexa Fluor 594 Anti-Rat or -Mouse IgG antibodies (1:500–1000, Life Technologies). Finally, cells were stained with DAPI (Vector Laboratories) and examined by TCS SP5 Confocal Laser Scanning Microscope (Leica Microsystems).
In vitro Glycosylation Assay
As a substrate, the pCAGGS-GRP78-WT-GST plasmid was expressed in HEK293 cell, and GST-tagged GRP78 protein was pulled down by Glutathione Sepharose 4B agarose (GE Healthcare). In parallel, pQCXIPG-GALNT6-6xHis plasmids (WT and H271D) were expressed in HEK293 cell, and His-tagged recombinant GALNT6 proteins (WT and H271D) were purified by Ni-NTA agarose (QIAGEN). Then in vitro glycosylation assay was performed as described previously [7].
Identification of O-glycosylation Sites in GRP78
For identification of O-glycosylation sites, HeLa-GALNT6 stable cells (WT and H271D) were transfected with pCAGGS-GRP78-WT-3xFlag expression vector and collected after 48 h incubation. Cells were lysed with 1% NP-40 lysis buffer and Flag-tagged GRP78 protein was immunoprecipitated with anti-Flag monoclonal antibody and Protein A agarose (Invitrogen). After five times washing with the lysis buffer, immunocomplexes were loaded in a SDS-PAGE gel and protein bands were visualized by CBB staining (Bio-Rad). The bands which indicated GRP78 were excised and proceed to analyze with QSTAR Elite QqTOF mass spectrometer (AB Sciex).
GALNT6 Knockdown by siRNA
We used two small interfering RNA (siRNA) (Sigma-Aldrich) for GALNT6 knockdown and SIC001 Mission siRNA Universal Negative Control (Sigma-Aldrich) as a control as previously described [9]. Briefly, GALNT6-siRNA or control siRNA was transfected into cells by using Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer's protocols. Seventy-two hours later, cells were collected for further study.
Reagents for Cell Treatment
To examine stability of GRP78 protein, cells were treated with 50 μg/ml of cycloheximide (Sigma-Aldrich) and incubated for various time points as indicated in the text. To induce ER stress, cells were treated with 2 μM of Thapsigargin (TG) for 4 h before immunocytochemistry.
Nutrition Deprivation by Treatment With EBSS
MCF7 stable cells were seeded in 6 cm or 10 cm dish. After 24 h, medium was changed and the cells were cultured in MEM with 10% FBS or washed by PBS for three times followed by culture in Earle's Balanced Salt Solution (EBSS) (Life Technologies) for 6 h. The cells were then collected for further study.
Results
Identification of GRP78 as a Substrate of GALNT6
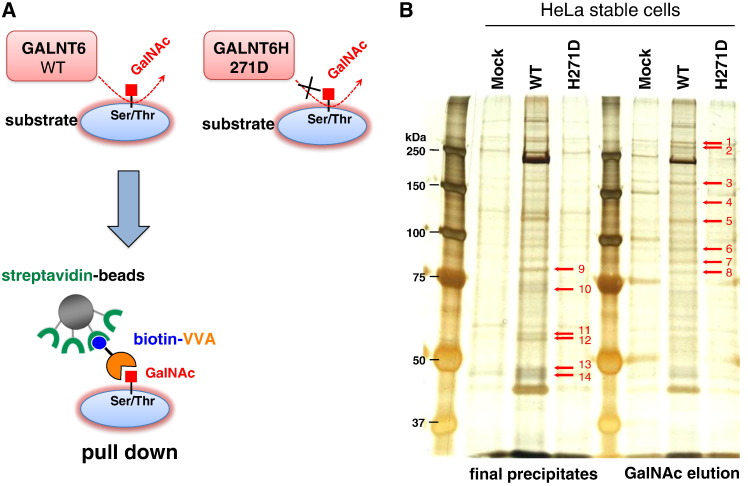
To investigate direct O-glycan substrates of GALNT6, we first prepared three HeLa cell-derived cell lines, each of which was designed to stably express HA-tagged wild-type GALNT6 protein (HeLa-GALNT6-WT) or HA-tagged enzyme-dead H271D-substituted GALNT6 protein (HeLa-GALNT6-H271D), or was transfected with mock vector (HeLa-Mock) [7]. Using these three cell lines, we performed pull-down assay using biotin-conjugated VVA-lectin (specific to GalNAc-Ser/Thr, also called Tn-antigen) and streptavidin-conjugated agarose (Figure 1A) with two elution methods at the final step, followed by silver staining (Figure 1B). Consequently, we identified more than ten protein bands showing significantly higher intensities in HeLa cells stably expressing wild-type GALNT6 protein, compared with HeLa-Mock cells or HeLa-GALNT6-H271D cells (Figure 1B). We excised these bands and performed mass spectrometry analysis to characterize the possible interacting proteins (Supplementary Figure 1, no candidate protein was identified from band 3 (~100 kDa), 13 and 14 (~50 kDa)). This analysis suggested 15 candidate proteins from 14 excised protein bands (Supplementary Table 3). We subsequently examined expression levels and possible biological functions of 15 genes encoding the candidate proteins by semi-quantitative RT-PCR and literature search, respectively (Supplementary Figure 2). In this study, we selected GRP78 (Band 10 in Figure 1B, gene name: HSPA5 in Supplementary Figure 2) for further characterization due to its high expression in cancer cells and its possible oncogenic functions.
Figure 1.
Screening of GALNT6 substrates. (A) A pictorial schema for identification of GALNT6 substrates by using VVA-lectin pull down assay. (B) Silver staining for finally precipitated proteins and protein fraction eluted by GalNAc after VVA-lectin pull down using three HeLa cell-derived cell lines stably expressing empty vector (Mock), HA-tagged GALNT6 wild-type (WT), and enzyme activity-depleted GALNT6 (H271D). Differential protein bands indicated as arrows were excised and analyzed by mass spectrometry.
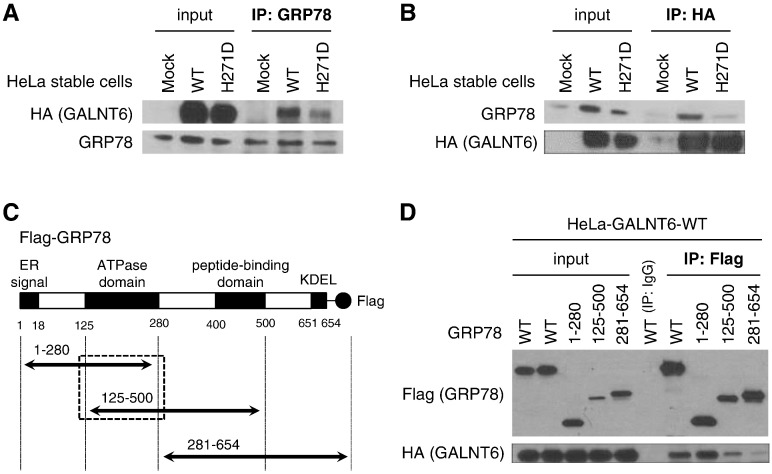
To validate the mass spectrometry result, we performed immunoprecipitation (IP)-western blot analysis by using anti-GRP78 antibody in HeLa-Mock, HeLa-GALNT6-WT, and HeLa-GALNT6-H271D stable cells, and found that HA-tagged GALNT6 protein was co-precipitated with endogenous GRP78 in HeLa-GALNT6-WT stable cells (Figure 2A). Concordantly, endogenous GRP78 was pulled down when the HA-tagged GALNT6 protein was immunoprecipitated (Figure 2B), suggesting the interaction of GALNT6 and GRP78 proteins. The binding affinity of GALNT6-H271D to GRP78 protein was weaker than that of GALNT6-WT. To define a GALNT6-binding region(s) in the GRP78 protein, we constructed expression vectors for Flag-tagged full-length wild-type GRP78 (GRP78-WT) and three Flag-tagged partial GRP78 fragments (Figure 2C). These fragments contained an ATPase domain (GRP78–1-280), an ATPase and peptide-binding domains (GRP78–125-500), or a peptide-binding domain (GRP78–281-654). We then transfected these four GRP78 expression vectors into HeLa-GALNT6-WT stable cells followed by IP-western blot analysis, and found that HA-tagged GALNT6 was co-immunoprecipitated with GRP78-WT, GRP78–1-280 and GRP78–125-500, all of which included the ATPase domain (Figure 2D), but the binding between GALNT6 and GRP78–281-654 lacking of an ATPase domain was significantly reduced. These results indicated that the ATPase domain of GRP78 was essential for the interaction with GALNT6.
Figure 2.
GRP78 interacts with GALNT6. (A and B) HA-tagged GALNT6 was co-immunoprecipitated with endogenous GRP78 in HeLa-GALNT6-WT and -H271D stable cells, but not in -mock stable cells. However, interaction of GRP78 with GALNT6-H271D was much weaker than that with GALNT6-WT. (A) Endogenous GRP78 was pulled down using anti-GRP78 antibody, followed by western blots for HA-tagged GALNT6 and endogenous GRP78. (B) HA-tagged GALNT6 was pulled down by anti-HA, and then endogenous GRP78 and HA-tagged GALNT6 were detected by western blot. (C) To map the binding domain on GRP78, four expressing vectors for Flag-tagged full-length wild-type GRP78 (WT) and three Flag-tagged partial GRP78 fragments with or without ATPase domain were generated. (D) Forty-eight hours after transfection with these four vectors in HeLa-GALNT6-WT stable cells, immunoprecipitation with anti-Flag antibody was performed followed by western blots.
Overexpression of GRP78 Drives Golgi-to-ER Relocation of GALNT6
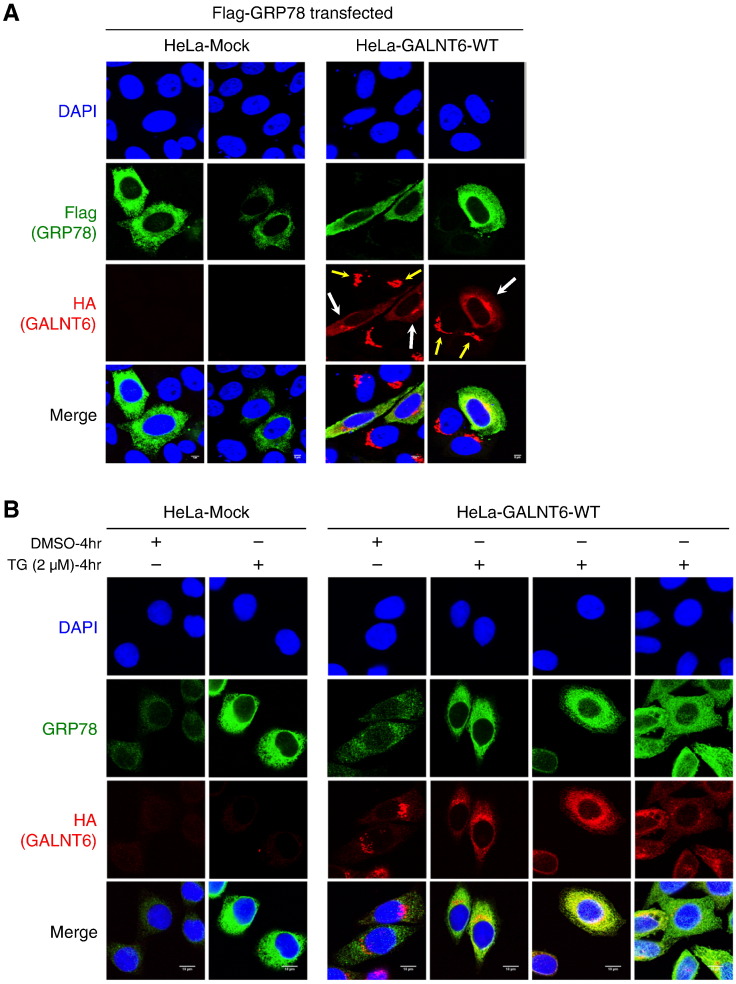
Subsequently we performed immunocytochemical analysis of GALNT6 and GRP78 using the HeLa-GALNT6-WT and HeLa-Mock stable cells. The HA-tagged GALNT6 was detected in the Golgi (Figure 3A, yellow arrows) when Flag-tagged GRP78 was absent, but in the presence of high level of Flag-tagged GRP78, HA-tagged GALNT6 was mainly located in ER of HeLa-GALNT6-WT cells (Figure 3A, white arrows), suggesting that Golgi-to-ER translocation of GALNT6 occurred through the interaction with GRP78. To further validate this hypothesis, we examined the subcellular localization of GALNT6 with endogenous GRP78 protein that could be activated by an ER stress activator, Thapsigargin (TG). As expected, we observed significant increase of endogenous GRP78 protein by treatment with TG, which also drove the Golgi-to-ER translocation of HA-tagged GALNT6 in HeLa-GALNT6-WT cells (Figure 3B). Although the TG treatment itself might activate other molecules involved in the ER stress response, this result further supported that high level of GRP78 protein could drive Golgi-to-ER relocation of GALNT6, as clearly shown in Figure 3A.
Figure 3.
Overexpression of GRP78 drives Golgi-to-ER relocation of GALNT6. Immunocytochemistry was performed in the HeLa-Mock and -GALNT6-WT stable cells. (A) Forty-eight hours after transfection of Flag-tagged GRP78 expression vectors into the HeLa stable cells, immunocytochemistry was performed by using anti-Flag and anti-HA antibodies. Flag-tagged GRP78 was shown in green color and HA-tagged GALNT6 was in red color. We observed that HA-tagged GALNT6 located in Golgi if exogenous GRP78 was not expressed (yellow arrows), but a large amount of Flag-tagged GRP78 drove Golgi-to-ER relocation of GALNT6 in the HeLa-GALNT6-WT stable cells (white arrows). Two fields were taken for each condition. (B) The HeLa stable cells were treated with DMSO (control) or 2 μM of an ER stress activator TG for 4 h, and immunocytochemistry was performed by using anti-GRP78 and anti-HA antibodies. Endogenous GRP78 was shown in green color and HA-tagged GALNT6 was in red color. After ER stress by TG treatment, endogenous expression of GRP78 was significantly increased, and drove Golgi-to-ER relocation of GALNT6. Three fields were taken for TG-treated HeLa-GALNT6-WT stable cells.
GALNT6 O-glycosylates GRP78
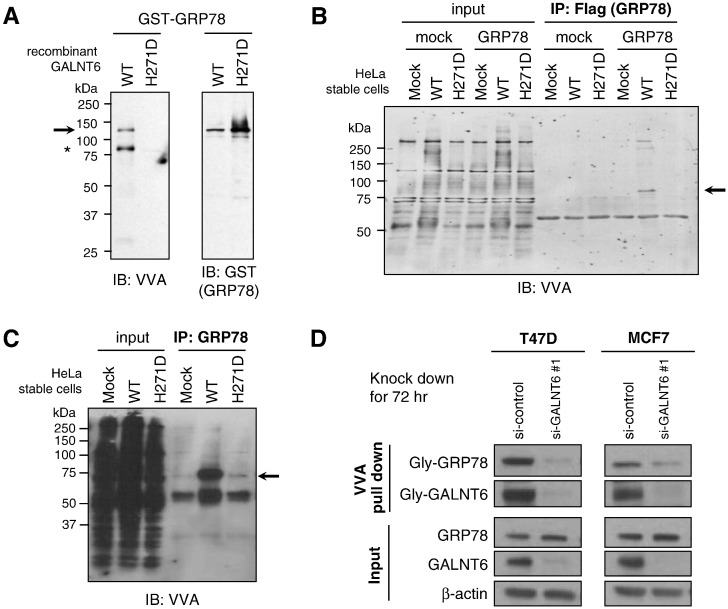
To examine the biological significance of O-glycosylation in GRP78 protein by GALNT6, we first generated GST-tagged recombinant GRP78 protein (GST-GRP78) and His-tagged recombinant GALNT6 (His-GALNT6-WT or -H271D) proteins, which were individually expressed and purified using HEK293 cells. Then we performed an in vitro O-glycosylation assay using the GST-GRP78 protein as a substrate, UDP-GalNAc as a sugar donor, and His-GALNT6-WT or -H271D protein as a source of O-type glycosyltransferase, followed by western blot analysis. Our results showed that GST-GRP78 protein was O-glycosylated by His-GALNT6-WT, but not by GALNT6-H271D (Figure 4A). We also observed auto-O-glycosylation in His-GALNT6-WT, but not in -H271D protein. In a cell-based assay, we found that both exogenous (Figure 4B) and endogenous (Figure 4C) GRP78 proteins were highly O-glycosylated in HeLa-GALNT6-WT stable cells, but not in HeLa-Mock and HeLa-GALNT6-H271D stable cells. We then investigated which site(s) of GRP78 was O-glycosylated by GALNT6. Exogenously expressed Flag-tagged GRP78 proteins in HeLa-GALNT6-WT or -H271D stable cells were immunoprecipitated with anti-Flag antibody followed by CBB staining. Protein bands corresponding to GRP78’s molecular weight were cut and submitted for mass spectrometry analysis, which consequently identified five O-glycosylation candidate sites (T85, T151, T166, T184, and T203) in GRP78 (Supplementary Table 4). Except T85, the later four sites were located in the ATPase domain. In addition, we found the O-glycosylation level of endogenous GRP78 protein was significantly reduced after GALNT6 knockdown in MCF7 and T47D breast cancer cells, suggesting that GALNT6 was critical for O-glycosylation of GRP78 in these two cancer cells (Figure 4D).
Figure 4.
GALNT6 O-glycosylates GRP78. (A) GST-tagged recombinant GRP78 protein (GST-GRP78) was incubated with UDP-GalNAc (sugar donor) and His-tagged recombinant wild-type GALNT6 (GALNT6-WT) or enzyme-dead GALNT6 (GALNT6-H271D) proteins. O-glycosylated proteins and GST-tagged GRP78 protein were detected by western blot analysis with VVA lectin and anti-GST antibody, respectively. GST-GRP78 was O-glycosylated by GALNT6-WT but not by enzyme activity-depleted GALNT6-H271D. We also observed that GALNT6-WT itself was auto-O-glycosylated (indicated by asterisk). In cell-based assay, we pulled down exogenous (B) or endogenous GRP78 (C), and detected its O-glycosylation level. GRP78 proteins had much higher level of O-glycosylation in HeLa-GALNT6-WT stable cells, when compared with -mock and -H271D stable cells (B and C). (D) GALNT6 was knocked down by using siRNA in T47D and MCF7 cells for 72 h, then VVA pull down assay and western blot were performed. The O-glycosylation level of GRP78 was significantly reduced after GALNT6 knockdown, suggesting that GALNT6 was critical for O-glycosylation of GRP78 in these cells.
O-glycosylation Stabilizes GRP78 Protein
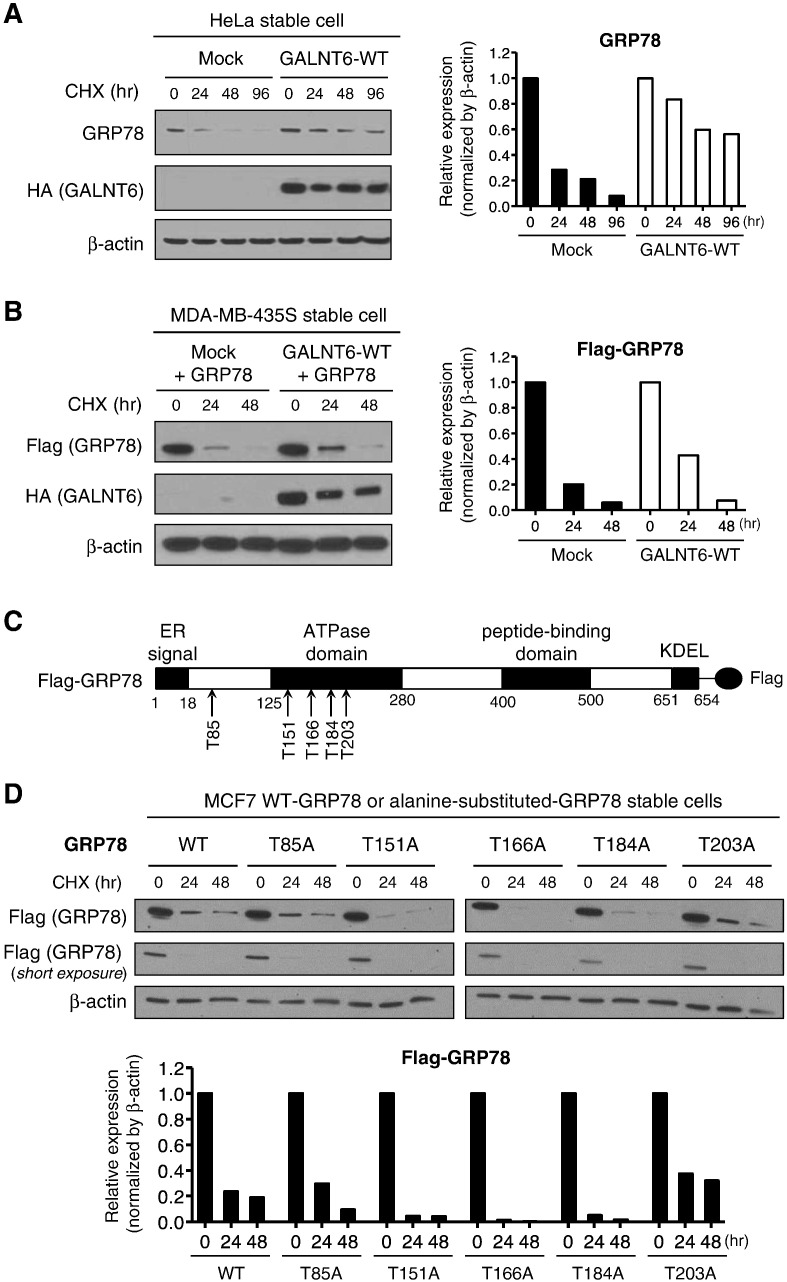
Because O-glycosylation was suggested to play important roles in maintaining protein stability [7], [8], we investigated the effect of O-glycosylation on the stabilization of GRP78 protein. After treatment with a protein synthesis inhibitor, cycloheximide (CHX), we examined the stability of GRP78 protein at different time points. We found endogenous (Figure 5A) and exogenous GRP78 proteins (Figure 5B) were more stable in GALNT6 overexpressing cells (HeLa- or MDA-MB-435S-derived cells in which GALNT6-WT protein was stably expressed) than the cells harboring mock vector (HeLa- or MDA-MB-435S-Mock). To further verify these candidate O-glycosylation sites and investigate the role of O-glycosylation on GRP78, we generated Flag-tagged constructs in which each of candidate O-glycosylation sites was substituted with an alanine residue (T85A, T151A, T166A, T184A, or T203A) (Figure 5C), and then transfected these constructs as well as Flag-tagged GRP78-WT protein into MCF7 cells and established stably-expressing cell lines. Western blot analysis after CHX treatment of these established cell lines showed rapid decrease of Flag-tagged GRP78 proteins in cells with T151A, T166A and T184A (Figure 5D), indicating that O-glycosylation at T151, T166 and T184 sites might be important for GRP78 protein stability. Taken together, these results have suggested that O-glycosylation at specific sites by GALNT6 is likely to be important for stabilization of the GRP78 protein.
Figure 5.
GALNT6 stabilizes GRP78 through O-glycosylation. (A) HeLa stable cells were treated with CHX (cycloheximide) prior to western blot analysis. Endogenous GRP78 was more stable in HeLa-GALNT6-WT stable cells when compared with -mock stable cells. (B) MDA-MB-435S-Mock and -GALNT6-WT stable cells were transfected with Flag-tagged GRP78 and treated with CHX prior to western blot. Overexpression of GALNT6 stabilized exogenously expressed GRP78 in MDA-MB-435S-GALNT6-WT stable cells. (C) We generated Flag-tagged constructs in which each of candidate O-glycosylation sites was substituted with an alanine residue (T85A, T151A, T166A, T184A, or T203A). (D) We established MCF7-derived cell lines that stably expressing each of Flag-tagged alanine-substituted GRP78 proteins, as well as Flag-tagged GRP78-WT protein. These stable cells were treated with CHX at different time points followed by western blot. Lower graph indicated Flag-tagged GRP78 protein intensities from western blot, which showed rapid reduction of GRP78 proteins harboring T151A, T166A, and T184A substitutions.
O-glycosylation Deficiency Causes ER-to-cytoplasm Translocation of GRP78
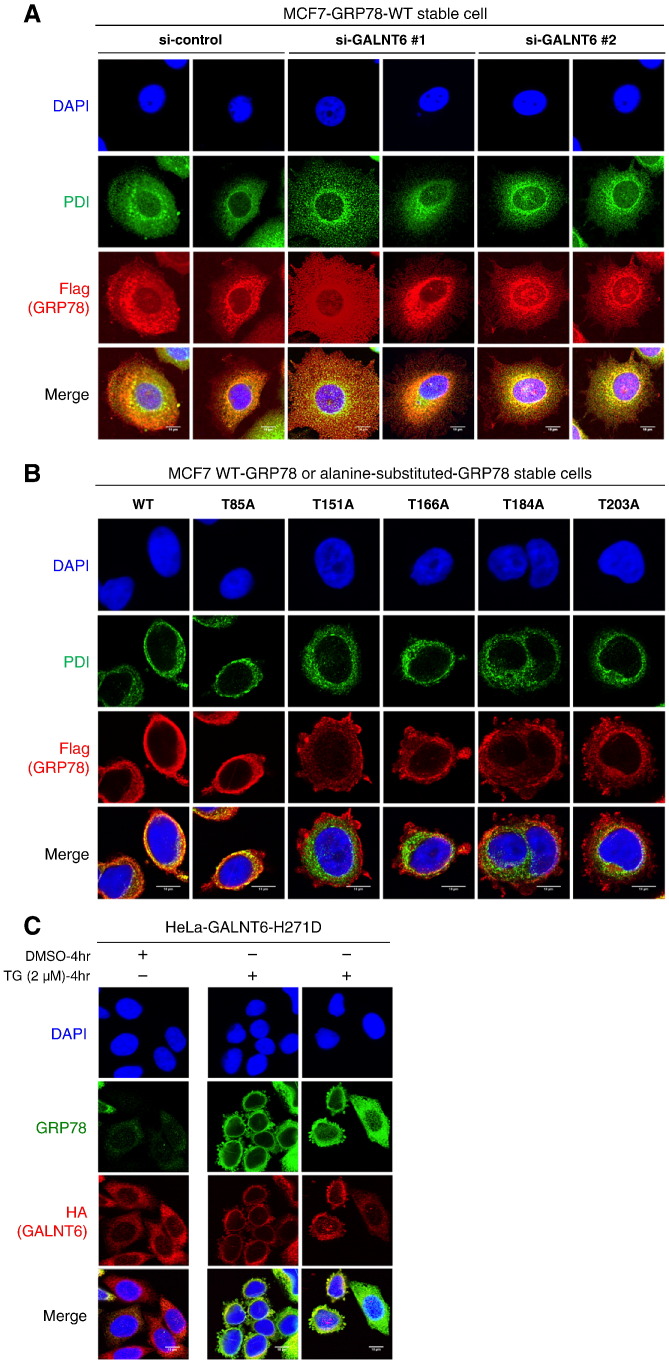
To investigate the effect of O-glycosylation on the subcellular localization of GRP78, we knocked down GALNT6 in MCF7 cells. We observed both Flag-tagged GRP78 and endogenous PDI (prolyl 4-hydroxylase subunit beta, P4HB), a classic ER marker for clarification of ER structure, were located at ER in MCF7 cells treated with control siRNA, while Flag-tagged GRP78 was diffusely distributed and cell shape became irregular when GALNT6 was knocked down (Figure 6A, and Supplementary Figure 3A). To further figure out whether the subcellular localization of GRP78 was changed or not, we examined Flag-tagged GRP78 proteins in six cell lines expressing Flag-tagged GRP78-WT or alanine-substituted GRP78. We found that Flag-tagged GRP78 protein was diffusely distributed in four cell lines harboring alanine-substitutions at O-glycosylation sites in the ATPase domain (T151A, T166A, T184A, or T203A) (Figure 6B, and Supplementary Figure 3B). In addition, irregular subcellular localization of endogenous GRP78 was also observed in HeLa-GALNT6-H271D stable cells after ER stress by TG treatment (Figure 6C), but not in Hela-GALNT6-WT stable cells (Figure 3B). Collectively, these results indicated that O-glycosylations might be important for subcellular localization of GRP78 in ER.
Figure 6.
O-glycosylation deficiency causes GRP78 ER-to-cytoplasm translocation. (A) GALNT6 was knocked down by siRNA for 72 h in MCF7-GRP78-WT stable cells, and then immunocytochemistry was performed. Flag-tagged GRP78 showed a diffuse and irregular distribution pattern in the cells in which GALNT6 was knocked down. Two fields were taken for each condition. (B) Immunocytochemistry was performed in MCF7-GRP78-WT and alanine-substituted GRP78 stable cells. We found that Flag-tagged GRP78 protein irregularly distributed out of ER in four cell lines harboring alanine-substitutions at O-glycosylation sites in the ATPase domain (T151A, T166A, T184A, or T203A). (C) HeLa-GALNT6-H271D stable cells were treated with DMSO or TG for 4 h, followed by immunocytochemistry. Irregular subcellular localization of endogenous GRP78 was also frequently observed in HeLa-GALNT6-H271D stable cells after induction of ER stress by TG treatment. Two fields were shown from TG-treated HeLa-GALNT6-H271D stable cells. More images from different fields for Figure 6, A and B, are provided in Supplementary Figure 3.
O-glycosylation of GRP78 is Critical for Cell Survival Under the Nutrition Deprivation Condition
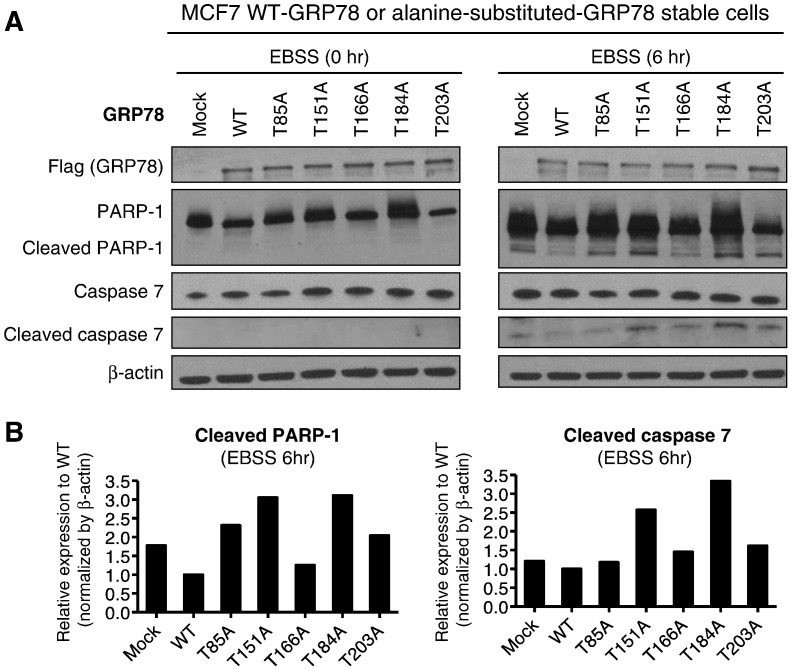
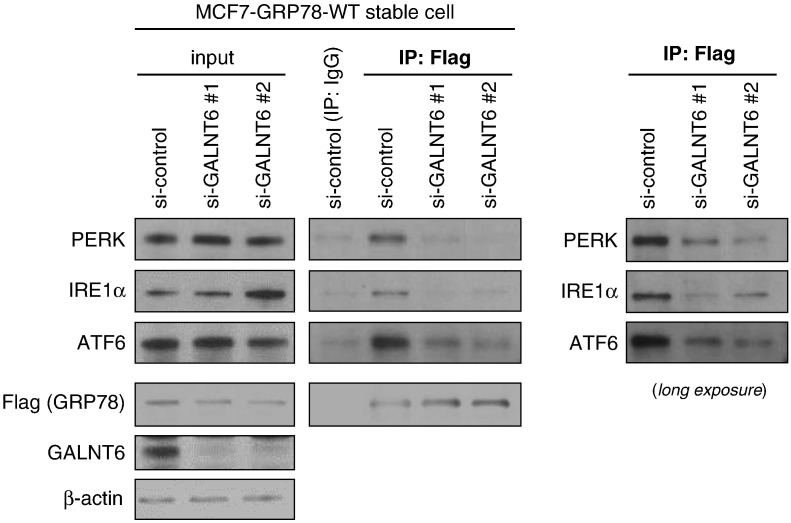
Since GRP78 was reported to play important roles in cancer cell survival under stress conditions [13], [14], [15], we tested MCF7 cells, in which GRP78-WT or each of five alanine-substituted-GRP78 proteins (GRP78-T85A, −T151A, −T166A, −T184A, or -T203A) was stably expressed, under a nutrition deprivation condition by treatment with Earle's Balanced Salt Solution (EBSS) (Figure 7, A and B). Compared with MCF7 cells with GRP78-WT, the cells with each of alanine-substituted GRP78 proteins were more susceptible to apoptosis under the stress condition, showing higher level of apoptosis markers, such as cleaved poly (ADP-ribose) polymerase 1 (PARP-1) and cleaved caspase 7 (CASP7). We could not examine cleavage of caspase 3 because MCF7 cells lacked its expression [19], [20]. Collectively, these data indicated that O-glycosylation deficient GRP78 lost its anti-apoptotic function. Since the anti-apoptotic roles of GRP78 in the ER stress condition was suggested to go through its interaction with PERK (eukaryotic translation initiation factor 2 alpha kinase 3), IRE1α (endoplasmic reticulum to nucleus signaling 1) and ATF6 (activating transcription factor 6) proteins in ER [10], [11], [12], [21], we hypothesized that O-glycosylation deficiency might affect these interactions. We observed the binding ability of Flag-tagged GRP78 with endogenous PERK, IRE1α and ATF6 proteins was significantly reduced after GALNT6 knockdown in MCF7-GRP78-WT stable cells (Figure 8). Taken together, these findings suggested that O-glycosylation of GRP78 was critical for the interaction with PERK, IRE1α and ATF6 to prevent apoptosis of cancer cell under the ER stress.
Figure 7.
GRP78 O-glycosylation is critical for cell survival under the nutrition deprivation condition. (A) MCF7-GRP78-WT and -alanine-substituted GRP78 stable cells were cultured in MEM with 10% FBS or after washed by PBS, and then in EBSS for additional 6 h, followed by western blot. Compared with MCF7-GRP78-WT cells, MCF7-alanine-substituted-GRP78 cells were more susceptible to apoptosis under the stress condition, showing higher level of apoptosis markers, such as cleaved PARP-1 and cleaved caspase 7. Nutrition deprivation treatment: EBSS medium treatment. (B) Graphs indicated protein band intensities of cleaved PARP-1 (left) or cleaved caspase 7 (right).
Figure 8.
O-glycosylation is critical for the interaction of GRP78 with PERK, IRE1α and ATF6. GALNT6 was knocked down by siRNA for 72 h in MCF7-GRP78-WT stable cells, and then Flag-tagged GRP78 was pulled down by using anti-Flag antibody followed by western blot. The binding ability of Flag-tagged GRP78 with endogenous PERK, IRE1α and ATF6 was significantly reduced after GALNT6 knockdown.
Discussion
Important roles of each of GALNT6 and GRP78 in carcinogenesis have been reported [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], but their interaction and the biological significance of O-glycosylated GRP78 have never been investigated. In this study, we have demonstrated that GRP78 is likely to be a substrate of GALNT6 and evidences showing that GALNT6 binds to the ATPase domain of GRP78 and stabilizes the GRP78 protein through O-glycosylations, which also contributed to proper subcellular localization, interaction and function of GRP78 in ER.
In this study, we firstly identified GRP78 as a novel substrate of GALNT6 and showed that the ATPase domain of GRP78 would be important for its binding with GALNT6 and O-glycosylations. Furthermore, we identified five glycosylation sites on GRP78 protein by GALNT6 using mass spectrometry analysis; among them, a glycosylation site T203 was also indicated by others [22]. In addition, when we knocked down GALNT6 in MCF7 and T47D breast cancer cells, the O-glycosylation level of endogenous GRP78 was significantly decreased, suggesting that GALNT6 was critical for O-glycosylation of GRP78 in these two cancer cells. A previous study suggested that GRP78 might be a glycoprotein based on mass spectrometry analysis [22], but it has been unknown which O-type glycosyltransferase glycosylates GRP78. To the best of our knowledge, we demonstrate for the first time that GALNT6 O-glycosylates GRP78, which may add a new insight for the function of GRP78 protein.
Secondly, GALNT6 was likely to stabilize GRP78 protein through O-glycosylations. In the presence of high level of GALNT6 protein, the half-life of GRP78 protein seemed to be significantly increased. Among five glycosylation-site-substituted GRP78 proteins, in which either T85, T151, T166, T184, or T203 was substituted to an alanine residue, three proteins (T151A, T166A and T184A) that have substitutions in the ATPase domain showed rapid degradation of GRP78 protein, suggesting that O-glycosylation by GALNT6 stabilized GRP78. The biological significance of GRP78 stabilization might be to prolong its oncogenic functions in cancer cells.
Thirdly, O-glycosylations also contributed to the proper subcellular localization, interaction and function of GRP78 in ER. O-glycosylation deficient GRP78 could not be located in ER, but was diffusely distributed in cytoplasm, and reduced the binding affinity with PERK, IRE1α and ATF6. As consequence, these cells failed to respond to the ER stress condition and resulted in cell death. Previous studies suggested promising anti-tumor effects by targeting GRP78, regardless to GRP78 knockdown or its functional inactivation [21]. In our study, O-glycosylation deficient GRP78 seemed to lose its anti-apoptotic function, and therefore, targeting GALNT6-GRP78 pathway might be a promising strategy to induce cancer cell death.
Lastly, GRP78 activation could drive Golgi-to-ER relocation of GALNT6 protein. Previous studies indicated that activation of growth factor receptors (e.g. EGFR, PDGFR) or Src protein kinase caused Golgi-to-ER relocation of GalNAc-Ts (e.g. GALNT1 and GALNT2), which in turn enhanced cancer cell invasiveness [23], [24]. Similar to these examples, we found that high level of GRP78 protein either by overexpression or ER stress condition induced the Golgi-to-ER relocation of GALNT6, where GALNT6 might O-glycosylate GRP78 as well as other proteins possibly involved in carcinogenesis. GRP78 is known to be up-regulated in many types of cancer and often induced by diverse cellular stresses in the tumor microenvironment, such as hypoxia, acidosis, starvation, as well as exogenous stresses including therapeutic interventions [10]. Under these conditions, Golgi-to-ER relocation of GALNT6 might frequently occur and then trigger O-glycosylation of diverse substrates at ER.
In summary, we found that GALNT6-induced O-glycosylation is critical for the stability, subcellular localization, and anti-apoptotic function of GRP78 protein in cancer cells. We also suggest that GRP78 might enhance the activity of GALNT6 in carcinogenesis through driving Golgi-to-ER relocation of GALNT6. These findings may provide a new insight in the GALNT6-GRP78 pathway and indicate that targeting this pathway is a promising approach for development of a novel class of anti-cancer therapies.
Disclosure of Potential Conflicts of Interest
Y.N. is a stock holder and a scientific advisor of OncoTherapy Science, Inc. J.P. is a scientific advisor of OncoTherapy Science, Inc.
Acknowledgements
We thank Kyoko Adachi and Yasuo Mochizuki for technical supports, Rui Wang for cell culture, and Toyomasa Katagiri for helpful discussion.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2016.11.007.
Appendix A. Supplementary data
. Supplementary Figure 1. Mass spectrometry analysis identified 15 proteins after the VVA-lectin pull-down assay. Peptide finger-printing results were shown with expected matching scores from the Mascot database search. No candidate protein was identified from band 3 (~100 kDa), 13 and 14 (~50 kDa). Supplementary Figure 2. Semi-quantitative RT-PCR for candidate substrates of GALNT6. Expression levels of total 15 genes encoding GALNT6’s candidate substrates were examined by semi-quantitative RT-PCR in breast cancer cells and HeLa-GALNT6 stable cells. Functional relationship of each gene with glycosylation and cancer was examined by literature search in PubMed (http://www.ncbi.nlm.nih.gov/pubmed). N: no report; Y: already reported. Supplementary Figure 3. Immunocytochemistry analysis for GRP78 ER-to-cytoplasm translocation. More figures were provided for Figure 6. Supplementary Figure 3A and B were corresponding to Figure 6, A and B, respectively.
Supplementary tables
References
- 1.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 2.Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7(6):599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan P, Dabelsteen S, Madsen FB, Francavilla C, Kopp KL, Steentoft C, Vakhrushev SY, Olsen JV, Hansen L, Bennett EP. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111(39):E4066–E4075. doi: 10.1073/pnas.1406619111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks SA, Carter TM, Bennett EP, Clausen H, Mandel U. Immunolocalisation of members of the polypeptide N-acetylgalactosaminyl transferase (ppGalNAc-T) family is consistent with biologically relevant altered cell surface glycosylation in breast cancer. Acta Histochem. 2007;109(4):273–284. doi: 10.1016/j.acthis.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Freire T, Berois N, Sonora C, Varangot M, Barrios E, Osinaga E. UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a potential new marker for detection of bone marrow-disseminated breast cancer cells. Int J Cancer. 2006;119(6):1383–1388. doi: 10.1002/ijc.21959. [DOI] [PubMed] [Google Scholar]
- 6.Patani N, Jiang W, Mokbel K. Prognostic utility of glycosyltransferase expression in breast cancer. Cancer Genomics Proteomics. 2008;5(6):333–340. [PubMed] [Google Scholar]
- 7.Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, Hirata K, Nakamura Y, Katagiri T. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70(7):2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, Katagiri T, Chung S, Kijima K, Nakamura Y. Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia. 2011;13(4):320–326. doi: 10.1593/neo.101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarhan YE, Kato T, Jang M, Haga Y, Ueda K, Nakamura Y, Park JH. Morphological Changes, Cadherin Switching, and Growth Suppression in Pancreatic Cancer by GALNT6 Knockdown. Neoplasia. 2016;18(5):265–272. doi: 10.1016/j.neo.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67(8):3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 11.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 12.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32(7):805–818. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook KL, Shajahan AN, Warri A, Jin L, Hilakivi-Clarke LA, Clarke R. Glucose-regulated protein 78 controls cross-talk between apoptosis and autophagy to determine antiestrogen responsiveness. Cancer Res. 2012;72(13):3337–3349. doi: 10.1158/0008-5472.CAN-12-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grkovic S, O'Reilly VC, Han S, Hong M, Baxter RC, Firth SM. IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene. 2013;32(19):2412–2420. doi: 10.1038/onc.2012.264. [DOI] [PubMed] [Google Scholar]
- 15.Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, Mao C, Ye R, Wang M, Pen L. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68(2):498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 16.Jiang CC, Yang F, Thorne RF, Zhu BK, Hersey P, Zhang XD. Human melanoma cells under endoplasmic reticulum stress acquire resistance to microtubule-targeting drugs through XBP-1-mediated activation of Akt. Neoplasia. 2009;11(5):436–447. doi: 10.1593/neo.09208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Deng Z, Tanikawa C, Shuin T, Miki T, Matsuda K, Nakamura Y. Downregulation of the tumor suppressor HSPB7, involved in the p53 pathway, in renal cell carcinoma by hypermethylation. Int J Oncol. 2014;44(5):1490–1498. doi: 10.3892/ijo.2014.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JH, Rasch MG, Qiu J, Lund IK, Egeblad M. Presence of insulin-like growth factor binding proteins correlates with tumor-promoting effects of matrix metalloproteinase 9 in breast cancer. Neoplasia. 2015;17(5):421–433. doi: 10.1016/j.neo.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Twiddy D, Cohen GM, Macfarlane M, Cain K. Caspase-7 is directly activated by the approximately 700-kDa apoptosome complex and is released as a stable XIAP-caspase-7 approximately 200-kDa complex. J Biol Chem. 2006;281(7):3876–3888. doi: 10.1074/jbc.M507393200. [DOI] [PubMed] [Google Scholar]
- 20.Zoli W, Ulivi P, Tesei A, Fabbri F, Rosetti M, Maltoni R, Giunchi DC, Ricotti L, Brigliadori G, Vannini I. Addition of 5-fluorouracil to doxorubicin-paclitaxel sequence increases caspase-dependent apoptosis in breast cancer cell lines. Breast Cancer Res. 2005;7(5):R681–R689. doi: 10.1186/bcr1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14(4):263–276. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L. Precision mapping of the human O-GalNAc glycoproteome through Simple Cell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill DJ, Chia J, Senewiratne J, Bard F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010;189(5):843–858. doi: 10.1083/jcb.201003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill DJ, Tham KM, Chia J, Wang SC, Steentoft C, Clausen H, Bard-Chapeau EA, Bard FA. Initiation of GalNAc-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proc Natl Acad Sci U S A. 2013;110(34):E3152–E3161. doi: 10.1073/pnas.1305269110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. Supplementary Figure 1. Mass spectrometry analysis identified 15 proteins after the VVA-lectin pull-down assay. Peptide finger-printing results were shown with expected matching scores from the Mascot database search. No candidate protein was identified from band 3 (~100 kDa), 13 and 14 (~50 kDa). Supplementary Figure 2. Semi-quantitative RT-PCR for candidate substrates of GALNT6. Expression levels of total 15 genes encoding GALNT6’s candidate substrates were examined by semi-quantitative RT-PCR in breast cancer cells and HeLa-GALNT6 stable cells. Functional relationship of each gene with glycosylation and cancer was examined by literature search in PubMed (http://www.ncbi.nlm.nih.gov/pubmed). N: no report; Y: already reported. Supplementary Figure 3. Immunocytochemistry analysis for GRP78 ER-to-cytoplasm translocation. More figures were provided for Figure 6. Supplementary Figure 3A and B were corresponding to Figure 6, A and B, respectively.
Supplementary tables