Abstract
Aberrant activation of the receptor tyrosine kinase-mediated RAS signaling cascade is the primary driver of embryonal rhabdomyosarcoma (ERMS), a pediatric cancer characterized by a block in myogenic differentiation. To investigate the cellular function of activated RAS signaling in regulating the growth and differentiation of ERMS cells, we genetically ablated activated RAS oncogenes with high-efficiency genome-editing technology. Knockout of NRAS in CRISPR-inducible ERMS xenograft models resulted in near-complete tumor regression through a combination of cell death and myogenic differentiation. Utilizing this strategy for therapeutic RAS targeting in ERMS, we developed a recombinant oncolytic myxoma virus (MYXV) engineered with CRISPR/Cas9 gene-editing capability. Treatment of pre-clinical human ERMS tumor xenografts with an NRAS-targeting version of this MYXV significantly reduced tumor growth and increased overall survival. Our data suggest that targeted gene-editing cancer therapies have promising translational applications, especially with improvements to gene-targeting specificity and oncolytic vector technology.
Keywords: CRISPR, myxoma, rhabdomyosarcoma, oncolytic virus, cancer gene therapy, RAS
Introduction
Embryonal rhabdomyosarcoma (ERMS) is a devastating pediatric sarcoma characterized by a pathologic block in myogenic differentiation. The initiation and progression of ERMS are primarily driven by RAS-associated signaling pathways.1 Whether RAS signaling is involved in repressing myogenic differentiation in ERMS is currently unknown. There are also no effective therapies capable of directly targeting activated RAS proteins. In fact, many common cancer oncogenes are undruggable by modern therapeutic approaches.2, 3 Developing therapies to target these oncogenic drivers remains one of the primary goals of current cancer research. However, creation of new targeted cancer therapies is a time-consuming process, requiring substantial research and development of resources. Directly targeting the genomic DNA of essential cancer genes could overcome many of the challenges associated with developing precision cancer treatments. Advances in genome-editing technology now make it possible to target oncogenes with remarkable precision, paving the way for a new generation of gene-editing cancer therapies. Because genome-editing technology relies only on knowledge of cancer genetics, it can be rapidly customized to the vulnerabilities present in individual tumors. Development of a programmable cancer therapy of this kind could profoundly impact cancer treatment by providing the flexibility to rapidly target a wide range of essential cancer genes in a variety of cancer types.
Although a growing number of studies have provided initial evidence that gene-editing technology could be used to target essential cancer genes,4, 5, 6, 7 several major obstacles remain before a cancer gene-editing therapy could be used in the clinic. The first is the development of high-efficiency gene-editing approaches that can target essential cancer genes with therapeutic efficiency and specificity. Cancer gene-editing therapies will also require the development of an effective gene therapy delivery system to facilitate treatment of cancer patients.
Our study used high-efficiency gene-targeting approaches to investigate the dependency of ERMS cells on activated RAS signaling, identifying RAS as a dominant repressor of myogenic differentiation in ERMS. Using this targeting strategy, we developed a novel oncolytic myxoma viral (MYXV) gene-editing vector system to directly target activated RAS genes in ERMS tumors. This study evaluates the therapeutic potential of this recombinant gene-editing oncolytic virus, providing important insights into the emerging field of gene-editing cancer therapy and establishing a framework for further development of this innovative technology.
Results
Targeting Activated RAS Inhibits Tumor Growth and Induces Myogenic Differentiation of ERMS Cells
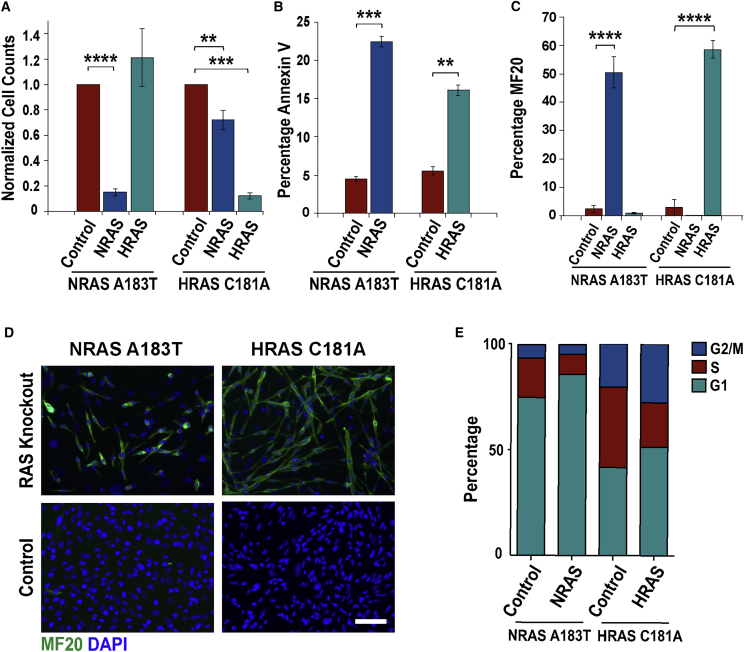
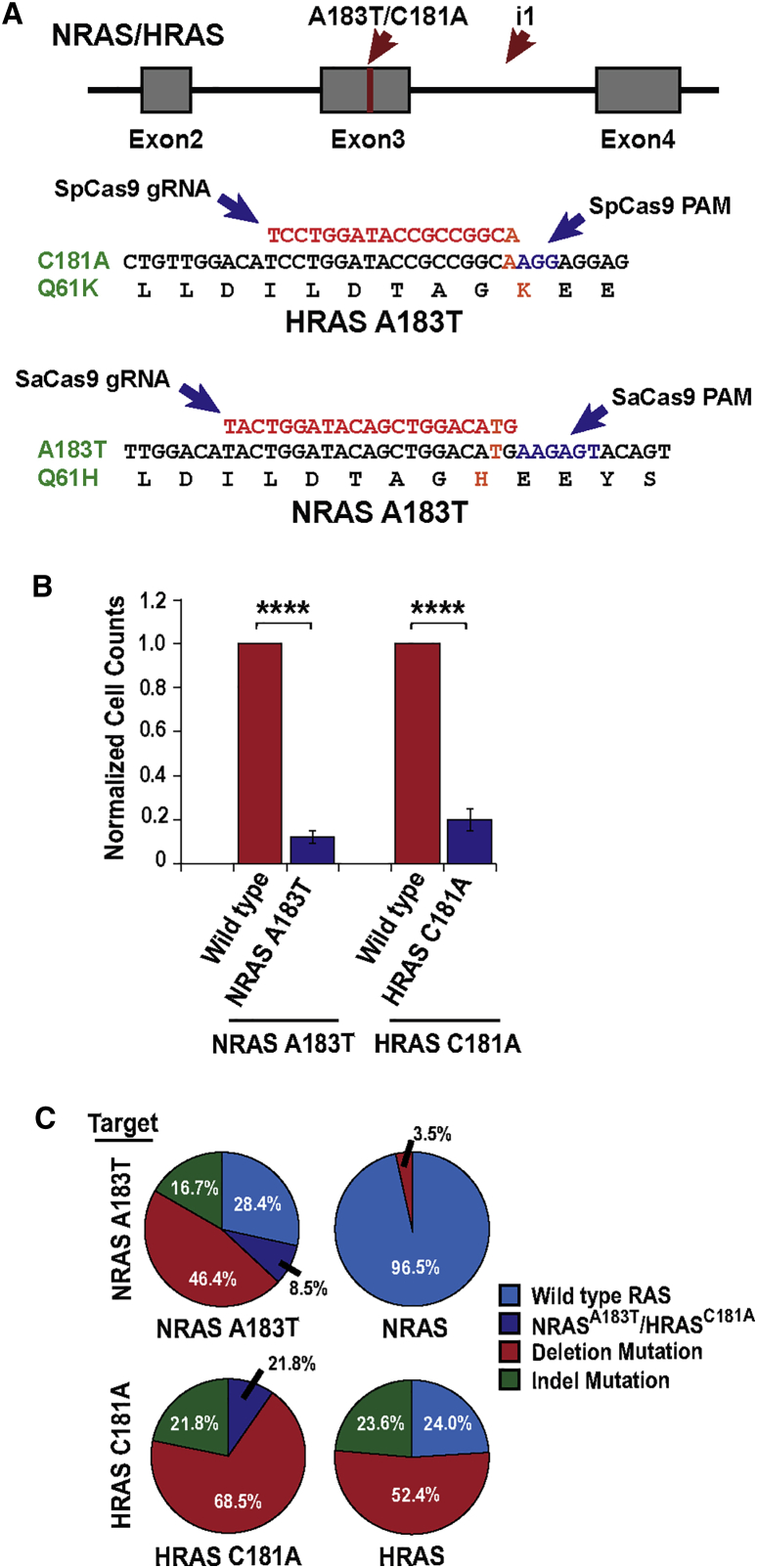
RAS-associated signaling pathways are responsible for ERMS initiation and progression. However, the cellular mechanisms by which RAS signaling promotes ERMS tumor growth are unknown. Therefore, we sought to disrupt RAS signaling in RAS mutant ERMS cells using CRISPR/Cas9 gene-editing approaches. Because RAS signaling was essential for the proliferation of ERMS cells, gene knockout required high-efficiency gene-editing approaches that can effectively target the oncogene in all cancer cells. Despite several improvements to CRISPR/Cas9 single guide RNA (sgRNA) design,8 the insertion and deletion (indel) mutation efficiency of a sgRNA is not sufficient for most cancer therapeutic applications. We previously observed that simultaneous targeting of genes with multiple gRNAs resulted in high-efficiency deletions between sgRNAs that were up to two to three times the average indel efficiency for sgRNAs (i.e., 40%–60%).7 We therefore adapted a similar high-efficiency gene-editing strategy to examine the phenotypic response of ERMS cells to ablation of RAS genes. To target RAS in ERMS, RAS mutant cancer cells were co-transduced with high-titer lentiviruses expressing Streptococcus pyogenes Cas9 (SpCas9) and dual gRNAs targeting exon 3 of the NRAS or HRAS genes (NRAS A183T mutant 381T and HRAS C181A mutant SMS-CTR ERMS cells, respectively). NRAS and HRAS knockout in RAS mutant ERMS cells resulted in a significant reduction in viability 5 days post-gene targeting (Figure 1A). We also observed a small but significant reduction in cell viability with NRAS targeting in HRAS-driven ERMS cells (Figure 1A), suggesting a potential role for wild-type RAS signaling in modulating growth of some ERMS cells. Flow cytometry-based annexin V assay for apoptosis revealed that the reduction in ERMS cell growth after RAS targeting was due to a modest but significant increase in cell death (Figure 1B). The vast majority of NRAS- and HRAS-targeted cells underwent terminal myogenic differentiation, as shown by a significant increase in myosin heavy chain positive-multinucleated myotubes (Figures 1C and 1D). Cell-cycle analysis of ERMS cells with NRAS A183T and HRAS C181A knockout revealed a decrease in the percentage of S phase cells compared with controls because of an increase in the percentage of cells arrested in the G1 phase of the cell cycle (NRAS: S, p = 0.005; G1, p = 0.002; HRAS: S, p < 0.0001; G1, p = 0.0005; Figure 1E).
Figure 1.
High-Efficiency Knockout of RAS Induces Differentiation of ERMS Cells
(A) RAS targeting reduces viability in ERMS cells harboring NRAS A183T or HRAS C181A activating mutations. Annexin V apoptosis (B) and MF20 (C), myosin heavy chain immunostaining analysis, after NRAS or HRAS knockout. (D) Fluorescent images showing myogenic differentiation of ERMS cells after RAS targeting (MF20, green; nuclei, blue; scale bar: 100 μm). (E) Changes in cell cycle in RAS knockout (KO) ERMS cells (n = 3). Cell viability (A) and MF20 (C) data were analyzed 6 days post-RAS targeting, and cell death (B) and cell-cycle analysis (E) were taken 4 days post-targeting, prior to myogenic differentiation. Data in (A)–(C) represent the average of four biological replicates with error bars ± SEM. **p < 0.01; ***p < 0.001; ****p < 0.0001.
To further assess the response of ERMS cells to the loss of RAS signaling, we engineered tamoxifen-inducible NRAS-targeting ERMS cells, using a system previously developed in our laboratory.7 We first compared the global gene expression profile of the tamoxifen-induced NRAS knockout ERMS cells with non-induced ERMS control cells prior to terminal myogenic differentiation (day 2 after NRAS knockout) to gain insight into genes and pathways regulated by NRAS. Disruption of NRAS resulted in a decrease in the expression of key cell-cycle and DNA replication genes and an increase in the expression of myogenic differentiation genes (Figure S1). Taken together, our loss-of-function studies and gene expression analysis demonstrate that RAS signaling is required for sustaining ERMS cell proliferation, in part, through its repression of the myogenic program.
NRAS Knockout Regresses ERMS Tumor Xenografts In Vivo
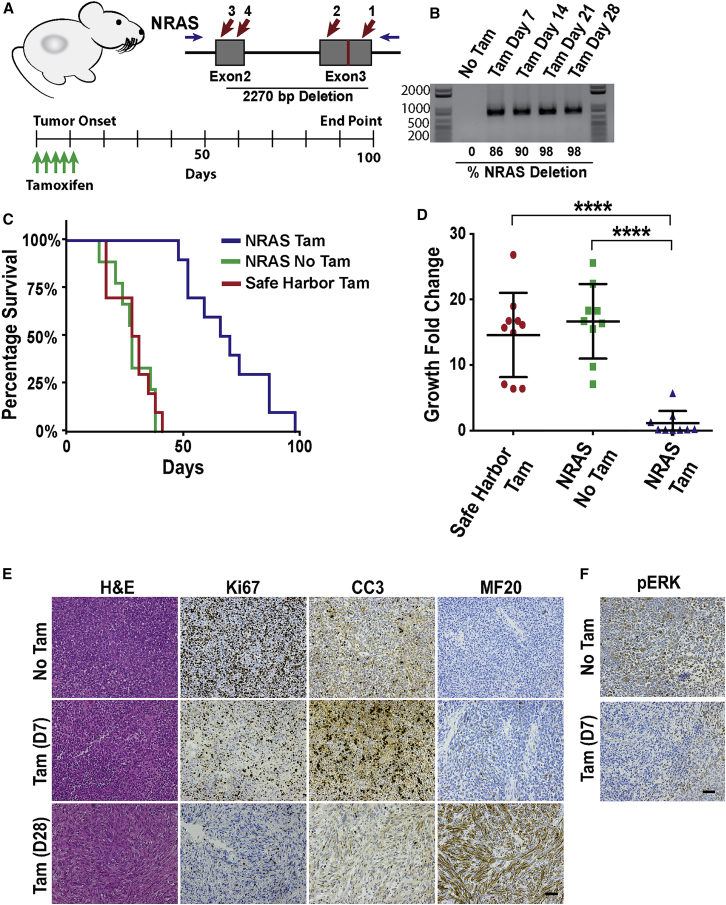
To examine the response of ERMS tumors to loss of RAS signaling in vivo, we established tamoxifen-inducible NRAS-targeting tumor xenografts in immunocompromised NOD-SCID Il2rg−/− (NSG) mice (Figure 2A). The growth of NRAS-targeted xenografts was compared with non-induced or control tumors targeted at a safe harbor region of chromosome 4 (58110237–58110808; GRCH38.p2).7 Treatment of tumor xenografts with tamoxifen resulted in extensive gene editing with complete NRAS knockout within a week of treatment (Figure 2B). The median survival of mice with NRAS-targeted tumors increased by approximately 2-fold (i.e., 38.5 days) over non-targeted or safe harbor control-targeted tumors (Figure 2C). NRAS knockout resulted in the regression of ERMS tumors, most of which became undetectable around 10 days after tamoxifen treatment (Figure 2D). There was a significant difference in tumor volume change, comparing NRAS-targeted tumor xenografts with non-treated and tamoxifen-treated control tumors (p < 0.0001; Figure 2D).
Figure 2.
Targeting NRAS in ERMS Xenografts Inhibits Tumor Growth and Induces Myogenic Differentiation
(A) Design of tamoxifen-inducible NRAS knockout experiment with NRAS gene-targeting strategy showing gRNA target sites (red arrows) and the activating A183T mutation site (red vertical bar). (B) Inducible gene editing in tumor xenografts as shown by the presence of deletion mutations in tamoxifen-treated tumors after PCR amplifying the deletion mutations with primers flanking the gRNA target sites. Efficiency of editing was estimated by combining deletion efficiencies between gRNA target sites. (C) Mouse survival after tamoxifen (Tam) induced knockout of NRAS in tumor xenografts (n = 10). (D) Analysis of changes in tumor volume over 21 days with mean ± SD of each treatment group. (E) Histological (H&E) and immunohistochemistry (IHC) analysis of NRAS knockout tumors prior to tamoxifen treatment and at 7 and 28 days after treatment. IHC analysis was used to determine the extent of cellular proliferation (Ki67), apoptosis (CC3), and myogenic differentiation (myosin heavy chain, MF20) of tumor samples. (F) Phosphorylated ERK (pERK) immunostaining in tumor xenografts 7 days after tamoxifen treatment. Scale bars represent 100 μm. ****p < 0.0001.
The cellular response of ERMS tumors to NRAS targeting was monitored every 7 days by analyzing changes in cell morphology (H&E), cellular proliferation (Ki67), cell death (cleaved caspase 3 [CC3]), and myogenic differentiation (MF20). NRAS knockout resulted in an initial decrease in cellular proliferation and increase in cell death (day 7), followed by myogenic differentiation throughout most of the tumor mass (day 28; Figure 2E). This was associated with a decrease in phosphorylated extracellular signal-regulated kinase, aka MAPK (pERK), signaling following NRAS targeting in ERMS tumors (Figure 2F). These findings indicate that RAS is essential for sustaining tumor growth, as well as repressing myogenic differentiation in ERMS.
Relapse of NRAS-Targeted Tumors
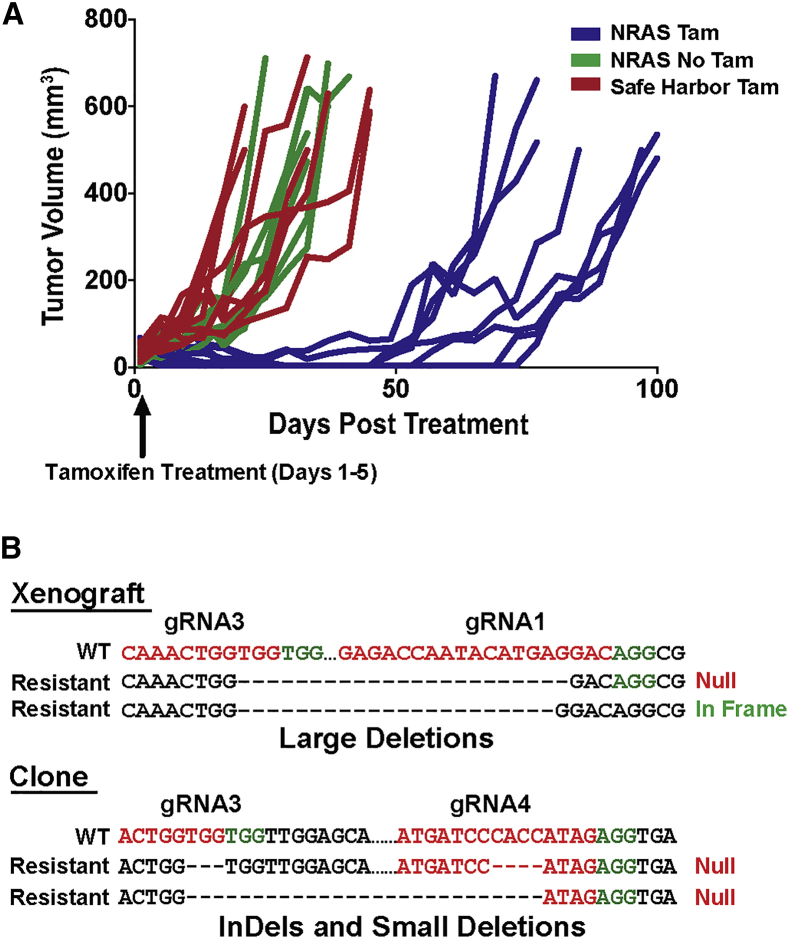
Despite effective NRAS knockout in ERMS tumor xenografts (Figure 2B), relapsed tumor growth eventually occurred in all mice (Figure 3A). The primary genotype found in NRAS-resistant tumors was a large gDNA deletion (2,270 bp) between the outermost gRNA target sites (gRNA3 and gRNA1; Figure 2A), resulting in the loss of 67 amino acids (aa) (35% of the protein coding sequence). Consistent with the high fidelity of non-homologous end joining (NHEJ) DNA repair, the majority of deletion mutations were direct fusions between the gRNA cut sites, resulting in frameshift mutations (Figure 3B). In-frame deletion mutations were also identified (Figure 3B), but the loss of the NRAS core region, including the activating mutation site, likely resulted in a non-functional protein. There was a very small percentage of non-targeted xenograft cells with compromised gene editing, but these cells did not represent the majority of the relapsed tumor mass. Indel mutations and small deletions that resulted in truncating frameshift mutations were also identified (Figure 3B). The findings suggest that relapsed tumor growth most likely occurred secondary to the resistance of tumor cells to NRAS gene targeting rather than the outgrowth of non-targeted cells.
Figure 3.
Relapse of ERMS Tumors following NRAS Targeting
(A) Change in tumor volume over time in NRAS-targeted tumors compared with controls. (B) Example of common NRAS mutations found in relapsed tumor xenografts and an isolated NRAS-targeting-resistant clone.
Development of Recombinant Gene-Editing Myxoma Virus for RAS-Targeted Oncolytic Viral Therapy
The pre-clinical RAS knockout studies highlighted the potential for exploiting the myogenic commitment of ERMS cancer cells for differentiation therapy. Because there are currently no viable RAS-targeted therapies, we sought to test the effectiveness of high-efficiency CRISPR/Cas9 technology for therapeutic targeting of the NRAS oncogene in ERMS. We recognized that delivery would be a major obstacle for any cancer gene-editing therapy. Our initial attempts to use traditional non-replicating lentiviral and adeno-associated viral gene therapy vectors failed because of low transduction efficiency in ERMS tumors. We therefore chose to investigate the replicating oncolytic MYXV from the Leporipoxvirus genus9, 10 as a potential delivery vector for the NRAS gene-editing cancer therapy. MYXV causes myxomatosis in European rabbits but is completely non-pathogenic to humans.9, 10 However, the virus is capable of replicating selectively in many cancer cells,11 including ERMS cells,12 and is currently being developed for a variety of clinical applications. The large genome size (161.8 kb) of MYXV is also capable of packaging all CRISPR/Cas9 gene-editing components into a single viral genome,10 making MYXV an ideal candidate for a therapeutic CRISPR/Cas9 gene-editing vector.
We confirmed the replication efficiency of recombinant MYXV in ERMS cells using a reporter virus that expressed both a constitutive vaccinia virus synthetic early and late promoter (vvSEL)-driven GFP cassette along with a replication-dependent late promoter (p11) expressing RFP12 (Figure S2A). Incubation of ERMS cells with low-titer MYXV resulted in effective viral infection and replication in vitro as shown by the expression of both GFP and RFP in infected cells (Figure S2A). Replication-competent MYXV virus could also be serially passaged in ERMS cells through the transfer of conditioned media or cellular lysate (Figure S2A). Recombinant MYXV exhibited active viral replication (expression of GFP and RFP) in ERMS tumors xenografts when administered by direct intra-tumoral injection (Figure S2B). Despite efficient viral replication, the spread of the recombinant myxoma viral vector was restricted to regional areas within the tumor mass (Figure S2B). This pattern of viral infection is likely reflective of the plaque-forming nature of MYXV infection, as well as the tight cellular junctions present in ERMS tumors. Although the myxoma virus (MYXV) was not able to infect all ERMS cells, we hypothesized that ablation of essential oncogenes could promote viral spread by facilitating the release of viral particles to surrounding cells. Similarly, in therapeutic applications, stimulated immune responses may facilitate the killing of RAS knockout cancer cells in the absence of direct infection. Unfortunately, there are currently no immune-competent, RAS-driven mouse models of ERMS. Therefore, we employed ERMS tumor xenografts to test the ability of engineered recombinant gene-editing MYXV to regress ERMS tumors independent of an active immune system.
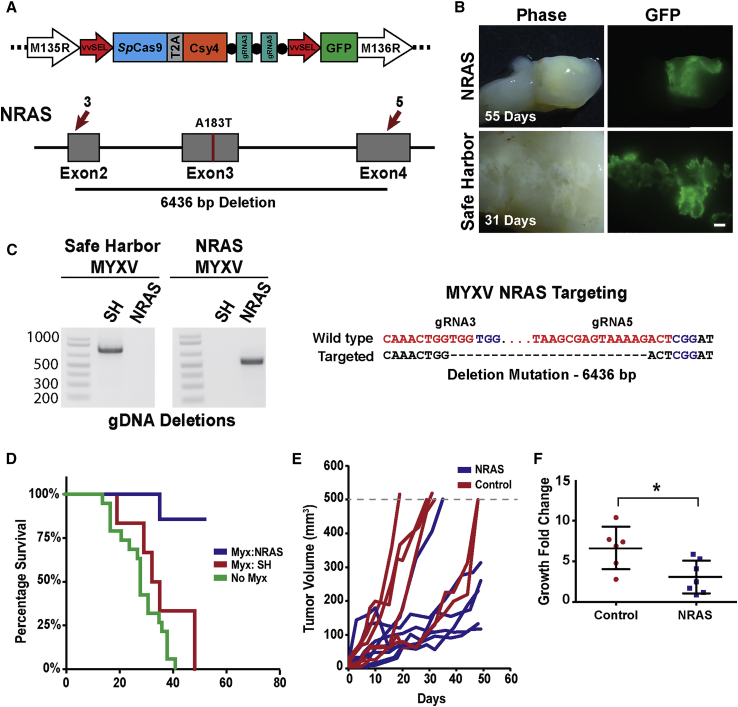
To target the NRAS gene in ERMS cells, we engineered a recombinant gene-editing MYXV containing an additional open reading frame expressing the SpCas9 CRISPR components from a single viral transcript (Figure 4A). This approach facilitated the use of high-expressing viral promoters (i.e., vvSEL) to ensure efficient gene editing when transcribed from the cytoplasmic MYXV. Our MYXV gene-editing system consisted of an SpCas9-2A-Csy4 cassette for simultaneous expression of SpCas9 and the CRISPR ribonuclease Csy4 (Figure 4A). The Csy4 ribonuclease is capable of cleaving CRISPR gRNAs at precise recognition sequences to split gRNAs from the 3′ end of mRNA transcripts.13 To facilitate targeted gene editing, we inserted dual gRNAs targeting the genes of interest into the 3′ end of the SpCas9-2A-Csy4 open reading frame separated by Csy4 cleavage sequences (Figure 4A). Human NRAS-driven ERMS tumor xenografts established in immunocompromised NSG mice were then injected intratumorally with four doses of the recombinant gene-editing MYXV (1 × 107 plaque-forming units [PFU]/100 μL) targeting the NRAS oncogene or a safe harbor locus in chromosome 4 (Figure 4B). Both gene-editing MYXVs effectively transduced ERMS tumor xenografts with no obvious visual difference in the percentage of infected cells between control and NRAS-targeted tumors (Figure 4B). This suggested that at least for ERMS tumors, NRAS targeting alone did not significantly influence the spread of MYXV viral vectors (Figure 4B). In vivo MYXV-mediated targeting of exons 2 and 4 of the human NRAS gene resulted in the removal of 52% of the core region of the NRAS protein (98 aa, xenograft genomic DNA deletion of 6,436 bp; Figure 4C). Although accurate quantification of gene-editing efficiency is not possible in tumor xenografts, due to incomplete viral infection, in vitro infection of ERMS cells with the gene-editing MYXV vectors resulted in over 70% deletion mutations at the target locus (Figure S2C), which is significantly higher than the editing efficiency observed with typical CRISPR lentiviral vectors.7 MYXV-mediated targeting of the NRAS oncogene in ERMS tumors significantly increased mouse survival compared with controls (p = 0.0037; Figure 4D) and resulted in a significant reduction in tumor growth in the majority of xenografts (Figures 4E and 4F). However, the effect of MYXV gene-editing therapy did not provide sustained therapeutic benefit because all tumors relapsed during treatment. This may be due to a number of factors, including incomplete viral infection and/or tumor cell resistance to NRAS targeting. Histological and immunohistochemical (IHC) analysis of MYXV-infected tumors revealed modest myogenic differentiation in NRAS-targeted tumors, with the majority of differentiated cells located in areas of low viral load outside the primary infection sites (Figure S3). This suggests that MYXV infection restricted the ability of ERMS cells to undergo myogenic differentiation.
Figure 4.
Myxoma Oncolytic Gene-Editing Viral Therapy Significantly Reduces ERMS Tumor Growth
(A) Schematic of the engineered gene-editing oncolytic MYXV and NRAS-targeting strategy with gRNA target sites (red arrows). (B) Phase and GFP fluorescent images showing recombinant MYXV-infected areas in ERMS tumors (scale bar: 1 mm). (C) High-efficiency gDNA deletions detected at the safe harbor (control virus) and NRAS target sites in representative MYXV-treated ERMS tumors. Sequencing of the most common NRAS deletion mutation is shown. (D) Mouse survival over time compared between NRAS-targeted (blue), safe-harbor-targeted (red), and wild-type uninfected ERMS tumors (n = 6, n = 6, and n = 20, respectively). Tumor growth over time (E) and change in tumor volume (3 weeks) (F) for NRAS- and safe-harbor-targeted tumors. Error bars represent mean ± SD. *p < 0.05.
Tumor-Specific RAS Targeting in ERMS
Although we demonstrate that gene-editing technology is capable of enhancing the capacity of MYXV vectors to kill cancer cells, our initial therapeutic strategy was not selective for cancer cells and could result in off-target editing of the NRAS gene in non-cancer cells. Future cancer gene-editing therapies would likely benefit from selective targeting of cancer cells, and therefore we examined the ability of CRISPR gene-editing technology to selectively target activating point mutations in RAS genes. Developing high-efficiency mutation-specific targeting strategies, however, poses significant challenges for many driver oncogenes because of restrictions on the placement of CRISPR gRNA target sites. Although several previous studies have suggested that tumor point mutations could be targeted with a sgRNA targeting the mutation site,14, 15 our previous experience with CRISPR/Cas9 gene editing suggests that the efficiency of NHEJ repair at a sgRNA site does not allow for the efficiencies necessary for therapeutic applications.
To develop a therapeutic gene-editing strategy capable of specifically targeting the activating mutations in RAS, we sought to identify CRISPR gRNA target sites that could specifically recognize the mutations present in ERMS. Unfortunately, the RAS mutations found in ERMS cells did not introduce new CRISPR protospacer adjacent motifs (PAMs) that could specify target sites unique to ERMS cells. We therefore looked for gRNA target sites that contained the activating point mutations in the seed region of the gRNAs immediately adjacent to the PAM recognition sequence (Figure 5A). Mutations in the first 7 bp adjacent to the PAM sequence, termed the seed region, are often not tolerated by the CRISPR system, facilitating the establishment of mutation-specific gRNAs.16 We have previously shown that mutations in this gRNA seed region blocks Cas9 activity in RMS cancer cells.7 Although an SpCas9 PAM recognition sequence was in close proximity to the HRAS C181A mutation site, no SpCas9 target sites were found near the activating NRAS A183T mutation site (Figure 5A). In contrast, the PAM recognition sequence for the CRISPR/Cas9 system isolated from Staphylococcus aureus (SaCas9) was located adjacent to the NRAS A183T mutation site in ERMS cells (Figure 5A). Therefore, we designed gRNA sequences using both the SpCas9 and SaCas9 systems to facilitate gene editing of the HRAS C181A and NRAS A183T mutation sites (Figure 5A). To enhance gene-editing efficiency, we placed a second gRNA target site in the third intron of the HRAS and NRAS genes, immediately adjacent to exon 3 containing the activating RAS mutations (i.e., gRNA i1; Figure 5A). This targeting strategy was designed to introduce harmless indel mutations in the third intron of the RAS gene in normal cells while creating non-functional indel mutations and high-efficiency deletions at the RAS mutation site in ERMS cells (Figure 5A).
Figure 5.
Tumor Mutation-Specific Targeting of NRAS and HRAS Oncogenes in ERMS
(A) RAS tumor mutation-specific targeting strategy with locations of the mutation site (red bar) and targeting gRNAs (red arrows). The placement of the mutation-specific gRNAs is identified. (B) Cell viability after treatment of ERMS cells with RAS mutation-specific lentivirus. Data represent mean ± SEM of three biological replicates. (C) Sequencing analysis of the NRAS and HRAS target site after treatment of NRAS A183T and HRAS C181A mutant and non-mutant cells with tumor-specific targeting virus. ****p < 0.0001.
To validate the specificity and assess off-target editing of these RAS gene-editing strategies, lentiviral CRISPR/Cas9 vectors targeting the activating point mutations in HRAS and NRAS were transduced into ERMS cells harboring either wild-type or mutant RAS. As expected, targeting of these RAS mutation sites led to a significant reduction in cell viability, similar to that observed with general targeting of NRAS and HRAS (Figure 5B). Next-generation sequencing analysis of the HRAS and NRAS target sites revealed high gene-editing efficiency from both the SpCas9 and SaCas9 systems, with 90.3% and 88.1% of the mutant alleles targeted, respectively (i.e., combination of deletion and indel mutations; Figure 5C). Both ERMS cell models used in our study exhibited chromosomal abnormalities at the RAS target sites, with NRAS A183T cells having at least two copies of the NRAS A183T allele1 and HRAS C181A mutant cells exhibiting loss of the wild-type HRAS allele.17 SaCas9 targeting was highly specific with no indel mutations detected at the NRAS A183T target site in non-mutant cells (Figure 5C). However, a small percentage of deletion mutations between the gRNA target sites were identified in wild-type cells (3.5%; Figure 5C). In contrast, the chosen HRAS C181A mutation-specific gRNA was highly promiscuous, targeting both mutant and wild-type alleles with high efficiency (wild-type allele targeting 76%; Figure 5C). The observed differences in off-target editing are likely not reflective of the fidelity of each Cas9 variant but instead signify the specificity inherent in individual CRISPR gRNA target sites.16 These results highlight some limitations in selectively targeting oncogenic point mutations because off-target editing could potentially result in the introduction of new oncogenic mutations in normal cells.
Discussion
This study establishes a platform for the development of future gene-editing cancer therapies by identifying core RAS vulnerabilities in ERMS cancer cells along with high-efficiency oncogene-targeting strategies. The results provide a unique window into the role of RAS oncogenes in promoting ERMS tumor progression by repressing myogenic differentiation. Our data suggest that ERMS cancer cells are pre-programmed myogenic cells that may respond well to targeted differentiation therapies. Even though the impaired differentiation status of RMS tumors is well established,18, 19 the molecular mechanisms restricting myogenic differentiation have not been fully explored. We have previously demonstrated that the HDAC3/NCOR epigenetic repressor complex restricts myogenic differentiation in RMS by blocking MYOD1-mediated transcriptional activation.7 The striking phenotypic similarities between RAS knockout and disruption of the HDAC3/NCOR epigenetic repressor complex suggest a role for the HDAC3/NCOR complex as a downstream mediator of RAS signaling, likely through direct interaction with the master myogenic regulatory factor MYOD1.7, 20 Despite promising data suggesting the importance of these factors for ERMS tumor growth, more research is needed to understand the connection between RAS signaling, MYOD1 regulation, and other additional factors, including the HDAC3/NCOR epigenetic repressor complex, in controlling myogenic differentiation in ERMS. Importantly, many of the driver genes in RMS are currently undruggable, and new inhibitor-based targeted therapies will take significant time and resources to develop. Our study highlights the use of recombinant gene-editing virotherapy as a promising alternative to target non-druggable driver genes in cancer.
In the absence of effective anti-RAS therapeutic agents, we examined the potential of using high-efficiency gene-editing approaches to directly target NRAS and HRAS in ERMS. Although several cancer therapeutic gene-editing studies have observed significant effects from the use of single gRNAs,6, 14, 15, 21 we found that higher gene-editing efficiency was required for therapeutic applications in ERMS.7 To establish an effective gene-editing system for ERMS tumors, we employed a multiplex gene-targeting approach to simultaneously introduce multiple disrupting indel mutations along with high-efficiency deletion mutations in ERMS cells.
In addition to establishing high-efficiency gene-targeting strategies, effective therapeutic delivery systems are critical for implementing this innovative technology for clinical use. Although previous reports have used non-replicating gene therapy vectors,6, 15, 21 we found that the transduction efficiency of these vectors was extremely limited in ERMS tumors. This led us to develop an oncolytic MYXV vector capable of replicating within ERMS cells. We demonstrated that this recombinant MYXV system can induce high-efficiency editing of endogenous human genes using a single SpCas9-Csy4 transcript with cleavable gRNAs. This approach not only facilitated the use of high-expression cytoplasmic viral promoters but can also be used to express large numbers of gRNAs, targeting multiple essential cancer genes. The versatility of this technology facilitates the rapid targeting of a wide range of genetic drivers or cancer types, making it customizable to individual cancer patients. The use of a non-pathogenic, rabbit-derived MYXV also has significant therapeutic potential because the vector is non-integrating and has no acquired immunity in the human population.11
It is important to note that direct cancer gene-editing therapy has only recently been proposed as a potential therapeutic option for targeting undruggable cancer genes. Although significant obstacles need to be overcome prior to the development of this technology for therapeutic applications, our data suggest that arming oncolytic viruses with gene-editing technology could have significant therapeutic advantages. There are several alternative oncolytic viruses currently used for cancer treatment,22 which may benefit from integrating CRISPR/Cas9 gene-editing technology into their therapeutic approaches. The primary obstacle to the development of a cancer gene-editing therapy is the inability of most oncolytic viruses to infect all cancer cells. Future oncolytic viral technology will need to overcome this limitation. Therefore, utilizing gene-editing technology to enhance the spread of oncolytic viruses by targeting antiviral genes or to induce potent cancer immune responses may hold more therapeutic potential than directly targeting cancer oncogenes. More research is also needed to understand how the immune system will impact oncolytic gene-editing therapy, using immunocompetent models. Although an active immune system could hamper the spread of oncolytic viruses, ablating essential cancer genes could trigger pathways that downregulate immune evasion or stimulate effective anti-cancer immune responses. The technological approach employed in this research could also be used to directly engineer the expression of immune-stimulatory factors to promote clearing of cancer cells.
Although our data support previous claims that gene-editing technology can be used to edit cancer-specific mutations,6, 15, 21 we find that targeting single-nucleotide variants is prone to low levels of off-target gene editing. These off-target effects could be reduced with thorough gRNA optimization; however, restrictions on the placement of tumor-specific gRNA target sites make finding efficient, high-fidelity gRNAs for many oncogenes a significant challenge. Because the target sites for these mutations likely lie in critical regions of oncogenes, such as RAS, the potential to create new oncogenic mutations in healthy tissues is also a major concern.15 The use of high-fidelity Cas9 variants may help overcome this limitation in gRNA specificity, but off-target gene editing still remains a challenge with all current CRISPR systems.16 Alternative methods of establishing cancer-targeting specificity could provide a more viable therapeutic strategy that does not rely on directly editing cancer mutations. This may include the use of replication-dependent viral promoters such as the late viral promoter, p11, to restrict expression of the CRISPR components to cancer cells.23 The enhanced flexibility of this approach would enable a variety of cancer gene-editing strategies, such as targeting non-mutant essential cancer genes, or multi gene-editing applications designed to reduce the probability of resistance. This is particularly relevant given our results with inducible NRAS knockout, which identified a sub-population of ERMS cancer cells that were resistant to NRAS targeting. RAS-resistant cells are not unique to ERMS and have been isolated after KRAS knockout in pancreatic cancer cells.24
Our study provides valuable insight into the use of gene-editing technology as a potential targeted therapy application in cancer treatment. We have established strong evidence that arming oncolytic viruses with gene-editing capability has significant therapeutic potential for targeting undruggable driver genes in cancer. This study provides an important framework for further development of gene-editing oncolytic virotherapy as a new approach to precision cancer treatment.
Materials and Methods
Gene-Editing Vectors
Gene-editing vectors were created to express either SpCas9 or SaCas9 from a core elongation factor 1a promoter. Dual gRNAs targeting the genes of interest were expressed from tandem human U6 promoters on independent viral vectors (SpCas9) or integrated into Cas9 expression vectors (SaCas9). All gRNA target sites were selected using ChopChopv2 software,25 and the gRNA sequences used in this study are listed in Table S1. The tamoxifen-inducible CRISPR/Cas9 gene-editing system has been previously described.7 The key gene-editing vectors used in the research can be found on Addgene (Cambridge, MA, USA). Validation of effective gene editing in target cells was analyzed by PCR amplification of expected deletions created by the targeting vectors. Gene-editing efficiency was estimated in inducible tumor xenografts and cultured cells by analyzing the band intensity of deletion mutations compared with amplification of non-targeted regions of the genome (total gDNA). These efficiency estimates are likely underestimates because we did not analyze the efficiency of indel mutations that occur at each independent gRNA target site. For complete analysis of gene-editing efficiency after tumor-specific RAS targeting, the edited regions of both NRAS and HRAS were submitted for next-generation amplicon sequencing using 150-bp paired-end reads (greater than 500× coverage; Genewiz, South Plainfield, NJ, USA) to quantify all gene-editing events (deletions and indel mutations).
Cell-Based Assays
The cell lines used in this study harbored activating NRAS A183T (381T) or HRAS C181A (SMS-CTR) point mutations. These cells lines were validated by short tandem repeat profiling performed at the cell line validation core facility at Dana Farber Cancer Institute or from the American Type Culture Collection (ATCC). The characteristic RAS mutations present in each line were also identified using next-generation sequencing. The cells were also confirmed mycoplasma-free prior to experimentation. All cells were cultured under standard high serum growth conditions or low serum differentiation conditions, as described.7 Cell viability was assessed with relative cell counts, and myogenic differentiation was analyzed with myosin heavy chain immunofluorescence (MF20, DSHB), to determine the percentage of myosin-positive cells. Apoptosis was measured using an annexin V fluorescent flow cytometry assay (Thermo Fisher Scientific). All cell culture experiments were performed in technical triplicate with the results averaged over three to four independent biological replicates as indicated in the figure legend.
Recombinant Gene-Editing MYXV
To engineer a recombinant gene-editing MYXV, a transgenic cassette expressing the SpCas9 and Csy4 CRISPR components was integrated by site-directed recombination between genes 135R and 136R of the MYXV genome (Figure 4A). This design enabled high-efficiency gene editing from a single cytoplasmic mRNA transcript (Figure 4A). Because MYXV is a non-integrating cytoplasmic virus, all CRISPR/Cas9 components were driven by a single vaccinia vvSEL to facilitate high levels of cytoplasmic expression. In addition to the CRISPR components, the MYXV vectors also expressed a separate vvSEL-GFP cassette to track transduction efficiency in vivo (Figure 4A). The MYXV used in the research was purified and concentrated as previously described.26
Xenografts
Immunodeficient NSG mice were implanted with a single subcutaneous ERMS tumor xenograft on the left flank. After tumor onset (10 mm3), the tumor volume was measured every 2–3 days until endpoint (500 mm3). Treatment groups were split into RAS knockout tumors and control tumors targeted at a safe harbor region of the genome. This safe harbor control was preferred over non-targeting controls to account for any non-specific DNA damage response that may occur in CRISPR-targeted cells. Randomization was not used in grouping of the isogenic mouse strains; however, male and female mice were distributed equally across the treatment groups. Because animals required multiple, sequential experimental treatments, blinding was not performed. For tamoxifen-inducible NRAS knockout studies, gene editing was induced with five intraperitoneal injections of tamoxifen over a 10-day period after the tumor had reached a volume of 10 mm3. For the time-course xenograft experiments, tamoxifen injections were performed on 100-mm3 tumors to facilitate subsequent sampling of the xenografts at multiple time points during tumor regression. For MYXV gene-editing studies, four doses of engineered MYXV virus (1 × 107 PFU/100 μL) were intratumorally administered to 10-mm3 xenografts over 8 days, and changes in tumor volume were analyzed until tumors reached endpoint or 50 days post-viral administration, whichever came first. Histological and immunohistochemical analysis of tumors was performed by the University of Washington histology and imaging core. The antibody against phospho-Erk1/2 (p44/42 mitogen-activated protein kinase [MAPK]) was obtained from Cell Signaling. All treatment groups were maintained in identical laboratory conditions, with experiments carried out under approved protocols from the University of Washington Institutional Animal Care and Use Committee (IACUC) and the office of animal welfare. Mouse numbers were determined from pilot experiments, using a power analysis with an expected large effect size.
Statistics and Data Reporting
Experimental data were analyzed with a two-tailed t test or ANOVA statistical test based on the number of treatment groups. Bonferroni corrections were made to the p value, based on the number of statistical comparisons. The mean difference between treatment groups was used for statistical comparisons except for mouse xenograft studies that analyzed changes in median survival between targeted and control mice. All data were analyzed with Prism 6, GraphPad software. The numbers of biological replicates are reported in the figure legends. Data are presented as mean ± SEM with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Transcriptome Analysis and Data Availability
Expression profiling was performed on NRAS-targeted and control cells 2 days after gene editing to identify genes immediately impacted by NRAS knockout, prior to terminal myogenic differentiation. RNA-sequencing analysis was performed by the Fred Hutchinson Cancer Research Center, Genomics Shared Resource Core using protocols previously identified.7 All transcriptome data produced during this research is available at NIH GEO: GSE118939.
Author Contributions
M.P.P. and E.C. conceived, designed, and oversaw the study; analyzed the data; and wrote the manuscript. M.P.P., H.Y., and S.P. performed most of the experiments and produced all lentiviral, inducible, and MYXV gene-editing vectors. M.M.R. and G.M. were instrumental with the design of MYXV studies and produced the recombinant MYXV.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We thank Terra Vleeshouwer-Neumann and Thao Pham for their assistance with some of the cell-based assays and critical reading of the manuscript. We also thank the University of Washington histology and imaging core for their histopathology expertise. The study is funded by NIH NCI grant 1R01CA196882-01A1 and the Rally foundation.
Footnotes
Supplemental Information includes three figures and one table and can be found with this article online at https://doi.org/10.1016/j.omto.2018.09.001.
Supplemental Information
References
- 1.Shern J.F., Chen L., Chmielecki J., Wei J.S., Patidar R., Rosenberg M., Ambrogio L., Auclair D., Wang J., Song Y.K. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papke B., Der C.J. Drugging RAS: know the enemy. Science. 2017;355:1158–1163. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 3.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 4.Valletta S., Dolatshad H., Bartenstein M., Yip B.H., Bello E., Gordon S., Yu Y., Shaw J., Roy S., Scifo L. ASXL1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts. Oncotarget. 2015;6:44061–44071. doi: 10.18632/oncotarget.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang X., Zhuang C., Zhuang C., Xiong T., Li Y., Gui Y. An enhanced hTERT promoter-driven CRISPR/Cas9 system selectively inhibits the progression of bladder cancer cells. Mol. Biosyst. 2017;13:1713–1721. doi: 10.1039/c7mb00354d. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y., Sassi S., Shen J.K., Yang X., Gao Y., Osaka E., Zhang J., Yang S., Yang C., Mankin H.J. Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system. J. Orthop. Res. 2015;33:199–207. doi: 10.1002/jor.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phelps M.P., Bailey J.N., Vleeshouwer-Neumann T., Chen E.Y. CRISPR screen identifies the NCOR/HDAC3 complex as a major suppressor of differentiation in rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA. 2016;113:15090–15095. doi: 10.1073/pnas.1610270114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan W.M., Rahman M.M., McFadden G. Oncolytic myxoma virus: the path to clinic. Vaccine. 2013;31:4252–4258. doi: 10.1016/j.vaccine.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron C., Hota-Mitchell S., Chen L., Barrett J., Cao J.-X., Macaulay C., Willer D., Evans D., McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 11.Lun X., Yang W., Alain T., Shi Z.-Q., Muzik H., Barrett J.W., McFadden G., Bell J., Hamilton M.G., Senger D.L., Forsyth P.A. Myxoma virus is a novel oncolytic virus with significant antitumor activity against experimental human gliomas. Cancer Res. 2005;65:9982–9990. doi: 10.1158/0008-5472.CAN-05-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinn V.G., Hilgenberg V.A., MacNeill A.L. Myxoma virus therapy for human embryonal rhabdomyosarcoma in a nude mouse model. Oncolytic Virother. 2016;5:59–71. doi: 10.2147/OV.S108831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koo T., Yoon A.-R., Cho H.-Y., Bae S., Yun C.-O., Kim J.-S. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017;45:7897–7908. doi: 10.1093/nar/gkx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W., Lee S., Kim H.S., Song M., Cha Y.H., Kim Y.-H., Shin J., Lee E.S., Joo Y., Song J.J. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res. 2018;28:374–382. doi: 10.1101/gr.223891.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinson A.R.P., Jones R., Crose L.E.S., Belyea B.C., Barr F.G., Linardic C.M. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front. Oncol. 2013;3:183. doi: 10.3389/fonc.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tapscott S.J., Thayer M.J., Weintraub H. Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science. 1993;259:1450–1453. doi: 10.1126/science.8383879. [DOI] [PubMed] [Google Scholar]
- 19.Keller C., Guttridge D.C. Mechanisms of impaired differentiation in rhabdomyosarcoma. FEBS J. 2013;280:4323–4334. doi: 10.1111/febs.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacQuarrie K.L., Yao Z., Fong A.P., Tapscott S.J. Genome-wide binding of the basic helix-loop-helix myogenic inhibitor musculin has substantial overlap with MyoD: implications for buffering activity. Skelet. Muscle. 2013;3:26. doi: 10.1186/2044-5040-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhen S., Hua L., Takahashi Y., Narita S., Liu Y.-H., Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Wennier S., Reinhard M., Roy E., MacNeill A., McFadden G. Myxoma virus expressing interleukin-15 fails to cause lethal myxomatosis in European rabbits. J. Virol. 2009;83:5933–5938. doi: 10.1128/JVI.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opgenorth A., Graham K., Nation N., Strayer D., McFadden G. Deletion analysis of two tandemly arranged virulence genes in myxoma virus, M11L and myxoma growth factor. J. Virol. 1992;66:4720–4731. doi: 10.1128/jvi.66.8.4720-4731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muzumdar M.D., Chen P.-Y., Dorans K.J., Chung K.M., Bhutkar A., Hong E., Noll E.M., Sprick M.R., Trumpp A., Jacks T. Survival of pancreatic cancer cells lacking KRAS function. Nat. Commun. 2017;8:1090. doi: 10.1038/s41467-017-00942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44(W1):W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smallwood S.E., Rahman M.M., Smith D.W., McFadden G. Myxoma virus: propagation, purification, quantification, and storage. Curr. Protoc. Microbiol. 2005;17:14A.1.1–14A.1.20. doi: 10.1002/9780471729259.mc14a01s17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.