Abstract
MicroRNA (miR) sponges containing miR binding sequences constitute a potentially powerful molecular therapeutic strategy. Recently, naturally occurring circular RNAs (circRNAs) were shown to function as efficient miR sponges in cancer cells. We hypothesized that synthetic circRNA sponges could achieve therapeutic loss-of-function targeted against specific miRs. Linear RNA molecules containing miR-21 binding sites were transcribed in vitro; after dephosphorylation and phosphorylation, circularization was achieved using 5′-3′ end-ligation by T4 RNA ligase 1. circRNA stability was assessed using RNase R and fetal bovine serum. Competitive inhibition of miR-21 activity by a synthetic circRNA sponge was assessed using luciferase reporter, cell proliferation, and cell apoptosis assays in three gastric cancer cell lines. circRNA effects on downstream proteins were also delineated by Tandem Mass Tag (TMT) labeling (data available via ProteomeXchange identifier PRIDE: PXD008584), followed by western blotting. We conclude that artificial circRNA sponges resistant to nuclease digestion can be synthesized using simple enzymatic ligation steps. These sponges inhibit cancer cell proliferation and suppress the activity of miR-21 on downstream protein targets, including the cancer protein DAXX. In summary, synthetic circRNA sponges represent a simple, effective, convenient strategy for achieving targeted loss of miR function in vitro, with potential future therapeutic application in human patients.
Keywords: synthetic circular RNA, microRNA sponge, microRNA loss-of-function, gastric carcinoma, molecular therapy
Introduction
Gastric carcinoma (GC) remains the fifth most common cancer and the third highest cause of cancer death worldwide, with 984,000 new cases and 841,000 deaths in 2013.1 Most GC cases are diagnosed at late stages with consequent poor outcomes, for which treatment is largely restricted to cytotoxic chemotherapy. Fortunately, as our understanding of gastric molecular carcinogenesis deepens,2 novel potential therapeutic strategies based on molecular target therapies are now being discovered.
MicroRNAs (miRs) are small non-coding RNAs that mediate post-transcriptional silencing of 30% of protein-coding genes by base pairing between the miR seed region (2–8 nucleotides) and target 3′ UTRs of mRNAs.3 Dysregulated miRs can act as oncogenes (oncomiRs) or tumor suppressor genes (ts-miRs), and play important roles in tumorigenesis and progression.4, 5, 6, 7 miR-21, one of the most well-characterized oncomiRs, is overexpressed in GC.8, 9 Elevated expression of miR-21 has been implicated in critical GC-related processes, including proliferation, invasion, and apoptosis, by inhibiting tumor suppressor genes such as PDCD4 and RECK.10, 11 Thus, miR-21 represents an appealing therapeutic target. Moreover, linear sponge RNAs containing binding sites for specific miRs have been shown to induce miR loss-of-function.12, 13
Circular RNAs (circRNAs) are a new class of unique RNAs with single-stranded, covalently closed, continuous loop structures.14 circRNAs are produced by “backsplicing” or “head-to-tail splicing” of linear RNAs.15 Due to their circularity, lack of 5′ to 3′ polarity, and absence of polyadenylated tails, circRNAs are more stable and resistant to exonuclease degradation than linear RNAs.16 Naturally occurring endogenous circRNAs can contain selectively conserved miR binding sites, thereby functioning as efficient miR “sponges” and counteracting miRs.16, 17, 18 Thus, recent circRNA research has provided promising new insights into miR regulation. However, to our knowledge, the construction and use of synthetic artificial circRNAs targeting specific oncomiRs (i.e., synthetic circRNA sponges) have not yet been explored.
In the current study, we hypothesized that a synthetic circRNA could function as a miR-21 sponge to suppress GC proliferation. We used enzymatic ligation to synthesize a circRNA containing several miR-21 bulged (i.e., partially mismatched) binding sites in vitro. This circRNA was shown to be more resistant to nuclease degradation than its linear counterpart RNA. We then assessed competitive inhibition of miR-21 activity by the circRNA, as well as its inhibition of GC cell proliferation, dysregulation of global protein expression, and upregulation of a known miR-21 tumor suppressor gene target, DAXX. We conclude that synthetic circRNA sponges represent a rapid, convenient, effective strategy for achieving miR loss-of-function in vitro, with substantial potential for future application in human gastric cancer patients.
Results
miR-21 Is Overexpressed in The Cancer Genome Atlas Patients
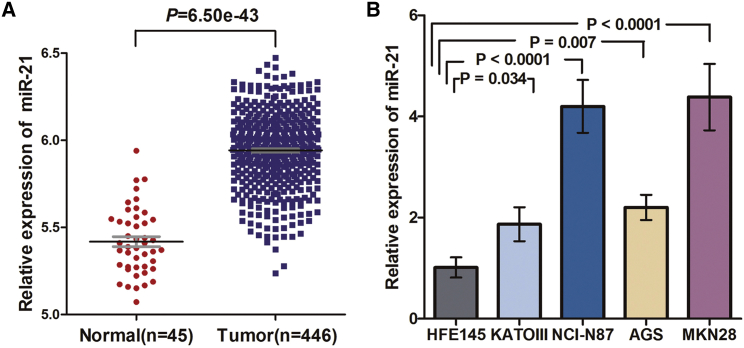
Based on miR sequence data from 446 GC and 45 normal samples within The Cancer Genome Atlas (TCGA), 185 aberrantly expressed miRs were identified, with 137 (74.1%) upregulated and 48 (25.9%) downregulated. miR-21, our miR of interest, was upregulated 1.76-fold in GC versus normal samples (p = 6.50e−43; Figure 1A).
Figure 1.
miR-21 Is Overexpressed in The Cancer Genome Atlas Patients and Also Overexpressed in GC Cell Lines
(A) Significant upregulation of miR-21 in GC versus normal samples. (B) Comparison of relative expression of miR-21 between NCI-N87, KATOIII, AGS, MKN28, and the normal gastric epithelial cell line HFE145, as determined by TaqMan qRT-PCR. Means ± SD are shown.
miR-21 Is Aberrantly Overexpressed in Gastric Cancer Cells
Using TaqMan qRT-PCR analysis, relative to the normal gastric epithelial cell line HFE145, we found significant overexpression of miR-21 in NCI-N87 (fold changes 4.2; p < 0.0001), KATOIII (fold changes 1.9; p = 0.034), AGS (fold changes 2.2; p = 0.007), and MKN28 (fold changes 4.4; p < 0.0001) (Figure 1B). We chose NCI-N87, AGS, and MKN28 for subsequent studies because miR-21 was expressed at higher levels than in KATOIII cells.
circRNA Sponges Can Be Correctly and Successfully Synthesized In Vitro
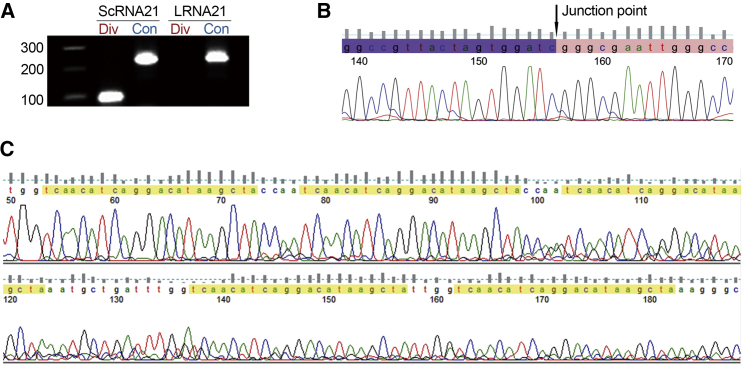
Our strategy for RNA circularization in vitro used enzymatic ligation of synthetic linear RNA containing five repeated miR bulged binding sites (Figure 2).12, 19 Accurate construction of circRNA sponges was validated by RT-PCR and Sanger sequencing. RT-PCR results confirmed that divergent primers generated the expected 100-nt product only from the circular (scRNA21) template RNA, but not from the corresponding linear (LRNA21) template RNA. In contrast, convergent primers generated the expected product from both circular and linear RNA templates (Figure 3A). Moreover, the head-to-tail circRNA junction point (a unifying characteristic of all circRNAs) was validated only in divergent PCR products generated from scRNA21 (Figure 3B). Five repeated miR-21 bulged (mismatched) binding sites were verified in convergent PCR products generated from both LRNA21 and scRNA21 sponges (Figure 3C). The head-to-tail junction and five repeated scrambled sites were also confirmed in a negative control scrambled circRNA.
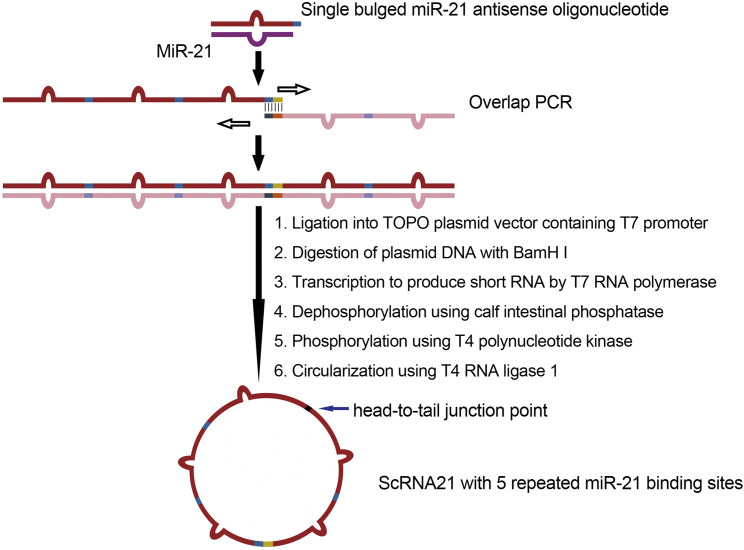
Figure 2.
Workflow for Producing Synthetic miR-21 circRNA Sponge
Each binding site sequence is perfectly complementary to the miR-21 seed region, but contains a bulge at positions 9–12 to prevent RNAi-type cleavage and degradation. One PCR cycle facilitates synthesis to generate a double-stranded DNA PCR fragment, which is then cloned into the TOPO PCR cloning vector (Invitrogen). The T7 RNA polymerase binding site located just 5′ to the PCR insert is used to generate, via T7 RNA polymerase, large quantities (150 μg) of linear RNA containing the miR sponge sequence described above. Calf intestinal phosphatase dephosphorylates the 5′ end of the RNA transcript, and T4 polynucleotide kinase (in the presence of ATP) generates RNA molecules suitable for ligation. Incubation with T4 RNA ligase results in RNA circularization.
Figure 3.
Validation of Synthetic Circular Sponge Construction
(A) RT-PCR of scRNA21 and LRNA21 using either convergent (Con) or divergent (Div) primers. Only scRNA21 could be amplified using divergent primers. (B) Results of sequencing of divergent PCR products generated from scRNA21 confirmed the head-to-tail junction point. (C) Results of sequencing of convergent PCR products generated from scRNA21 confirmed the five repeated miR-21 bulged binding sites (yellow highlighting).
A circRNA Sponge Is More Resistant to Nuclease Degradation Than Its Linear Counterpart RNA
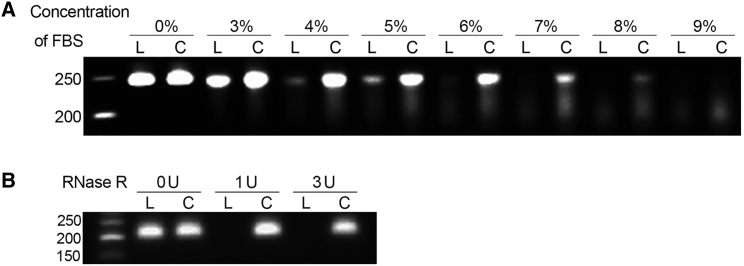
Resistance to nuclease-mediated degradation of the scRNA21 sponge was compared with that of its linear counterpart LRNA21, first using fetal bovine serum (FBS)-mediated degradation. LRNA21 was 92% degraded in 4% FBS after 30 min (Figure 4A). In contrast, the scRNA21 was significantly more stable, being only 9% degraded after 30 min (p < 1 × 10−4). In fact, the scRNA21 was degraded only in FBS at concentrations of 7% or higher. Stabilities of the circular and linear RNAs were then compared after digestion with the specific exonuclease, RNase R. LRNA21 was 94% or 97% degraded with 1 or 3 U RNase R/1 μg RNA, respectively; in contrast, the scRNA21 was only 9% or 14% degraded (p = 8 × 10−4 and p = 1.2 × 10−3, respectively; Figure 4B).
Figure 4.
Resistance of Synthetic circRNA scRNA21 to Nuclease Digestion
(A) scRNA21 (C) was more resistant to degradation by increasing concentrations of fetal bovine serum (FBS) than its linear counterpart LRNA21 (L) at 37°C × 30 min. (B) scRNA21 (C) was more resistant to RNase R degradation than its linear counterpart LRNA21 (L). Treatment ratios were 1 U RNase R/1 μg RNA and 3 U RNase R/1 μg RNA, respectively.
Functional Efficacy of a Synthetic circRNA miR Sponge
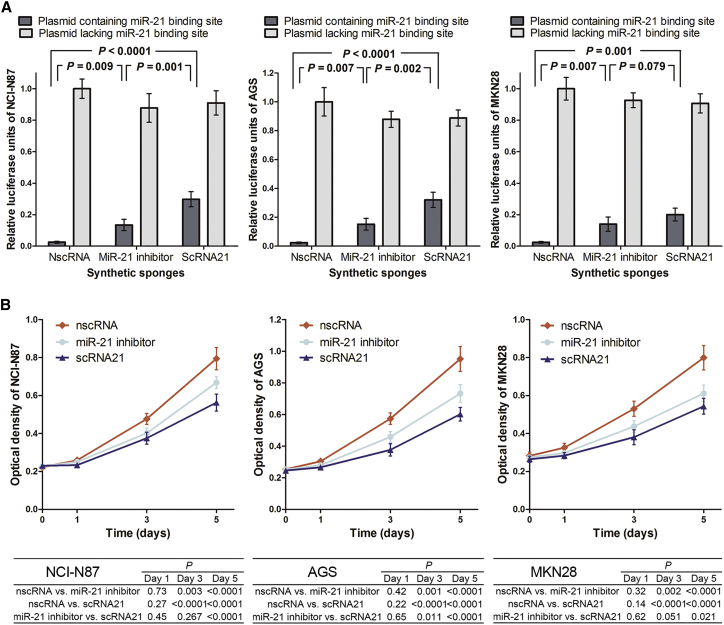
To assess the functional effectiveness of our synthetic circular miR-21 sponge, we assayed normalized firefly luciferase activity (i.e., firefly luciferase activity/Renilla luciferase activity) in NCI-N87, AGS, and MKN28 GC cells 24 hr after co-transfection. Relative to a luciferase reporter plasmid lacking any miR-21 binding sites, decreased normalized firefly luciferase activity was observed in all three cell lines upon co-transfection of a luciferase reporter plasmid containing miR-21 binding sites with a negative control scrambled circRNA (Figure 5A). Firefly luciferase activity was restored when the luciferase reporter plasmid containing multiple miR-21 binding sites was co-transfected with a conventional linear miR-21 inhibitor. Notably, even greater restoration of luciferase activity was observed when cells were co-transfected with our scRNA21 (Figure 5A). These findings confirmed the functional effectiveness of scRNA21 as a miR-21 sponge.
Figure 5.
Functional Effectiveness of scRNA21 as a miR-21 Sponge
(A) Dual-luciferase reporter assay results. After co-transfection with a negative control scrambled circRNA, a miR-21 inhibitor, or scRNA21 plus a luciferase reporter plasmid containing miR-21 binding sites, normalized luciferase activities were 0.0256, 0.1347, and 0.2981, respectively, in NCI-N87 cells (left); 0.0236, 0.1518, and 0.3214, respectively, in AGS cells (middle); and 0.0250, 0.1400, and 0.2004, respectively, in MKN28 cells (right). There was no significant difference between MKN28 cells co-transfected with miR-21 inhibitor versus scRNA21. (B) Significant inhibition of cancer cell proliferation by synthetic anti-oncomiR-21 circRNA (scRNA21). Cell proliferation was assessed by WST-1 assay. Relative to negative control scrambled circRNA or miR-21 inhibitor at day 5, scRNA-21 significantly inhibited proliferation in NCI-N87 cells (left) (p < 1 × 10−4 or p < 1 × 10−4, respectively), AGS cells (middle) (p < 1 × 10−4 or p < 1 × 10−4, respectively), and MKN28 cells (right) (p < 1 × 10−4 or p = 0.021, respectively). Each experiment was repeated independently three times.
Subsequently, we evaluated the effects of scRNA21 on proliferation in NCI-N87, AGS, and MKN28 GC cells by WST-1 assays. On day 5, a significantly decreased cell proliferation was observed in all three cell lines after transfection with our scRNA21, relative to either a scrambled control circRNA or a conventional miR-21 inhibitor (Figure 5B).
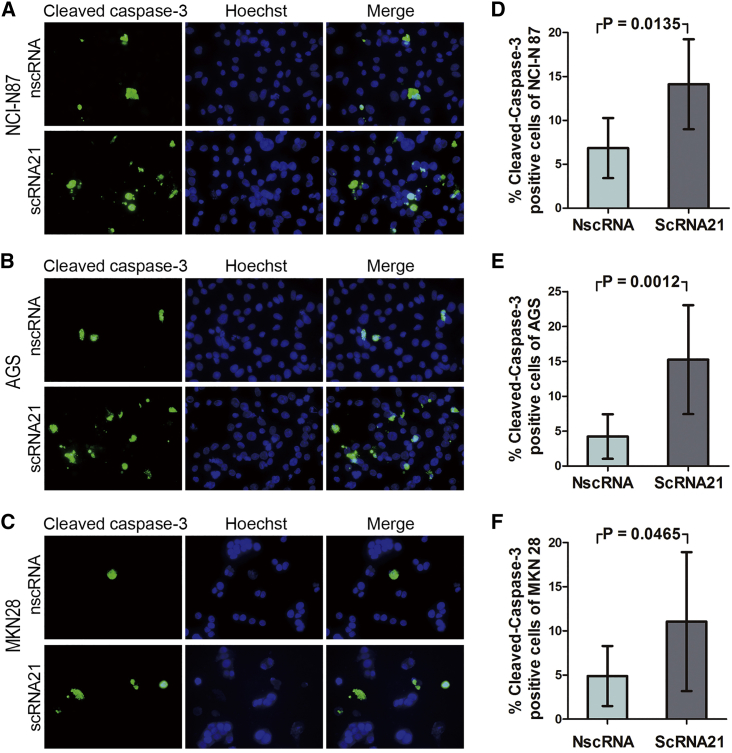
Next, we assessed the functional effects of scRNA21 on apoptosis. 24 hr after transfection of either negative control scrambled circRNA or scRNA21, we measured cleaved caspase-3 levels in NCI-N87, AGS, and MKN28 GC cells. scRNA21 induced increased apoptosis, evidenced by significantly higher cleaved caspase-3 levels than with negative control scrambled circRNA in all three GC cell lines (Figure 6).
Figure 6.
Induction of Apoptosis by scRNA21
(A–C) Relative to transfection with a negative control scrambled circRNA, significantly higher cleaved caspase-3 levels occurred after transfection with scRNA21 in NCI-N87 (A), AGS (B), and MKN28 (C) cells. (D) Mean cleaved caspase-3 levels in NCI-N87 cells were 6.84% and 14.11%, respectively (p = 0.0135). (E) Mean cleaved caspase-3 levels in AGS cells were 4.24% and 15.25%, respectively (p = 0.0012). (F) Mean cleaved caspase-3 levels in MKN28 were 4.89% and 11.05%, respectively (p = 0.0465). The number of cleaved caspase-3-positive cells and total cells was counted using ImageJ software, and ratios were calculated. Data are presented as mean; error bars indicate SD of three slides from each group of three independent experiments.
Downstream Proteomic Effects of a Synthetic Circular miR Sponge
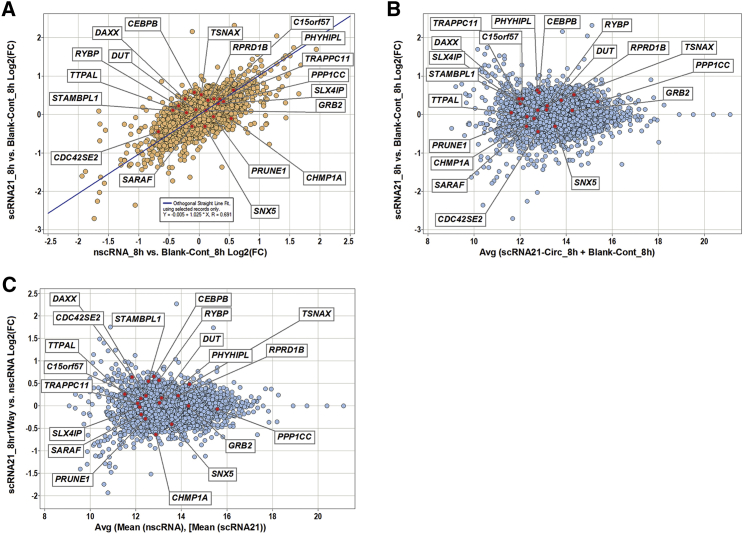
Effects of our scRNA21 on global protein expression were then comprehensively analyzed. Because oncomiRs exert at least some effects on target mRNAs by altering their translation into proteins,6, 13, 20 we performed pan-proteomic screening using Tandem Mass Tag (TMT) labeling to delineate proteins whose levels were altered by scRNA21. These experiments generated a list of 6,314 individual proteins. According to target prediction by Ingenuity Pathway Analysis, 235 of these 6,314 proteins represented putative direct binding targets of miR-21. 19 of these 235 proteins increased by a fold change at least two SDs above control levels (Table S1). Protein levels of these predicted direct miR-21 targets, which should be downregulated by miR-21, were restored after scRNA21 treatment (which is expected to antagonize miR-21; Figures 7A–7C). We then further explored these 19 proteins in the COSMIC consensus cancer gene database21: only 1 of these 19, the death domain-associated tumor suppressor protein DAXX, was found in the COSMIC consensus cancer gene list. Therefore, Daxx was selected for further study, due to its known tumor-suppressive function22, 23, 24, 25 and based on its fold change increase of >2 SDs 8 hr after scRNA21 treatment.
Figure 7.
Global Proteomic Analysis following Synthetic Circular RNA Treatment
(A) Quantile plot of TMT global proteomic analysis of differentially expressed proteins 8 hr after scRNA21 transfection. Dots represent proteins showing >2 SD differential expression in scRNA21-treated versus blank control samples, but <2 SD differential expression in negative control scrambled circRNA-treated versus blank control samples. 19 of these significantly differentially expressed proteins were predicted putative direct binding targets of miR-21 by Ingenuity Pathway Analysis. (B) MvA plot of TMT global proteomic analysis of differentially expressed proteins 8 hr after scRNA21 transfection (scRNA21 versus blank control). y axis: log2 fold change; x axis: average log2 signal. The term “MvA” is standard shorthand for minus versus average, a standard graphic plot to depict expression comparisons for amino or nucleic acid analyses. Minus is the difference between two biological conditions determined by subtracting (division in log world) the control signal from the case signal. Average is the geometric mean of the case’s and control’s log signals. In this case, our signals were the quantile-normalized log2 values derived from mass spectrometric analysis software (Proteome Discoverer) output. In practice, the MvA y axis is the log2 fold change between the two biological conditions, in this case the samples’ underlying genotypes. It is also called the log ratio. Thus, the zero value for log2 represents no difference. The x axis represents the average of both case’s and control’s normalized log2 signals in simple log2 notation. This can be thought of as depicting the full dynamic range of the underlying mass spectrometry platform. (C) MvA plot of TMT global proteomic analysis of differentially expressed proteins after 8 hr of scRNA21 treatment (scRNA21 versus negative control scrambled circRNA).
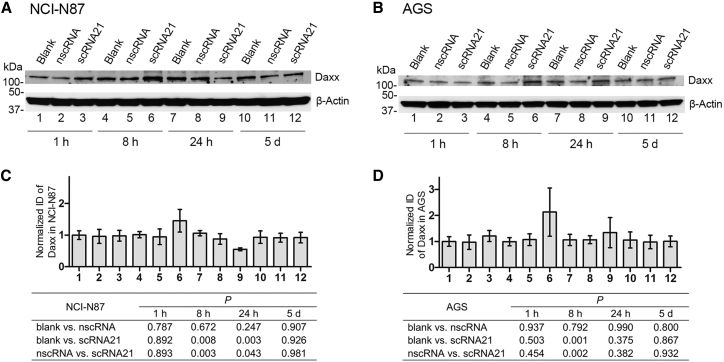
To validate our TMT results, we then performed western blotting using an anti-DAXX antibody. These experiments confirmed restoration of DAXX protein levels 8 hr after scRNA transfection of two GC cell lines, NCI-N87 and AGS (Figure 8). These data demonstrate the functional activity of scRNA21 against a specific miR-21 downstream target tumor suppressor gene.
Figure 8.
Effect of scRNA Treatment on Daxx Protein Levels
(A and B) Western blotting of protein extracted from NCI-N87 (A) or AGS (B) cells transfected with blank control, negative control scrambled circRNA, or scRNA21. (C and D) Integrated density (ID) of western blotting bands was quantified and calculated by ImageJ software for each group of three independent experiments, normalized to blank control: NCI-N87 (C) and AGS (D). Daxx protein was upregulated by scRNA21 treatment relative to either negative control scrambled circRNA or blank control 8 hr after transfection in both NCI-N87 and AGS cells.
Discussion
miRs are important post-transcriptional regulators of gene expression. They exert their effects by cleavage or translational arrest of target mRNAs, through direct base pairing to the binding sequences of the mRNAs.26 Numerous studies, including our own, confirm that miR dysregulation occurs frequently and plays a critical role in human cancers.4, 5, 20, 27 Thus, there has been a growing interest in developing strategies to achieve miR loss-of-function. miR sponges are competitive inhibitors containing multiple tandem binding sites complementary to a miR of interest.12 The metastasis of highly malignant breast cancer cells was suppressed via inhibition of miR-9, using an artificial linear miR-9 sponge.13 Similarly, inhibition of miR-135b by a linear miR-135b sponge suppressed cancer cell invasion, tumor growth, and metastasis in a mouse orthotopic lung tumor model.28 However, to our knowledge, there have been no publications describing miR inhibition by synthetic circular miR sponges.
Recently, circRNAs have drawn widespread attention as an important category of endogenous RNA. Two features of naturally occurring circRNAs, their resistance to exonucleases and their harboring of conserved miR binding sites, make them appealing tools for achieving miR loss-of-function.14, 16 Accumulating studies have revealed that several naturally occurring circRNAs can act as efficient miR sponges and play important roles in the pathogenesis of cancer. For example, overexpression of endogenous ciRS-7, which contains 63 conserved miR-7 target sites, was associated with inhibition of miR-7 and downstream activation of the oncogenes EGFR and RAF1, resulting in a more aggressive phenotype in colorectal cancer.29 circCCDC66, circFoxo3, circHIPK3, and circPVT1 represent additional naturally occurring circRNAs that harbor miR-binding sites and function as miR sponges in cancers.30, 31, 32, 33 However, to our knowledge, the construction or use of artificial circRNAs to achieve miR loss-of-function has not yet been reported.
We combined RNA circularization by enzymatic ligation with RNA synthesis containing multiple bulged miR binding sites to achieve artificial circRNA sponge synthesis in vitro, based on new permutations of previous methods.12, 19 In vitro transcribed linear RNA molecules, after dephosphorylation by calf intestinal phosphatase and phosphorylation by T4 polynucleotide kinase, were successfully circularized by 5′-3′ end ligation using T4 RNA ligase 1. Our results suggest that due to their bulged miR binding sites and closed-loop structures, with the absence of free 5′ and 3′ ends, synthetic circular miR sponges are more resistant to nuclease digestion and exert effective inhibition of miR activity. The marked observed stability of synthetic circRNA against two different nuclease digestion regimens emphasizes this key theoretical advantage of synthetic circRNAs as potential therapeutic agents. We also found that gastric carcinoma cell proliferation was inhibited, and that expression of DAXX, a key tumor suppressor gene that is also a known direct miR-21 target,22, 23, 24, 25, 34 was upregulated by a synthetic circRNA sponge. The inhibition of GC cell proliferation we observed could have been due, at least in part, to augmented protein levels of the miR-21 target gene Daxx. These results further establish the functional efficacy of synthetic circRNA sponges. Additional proteins, including GRB2, PPP1CC, CDC42SE2, SARAF, DUT, and RPRD1B, were increased at 24 hr post-scRNA-21 transfection: these additional proteomic changes further support the substantial biologic effects of this synthetic circRNA. These additional proteins merit further investigation in future circRNA experiments but are beyond the scope of the current initial exploratory study.
We draw several conclusions from the current study. First, to our knowledge, we have directly created the first artificial circRNAs containing multiple bulged oncomiR miR-21 binding sites by enzymatic ligation and synthesis in vitro. Second, we show that this synthetic circRNA is more resistant to nuclease degradation than its linear counterpart RNA. Third, we demonstrate that this synthetic circRNA functions actively as a miR-21 sponge, suppressing the posttranscriptional regulatory activity of miR-21 and increasing the expression of miR-21 downstream proteins, while also inhibiting GC cell proliferation. Notably, circRNA sponges can be synthesized in large quantities using simple and easily controllable steps. However, several potential drawbacks still exist, and additional troubleshooting may be required for broader application of this strategy. For example, we used enzymatic ligation for circularization of short RNAs (<500 nucleotides); circularization of larger RNAs may be challenging. Second, due to varying base composition and the terminal heterogeneity associated with in vitro transcription, the yield of ligation for circularization may vary for each gene. To achieve higher circRNA purity, RNase R digestion followed by gel purification of RNA may be useful. Third, RNA transfection protocols, such as the RNA-plasmid co-transfection steps, need to be further optimized. Fourth, we and others35 suggested that when using luciferase reporter assays to determine sponge effectiveness, it may be preferable to design two to three different oligonucleotide single strands containing different base compositions and to then choose the strand exhibiting the highest effectiveness. In future studies, we will confirm effects of scRNA21 on tumor growth, investigate mechanisms underlying scRNA21 activity, and evaluate scRNA21 toxicity in animal models. Notwithstanding these potential pitfalls, and considering the crucial role of oncomiRs in tumorigenesis and tumor progression, these results for the first time, to our knowledge, reveal enormous promise of synthetic circRNA sponges as novel tools for anticancer research and future molecular therapy.
Materials and Methods
Differential miR-21 Expression in TCGA Patients
446 GC and 45 normal samples with miR sequence data were retrieved from the TCGA data portal. After the data were combined and data without expression were filtered out, differential miR expression analysis was carried out using the Bioconductor package EdgeR, based on R Project version 3.5.1.36 Read counts were normalized, and miRs with fold changes >1.5 and p < 0.05 were selected.
Cell Lines
The human GC cell lines NCI-N87, KATOIII, AGS, and MKN28 were purchased from the American Type Culture Collection (Manassas, VA, USA). The human immortalized normal gastric epithelial cell line HFE145 was obtained from Dr. Hassan Ashktorab (Department of Medicine and Cancer Center, Howard University, Washington, DC, USA) and Dr. Duane Smoot (Department of Medicine, Meharry Medical College, Nashville, TN, USA).37 All five cell lines were cultured in DMEM (GIBCO, Grand Island, NY, USA) supplemented with 1% penicillin-streptomycin (GIBCO) and 10% FBS (GIBCO).
RNA Extraction
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA).
qRT-PCR Analysis of miR-21 Expression Assays
TaqMan miR assays, human (Applied Biosystems, Foster City, CA, USA) were used to assess miR-21 expression. Threshold cycle (Ct) was normalized to RNU6B expression. Fold change in miR-21 level was calculated using the 2−ΔΔCt method. All qRT-PCR assays were performed in duplicate.
Circular Sponge Design
To engender resistance to Argonaute 2-mediated endonucleolytic cleavage, we designed each miR-21 binding site as perfectly complementary to the miR-21 seed region, but with a bulge (mismatch) at positions 9–12 caused by deleting one nucleotide.12 We incorporated a bulge because linear bulged miR sponges are more effective than perfectly complementary sponges in competing with regulation of natural miR targets.12, 26, 38 The sequence of each miR-21 binding site was 5′-TCAACATCAGGACATAAGCTA-3′. A short 4-nt sequence (“spacer”) was inserted to separate the two miR-21 binding sites.
To produce linear RNA molecules, two sense and antisense DNA oligonucleotides, each containing three or two miR-21 bulged binding sites, respectively, separated by a “spacer,” were first ordered (IDT, Coralville, IA, USA). These oligonucleotides each contained a 19- to 21-nt region at their respective 3′ ends that allowed them to hybridize to one another after heat denaturation. The sequences of the two oligonucleotides were 5′-GATTCCTTGGTCAACATCAGGACATAAGCTACCAATCAACATCAGGACATAAGCTACCAATCAACATCAGGACATAAGCTAAATGCTGAT-3′ (sense) and 5′-TAGCTTATGTCCTGATGTTGACCAATAGCTTATGTCCTGATGTTGACCAAATCAGCATTTAGCTTATGTCC-3′ (antisense).
As a control, a sponge containing a scrambled seed sequence was also designed and constructed. The sequence of each miR-21 scrambled site was 5′-ACTTAGTGTCTATGCGTATGT-3′. This sequence was predicted by BLAST to not bind to any known human miR (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of the two scrambled oligonucleotides were 5′-ACTTAGTGTCTATGCGTATGTCCAAACTTAGTGTCTATGCGTATGTCCAAATCAGCATTACTTAGTGTC-3′ (sense) and 5′-GATTCCTTGGACATACGCATAGACACTAAGTCCAAACATACGCATAGACACTAAGTCCAAACATACGCATAGACACTAAGTAATGCTGAT-3′ (antisense).
A specific synthetic circRNA directed against miR-93 was also designed and constructed as another experimental control. The sequences of the two oligonucleotides were 5′-TTCCTTGGCTACCTGCACGTGTGCACTTTGCCAACTACCTGCACGTGTGCACTTTGCCTTCTACCTGCACGTGTGCACTTTGAATGCTGA-3′ (sense) and 5′-CAAAGTGCACACGTGCAGGTAGCCAACAAAGTGCACACGTGCAGGTAGCCAATCAGCATTCAAAGTGCACACG-3′ (antisense).
Circular Sponge Synthesis
To synthesize the scRNA21, we generated a double-stranded DNA PCR product by one cycle of overlap PCR.39 Overlapping, complementary 3′ ends allowing the two oligomers to anneal to one another after heating were used as template DNA for PCR to create the full-length product. The product was then cloned into the pCR 4-TOPO TA vector (Thermo Fisher Scientific, Carlsbad, CA, USA) and transformed into Escherichia coli (Thermo Fisher Scientific). Plasmid DNA was extracted using QIAprep Spin Miniprep Kits (QIAGEN, Valencia, CA, USA). Plasmid DNA was subsequently digested with BamHI (New England Biolabs, Ipswich, MA, USA) and purified by QIAprep Spin Miniprep Kit (QIAGEN). Large quantities of 5′-monophosphorylated linear RNA were synthesized by T7 RNA polymerase using HiScribe T7 High Yield RNA Synthesis Kits (New England Biolabs): approximately 150 μg of RNA could be transcribed from 1 μg of template DNA. After purification by RNeasy MinElute Cleanup Kits (QIAGEN), the linear RNA was treated with calf intestinal alkaline phosphatase (New England Biolabs) and then by T4 polynucleotide kinase (New England Biolabs) in the presence of ATP, to provide a phosphorylated 5′ terminus (required for ligation). Finally, the linear RNA was circularized using T4 RNA ligase 1 (New England Biolabs). A scrambled control circRNA (negative control scrambled circRNA) (described above) and a specific synthetic circRNA directed against miR-93 were also synthesized using the same methods.
RT-PCR and Sanger Sequencing of Synthetic circRNA Sponges
After reverse transcription, divergent primers, 5′-AGCACACTGGCGGCCGTTAC-3′ (sense) and 5′-ATATCCATCACACTGGCGG-3′ (antisense), were used to amplify a DNA PCR fragment containing the head-to-tail circRNA junction point. Convergent primers, 5′-AATTGGGCCCTCTAGATGCA-3′ (sense) and 5′-ACTAGTAACGGCCGCCAGTG-3′ (antisense), were used to amplify a DNA PCR fragment containing the five repeat miR binding sites. PCR products were then cloned and subsequently sequenced.
Stability Assays of Synthetic circRNA Sponges
Increasing Concentrations of FBS Digestion
Because several types of RNA exonucleases and endonucleases are enriched in serum,40 and because a future goal is to administer circRNAs intravenously, we compared the stability of scRNA21 and its sequence-identical LRNA21 by incubating 0.5 μg of each RNA with increasing concentrations of FBS (from 0% to 9%) at 37°C for 30 min.
RNase R Digestion
To evaluate circRNA resistance to a specific ribonuclease, we incubated scRNA21 and LRNA21 with or without RNase R (Epicenter Biotechnologies) at 37°C for 10 min. Ratios after incubation with either 1 U RNase R/1 μg RNA or 3 U RNase R/1 μg RNA were then calculated. Finally, RNAs were analyzed by 2.0% TAE-agarose gel electrophoresis; band intensities were determined using ImageJ software, and relative ratios were calculated.
Luciferase Reporter Assays
A luciferase reporter vector containing the miR-21 binding site was constructed by inserting annealed miR-21 binding site (IDT) into the pmirGLO vector (Promega, Madison, WI, USA), using PmeI and XbaI restriction enzymes (New England Biolabs). At 70%–80% confluence, NCI-N87, AGS, and MKN28, which express high endogenous levels of miR-21 based on our qRT-PCR results (described in Results), were co-transfected using Lipofectamine 2000 (Invitrogen) with 15 nM of either negative control scrambled circRNA or miR-21 inhibitor (MH10206; Thermo Fisher Scientific) or scRNA21, together with 80 ng of either luciferase reporter plasmid containing the miR-21 binding site, or empty pmirGLO vector per well of a 96-well plate. 24 hr after transfection, cells were analyzed for luciferase activity using a Dual-Glo Luciferase Assay Kit (Promega) and VICTOR2 fluorometry (Perkin Elmer, Waltham, MA, USA). Firefly luciferase activity was normalized to that of Renilla luciferase. All transfections were performed in triplicate, whereas luciferase activity was averaged from duplicate experiments.
Transfection of Synthetic circRNAs and Cell Proliferation Assays
NCI-N87, AGS, and MKN28 were transfected using Lipofectamine RNAiMAX (Invitrogen) with 15 nM of either negative control scrambled circRNA or miR-21 inhibitor or scRNA21 per well of a six-well plate. Cells were re-seeded onto 96-well plates at a density of 1,000 cells/well at day 0 (24 hr after RNA transfection). Cell proliferation was assessed at days 0, 1, 3, and 5 using the cell proliferation reagent WST-1 (Roche, Mannheim, Germany).
Immunofluorescence Staining
NCI-N87, AGS, and MKN28 were transfected using Lipofectamine RNAiMAX with 15 nM of either negative control scrambled circRNA or scRNA21. 24 hr after transfection, cells were fixed in acetone. Primary antibodies were rabbit anti-cleaved caspase-3 (1:400; #9661; Cell Signaling Technology), and secondary antibodies were Alexa Fluor 488 goat anti-rabbit IgG (1:500; A-11034; Invitrogen). Nuclei were counterstained with Hoechst 33342 (Thermo Fisher Scientific).
TMT Labeling and Proteomics Analyses
NCI-N87 cells were transfected using Lipofectamine RNAiMAX (Invitrogen) in six-well plates with 15 nM per well of scRNA21, negative control scrambled circRNA, or PBS blank control; a specific synthetic circRNA directed against miR-93 was also assayed as an experimental control. Because we reasoned that some downstream proteomic effects of scRNAs would occur shortly after treatment, whereas others might take longer to become apparent, total protein was extracted from treated and control cells at three different treatment time intervals: at 1, 8, and 24 hr after transfection.
TMT labeling, liquid chromatography-tandem mass spectrometry, and proteomics analyses were performed according to the manufacturer’s instructions. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD008584.
Bioinformatics Analyses
Proteomic data derived from a 10-plex TMT labeling Mass-Spec analysis using the Proteome Discoverer platform (Thermo Fisher Scientific, Waltham, MA, USA) were imported into Partek Genomics Suite v6.6 (Partek, St. Louis, MO, USA) for protein annotation and further analysis. One or more mass spectra were assigned to each NCBI gi identifier, which represented a single protein, and for each protein the median of these multiple spectra values was calculated to produce a single value representing that protein. These protein identifiers were mapped to their cognate NCBI Entrez genes to facilitate downstream analyses. Only those spectra with a Proteome Discoverer Isolation Interference value <30% were accepted for further evaluation. The values of these human proteins were converted to log2 notation for statistical analysis and quantile-normalized to minimize possible noise and experimental variation across the 10 experimental samples. For quality control (QC), Partek Genomics Studio was used to create boxplots and principal components analysis (PCA) plots to detect possible outliers. Different pairs of biological samples’ values were compared, yielding linear and log2 fold change values for each protein and its associated gene counterpart.
To examine differential protein expression further, these results were imported into Spotfire DecisionSite with Functional Genomics v9.1.2 (TIBCO Spotfire, Boston, MA, USA). To characterize the proteins’ fold changes, their log2 values underwent SD analyses, allowing us to set appropriate fold change thresholds of differential expression.
Western Blotting
Total protein was extracted from NCI-N87 and AGS, after transfection with either PBS blank control, negative control scrambled circRNA, or scRNA21, at four different treatment time intervals: 1 hr, 8 hr, 24 hr, and 5 days. Mouse anti-Daxx (H-7; Santa Cruz Biotechnology) and anti-β-actin peroxidase (AC-15; Sigma-Aldrich), horseradish-peroxidase-conjugated goat anti-mouse IgG (Invitrogen), and ECL Western Blotting detection kits (Amersham-Pharmacia Biotech) were used.
Statistical Analyses
Means and SD were displayed as representative values for data in the figures. ANOVA with post hoc test of means and Student’s unpaired t test were used to assess statistical significance, which was done with SPSS (version 14.0; SPSS, Chicago, IL, USA), unless otherwise noted. p < 0.05 was regarded as significant.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD008584.
Author Contributions
Conception and Design: S.J.M., X.L., J.M.A., and Z.L. Circular RNA Sponge Synthesis and Functional Verification: X.L., Y.C., and J.M.A. RNA Extraction: Zhixiong Wang and Zhe Wang. Technical Support: Y.C., J.M.A., G.Z., D.T.S., H.A., and S.J.M. Tandem Mass Tag (TMT) Labeling and Proteomics Analysis: R.N.C., T.N.B., and L.D.V. Bioinformatics Analysis and Interpretation of Data: C.J.T. Writing and Revision of Manuscript: S.J.M. and X.L. Study Supervision: S.J.M.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
S.J.M. was supported by NIH grants CA190040, CA211457, and DK118250 and the Emerson Research Foundation; X.L. is supported by the Key Science and Technology Program of Shaanxi Province, China grant 2015SF128; G.Z. is supported by the International Scientific and Technological Cooperation and Exchange Program of Shaanxi Province, China grant 2015KW-030; and R.N.C. and T.N.B. were supported by Hopkins Digestive Diseases Basic and Translational Research Core Center (NIDDK center) grant P30 DK089502. S.J.M. is the Harry and Betty Myerberg Professor of Gastroenterology and American Cancer Society Clinical Research Professor.
Footnotes
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.010.
Supplemental Information
References
- 1.Fitzmaurice C., Dicker D., Pain A., Hamavid H., Moradi-Lakeh M., MacIntyre M.F., Allen C., Hansen G., Woodbrook R., Wolfe C., Global Burden of Disease Cancer Collaboration The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katona B.W., Rustgi A.K. Gastric cancer genomics: advances and future directions. Cell. Mol. Gastroenterol. Hepatol. 2017;3:211–217. doi: 10.1016/j.jcmgh.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura A., Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farazi T.A., Spitzer J.I., Morozov P., Tuschl T. miRNAs in human cancer. J. Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selaru F.M., Olaru A.V., Kan T., David S., Cheng Y., Mori Y., Yang J., Paun B., Jin Z., Agarwal R. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49:1595–1601. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X., Meltzer S.J. Gastric cancer in the era of precision medicine. Cell. Mol. Gastroenterol. Hepatol. 2017;3:348–358. doi: 10.1016/j.jcmgh.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueda T., Volinia S., Okumura H., Shimizu M., Taccioli C., Rossi S., Alder H., Liu C.G., Oue N., Yasui W. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Zhang Y., Zhang Y., Ding J., Wu K., Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Li Z., Gao C., Chen P., Chen J., Liu W., Xiao S., Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 11.Motoyama K., Inoue H., Mimori K., Tanaka F., Kojima K., Uetake H., Sugihara K., Mori M. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int. J. Oncol. 2010;36:1089–1095. doi: 10.3892/ijo_00000590. [DOI] [PubMed] [Google Scholar]
- 12.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., Teruya-Feldstein J., Reinhardt F., Onder T.T., Valastyan S. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 16.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Chen L., Zhang S., Wu J., Cui J., Zhong L., Zeng L., Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 19.Petkovic S., Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan T., Sato F., Ito T., Matsumura N., David S., Cheng Y., Agarwal R., Paun B.C., Jin Z., Olaru A.V. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C.W., Wang L.K., Wang S.P., Chang Y.L., Wu Y.Y., Chen H.Y., Hsiao T.H., Lai W.Y., Lu H.H., Chang Y.H. Daxx inhibits hypoxia-induced lung cancer cell metastasis by suppressing the HIF-1α/HDAC1/Slug axis. Nat. Commun. 2016;7:13867. doi: 10.1038/ncomms13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papagiannakopoulos T., Shapiro A., Kosik K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 24.Li Q., Wang X., Wu X., Rui Y., Liu W., Wang J., Wang X., Liou Y.C., Ye Z., Lin S.C. Daxx cooperates with the Axin/HIPK2/p53 complex to induce cell death. Cancer Res. 2007;67:66–74. doi: 10.1158/0008-5472.CAN-06-1671. [DOI] [PubMed] [Google Scholar]
- 25.Ma X., Choudhury S.N., Hua X., Dai Z., Li Y. Interaction of the oncogenic miR-21 microRNA and the p53 tumor suppressor pathway. Carcinogenesis. 2013;34:1216–1223. doi: 10.1093/carcin/bgt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tay F.C., Lim J.K., Zhu H., Hin L.C., Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug Deliv. Rev. 2015;81:117–127. doi: 10.1016/j.addr.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Song J.H., Meltzer S.J. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35–47.e2. doi: 10.1053/j.gastro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Lin C.W., Chang Y.L., Chang Y.C., Lin J.C., Chen C.C., Pan S.H., Wu C.T., Chen H.Y., Yang S.C., Hong T.M., Yang P.C. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat. Commun. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 29.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiao K.Y., Lin Y.C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W., Du W.W., Li X., Yee A.J., Yang B.B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Pan X., Wang Z.X., Wang R. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol. Ther. 2010;10:1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 35.Kluiver J., Slezak-Prochazka I., Smigielska-Czepiel K., Halsema N., Kroesen B.J., van den Berg A. Generation of miRNA sponge constructs. Methods. 2012;58:113–117. doi: 10.1016/j.ymeth.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smoot D.T., Allen C.R., Barnes P., Brown M., Phadnis S., Gold B., Ashktorab H. Human gastric epithelial cell lines derived from primary cultures of normal gastric epithelial cells. Gastroenterology. 2000;118:A540–A541. [Google Scholar]
- 38.Gentner B., Schira G., Giustacchini A., Amendola M., Brown B.D., Ponzoni M., Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 39.Heckman K.L., Pease L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 40.Choung S., Kim Y.J., Kim S., Park H.O., Choi Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PRIDE: PXD008584.