Abstract
Bright-field transmission electron microscopy (TEM), TEM-negative staining technique, resin-embedding and ultramicrotomy, scanning TEM, scanning electron microscopy, atomic force microscopy and cryoelectron microscopy are imaging techniques used for describing giant viruses, their cycle and ultrastructure. Here we used the SECOM system, an integrated correlative light and electron microscopy using light and electronic imaging without sample transfer, to study cells infected with giant viruses, as shown by Tupanvirus, the ultrastructure of which was successfully observed. An improvement of the SECOM system with an eye to its use in fundamental and clinical research could be considered in the field of microbiology.
Keywords: Amoeba, CLEM, giant viruses, SECOM system, Tupanvirus
Introduction
The discovery of giant viruses (GVs) has upended the definition of the virus by exceeding the barrier of particle size and genome [1], [2]. Tupanvirus is a GV with a ∼450 nm capsid containing fibrils and large cylindrical tail (∼550 × ∼450 nm including fibrils) attached to the base of the capsid that is the longest described in the virosphere. The average length of a complete virion is ∼1.2 μm, which makes them one of the longest viral particles [3].
Bright-field transmission electron microscopy (TEM), TEM-negative staining technique and resin embedding and ultramicrotomy have been used, respectively, to obtain nanometer-scale information about GVs [1], [2], [4], [5], [6], [7], to picture whole-GV morphology [2], [3], [7], [8], [9], to study the ultrastructure of GVs and to describe infection cycles over time [1], [2], [3], [4], [5], [8], [9], [10]. Scanning TEM [8], scanning electron microscopy (SEM) [11] and atomic force microscopy have been used to study sections of GV-infected cells [12] and to characterize GV morphology. Cryoelectron microscopy has been a popular technique because it enables the preservation of GV ultrastructure [9], [10], [12], [13], [14], [15].
Because the diameter of GVs is larger than the optical resolution limit, GVs are readily visible in bright-field transmitted or fluorescence light microscopy (LM) [2], [3], [10]. But despite this unique property, LM is not as often used as electron microscopy (EM) for characterizing GV morphology or infection cycle because of limitations in resolution. Correlative light and electron microscopy (CLEM) helps to bridge this gap between LM and EM of GV-infected cells with the combination of the specificity of fluorescent labeling and the high-resolution structural information of EM, making it the perfect tool to study the complex relationship between form and function in biology. The SECOM system is a system for integrated CLEM wherein light and electron imaging are performed in one system without the need for sample transfer [16], [17].
Here we report our study of cells infected with GVs using integrated CLEM with the goal of determining whether GVs such as Tupanvirus [3] could be detected using the SECOM system so we could study their unique ultrastructure.
Materials and methods
Acanthamoeba castellanii samples in periodic acid–Schiff medium were infected with purified Tupanvirus with a multiplicity of infection of 10 at 30°C for 18 hours and stained with FM4-64FX (aldehyde fixable; F34653, Thermo Fisher) for 30 minutes at 30°C in the dark, then fixed overnight at 4°C with paraformaldehyde 4% in sodium cacodylate 0.1 M buffer. After rinsing two times for 15 minutes each with a cacodylate 0.1 M/saccharose 0.2 M in water solution, cells were dehydrated with ethanol 50%, 70% and 96%, for 15, 30 and 30 minutes, respectively. Cells were then placed for 1 hour in a mix of LR-White resin 100% (Polysciences, Ref. 17411 MUNC-500) and ethanol 96% in a 2:1 ratio. After 30 minutes in pure 100% LR-White resin, cells were placed in 100% LR-White resin overnight at room temperature. The day after, cells were placed for 1 hour in 100% resin at room temperature. A total of 1.5 mL of Pure 100% LR-White resin was added on the cell pellet. Polymerization was achieved at 60°C for 3 days. Between all steps, the samples were ultracentrifuged at 5000 rpm, and the supernatant was discarded. Sections 70, 100 or 1000 nm thick were cut on a UC7 ultramicrotome (Leica). For TEM, 70 nm thick sections were deposited on 300 mesh copper/rhodium grids (Maxtaform HR25, TAAB). They were poststained with 5% uranyl acetate and lead citrate according to the Reynolds method [18]. Electron micrographs were obtained on a Morganii 268D (Philips); TEM was operated at 80 keV and was equipped with a 1024 × 1024 pixel MegaView3 camera. For fluorescence microscopy, 100 nm thick sections on grids were imaged with a confocal laser scanning AiryScan LS800 microscope (Zeiss). For the SECOM system, 100 nm thick uncontrasted sections were deposited on glass slides coated with a conducting layer of indium tin oxide, making it possible to image the sections with electrons while maintaining optical transparency for fluorescence imaging. The SECOM system was mounted on a Verios 460 (Thermo Fisher) SEM. The fluorescence images were obtained by excitation with a 467 nm light source.
Results and discussion
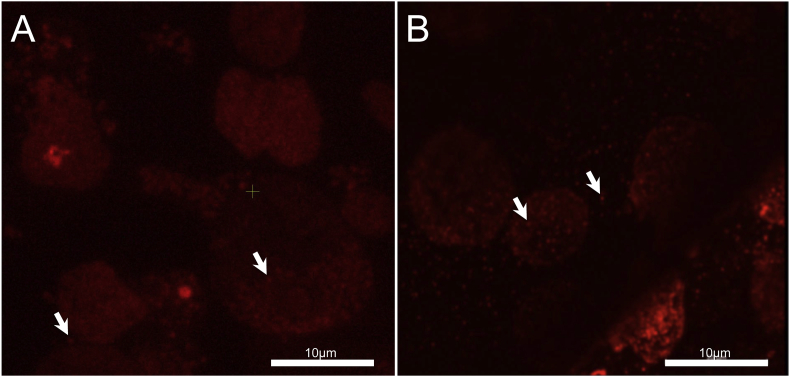
Thin sections containing whole infected cells were identified optically on the SECOM system using a low-magnification (40×) air objective lens, followed by imaging with a high NA (=1.2) 60× water immersion objective (Fig. 1(A)), where amoeba cells can clearly be identified.
Fig. 1.
(A) SECOM system fluorescence image of 100 nm thick ultrathin section on indium tin oxide slide using 60× water immersion objective with NA of 1.2 (B) Confocal laser scanning microscope reference image of 100 nm thick ultrasection on grid using 63× objective lens (Z maximal projection). White arrows point to intracellular or extracellular single Tupanvirus particles in (A) and (B).
EM images of the same (uncontrasted) sections were acquired on the Verios 460 SEM using the in-lens secondary electron detector in immersion mode. A 1 keV beam and 100 pA currents were used to obtain the high-resolution EM images which have sufficient contrast to enable the identification of virus particles as well as cell plasma membranes and intracellular organelles. Further, the high magnification SEM image of the virus (Fig. 2(A)) shows the possibility of locating and imaging an individual virus. A reference image of an EM-stained section acquired in a TEM is shown in Fig. 2(B).
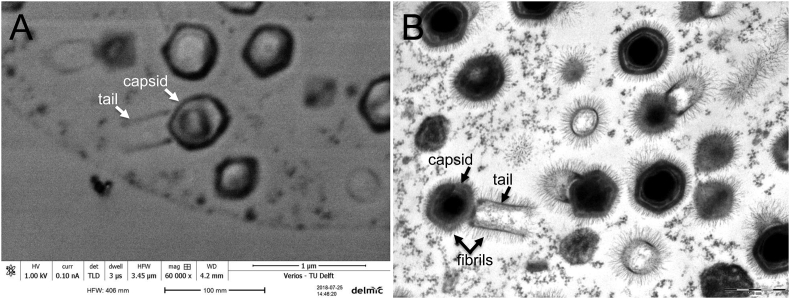
Fig. 2.
(A) High-magnification scanning electron microscopy image of 100 nm thick ultrathin section with no EM stain acquired with FEI Verios 460 (FEI Company) and showing individual tupanviruses; virus particle shows typical capsid and attached tail. (B) High-magnification transmission electron microscopy image of 70 nm thick ultrathin section with EM stain acquired at 80 keV on Morgagni 268D (Philips) transmission electron microscope.
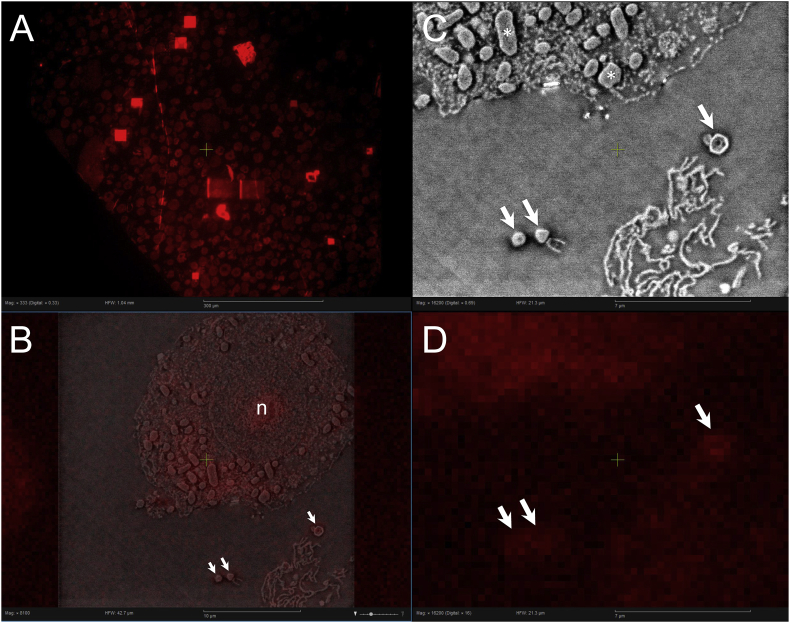
The optical and electron beam imaging parameters were optimized at a location far away from the region of interest. Then the fluorescence image was acquired, followed by the electron image. Finally the overlay procedure was performed by electron beam exposure of a grid of points, from which the cathodoluminescence was detected by the camera and used to align the objective lens with the electron beam axis. The correlative imaging including the overlay is fully automated when giving a high-resolution image, with two mixing ratios of optical and electron contrast. The Tupanvirus was successfully identified and imaged on the SECOM system (Fig. 3, Fig. 4). This also shows the optical LM, EM and LM/EM correlative image in the Odemis image acquisition interface.
Fig. 3.
Example of four-panel Odemis software interface of SECOM system with single and overlay channels. (A) Fluorescence light microscopy overview image of 100 nm thick ultrathin LR-white section containing Tupanvirus-infected cells. (B) Correlative light-electron microscopy overlay image of Tupanvirus particles (white arrows) located outside amoeba cell; n indicates amoeba nucleus. (C) Scanning electron microscopy of Tupanvirus particles depicted in (B). Depending on cutting plane of section, ultrastructural features such as capsid and tail can be seen. Mitochondria (asterisks) can be observed in amoeba cytoplasm. (D) Light microscopy of Tupanvirus particles (white arrows) depicted in (B) and detected by as fluorescent spots.
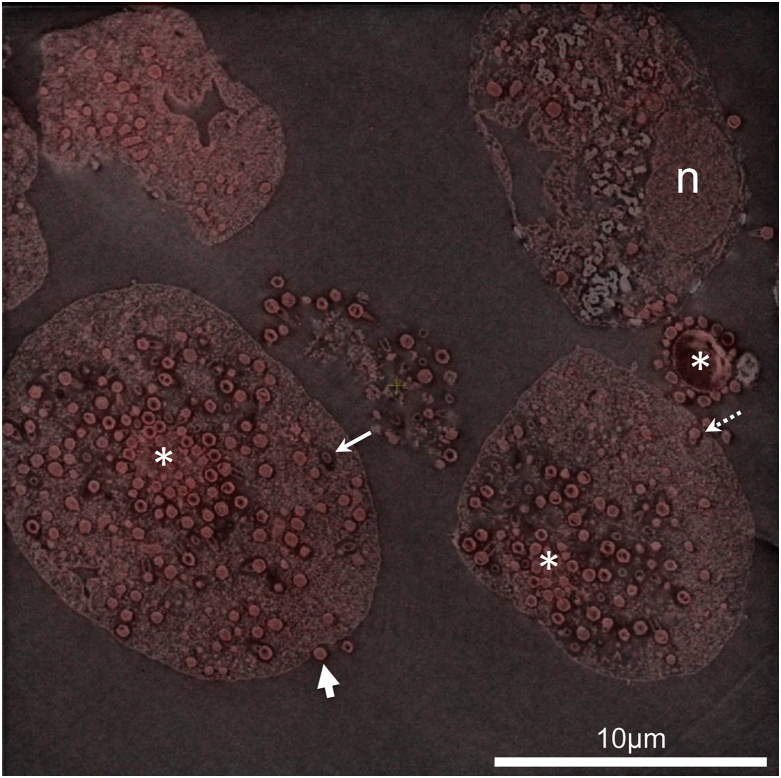
Fig. 4.
Example of correlative light-electron microscopy overlay image of Tupanvirus-infected amoeba cells acquired with SECOM system. Nucleus (n) in cell as well as virus factories (asterisks) with multiplying viruses are shown. Tupanvirus particles can be found inside cells cytoplasm as well as inside cellular vacuoles or plasma membrane invaginations (solid and dashed thin arrows), probably on their way to externalization (thick arrow).
Conclusion
The SECOM system can successfully detect GV particles by fluorescence LM while simultaneously providing information on the EM ultrastructure such as the Tupanvirus capsid and tail features. Although this ultrastructural information is not as high in resolution as that achieved in a conventional TEM (the fibrils were not seen in SEM), it is sufficient for determining the gross morphology of GV particles and their locations regarding cellular compartments. In the field of virology the SECOM system has the potential to be an efficient tool for screening suspicious samples and virus-infected cells at low and higher resolution. More broadly, the SECOM system has the potential to be useful for fundamental and clinical research in the field of microbiology.
Funding
This work was supported by the French Government under the “Investissements d’avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, French National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03).
Acknowledgements
We thank S. Aguy from Eden Instruments. We thank all the IHU staff members for providing an enthusiastic research atmosphere.
Conflict of interest
None declared.
References
- 1.La Scola B., Audic S., Robert C., Jungang L., de Lamballerie X., Drancourt M. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Colson P., La Scola B., Levasseur A., Caetano-Anollés G., Raoult D. Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nat Rev Microbiol. 2017;15:243–254. doi: 10.1038/nrmicro.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahão J., Silva L., Silva L.S., Khalil J.Y.B., Rodrigues R., Arantes T. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun. 2018;9:749. doi: 10.1038/s41467-018-03168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil J.Y., Andreani J., La Scola B. Updating strategies for isolating and discovering giant viruses. Curr Opin Microbiol. 2016;31:80–87. doi: 10.1016/j.mib.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Khalil J.Y., Langlois T., Andreani J., Sorraing J.M., Raoult D., Camoin L. Flow cytometry sorting to separate viable giant viruses from amoeba co-culture supernatants. Front Cell Infect Microbiol. 2017;6:202. doi: 10.3389/fcimb.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Temmam S., Monteil-Bouchard S., Robert C., Baudoin J.P., Sambou M., Aubadie-Ladrix M. Characterization of viral communities of biting midges and identification of novel thogotovirus species and rhabdovirus genus. Viruses. 2016;8:77. doi: 10.3390/v8030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somiya M., Liu Q., Kuroda S. Current progress of virus-mimicking nanocarriers for drug delivery. Nanotheranostics. 2017;1:415–429. doi: 10.7150/ntno.21723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutsafi Y., Shimoni E., Shimon A., Minsky A. Membrane assembly during the infection cycle of the giant Mimivirus. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003367. e1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao C., Chipman P.R., Battisti A.J., Bowman V.D., Renesto P., Raoult D. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 10.Schrad J.R., Young E.J., Abrahão J.S., Cortines J.R., Parent K.N. Microscopic characterization of the Brazilian giant samba virus. Viruses. 2017;9:E30. doi: 10.3390/v9020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertelli C., Mueller L., Thomas V., Pillonel T., Jacquier N., Greub G. Cedratvirus lausannensis—digging into Pithoviridae diversity. Environ Microbiol. 2017;19:4022–4034. doi: 10.1111/1462-2920.13813. [DOI] [PubMed] [Google Scholar]
- 12.Xiao C., Kuznetsov Y.G., Sun S., Hafenstein S.L., Kostyuchenko V.A., Chipman P.R. Structural studies of the giant Mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao C., Fischer M.G., Bolotaulo D.M., Ulloa-Rondeau N., Avila G.A., Suttle C.A. Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests novel assembly pathway for giant viruses. Sci Rep. 2017;7:5484. doi: 10.1038/s41598-017-05824-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zauberman N., Mutsafi Y., Halevy D.B., Shimoni E., Klein E., Xiao C. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto K., Miyazaki N., Song C., Maia F.R.N.C., Reddy H.K.N., Abergel C. Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy. Sci Rep. 2017;7:13291. doi: 10.1038/s41598-017-13390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Boer P., Hoogenboom J.P., Giepmans B.N. Correlated light and electron microscopy: ultrastructure lights up! Nat Methods. 2015;12:503–513. doi: 10.1038/nmeth.3400. [DOI] [PubMed] [Google Scholar]
- 17.Sueters-Di Meo J., Liv N., Hoogenboom J.P. Encyclopedia of analytical chemistry: applications, theory and instrumentation. 2016. Using advanced correlative microscopy to study complex biological samples. Available at: [DOI] [Google Scholar]
- 18.Reynolds E.S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]