Abstract
Macrophages play a crucial role in the biological performance of biomaterials, as key factors in defining the optimal inflammation-healing balance towards tissue regeneration and implant integration. Here, we investigate how different surface modifications performed on poly(L-lactic acid) (PLLA) films would influence the differentiation of human monocytes to macrophages. We tested PLLA films without modification, surface-modified by plasma treatment (pPLLA) or by combining plasma treatment with different coating materials, namely poly(L-lysine) and a series of proteins from the extracellular matrix: collagen I, fibronectin, vitronectin, laminin and albumin. While all the tested films are non-cytotoxic, differences in cell adhesion and morphology are observed. Monocyte-derived macrophages (MDM) present a more rounded shape in non-modified films, while a more elongated phenotype is observed containing filopodia-like and podosome-like structures in all modified films. No major differences are found for the expression of HLA-DR+/CD80+ and CD206+/CD163+ surface markers, as well as for the ability of MDM to phagocytize. Interestingly, MDM differentiated on pPLLA present the highest expression of MMP9. Upon differentiation, MDM in all surface modified films present lower amounts of IL-6 and IL-10 compared to non-modified films. After stimulating MDM with the potent pro-inflammatory agent LPS, pPLLA and poly(L-lysine) and fibronectin-modified films reveal a significant reduction on IL-6 secretion, while the opposite effect is observed with IL-10. Of note, in comparison to non-modified films, all surface modified films induce a significant reduction of the IL-6/IL-10 ratio, a valuable prognosticator of pro- versus anti-inflammatory balance. These findings give important insights about MDM-biomaterial interactions, while strengthening the need of designing immune-informed biomaterials.
Introduction
Biodegradable polymers are commonly used in biomedical applications, including the development of surgical implants, drug delivery systems and tissue engineering constructs.1 Materials targeting organ substitution with permanent function in the body are usually preferred to show high inertness, associated with the concept of biotolerability.2 Oppositely, biodegradable polymers aiming tissue regeneration must show an interplay with the implantation environment.3,4
One of the most important characteristics of biomaterials is their ability to trigger adequate levels of host inflammatory response. An exacerbated inflammatory response leads to abnormal tissue healing, characterized by chronic inflammation, fibrotic encapsulation, and scar tissue formation. These events ultimately promote the rejection of the implanted biomaterial.5,6 On the other hand, the innate inflammatory process followed by the implantation of a biomaterial is also a keystone for efficient tissue regeneration and repair.7,8 Therefore, designing biomaterial-based therapies that have the ability to modulate the inflammatory response must be accompanied by the understanding of biomaterial–immune system interactions.
Inflammation is an extremely complex multistage process involving numerous cell types and mediator signals.9 In particular, macrophages (Mϕ) play an important role in wound healing and biomaterial-mediated inflammation. Immediately after implantation, monocytes are recruited from peripheral blood to the implantation site where they differentiate into Mϕ. Inflammatory signals from the tissue injury around the implanted biomaterial mediate the differentiation of monocytes into inflammatory Mϕ via autocrine and paracrine signalling.10 In the early stages of inflammation, Mϕ attempt to remove the foreign body via phagocytosis and secrete large amounts of bioactive mediators, such as (i) reactive oxygen species to degrade the biomaterial, (ii) chemokines to direct additional inflammatory cells to the site of injury, and (iii) cytokines to further activate the surrounding inflammatory cells.11,12 If the inflammatory response is not excessive, Mϕ enable the injured site to move into healing phase by releasing growth factors that promote the proliferation of fibroblasts and blood vessel formation.13 Mϕ may present a wide plethora of phenotypes with inflammatory or anti-inflammatory/regeneration-adjuvant features.14 States of ‘functionally active’ Mϕ phenotypes, with upregulation of both pro- and anti-inflammatory markers have been reported as efficient in modulating the integration of biomaterials.15 The classic and alternative activation of Mϕ are also termed as M1 and M2 to mimic T helper (Th) cell nomenclature. While the classically activated (M1) enhances Th1 type inflammation,16 the alternatively activated (M2) enhances Th2 response and improve tissue healing.16,17 Physical properties such as substrate stiffness, topography, pore size and size of wear debris; chemical properties such as surface chemistry, ligand presentation and release of growth factors; and temporal properties, such as degradation rates, all influence the monocyte-to-macrophage differentiation and activation as well as their cytokine secretion.9,14,18–23 With this in mind, new generation of biomaterials are being designed in an attempt to control the immune response of the body after implantation. The idea is to tailor the phenotype and physiology of Mϕ, and thus ultimately guide them into an appropriate stimulus towards tissue regeneration.
PLLA is a biodegradable thermoplastic aliphatic polyester derived from the polymerization of L-lactide in lactic acid.24 Polyesters have been well documented for their excellent biodegradability, biocompatibility, nontoxicity and their biocompatible degradation products. However, poor hydrophilicity and the lack of natural recognition sites on polyester surfaces for covalent cell-recognition signal molecules to promote cell attachment are the main drawbacks of PLLA as tissue engineering biomaterials.25,26 Therefore, different techniques have been proposed to modify its surface to enhance cell-material interaction.
Plasma treatment is one of the most widely used treatment techniques for improving the surface properties of polymers for use in Tissue Engineering and Regenerative Medicine (TERM).27 The principle is based on the presence free electrons in the air, which are accelerated by a high voltage discharge and ionize the gas. As a result, chemical and physical modifications occur on the modified surfaces, creating reactive sites such as amine and carboxyl groups.24,28,29 Despite the use of polyester biomaterials in clinical practice, only a few studies addressed its immunomodulatory effect.9,30,31
In this context, we aim to study the interaction of surface-modified PLLA films with human monocyte-derived macrophages (MDM). We aim to assess how different surface modifications performed to PLLA films would influence the differentiation of human monocytes to Mϕ and their phenotypic polarization and activation profile. To this end, films were produced and tested without modification (PLLA) or after different surface modifications known to enhance cell adhesion. The surface of such films was modified by employing air-plasma technology (pPLLA) or by combining air-plasma with different coating materials, namely poly(L-lysine) (pPLLA-PLL) and various extracellular matrix (ECM)-derived proteins, namely collagen I (pPLLA-COLL I), fibronectin (pPLLA-FN), vitronectin (pPLLA-VTN), laminin (pPLLA-LAM) and albumin (pPLLA-ALB). MDM were characterized regarding their morphology, surface markers, and cytokines secretion. Moreover, an additional pro-inflammatory stimulus was added with lipopolysaccharide (LPS) and the resultant cytokine profile was evaluated.
Experimental
PLLA films production
PLLA films were produced by solvent casting methodology. Briefly, PLLA (5% w/v, Mw~1600-2400, 70% crystallinity, Polysciences) dissolved in methylene chloride (Fisher Chemical) was poured to a glass petri dish. Solvent was left to evaporate overnight at room temperature (RT) inside a fume hood. Afterwards, the produced PLLA films were cut into disks to obtain films with 1 cm2.
Plasma surface modification
The plasma-treatment of PLLA was performed according to our previous described protocol.32 Briefly, PLLA films were placed in a plasma reactor chamber (PlasmaPrep5, Gala Instrumente) fitted with a radio frequency generator. Air was used as the working atmosphere to generate a glow discharge plasma at 0.2 mbar and 30 V for 15 min. Films were placed vertically to allow plasma treatment on both sides.
Coating materials
The adsorption of the different coating materials was performed following the respective commercially available protocols. Following plasma treatment, PLLA films were immediately sterilized by immersion in 70% v/v ethanol for 2 h at RT. After washing with phosphate-buffered saline (PBS, Sigma-Aldrich), the films were immersed in the different coating solutions, namely collagen I (8 µg.cm-2, collagen from human placenta, Bornstein and Traub Type I, Sigma-Aldrich), poly(L-lysine) (2 µg.cm-2, poly-L-lysine hydrobromide, Mw=30000-70000, Sigma-Aldrich), fibronectin (3 µg.cm-2, human plasma fibronectin purified protein, Merk Millipore), vitronectin (0.1 µg.cm-2, vitronectin from human plasma, Sigma-Aldrich), laminin (2 µg.cm-2, laminin from human placenta, Sigma-Aldrich) and albumin (4 µg.cm-2, albumin solution human 30% in 0.85% sodium chloride, Sigma-Aldrich). PLLA films were incubated with each coating solution for 2 h at 37°C and then at 4°C overnight.
Isolation of blood-derived monocytes and in vitro differentiation into macrophages
Experiments were conducted using buffy coats from healthy donors (n=7) supplied by the Hospital of Braga, after approval of the Competent Ethics Committee (CEC). The human samples received were handled in accordance with the guidelines approved by the CEC. All the donors agreed and signed an authorized consent (ethical approval reference SECVF014/2015).
Monocytes were isolated from buffy coats of healthy donors by centrifugation on Histopaque®-1077 (Sigma-Aldrich) followed by immunomagnetic separation with human anti-CD14 purification kit (Miltenyi Biotec). The purity of the separation was always confirmed by flow cytometry and was superior to 95% (Figure SI1). Purified monocytes were differentiated in vitro into MDM in the different PLLA films, namely without modification, plasma-treated, or combining plasma treatment with various coating materials. Cells were cultured above the films in RPMI 1640 medium containing heat-inactivated fetal bovine serum (10%, FBS, Gibco), L-glutamine (2 mM, Gibco), penicillin (50 U.mL-1, Gibco), streptomycin (50 µg.mL-1, Gibco) and HEPES (10 mM, Gibco), and supplemented with human macrophage colony stimulating factor (20 ng.mL-1, M-CSF, Peprotech) for 7 days at 37°C in a humidified 5% CO2 air atmosphere. The medium was renewed on the third day of culture. MDM cultured in modified and non-modified PLLA films were supplemented with LPS (100 ng.mL-1, Sigma-Aldrich) for 24 hours at 37°C in a humidified 5% CO2 air atmosphere. Positive controls for surface marker analysis were obtained by polarizing macrophages with 10 ng.mL-1 of LPS (Sigma-Aldrich) plus 100 U.mL-1 of IFN-Υ (Biolegend) for M1 and with 20 ng.mL-1 of IL-4 (Biolegend) for M2.
Cell quantification
The amount of adhered cells after 7 days of culture at the surface of PLLA films was quantified by image analysis. Samples were fixed with 10% v/v formalin (BD Biosciences) for 30 min at RT. Upon PBS washing, 500 µL of PBS containing 0.5 µL DAPI (1 mg.mL-1, 4,6-diaminidino-2-phenylindole-dilactate, Sigma-Aldrich) was added to each sample. Ultimately, samples were washed with PBS and visualized by fluorescence microscopy (Axioimage RZ1M, Zeiss, Germany). Total cell number was quantified by counting the number of stained nuclei in PLLA films (for each biomaterial formulation, 3 replicates were analyzed per donor; n=4) using ImageJ software (NIH, USA). Five images per film were used for the quantification.
Phalloidin/DAPI fluorescence stainings
In order to observe cellular morphology, MDM were stained for actin and nuclei. After 7 days of culture, samples were fixed with 10% v/v formalin (BD Biosciences) for 30 min followed by permeabilization with 0.1% v/v Triton-X (Sigma-Aldrich) for 5 min, both at RT. Upon PBS washing, 500 µL of PBS containing 10 µL phalloidin (50 µg.mL-1, phalloidin tetramethylrhodamine B isothiocyanate, Sigma-Aldrich) was added to each sample. After 1h at RT, samples were washed with PBS and counterstained with DAPI by immersing the samples in 500 µL of PBS containing 0.5 µL DAPI (1 mg.mL-1, 4,6-diaminidino-2-phenylindole-dilactate, Sigma-Aldrich). Ultimately, samples were washed with PBS and visualized by fluorescence microscopy (Axioimage RZ1M, Zeiss, Germany). Z-stack mode was used with a resolution of 5 µm between slides by using the AxioVision software.
Live-dead assay
To assess the viability of MDM in contact with PLLA films, a live-dead assay was performed using calcein-AM and propidium iodide dyes. Briefly, 1 mL of PBS containing 2 µL of calcein-AM (1 mg.mL-1, Invitrogen) and 1 µL of PI (1 mg.mL-1, Invitrogen) was added to each well. Samples were then incubated at 37 °C for 10 min protected from light. Afterward, samples were washed with PBS and immediately visualized by fluorescence microscopy (Axioimage RZ1M, Zeiss, Germany).
Scanning electron microscopy
The morphology of the MDM adhered at the surface of the PLLA films was analyzed by scanning electron microscopy (SEM). After 7 days of culture, samples were fixed with 10% v/v formalin (BD Biosciences) for 30 min and subsequently dehydrated using sequential ethanol series (60, 70, 80, 90, 96, and 100%, 10 min each). Samples were gold-sputtered and visualized (Leica Cambridge S-360) operating at 15.0 kV accelerating voltage.
Surface staining and phagocytic activity of monocyte-derived macrophages
For analysis, MDM were detached from the PLLA films by incubation with TrypLE™ Express solution (Life Technologies) at 37°C for 10 min. For the analysis of surface markers, MDM were incubated for 20 minutes with saturating concentrations of monoclonal antibodies against HLA-DR (clone L243, Biolegend), CD206 (clone 15-2, Biolegend) and CD163 (clone GHI/61, Biolegend). To assess phagocytic activity, fluorescent yellow-green latex beads (1.0 µm mean particle size, Sigma-Aldrich) were incubated with Mϕ at a 1:10 (cell/beads) ratio for 45 min or 4 hours at 37°C. Before acquisition, cells were washed with FACS buffer (PBS with 2% FBS) to remove non-phagocytized beads. Samples were acquired on a LSRII flow cytometer with FACS Diva software (BD Biosciences). All data were analyzed using FlowJo v10 software (TreeStar Inc., Ashland, USA). Figure SI2 depicts the gating strategy for the characterization of macrophage surface markers.
Protein quantification
TNF-α, IL-6, and IL-10 concentration in the supernatants of differentiated and/or LPS-treated MDM were determined by ELISA using commercially available kits (Biolegend) according manufacturer’s instructions. Results are shown as arbitrary units (AU), calculated as follow: [cytokine (pg.µL-1) / number of cells] x 1000.
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA from cultured macrophages was extracted with TripleXtractor reagent (Grisp) according manufacturer’s instructions. cDNA was synthesized using GRS cDNA Synthesis MasterMix (Grisp) for reverse-transcription PCR. Target gene mRNA expression was quantified in 20 ng of cDNA by real-time PCR (Bio-Rad CFX96 Real-Time System C1000 Thermal Cycler) using a KAPA SYBR® FAST qPCR kit Master Mix (KAPA Biosystems) and normalized to Gapdh and β-actin mRNA levels. Specific oligonucleotides for Mmp9 are (forward) TGT ACC GCT ATG GTT ACA CTC G, (reverse) GGC AGG GAC AGT TGC TTC T; for Mmp12 are (forward) GAT CCA AAG GCC GTA ATG TTC C, (reverse) TGA ATG CCA CGT ATG TCA TCA G; for Gapdh are (forward) AAG GTG AAG GTC GGA GTC AAC, (reverse) GGG GTC ATT GAT GGC AAC AAT A; and for β-actin are (forward) GCC GTC TTC CCC TCC ATC GTG, (reverse) GGA GCC ACA CGC AGC TCA TTG TAG A.
Statistical analysis
Data are shown as mean ± standard deviation or mean ± SEM from at least four donors. Statistical analyses were performed using one-way ANOVA test with a Tukey’s multiple-comparison post-test for multiple group comparisons.
Results and discussion
It is well established that microenvironmental cues presented by biomaterials play a crucial role in modulating immune cell response.33 While much progress has been made in understanding these effects on both somatic34 and stem34,35 cells, the effect of such biophysical and biochemical signals on immune cells, specifically Mϕ, is less well known. To fulfill their plethora of functions, Mϕ exhibit a spectrum of transient polarization states that are influenced by varying microenvironmental cues, some of which may be biomaterial-based.14 This deficit in understanding macrophage-biomaterials interaction led us to study the interplay between human monocytes during their differentiation process into macrophages with PLLA, a widely used biomaterial in TERM applications.
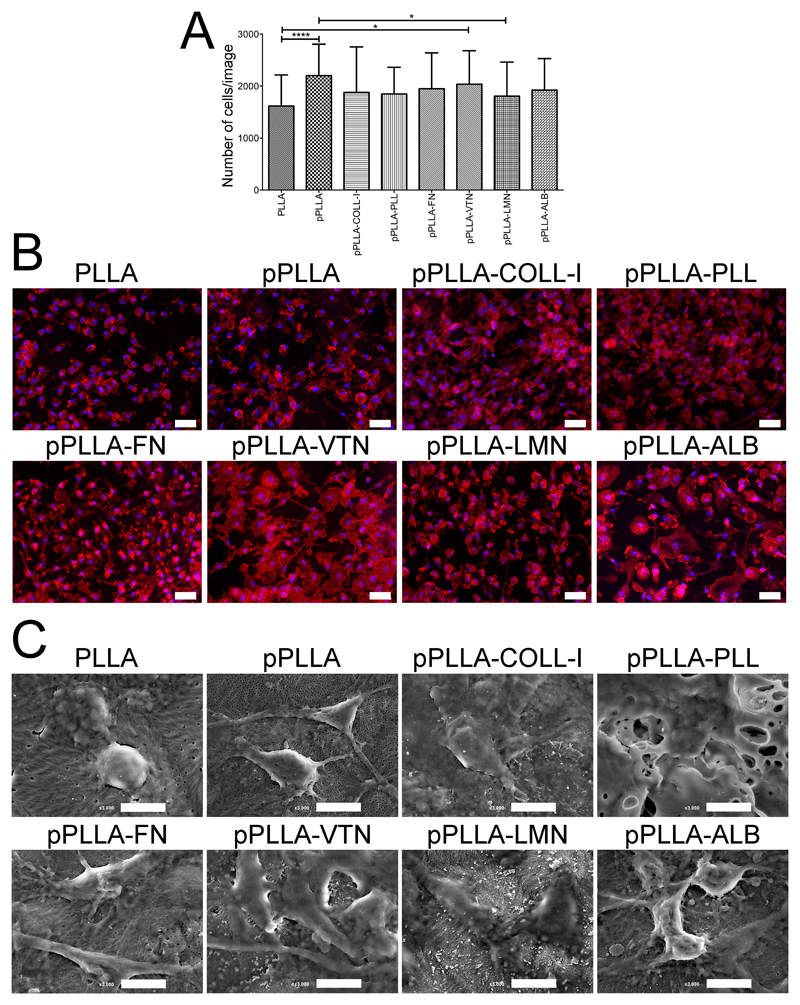
After isolation, monocytes were immediately cultured on the surface of PLLA films with or without different surface modifications. After 7 days of differentiation, the amount of adhered Mϕ per film was quantified. As shown in Figure 1A, PLLA films without modification presented the lowest number of cells. This difference was highly significant when comparing non-modified with plasma-modified films (pPLLA). Although with lower significance, plasma-modified films combined with vitronectin (pPLLA-VTN) or laminin (pPLLA-LMN) coatings also showed significantly lower number of adhered cells compared to pPLLA. In fact, the low hydrophilicity and lack of natural recognition sites for cell adhesion are well-known major drawbacks of PLLA for TERM applications.25,26 The treatment of the PLLA surface with plasma modification allows increasing the hydrophilicity of PLLA.36 Consequently, the binding strength and structural arrangement of water molecules at the surface of PLLA is increased, affecting protein–surface interactions.24 Indeed, protein adsorption to surfaces with different chemistries has been shown to affect Mϕ adhesion and morphology.22 Therefore, the adsorption of serum proteins from the culture medium to the surface of pPLLA might have also contributed to the highest MDM adhesion. The presence of serum in culture medium also allowed studying MDM behavior in a closer environment to what occurs in vivo following implantation, in which serum proteins adsorb to the surface of the implant within seconds. A transient surface matrix is then formed, and, after activation of the coagulation cascade and complement systems, different cell populations are activated following thrombus formation.[37] Among a series of cellular events, Mϕ cells adhere to the surface of the implant and develop a response.12,38 Additionally, coating PLLA films with PLL, well known to improve cell adhesion, or with ECM-derived proteins also contributed to higher MDM adhesion compared to non-modified PLLA, since their presence improve the surface recognition by MDM.39,40 Importantly, all tested PLLA surfaces were non-toxic, as shown by the higher ratios of living cells in the live-dead staining assay (Figure SI3). The morphologies of MDM in contact with different PLLA surfaces were further observed by F-actin staining (Figure 1B). Fluorescence microscopy observations demonstrated distinct MDM morphologies upon differentiation in the presence of different PLLA surfaces. While non-modified PLLA films revealed the presence of rounded MDM, in the other formulations a heterogenic population composed by more elongated MDM with higher surface area was observed. Additionally, in all surface modified films, MDM exhibited more filopodia-like and podosome-like structures, which are involved in cell motility and adhesion, respectively, compared to non-modified PLLA. A more detailed observation of the MDM morphologies could be confirmed by SEM analysis, corroborating the actin-staining results (Figure 1C). Overall, these results show that the different surface modifications of PLLA films affected MDM adhesion and morphology during monocyte-to-macrophage differentiation.
Figure 1.
Quantification and morphology of human monocyte-derived macrophages (MDM) differentiated in PLLA films after 7 days of culture. Cells were differentiated in films without treatment (PLLA), submitted to air-plasma treatment (pPLLA) or with additional coatings, namely collagen I (pPLLA-COLL-l), poly(L-lysine) (pPLLA-PLL), fibronectin (pPLLA-FN), vitronectin (pPLLA-VTN), laminin (pPLLA-LMN), and albumin (pPLLAALB). (A) Quantification of cells nuclei by image analysis after DAPI staining. Results are shown as mean ± SD (*p<0.05 and ****p<0.0001). (B) Actin polymerization by phalloidin fluorescence staining. Cells nuclei are counterstained in blue by DAPI. Scale bars are 50 µm. (C) SEM images evidencing the different morphologies of MDM in contact with the different surfaces of PLLA films. Scale bars are 10 µm.
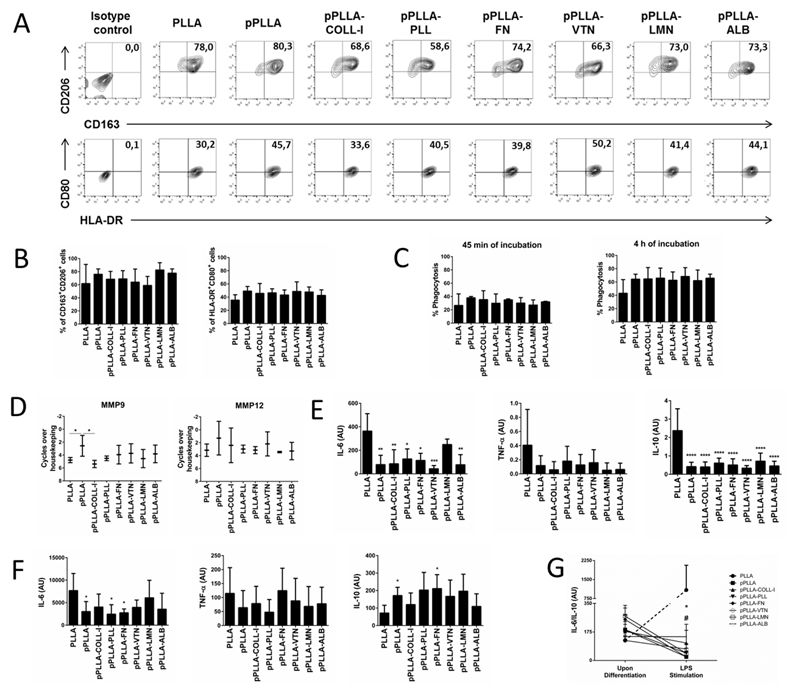
The expression profile of surface markers and cytokine secretion of MDM differentiated in different surface-modified PLLA films was assessed. The phenotypical profile of MDM differentiated in non-modified PLLA films or after surface modification by plasma treatment alone or combined with different coating materials was characterized by the expression of specific cell surface markers by flow cytometry. No major differences were found in the expression of the well-known classically activated (M1) markers HLA-DR+CD80+, when comparing the percentage of positive cells in the several conditions of differentiation (Figure 2A and B). Similar results were observed for the proportion of the alternatively activated (M2) markers CD163+CD206+ (Figure 2A, 2B).41 Moreover, no differences were observed when comparing the mean fluorescence intensity (MFI) of each one of the cell surface markers in MDM differentiated in the diverse PLLA films (Table 1). These results indicate that the different surface modifications performed in PLLA films did not significantly impact the phenotype of Mϕ during the differentiation process.
Figure 2.
(A and B) Cell surface expression of CD163, CD206, HLA-DR, and CD80 in human monocyte-derived macrophages (MDM) differentiated in films without modification (PLLA), modified by plasma treatment (pPLLA), or modified combining plasma treatment with different coating materials, namely collagen I (pPLLA-COLL-I), poly(L-lysine) (pPLLA-PLL), fibronectin (pPLLA-FN), vitronectin (pPLLA-VTN), laminin (pPLLA-LMN), or albumin (pPLLA-ALB). (C) Phagocytic activity of MDM differentiated in each film evaluated by the co-incubation with fluorescent latex-beads for 45 minutes or 4 hours. The percentage of phagocytized beads was quantified by flow cytometry. (D) qPCR quantification of transcriptional levels of MMP9 and MMP12. The nomenclature “cycles over housekeeping” corresponds to the difference between the cycle threshold (CT) of the tested gene and the housekeeping gene. (E) IL-6, TNF-α and IL-10 secretion quantification by ELISA assay. (F) IL-6, TNF-α and IL-10 secretion quantification by ELISA assay after an additional stimulation for 24h with LPS. (G) IL-6/IL-10 ratio before and after LPS stimulation. In (E) and (F), statistical analysis is relative to non-modified films (PLLA condition) and cytokine quantifications are shown as arbitrary units (AU). In (G) the symbol # represents significant differences p<0.005 between PLLA and pPLLA or PLLA and pPLLA-PLL. The **p<0.05 relates to the significant difference between PLLA and all the other tested conditions. Mean ± SD are from at least four different donors (*p<0.05, **p<0.005, ***p<0.001 and ****p<0.0001).
Table 1.
Mean Fluorescence Intensity (MFI) of surface markers CD163, CD206, HLA-DR and CD80 before differentiation (monocytes) or after differentiation in macrophages in the presence of the films without treatment (PLLA), treated with plasma (pPLLA), or treated with plasma combined with treatments with collagen I (pPLLA-COLL-I), poly(L-lysine) (pPLLA-PLL), fibronectin (pPLLA-FN), vitronectin (pPLLA-VTN), laminin (pPLLA-LMN) or albumin (pPLLA-ALB). MFI of macrophages polarized to a M1 or M2 phenotypes were also included. Values are shown as mean ± SEM for at least four different donors.
| Surface Markers |
Monocytes (MFI) |
Macrophages (MFI) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | PLLA | pPLLA | pPLLA-COLL-I | pPLLA-PLL | pPLLA- FN |
pPLLA-VTN | pPLLA-LMN | pPLLA-ALB | |||
| CD163 | Mean SEM |
354.50 33.50 |
281.75 60.01 |
2154.17 178.44 |
1424.25 200.57 |
2099.25 88.26 |
1750.25 144.98 |
1841.25 231.76 |
1653.00 115.90 |
1867.75 173.93 |
2113.00 452.44 |
1586.00 322.43 |
| CD206 | Mean SEM |
127.00 4.00 |
3070.68 524.78 |
2747.84 517.75 |
2985.67 349.61 |
2705.00 365.44 |
2671.33 347.30 |
2761.67 482.65 |
3123.67 469.41 |
2758.67 323.37 |
2462.00 528.95 |
2317.67 342.52 |
| HLA-DR | Mean SEM |
376.00 93.00 |
2843.60 218.51 |
2219.60 131.44 |
2124.00 435.01 |
2588.00 340.11 |
2244.67 307.09 |
1809.67 266.02 |
1993.67 136.94 |
2804.67 350.79 |
2681.00 450.65 |
2386.33 299.60 |
| CD80 | Mean SEM |
8.78 0.96 |
1049.80 108.13 |
191.61 24.73 |
111.40 12.93 |
146.00 11.55 |
139.33 19.91 |
142.67 20.02 |
134.33 10.41 |
150.00 14.15 |
139.00 12.17 |
131.33 16.76 |
In order to dissect potential functional alterations in Mϕ due to the biomaterials composition, we further characterized the effector functions of these cells. The percentage of phagocytosis was evaluated by flow cytometry in two distinct periods of contact with the latex beads. We observed that MDM maintained the same ability to phagocytize, since the percentage of phagocytized beads is similar in all conditions, independently of the period of contact (Figure 2C). We further evaluated the expression of genes involved in ECM organization such as matrix metalloproteinases (MMPs) as this family of enzymes is known to regulate various inflammatory and repair processes.42 In particular, MMP9 is known to induce extracellular matrix remodelling,43 while MMP12 is expressed by pro-healing macrophages.44 We observed that Mϕ differentiated in films modified with plasma (pPLLA) had the highest expression of MMP9, with a significant difference when compared with those differentiated in non-modified PLLA films and with collagen I-coated films (pPLLA-COLL-I). No significant alterations between the expression levels of MMP12 were observed (Figure 2D). This result in part reveals the specificity of surface modifications on films in modulating macrophage MMP’s expression, confirming previous studies that demonstrate biomaterial-dependent effect on MMPs.45,46 Although significance was restricted to pPLLA, the increase in MMP9 expression can reflect an effect of plasma treatment alone to predispose MDM towards alternative activation given that the production and activity of MMP9 was described higher in M2 macrophages.47 Moreover, MMP9 was shown to be required for tissue remodeling in distinct settings,48–50 acting also as a strong angiogenic factor.43 Our data suggests that the distinct coating materials may hinder the transcription upregulation conferred by plasma treatment alone. Future studies will shed light on correlation between increasing MMP9 transcript levels and surface modifications upon plasma treatment.
It is well described in the literature the ability of Mϕ to secrete a wide array of inflammatory mediators in response to external signals such as cytokines.9,21,22,51,52 We determined the cytokine profile of MDM during differentiation when cultured in contact with different surface modified PLLA films. The secretions of tumor necrosis factor (TNF)-α, and interleukin (IL)-6 and -10 were analyzed. TNF-α and IL-6 are pro-inflammatory cytokines commonly analyzed when studying the inflammatory response induced by biomaterials.9,53 IL-10 is one of the most studied anti-inflammatory cytokine, and is crucial in restraining inflammation.54 Results show that MDM differentiated in almost all surface-modified films produced significantly lower amounts of pro-inflammatory cytokines IL-6 but also of anti-inflammatory cytokine IL-10, when compared to non-modified PLLA films. The secretion profile of TNF-α followed the same trend, although without statistical significance. Exceptionally, no significant differences were found in the secretion profile of IL-6 secretion when comparing laminin-modified films (pPLLA-LMN) with non-modified films. These results evidenced the tailoring role of surface modified films, leading to lower secretions of pro- and anti-inflammatory cytokines compared to non-modified films (Figure 2E).
To further understand if the differentiated response observed using different surface modified PLLA films would be maintained after stimulation with external factors, MDM were treated with LPS, a well-known pro-inflammatory stimulus. Subsequently, the secretions of TNF-α, IL-6, and IL-10 were again quantified.
Our results suggest that MDM stimulated in several surface modified films showed a lower IL-6 secretion when compared with non-modified films (Figure 2F). In particular, pPLLA, pPLLA-PLL, and pPLLA-FN films showed a significant lower IL-6 secretion when compared with non-modified films. Remarkably, after the pro-inflammatory LPS stimulus, the IL-10 secretion in all surface-modified films was higher compared to non-modified films. These findings were statistically significant for pPLLA and pPLLA-FN films. Again, no significant differences could be observed for TNF-α secretion.
The IL-6/IL-10 ratio has been used in distinct experimental settings as a valuable prognosticator of pro- versus anti-inflammatory balance.55–58 Increased IL-6/IL-10 ratio was significantly associated in favor of pro-inflammatory responses even in cases with similar TNF-α and IL-1β levels.55,58 Remarkably, while the IL-6/IL-10 ratio (Figure 2G) remained similar upon Mϕ differentiation in all the films tested, a striking decrease could be observed upon LPS stimulus in all surface-modified films. This was in clear contrast with Mϕ differentiated in non-modified films, in which a 9-fold increase of IL-6/IL-10 ratio was observed. Further experimental evidences support the anti-inflammatory potential of MDM differentiated in surface-modified films. After evaluation of MFI levels of classical and alternative markers, recent studies demonstrate that M2 macrophages present high surface levels of CD163, while maintaining the CD80 expression low. An opposite trend was observed for M1 macrophages.41,59 Accordingly, Mϕ differentiated in all surface-modified films presented a phenotype closer to M2 macrophages in terms of CD163 and CD 80 surface MFI, although in any significant differences were found for all the films tested (Table 1). In agreement with our data, previous studies studying macrophage-biomaterials interaction have relied on the release of the immunoregulatory cytokine IL-10 and CD163 expression, to specifically characterize an M2 and anti-inflammatory polarization.60–62 Overall, the study of cytokines and surface markers profile in the present work suggests that all surface-modified films contributed to a lower inflammatory response of MDM in comparison to non-modified PLLA films. This ability was particularly distinct after the pro-inflammatory LPS stimulus. These data provides important insights about the interaction of MDM with different surface-modified PLLA films. Most importantly, this study also reinforced the idea that Mϕ phenotype occupies a continuum between M1 and M2 designations, thus expressing and releasing both pro- and anti-inflammatory markers and cytokines, as it has been suggested in other studies.9,37,63,64
Therefore, more than classifying monocytes/macrophages populations in contact with biomaterials in a “bipolar classification”, the well-known M1/M2 paradigm, our focus was mainly to understand which functional responses of MDM in their differentiation were being stimulated by their interaction with different surface-modified PLLA films. The induction of “functional” phenotypes of Mϕ, which do not fit in the conventional M1 or M2 phenotype classifications, has been reported in different sites of the fibrotic capsules elicited by the implantation of biomaterials with different pore sizes.15 One may speculate that such intermediate Mϕ phenotypes may be valuable to induce a balanced regeneration of tissues, avoiding the triggering of excessive implant rejection associated with M1 phenotype or excessive angiogenesis promoted by M2 phenotype. These results reinforce the need to study the role of these simultaneously pro- and anti-inflammatory Mϕ in the healing process.
The study presented herein rendered valuable conclusions about the modulation effect of different surface-modified PLLA films on the differentiation of MDM. Most importantly, plasma-modification alone or combined with various coating materials showed to be able to restrain the pro-inflammatory character of non-modified films. Additionally, although all coating materials showed anti-inflammatory potential, they also did not elicit the secretion of pro-inflammatory cytokines. This highlights the potential of such surface modifications to be used as pro-healing biomaterials. The surface of biomaterials is commonly modified with ECM-derived proteins to improve not only cell adhesion but also to function as biomimetic substrates. For example, the use of collagen I has been reported for bone regeneration strategies,65 and laminin for neural tissue engineering66 or combined with vitronectin for human pluripotent stem cells expansion.67 The integration of the output of these studies with the data presented herein suggests that the proposed surface modifications of PLLA films are amenable to be used as biomimetic coatings, while avoiding an excessive pro-inflammatory response of MDM. Therefore, it may be a promising strategy to extrapolate such surface modifications to other biomaterials, seeking the improvement of their biotolerability.
The findings obtained within this work contributed to increase the knowledge about the interaction of human Mϕ with the widely used PLLA biomaterial, and also how different surface modifications can affect this dynamic interaction.
Conclusions
Insights into macrophage-biomaterial biology and an improved understanding of other components of the immune system are required to establish a set of design principles that aid in the engineering of a new generation of immuno-informed biomaterials that can actively direct the innate immune system. With this in mind, we demonstrated how human MDM interact with PLLA films presenting different surface modifications. Overall, surface-modified PLLA films disrupt the balance of macrophage polarization towards a favorable anti-inflammatory profile, particularly when facing an inflammatory stimulus. These results clearly demonstrate the complexity of the interplay of biomaterials and the immune response. This urge for the performance of more studies addressing this dynamic interaction, which are expected to provide insight in the selection and design of biomaterials based on how their chemistry, surface properties, geometry, and other features will impact on the know-how about immunological system response. The “ideal” response and cytokine environment remains uncertain, and more complex studies are required to mimic more closely an implantation scenario (e.g. different immune cells combined with other cell types involved in the healing process).
Supplementary Material
Electronic Supplementary Information (ESI) available: Figure SI1 - Histogram of CD14+ monocytes after immunomagnetic isolation, Figure SI 2 - Gating strategy for the evaluation of the surface markers for macrophages’ characterization, and Figure SI 3 - Live-dead assay. See DOI: 10.1039/x0xx00000x
Representative histogram of CD14+ monocytes after immunomagnetic isolation. The black line represents isotype control, whereas the red line represents CD14+ cells in the isolated sample.
Gating strategy for the evaluation of the surface markers for macrophages’ characterization. (A) Cell population was depicted by FSC and SSC analysis, in which (B) doublets were excluded using FSC-A and FSC-H. (C) Macrophages were selected by CD14+CD16high expression, and within this population (D) the M1 markers HLA-DR and CD80, and (E) the M2 markers CD163 and CD206 were analyzed.
Viability of human macrophages differentiated in PLLA membranes by live-dead assay after 7 days of culture. Cells were differentiated in membranes without treatment (PLLA), modified with plasma treatment (pPLLA), or modified by combining plasma treatment with multiple coating materials, namely collagen I (pPLLA-COLL I), poly(L-lysine) (pPLLA-PLL), fibronectin (pPLLA-FN), vitronectin (pPLLA-VTN), laminin (pPLLA-LMN), or albumin (pPLLA-ALB). Living cells are marked by green fluorescence (calcein-AM) and dead cells in red (propidium iodide). Scale bars are 200 µm.
Acknowledgements
This work was developed under the scope of the project NORTE-01-0145-FEDER-000023, supported by the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement through the European Regional Development Fund (FEDER). C.R. Correia and J.F. Mano acknowledge the funding from European Research Council for project ATLAS with the grant agreement number ERC-2014-ADG-669858. J. Gaifem, M.B. Oliveira and R. Silvestre acknowledge the Portuguese Foundation for Science and Technology (FCT) for the doctoral (PD/BD/106053/2015), post-doctoral (SFRH/BPD/111354/2015) and FCT Investigator (IF/00021/2014) grants, respectively. The authors also acknowledge Hospital de Braga for providing the buffy coats.
Notes and references
- 1.Tate MLK, Detamore M, Capadona JR, Woolley A, Knothe U. Biomaterials. 2016;95:35. doi: 10.1016/j.biomaterials.2016.03.038. [DOI] [PubMed] [Google Scholar]
- 2.Ratner BD. Journal of Cardiovascular Translational Research. 2011;4:523. doi: 10.1007/s12265-011-9287-x. [DOI] [PubMed] [Google Scholar]
- 3.Brown BN, Badylak SF. Acta Biomaterialia. 2013;9:4948. doi: 10.1016/j.actbio.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 4.Lutolf MP, Hubbel JA. Nature Biotechnology. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 5.Eming SA, Krieg T, Davidson JM. Journal of Investigative Dermatology. 2007;127:514. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 6.Landgraeber S, Jager M, Jacobs JJ, Hallab NJ. Mediators of Inflammation. 2014;2014 doi: 10.1155/2014/185150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown BN, Ratner BD, Goodman SB, Amar S, Badylak SF. Biomaterials. 2012;33:3792. doi: 10.1016/j.biomaterials.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mountziaris PM, Spicer PP, Kasper FK, Mikos AG. Tissue Engineering Part B. 2011;17:393. doi: 10.1089/ten.teb.2011.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida CR, Serra T, Oliveira MI, Planell JA, Barbosa MA, Navarro M. Acta Biomaterialia. 2014;10:613. doi: 10.1016/j.actbio.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 10.Italiani P, Boraschi D. Frontiers in Immunology. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eming SA, Hammerschmidt M, Krieg T, Roers A. Semininars in Cell and Developmental Biology. 2009;20:517. doi: 10.1016/j.semcdb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Anderson JM, Rodriguez A, Chang DT. Seminars in Immunology. 2008;20:86. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper RL, Segal RA, Diegelmann RF, Reynolds AM. Journal of Theoretical Biology. 2015;367:86. doi: 10.1016/j.jtbi.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Sridharan R, Cameron AR, Kelly DJ, Kearney CJ, O’Brien FJ. Materials Today. 2015;18:313. [Google Scholar]
- 15.Sussman EM, Halpin MC, Muster J, Moon RT, Ratner BD. Annals of Biomedical Engineering. 2014;42:1508. doi: 10.1007/s10439-013-0933-0. [DOI] [PubMed] [Google Scholar]
- 16.Brown BN, Valentin JE, Stewart-Akers AM, McCabe GP, Badylak SF. Biomaterials. 2009;30:1482. doi: 10.1016/j.biomaterials.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau SK, Chu PG, Weiss LM. American Journal of Clinical Pathology. 2004;122:794. doi: 10.1309/QHD6-YFN8-1KQX-UUH6. [DOI] [PubMed] [Google Scholar]
- 18.Wójciak-Stothard B, Curtis A, Monaghan W, Macdonald K, Wilkinson C. Experimental Cell Research. 1996;223:426. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs AK, Syrovets T, Haas KA, Loos C, Musyanovych A, Mailänder A, Landfester VK, Simmet T. Biomaterials. 2016;85:78. doi: 10.1016/j.biomaterials.2016.01.064. [DOI] [PubMed] [Google Scholar]
- 20.Garg K, Pullen NA, Oskeritzian CA, Ryan JJ, Bowlin GL. Biomaterials. 2013;34:4439. doi: 10.1016/j.biomaterials.2013.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf MT, Dearth CL, Ranallo CA, LoPresti ST, Carey LE, Daly KA, Brown BN, Badylak SF. Biomaterials. 2014;35:6838. doi: 10.1016/j.biomaterials.2014.04.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maciel J, Oliveira MI, Goncalves RM, Barbosa MA. Acta Biomaterialia. 2012;8:3669. doi: 10.1016/j.actbio.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Boersema GS, Grotenhuis N, Bayon Y, Lange JF, Bastiaansen-Jenniskens YM. Bioresearch Open Access. 2016;5:6. doi: 10.1089/biores.2015.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao YP, Cui FZ. Biomedical Materials. 2007;2:R24. doi: 10.1088/1748-6041/2/4/R02. [DOI] [PubMed] [Google Scholar]
- 25.Tjia J, Aneskievich B, Moghe P. Biomaterials. 1999;20:2223. doi: 10.1016/s0142-9612(99)00153-2. [DOI] [PubMed] [Google Scholar]
- 26.Antunes JC, Oliveira JM, Reis RL, Soria JM, Gómez-Ribelles JL, Mano JF. Journal of Biomedical Materials Research Part A. 2010;94A:856. doi: 10.1002/jbm.a.32753. [DOI] [PubMed] [Google Scholar]
- 27.Thakur S, Neogi S. Reviews of Adhesion and Adhesives. 2015;3:53. [Google Scholar]
- 28.Cheng Q, Lee BL, Komvopoulos K, Yan Z, Li S. Tissue Engineering Part A. 2013;19:1188. doi: 10.1089/ten.tea.2011.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song W, Veiga DD, Custódio CA, Mano JF. Advanced Materials. 2009;21:1830. [Google Scholar]
- 30.Saino E, Focarete ML, Gualandi C, Emanuele E, Cornaglia AI, Imbriani M, Visai L. Biomacromolecules. 2011;12:1900. doi: 10.1021/bm200248h. [DOI] [PubMed] [Google Scholar]
- 31.Pan H, Jiang H, Kantharia S, Chen W. Biomedical Materials. 2011;6 doi: 10.1088/1748-6041/6/6/065002. [DOI] [PubMed] [Google Scholar]
- 32.Correia CR, Reis RL, Mano JF. Biomacromolecules. 2012;14:743. doi: 10.1021/bm301833z. [DOI] [PubMed] [Google Scholar]
- 33.Adutler-Lieber S, Zaretsky I, Platzman I, Deeg J, Friedman N, Spatz JP, Geiger B. Journal of Autoimmunity. 2014;54:100. doi: 10.1016/j.jaut.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Discher DE, Janmey P, Wang Y-L. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 35.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Bei J, Wang S. Biomaterials. 2002;23:2607. doi: 10.1016/s0142-9612(01)00400-8. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Klein T, Murray RZ, Crawford R, Chang J, Wu C, Xiao Y. Materials Today. 2016;19:304. [Google Scholar]
- 38.Jenney CR, Anderson JM. Journal of Biomedical Materials Research. 2000;49:435. doi: 10.1002/(sici)1097-4636(20000315)49:4<435::aid-jbm2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 39.Turner NJ, Yates AJ, Jr, Weber DJ, Qureshi IR, Stolz DB, Gilbert TW, Badylak SF. Tissue Engineering Part A. 2010;16:3309. doi: 10.1089/ten.TEA.2010.0169. [DOI] [PubMed] [Google Scholar]
- 40.Kajahn J, Franz S, Rueckert E, Forstreuter I, Hintze V, Moeller S, Simon JC. Biomatter. 2012;2:226. doi: 10.4161/biom.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rőszer T. Mediators of Inflammation. 2015 doi: 10.1155/2015/816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parks WC, Wilson CL, Lopez-Boado YS. Nature Reviews Immunology. 2004;4:617. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 43.Jadhav U, Chigurupati S, Lakka SS, Mohanam S. International Journal of Oncology. 2004;25:1407. [PubMed] [Google Scholar]
- 44.Lee JT, Pamir N, Liu N-C, Kirk EA, Averill MM, Becker L, Larson I, Hagman DK, Foster-Schubert KE, van Yserloo B, Bornfeldt KE, et al. Endocrinology. 2014;155:3409. doi: 10.1210/en.2014-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones JA, McNally AK, Chang DT, Qin LA, Meyerson H, Colton E, Kwon ILK, Matsuda T, Anderson JM. Journal of Biomedical Materials Research Part A. 2008;84A:158. doi: 10.1002/jbm.a.31220. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira MI, Santos SG, Oliveira MJ, Torres AL, Barbosa MA. European Cells and Materials. 2012;24:136. doi: 10.22203/ecm.v024a10. [DOI] [PubMed] [Google Scholar]
- 47.Lolmede K, Campana L, Vezzoli M, Bosurgi L, Tonlorenzi R, Clementi E, Bianchi ME, Cossu G, Manfredi AA, Brunelli S, Rovere-Querini P. Journal of Leukocyte Biology. 2009;85:779. doi: 10.1189/jlb.0908579. [DOI] [PubMed] [Google Scholar]
- 48.Zambuzzi WF, Paiva KBS, Menezes R, Oliveira RC, Taga R, Granjeiro JM. Journal of Molecular Histology. 2009;40:301. doi: 10.1007/s10735-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 49.Delaissé J-M, Engsig MT, Everts V, Ovejero MC, Ferreras M, Lund L, Vu TH, Werb Z, Winding B, Lochter A, Karsdal MA, et al. Clinica Chimica Acta. 2000;291:223. doi: 10.1016/s0009-8981(99)00230-2. [DOI] [PubMed] [Google Scholar]
- 50.Corotti MV, Zambuzzi WF, Paiva KBS, Menezes R, Pinto LC, Lara VS, Granjeiro JM. Archives of Oral Biology. 2009;54:764. doi: 10.1016/j.archoralbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Reeves AR, Spiller KL, Freytes DO, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2015;73:272. doi: 10.1016/j.biomaterials.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiller KL, Nassiri S, Witherel CE, Anfang RR, Ng J, Nakazawa KR, Yu T, Vunjak-Novakovic G. Biomaterials. 2015;37:194. doi: 10.1016/j.biomaterials.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hunter CA, Jones SA. Nature Immunology. 2015;16:448. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 54.Banchereau J, Pascual V, O’Garra A. Nature Immunology. 2012;13:925. doi: 10.1038/ni.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Day NPJ, hien TT, Schollaardt T, Loc PP, Chuong LV, Chau TTH, Mai NTH, Phu NH, Sinh DX, White NJ, Ho M. The Journal of Infectious Diseases. 1999;180:1288. doi: 10.1086/315016. [DOI] [PubMed] [Google Scholar]
- 56.Jerin A, Požar-Lukanovič N, Sojar V, Stanisavljevič D, Paver-Eržen V, Osredkar J. Clinical Chemistry and Laboratory Medicine. 2005;41:899. doi: 10.1515/CCLM.2003.136. [DOI] [PubMed] [Google Scholar]
- 57.Sapan HB, Paturusi I, Jusuf I, Patellongi I, Massi MN, Pusponegoro AD, Arief SK, Labeda I, Islam AA, Rendy L, Hatta M. International Journal of Burns and Trauma. 2016;6:37. [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz S, Vardon-Bounes F, Merlet-Dupuy V, Conil J-M, Buléon M, Fourcade O, Tack I, Minville V. Intensive Care Medicine Experimental. 2016;4:22. doi: 10.1186/s40635-016-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rey-Giraud F, Hafner M, Ries CH. Plos One. 2012 doi: 10.1371/journal.pone.0042656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kajahn J, Franz S, Rueckert E, Forstreuter I, Hintze V, Moeller S, Simon JC. Biomatter. 2012;2:226. doi: 10.4161/biom.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rostam HM, Singh S, Salazar F, Magennis P, Hook A, Singh T, Vrana NE, Alexander MR, Ghaemmaghami AM. Immunobiology. 2016;221:1237. doi: 10.1016/j.imbio.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 62.Fuchs A-K, Syrovets T, Haas KA, Loos C, Musyanovych A, Mailänder V, Landfester K, Simmet T. Biomaterials. 2016;85:78. doi: 10.1016/j.biomaterials.2016.01.064. [DOI] [PubMed] [Google Scholar]
- 63.Martinez FO, Gordon S. F1000Prime Reports. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen JS, Fuentes-Duculan J, Suárez-Fariñas M, Pierson KC, Pitts-Kiefer A, Fan L, Belkin LDA, Wang CQF, Bhuvanendran S, Johnson-Huang LM, Bluth MJ, et al. Journal of Investigative Dermatology. 2011;131:1322. doi: 10.103/jid.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Correia CR, Pirraco RP, Cerqueira MT, Marques AP, Reis RL, Mano JF. Scientific Reports. 2016;6:21883. doi: 10.1038/srep21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu BF, Ma J, Xu QY, Cui FZ. Colloids and Surfaces B: Biointerfaces. 2006;53:175. doi: 10.1016/j.colsurfb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 67.Lam AT, Li J, Chen AK, Reuveny S, Oh SK, Birch WR. Stem Cells and Development. 2014;23:1688. doi: 10.1089/scd.2013.0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative histogram of CD14+ monocytes after immunomagnetic isolation. The black line represents isotype control, whereas the red line represents CD14+ cells in the isolated sample.
Gating strategy for the evaluation of the surface markers for macrophages’ characterization. (A) Cell population was depicted by FSC and SSC analysis, in which (B) doublets were excluded using FSC-A and FSC-H. (C) Macrophages were selected by CD14+CD16high expression, and within this population (D) the M1 markers HLA-DR and CD80, and (E) the M2 markers CD163 and CD206 were analyzed.
Viability of human macrophages differentiated in PLLA membranes by live-dead assay after 7 days of culture. Cells were differentiated in membranes without treatment (PLLA), modified with plasma treatment (pPLLA), or modified by combining plasma treatment with multiple coating materials, namely collagen I (pPLLA-COLL I), poly(L-lysine) (pPLLA-PLL), fibronectin (pPLLA-FN), vitronectin (pPLLA-VTN), laminin (pPLLA-LMN), or albumin (pPLLA-ALB). Living cells are marked by green fluorescence (calcein-AM) and dead cells in red (propidium iodide). Scale bars are 200 µm.