Abstract
The classical model of cytosine DNA methylation (5mC) regulation depicts this covalent modification as a stable repressive regulator of promoter activity. However, whole genome analysis of 5mC reveals more widespread tissue and cell type-specific patterns, and pervasive dynamics during mammalian development. Here, we review recent findings that delineate 5mC functions in developmental stages and diverse genomic compartments as well as discuss the molecular mechanisms that connects transcriptional regulation and 5mC. Beyond the newly appreciated dynamics, regulatory roles for 5mC have been suggested in new biological contexts such as learning and memory or aging. Utilizing new single-cell measurement techniques and precise editing tools will enable functional analyses of 5mC in gene expression, clarifying its role in various biological processes.
Introduction
DNA methylation (the presence of 5-methylcytosine, 5mC) is one of the best studied epigenetic systems in mammals with well-characterized mechanisms to write, read and perpetuate this covalent epigenetic mark (Fig. 1–2) (1, 2). Gene regulation by 5mC was initially studied by analyzing CpG islands (CGIs), which are stretches of DNA sequence enriched in CpG dinucleotides found at the majority of mammalian promoters (2). The presence of 5mC at CGI is usually associated with long term, stable gene repression (2). However, the pattern of 5mC outside of promoters and during dynamic biological processes was less understood prior to the development of epigenomic tools. Whole-genome 5mC profiling has uncovered dynamics of 5mC at enhancers, gene bodies and extended transcriptionally inactive partially methylated domains (PMDs), implicating regulatory roles of 5mC in development and disease (3–5). Although 5mC is broadly accepted as a repressive epigenetic mark, the mechanism of 5mC mediated transcription repression is still not fully understood (1, 2). Several studies suggest that gene regulation by 5mC may involve functional interaction between 5mC and sequence-specific transcription factors (TFs) (6–9). Here we review recent insights regarding the dynamics of 5mC in development and disease, discuss the utility of single-cell methods and new epigenome editing technologies for further analysis of 5mC function and discovery of additional DNA base modifications in mammalian genomes.
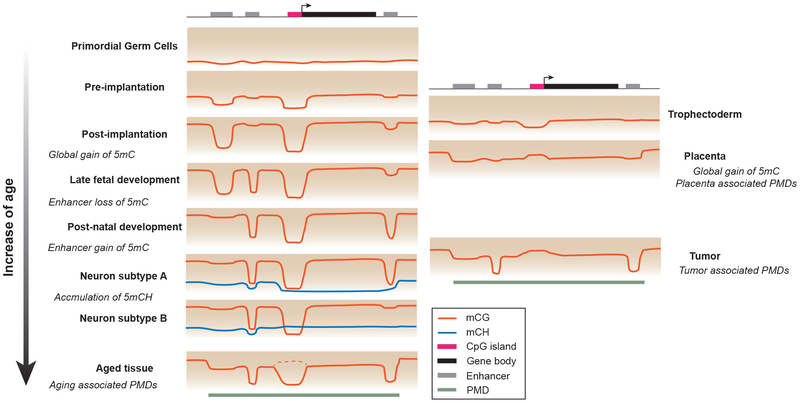
Figure 1.
DNA methylation signatures of developmental stages and tumors - primordial germ cells and preimplantation: low global 5mC level; postimplantation: global gain of 5mC. late fetal development: local demethylation at embryonically active regulatory sequences; postnatal development: gain of 5mC at embryonically active regulatory sequences, loss of 5mC at regulatory sequences active in differentiated tissues. neuron subtypes A and B: brain specific accumulation of mCH, which abundance is inversely correlated with transcription activity; aged tissues: PMDs in late-replication regions resulted from progressive methylation loss. Dashed line indicates a small percentage of CGI shows hyper-methylation in aged tissues (67). trophectoderm: low global 5mC level similar to preimplantation embryo; placenta and tumors: pervasive PMDs and CGI hypermethylation. The color gradient in the background indicates the increase of 5mC level from bottom to top of each panel.
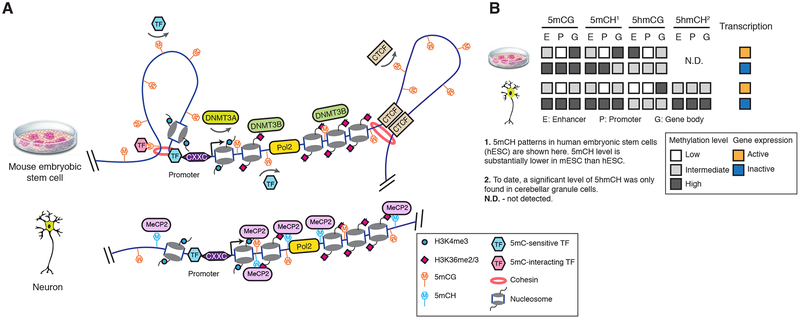
Figure 2.
The function and regulation of DNA methylation in diverse genomic contexts. (A) Gene promoter CpG islands are protected from 5mC by a CXXC domain containing proteins. De novo DNA methyltransferase DNMT3A activity is inhibited by H3K4me3. At distal regulatory elements, 5mC can either prohibit the binding of 5mC-sensitive TFs, or interact with TFs with preference for methylated binding sequence. DNA methylation at CTCF binding sites regulates chromatin conformation by modulating CTCF binding. In mouse embryonic stem cells (mESCs, top), DNMT3B is recruited to transcribed regions by H3K36me and establishes gene body methylation. In post-mitotic neurons (bottom), 5mCH is bound by MeCP2. (B) Transcription outputs are correlated with 5mC and 5hmC levels of enhancers, promoters and gene bodies.
Dynamic DNA methylation in development
In mammalian genome, 5mC is predominantly located within the CpG dinucleotide context. In most somatic tissues, the majority (>80%) of CpGs in the genome are methylated, with the exception of CGIs, and other gene regulatory sequences that show reduced 5mC levels (3). In fact, local 5mC depletion is a reliable signature of promoters and enhancers: CpG sites in promoter associated CGIs are often less than 10% methylated, while distal regulatory sequences such as enhancers are commonly marked by levels of 5mC ranging from 10% to 50% (6). CGI methylation shows few dynamics across healthy tissues. In contrast, aberrant CGI hypermethylation is commonly observed in cancers (2). With respect to methylation patterns outside CGIs, direct comparison of whole genome methylome profiles derived from numerous cell types has revealed that under normal physiological conditions 15%-21% of CpGs vary in methylation status between human tissues (4, 5). These dynamically methylated CpGs cluster into over 1 million tissue specific differentially methylated regions (DMRs). DMRs are predominantly distal to transcriptional start sites and overlap with enhancers. Intragenic DMRs that stretch for kilobases (large-DMRs) can overlap with tissue defining “signature genes” as well as with predicted super-enhancers, which are clusters of enhancers that function cooperatively (4, 10, 11). These tissue-type specific DMRs are defined by both lineage specific gain and loss of 5mC during embryogenesis. A study profiling 5mC dynamics during mouse late fetal development with daily sampling (12) revealed a temporal uncoupling of the loss and gain of 5mC during the development of all tissues analyzed: enhancers predominantly lose 5mC during fetal development whereas most de novo 5mC occurs after birth (Fig. 1) (12). In mouse brain cortex, synchronized de novo 5mC is observed during the second postnatal week, primarily targeting enhancers that are embryonically active (12). Although few analyses have been systematically performed across the lifespan of mammals, existing data suggests that, overall the methylome is stable beyond adolescence (13).
Contrary to what is observed in somatic cells, genome-wide changes of DNA methylation patterns occur during preimplantation mammalian development and in the early embryonic germ line (Fig. 1) (14–17). However, these global methylation changes are coordinated with genome-wide changes in histone modifications and reflect a global reprogramming of epigenetic information rather than locus specific transcriptional regulation. For example, global erasure of DNA methylation in primordial germ cells (PGCs) does not lead to widespread transcriptional activation (16, 17) and numerous genes are expressed during spermatogenesis despite high level methylation at their promoters (18).
In summary, 5mC patterns in differentiated tissues are highly specific and established primarily by site specific remodeling at regulatory sequences. Sequential waves of gain and loss of 5mC in late-fetal to early postnatal development contribute to the majority of 5mC diversity found in differentiated tissues.
Transcription factors direct local 5mC remodelling
The landscape of 5mC in mammalian genome is shaped by local depletion of 5mC at regulatory sequences (Fig. 2). The depletion of 5mC at CGIs is, in part, determined by the presence of CpG dinucleotide clusters. that are specifically recognized and protected from methylation by proteins containing a CXXC zinc finger domain (19). These proteins include components of the H3K4 methyltransferase complex (19). Promoter H3K4me3, in turn, inhibits the activity of the de novo DNA methyltransferase DNMT3a thus preventing methylation at CGIs (20). Beyond CXXC containing proteins, the depletion of 5mC at CGIs is also dependent on the binding of other specific TFs (19).
Unlike CGI promoters, enhancers have a comparable CpG density to the genomic average. Tissue or cell type-specific regulatory sequences can be effectively identified by DMRs and are enriched in recognition sites for TFs with tissue specific functions (4, 5, 21). We speculate that the large quantity of DMRs (20,000 – 100,000 per tissue type) reflects the collective local demethylation induced by specific TF binding. This model has been supported by experiments in mouse embryonic stem cell (mESC) using insulator protein CCCTC-binding factor (CTCF) and a transcriptional repressor of neural genes - RE1-Silencing Transcription factor (REST) (6). The presence of an intact CTCF motif is sufficient and necessary for local methylation depletion. The deletion of REST led to increased 5mC at lowly methylated regions normally bound by REST. Notably, such gain of 5mC can be reversed by re-introducing REST.
It has been postulated that ‘pioneer’ TFs, which can direct cellular reprogramming, are likely to induce local epigenetic remodeling (22). One example is ASCL1 whose forced expression directly converts fibroblasts to post-mitotic neurons and leads to reduced 5mC at most of its binding sites (22, 23). However, given the small number of TFs tested to date, it is unknown whether the ability to remodel 5mC is indeed restricted to TFs with ‘pioneer’ activity. In addition, the mechanism of TF induced local 5mC loss is not understood. TFs may directly recruit cytosine demethylation machinery containing Ten Eleven Translocation methylcytosine dioxygenase (TET – see below) (24), or alternatively involve intermediate steps of chromatin remodeling or histone modification.
Transcription factors read DNA methylation
The depletion of 5mC at active regulatory elements suggests that the absence of this repressive mark is essential for active transcription regulation. However, our understanding of the mechanism of how 5mC impacts transcription machinery remains incomplete. 5mC readers including proteins containing a methyl-CpG-binding domain such as MECP2 or MBD proteins can recognize 5mC and recruit co-regulators to methylated DNA (25). In addition, studies suggest that numerous transcription factors (TFs) can also directly sense the state of cytosine methylation through the modulation of their binding affinity and specificity (26).
The development of high-throughput assays for TF-DNA interaction has identified many transcription factors with binding specificity altered by the presence of 5mC in recognition sequences (26). A protein microarray study that interrogated more than 1,500 TFs found over 40 TFs that are able to bind methylated motifs (27). Pull down experiments of methylated and unmethylated random sequences using a recombinant TF protein followed by sequencing (methyl-SELEX) concluded that the binding of approximately 60% of TFs was influenced by 5mC, with the binding of comparable numbers of TFs enhanced or inhibited by 5mC (28). These studies unexpectedly uncovered that certain interactions between TFs and low affinity binding sites can be enhanced by the presence of 5mC. Similarly, DNA affinity purification sequencing (DAP-seq) of over 500 Arabidopsis (plant) TFs determined that >75% of TFs are sensitive to 5mC (29), suggesting the regulation of TF binding by 5mC is conserved across animal and plant kingdoms.
Although in vitro studies reveal that the majority of TF-DNA interaction can be modulated by 5mC, it is still uncertain how broadly this mechanism contributes to in vivo gene regulatory programs. There are only a handful of reports of 5mC regulating TF binding in a physiological context. In one well characterized example, aberrant gain of 5mC at the binding site of insulator protein CCCTC-binding factor (CTCF) is observed in the Igf2/H19 locus in Beckwith-Wiedemann (fetal overgrowth) syndrome, and in the PDGFRA locus of IDH mutant gliomas (8, 9). In both cases, hypermethylation of the binding site prevents CTCF binding and results in the disruption of chromatin topological domains leading to ectopic enhancer-promoter interactions. Intriguingly, 5mC only affects CTCF binding at a specific subset of recognition sequences (30). This selective 5mC sensitivity can be explained by the different biophysical property of CTCF’s 11 zinc fingers leading to its sensitivity to specific cytosine within its binding motif being methylated (31).
We speculate that the in vivo regulation of TF binding by 5mC is more common than what has been reported in the literature. However, such mechanisms may only affect TF function at a subset of sites showing developmental or disease related 5mC dynamics and thus might have been previously overlooked. In addition, the bi-directional regulation between TF binding and 5mC presence poses a challenge in determining whether TF binding is the cause or consequence of 5mC remodeling. We anticipate that new epigenetic editing tools for specific manipulation of 5mC (see below) may significantly facilitate functional studies of TF-5mC interactions.
Intragenic DNA methylation and transcription
Compared to its well-established repressive function at regulatory elements, less is known about 5mC regulation and function(s) at intragenic regions (Fig. 2). Studies with mESC and a heterologous yeast system found that the establishment of intragenic 5mC is a controlled process carried out by de novo DNA methyltransferase DNMT3B and co-transcriptionally regulated histone modification H3K36me (32, 33). Specifically, DNMT3B preferentially binds to H3K36me and deposits 5mC within gene bodies.
Although promoter 5mC shows a negative correlation with transcription, early studies of intragenic 5mC function posited the opposite finding (2). Indeed, for certain tissue types, active genes are more methylated than repressed genes. However, this correlation appears to be tissue specific. The interaction between DNMT3B and H3K36me leads to greater levels of intragenic 5mC for active genes in mESC (32). For some tissue types, the positive correlation between 5mC and transcription can be explained by the stratification of genes by chromatin states. At many developmental genes enriched in H3K27me3, intragenic 5mC antagonize polycomb repression and thus is positively correlated with transcription (34). In PMD associated tissues such cultured fibroblasts, genes located in PMDs are both lowly expressed and methylated (3). Many signature genes of human tissues overlap with intragenic large-DMRs, suggesting that intragenic 5mC may regulate tissue specific gene expression (4). In addition, intragenic 5mC has also been postulated to regulate alternative promoter usage, alternative splicing and prevent spurious internal transcription initiation (2, 35, 36).
Human embryonic stem cells (hESC) and mammalian brain tissues contain abundant non-CG methylation (5mCH, where H equals A, C or T) although the underlying three-base DNA sequence context (5mCAG vs. 5mCAC, respectively) is distinct (3, 13). Intragenic 5mCH is positively correlated with gene expression in hESC, but is negatively correlated with gene expression in the brain (13). In neurons, intragenic 5mCH quantitatively modulates gene expression by interacting with MECP2 and recruiting NCoR corepressor complex to chromatin (37–40). MECP2 is a methyl-CpG binding protein and mutations in MECP2 have been linked to Rett syndrome - an autism spectrum disorder (41). Independent studies have reported that MECP2 binds to both 5mCA and 5mCG sites with high affinity (37, 39, 40). CA sites are the most frequently methylated non-CG sites (13). MECP2 and 5mCA dependent repression is particularly critical for long genes (with a length greater than 100kb), many of which have neuronal functions (39).
In summary, intragenic 5mC (including 5mCH) performs fine-tuning and supportive roles in transcription regulation but is nevertheless functionally important as suggested by the disease links to intragenic 5mCA and MECP2 repression (39).
Functions of cytosine hydroxymethylation
The dynamic changes in 5mC pattern have raised the question regarding the possible mechanisms of 5mC removal. In this context, 5-hydroxymethylcytosine (5hmC) is the most abundant oxidative derivative of 5mC. 5hmC is generated by the Ten Eleven Translocation methylcytosine dioxygenase (TET) family of proteins and is an intermediate product of 5mC demethylation mediated by TET in combination with thymine DNA glycosylase (TDG)-mediated base excision repair (24). Although TET enzymes are highly expressed during the developmental periods of global DNA demethylation, their role in the observed methylation dynamics seems auxiliary and primarily antagonizes the activity of de novo methylation (17, 42). However, TETs are likely to play important roles in local 5mC dynamics at the regulatory sequences as TET binding is enriched in promoters and enhancers and found in complexes with transcription factors suggesting involvement in 5mC dynamics following TF binding (24).
Studies also suggest that the majority of 5hmC can be relatively stable arguing against 5hmC solely as a short lived demethylation intermediate and potentially pointing towards epigenetic/regulatory role for this modification (43). This is also supported by the abundance of 5hmC in the brain (up to 20% of 5mCG+5hmCG) (13, 40). Although sequencing-based approaches have only found 5hmC in CG dinucleotides in mESC and brain cortical tissue (24), 5hmCH accounted for 38% of 5mCH+5hmCH in cerebellar granule cells, suggesting that 5mCH is also targeted for TET mediated methylation turnover (40). Interestingly, 5hmCG and 5hmCA (the main context of non-CG methylation) interact with MECP2 differently and lead to opposite gene regulatory outputs. Both 5mCA and 5hmCA can be bound by MECP2 (39, 40), whereas the oxidation of 5mCG to 5hmCG abolishes the MECP2 interaction and disrupts the function of 5mCG as a repressive mark (Fig. 2B).
Aberrant methylation patterns in diseases and aged tissues
Global changes of methylation patterns are observed in pathology. Tumor methylomes are characterized by global hypomethylation and localized hypermethylation at specific CGIs, many of them associated with tumor suppressor genes (2). Analysis of the whole genome methylomes of 39 primary tumors in mouse and human shows a pronounced loss of methylation at nuclear lamina associated partially methylated domains (PMDs) located in late replicating parts of the genome (44). Intriguingly, limited PMD hypomethylation can be detected in healthy tissue and increases with age (44). This loss in DNA methylation observed during aging may be due to the insufficient maintenance of DNA methylation across an increasing number of cell divisions (44), which ties similar methylation changes to both cancer and in aging (44, 45). An increase in PMD abundance is also found during the early development of placenta, along with comparable CGI hypermethylation as also observed in cancers (46). The similarity between tumor and placenta methylomes suggests that tumorigenesis may represent a latent developmental epigenomic program.
Despite global DNA hypomethylation, a limited number of genomic loci gain DNA methylation during aging and in cancer. Interestingly, these are sequences that are transcriptionally repressed by the polycomb system (and lack DNA methylation) under normal physiological conditions (47). Similar gains of methylation at distinct polycomb repressed sequences has also been observed, although to a lesser degree, during placental development (46), suggesting an interesting and mechanistically important interplay between DNA methylation and polycomb regulated epigenetic systems.
In instances where DNA methylation is globally reduced as a part of normal developmental program, no aberrant transcriptional activation is observed, suggesting an alternative or compensatory gene expression regulatory mechanism. The situation is, however, different under pathological conditions where the global loss of DNA methylation is often associated with an aberrant transcriptional outcome.
Single-cell techniques uncover the diversity of DNA methylation patterns
Developing tissues contain heterogeneous cell populations derived from different cell lineages and exhibiting various degrees of maturation. Although computational approaches for tissue deconvolution are improving (48), bulk tissue analyses cannot easily distinguish molecular signatures contributed by distinct cell populations. Improvements in single cell methylome approaches now provide the power to distinguish cellular heterogeneity within a tissues/ cell population and identify epigenome fluctuations in a given cell type over time.
Since single-cell reduced representation bisulfite sequencing (scRRBS) and whole-genome single-cell methylome (scBS-seq) were first reported (49), methodological improvements have increased both throughput and resolution. Datasets containing thousands of single-cell methylomes, such as neuron classification in the human and mouse frontal cortices, have been generated with a well-based nuclei collection method, snmC-seq, or a combinatorial indexing-based method, sci-MET (10, 50). Unlike single cell RNA-seq which captures transcriptomic information, single nuclei methylome data allowed prediction of cell-type specific regulatory elements with 5mC signatures. Building upon single-cell methylome methods, single cell multi-omics approaches have been developed that are capable of analyzing both the transcriptomic and epigenomic profiles from a single cell (49). Studies using these techniques have identified cell-to-cell epigenetic variation in early mouse and human embryos (51, 52), demonstrating the enhancement of resolution enabled by multi-omic single-cell methods.
In addition to sequencing-based single-cell methylome strategies, a fluorescent in vivo reporter of genomic methylation (RGM) enables microscopic examination of the 5mC status at a single genomic locus with cellular resolution (53). In an alternative strategy, the fluorescent protein gene reporter can be directly inserted into the methylation sensitive locus which provides a direct readout of the methylation status in living cells or a whole animal (54). Although single-cell methods are just starting to be applied to analyzing 5mC functions, studies have already identified 5mC heterogeneity both across or within cell types, providing information that cannot be accessed with bulk level analyses.
Towards site-specific manipulation of DNA methylation
Functional analyses of 5mC have traditionally relied on pharmacological or genetic perturbations that affect 5mC levels genome-wide (30, 32). Although discoveries regarding 5mC function and regulation have been made with genome-wide perturbation strategies, these methods are associated with pleiotropic phenotypes and inevitably confound the interpretation of results (35). Modern genome engineering tools have opened up the possibility for a more precise site specific intervention that would potentially avoid pleiotropic effects of global 5mC disruption (55).
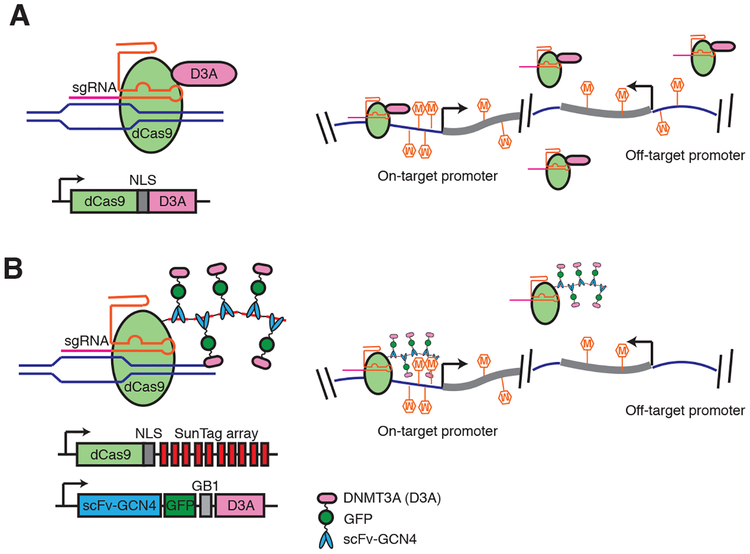
Strategies for targeting 5mC to specific genomic locations have utilized different programmable DNA-binding domain including zinc fingers, transcription activator-like effectors (TALEs) and cleavage deficient dCas9. dCas9-TET and dCas9-DNMT3a fusions have been shown to induce effective remodeling of 5mC (Fig. 3) (56). Most studies take a candidate approach to estimate the extent of off-target manipulation. However, off-target de novo methylation activity can be widespread across the entire genome (57), and not easily detected with steady-state 5mC measurements. Local amplification of regulator concentration through protein (SunTag) or RNA scaffolding (e.g. Casilio) has been incorporated into epigenetic editing to enhance the on/off target ratio (Fig. 3). This scaffold allows multiple copies of epigenetic effectors (e.g. DNMT3A catalytic domain) to be tethered to a genomic site, providing more robust on-target modulation and reduced non-specific effects (58).
Figure 3.
Comparison of direct fusion (A) and SunTag protein scaffold (B) based 5mC editing tools. SunTag system allows local amplification of effector (DNMT3A) concentration and independent control of dCas9 and DNMT3A expression levels to minimize off-target de novo methylation caused by un-tethered DNMT3A. Schematics were drawn on the basis of mC editing tools developed by Liu et al. (56), (A) and Pflueger et al. (56, 58) (B). NLS: nuclear localization sequences. GB1: solubility tag (protein G B1 domain). SunTag based mC editing tool (dCas9Sun-D3A) shows comparable on-target methylation at on-target promoters as the direct fusion (dCas9-D3A), but much lower off-target methylation.
Beyond cytosine base modifications: 6mA -the new kid on the block
5mC and its oxidation derivatives have been considered the only type of mammalian DNA modification until reports of N6-methyladenine (6mA) presence in DNA of higher eukaryotes (59). Compelling multimodal evidence has been presented for the existence of 6mA in mouse and human genomes. These findings are supported by the identification of both methyltransferase and demethylase for deposition and removal of 6mA, respectively (60, 61). Interestingly, the genome-wide distribution of 6mA is species-specific with lack of a conserved pattern across the eukaryotic species analyzed to date (59). Reported difference in 6mA distribution between mouse and human may represent tissue specific patterns rather than species differences, since the tissues used for the mouse and human studies were unique to each species (mESC and blood DNA, respectively (60, 61)). Functional analysis of 6mA in mammalian genomes may be most productive when applied to tissue with abundant 6mA, to be determined with surveys of 6mA across tissues and developmental stages.
Outlook
Recent studies have implicated intriguing, possibly regulatory, roles for 5mC dynamics in diverse biological processes and disease states. Animal models, such as mouse fear conditioning, suggests a role for 5mC in regulating learning and memory (62). Epigenome-wide association studies have associated 5mC signatures with many diseases (63). However, many studies use correlative analysis and report only moderate 5mC differences detected from complex tissues. Thus, while interesting, these studies should be considered as exploratory in nature. In addition, meta-analysis of 5mC microarray data has identified a small number (~300–400) of CpG sites that are associated with age across multiple tissues and the existence of an methylome-based clock (45). Further work is needed to link these clock methylation variants with other molecular information (e.g. gene expression) and aging related phenotypes.
In mitotically inactive mammalian tissues, moderate changes in 5mC dynamics likely reflect cellular heterogeneity either across cell types or within a cell type. In this context, we anticipate that single-cell multi-omic strategies will enhance the specificity and sensitivity allowing determination of cellular identity in tissues (cell atlas), and measuring alterations pertinent to experimental and environmental perturbations. Effectively 5mC dynamics of all cell-types in the tissue can be reconstructed with single-cell profiling without cell type labeling or enrichment strategies, contributing to organism cell reference atlas initiatives as well cell resolution profiling of disease tissues (64).
Site-specific methylome editing will serve as a key technique for the study of 5mC function, and should pave the way for the development of epigenetic-based strategies for cellular engineering and therapies. Specific editing tools will be crucial for moving beyond correlation allowing separation of cause from consequence. An important utility of methylome editing will be to understand the impact of 5mC on transcription and its relationship to other types of epigenetic modifications; a subject actively debated. For example, a recent methylation editing study demonstrating that deposition of 5mC at thousands of gene promoters led to only moderate transcriptional alterations (65, 66). With the availability of facile methods for single cell methylome profiling and editing tools that allow manipulation of 5mC in cells at the right place and time, we anticipate that, in the coming decade, discoveries of new biological functions (or debunking of proposed ones) for DNA methylation will proceed at an unprecedented pace.
Acknowledgements
We thank Drs. B.A. Wang, J.A. Law and J.R. Dixon (Salk Institute), S.C. Huang (New York University) and B. Ren (University of California San Diego) for insightful discussions. Work in the Hajkova lab is supported by MRC funding (MC_US_A652_5PY70) and by an ERC grant (ERC-CoG- 648879 - dynamic modifications). Research in the Ecker lab is supported by NIH awards 5R21HG009274, 5U24DK112348, 5R01ES025585, 5R21MH112161, 5R01MH112763, 1U19MH114831. Additional supports to J.R.E includes Office of Naval Research N00014-16-1-3159 and the Cure Alzheimer’s Fund. J.R.E is a investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Not applicable
References and notes
- 1.Schübeler D, Function and information content of DNA methylation. Nature. 517, 321–326 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet 13, 484–492 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Lister R et al. , Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz MD et al. , Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 523, 212–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziller MJ et al. , Charting a dynamic DNA methylation landscape of the human genome. Nature. 500, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadler MB et al. , DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 480, 490–495 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Domcke S et al. , Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 528, 575–579 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Bell AC, Felsenfeld G, Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 405, 482–485 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Flavahan WA et al. , Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo C et al. , Single Cell Methylomes Identify Neuronal Subtypes and Regulatory Elements in Mammalian Cortex. Science (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whyte WA et al. , Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Hariharan M, Gorkin DU, Dickel DE, Luo C, Spatiotemporal DNA Methylome Dynamics of the Developing Mammalian Fetus. bioRxiv (2017) (available at http://www.biorxiv.org/content/early/2017/07/21/166744.abstract). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R et al. , Global epigenomic reconfiguration during mammalian brain development. Science. 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith ZD et al. , A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 484, 339–344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith ZD et al. , DNA methylation dynamics of the human preimplantation embryo. Nature. 511, 611–615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seisenberger S et al. , The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill PWS et al. , Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature. 555, 392–396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammoud SS et al. , Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 15, 239–253 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Smith ZD, Meissner A, DNA methylation: roles in mammalian development. Nat. Rev. Genet 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Guo X et al. , Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 517, 640–644 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Hon GC et al. , Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet 45, 1198–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wapinski OL et al. , Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 155, 621–635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo C et al. , Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. bioRxiv (2018), p. 371427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Zhang Y, Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 156, 45–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Q, Luu P-L, Stirzaker C, Clark SJ, Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 7, 1051–1073 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Zhu H, Wang G, Qian J, Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet 17, 551–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu S et al. , DNA methylation presents distinct binding sites for human transcription factors. Elife. 2, e00726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y et al. , Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 356 (2017), doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Malley RC et al. , Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurano MT et al. , Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 12, 1184–1195 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto H et al. , Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol. Cell 66, 711–720.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baubec T et al. , Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 520, 243–247 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Morselli M et al. , In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. Elife. 4, e06205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H et al. , Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 329, 444–448 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neri F et al. , Intragenic DNA methylation prevents spurious transcription initiation. Nature. 543, 72–77 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Teissandier A, Bourc’his D, Gene body DNA methylation conspires with H3K36me3 to preclude aberrant transcription. EMBO J. 36, 1471–1473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo JU et al. , Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci 17, 215–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L et al. , MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl. Acad. Sci. U. S. A 112, 5509–5514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabel HW et al. , Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 522, 89–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellén M, Ayata P, Heintz N, 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci. U. S. A 114, E7812–E7821 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guy J, Cheval H, Selfridge J, Bird A, The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol 27, 631–652 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Amouroux R et al. , De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachman M et al. , 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem 6, 1049–1055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou W et al. , DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet 50, 591–602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvath S, Raj K, DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet 19, 371–384 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Smith ZD et al. , Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature. 549, 543–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlesinger Y et al. , Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet 39, 232–236 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Jaffe AE, Irizarry RA, Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelsey G, Stegle O, Reik W, Single-cell epigenomics: Recording the past and predicting the future. Science. 358, 69–75 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Mulqueen RM et al. , Highly scalable generation of DNA methylation profiles in single cells. Nat. Biotechnol 36, 428–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L et al. , Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. (2018), doi: 10.1038/s41556-018-0123-2. [DOI] [PubMed] [Google Scholar]
- 52.Rulands S et al. , Genome-Scale Oscillations in DNA Methylation during Exit from Pluripotency. Cell Syst. 7, 63–76.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stelzer Y, Shivalila CS, Soldner F, Markoulaki S, Jaenisch R, Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 163, 218–229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van de Pette M et al. , Visualizing Changes in Cdkn1c Expression Links Early-Life Adversity to Imprint Mis-regulation in Adults. Cell Rep. 18, 1090–1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pulecio J, Verma N, Mejía-Ramírez E, Huangfu D, Raya A, CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell. 21, 431–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu XS et al. , Editing DNA Methylation in the Mammalian Genome. Cell. 167, 233–247.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galonska C et al. , Genome-wide tracking of dCas9-methyltransferase footprints. Nat. Commun 9, 597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pflueger C et al. , A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. (2018), doi: 10.1101/gr.233049.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo G-Z, He C, DNA N6-methyladenine in metazoans: functional epigenetic mark or bystander ? Nat. Struct. Mol. Biol 24, 503–506 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Xiao C-L et al. , N6-Methyladenine DNA Modification in the Human Genome. Mol. Cell 71, 306–318.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Wu TP et al. , DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 532, 329–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Day JJ, Kennedy AJ, Sweatt JD, DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu. Rev. Pharmacol. Toxicol 55, 591–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paul DS, Beck S, Advances in epigenome-wide association studies for common diseases. Trends Mol. Med 20, 541–543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Science forum: the human cell atlas. Elife (2017) (available at https://cdn.elifesciences.org/articles/27041/elife-27041-v2.pdf). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ford EE, Grimmer MR, Stolzenburg S, Bogdanovic O, Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. bioRxiv (2017) (available at http://www.biorxiv.org/content/early/2017/08/17/170506.abstract). [Google Scholar]
- 66.Korthauer K, Irizarry RA, Genome-wide repressive capacity of promoter DNA methylation is revealed through epigenomic manipulation. bioRxiv (2018) (available at https://www.biorxiv.org/content/early/2018/08/01/381145.abstract). [Google Scholar]
- 67.Berdasco M, Esteller M, Hot topics in epigenetic mechanisms of aging: 2011. Aging Cell. 11, 181–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]