Abstract
Human brain connectivity is complex. Graph theory based analysis has become a powerful and popular approach for analyzing brain imaging data, largely because of its potential to quantitatively illuminate the networks, the static architecture in structure and function, the organization of dynamic behavior over time, and disease related brain changes. The first step in creating brain graphs is to define the nodes and edges connecting them. We review a number of approaches for defining brain nodes including fixed versus data-driven nodes. Expanding the narrow view of most studies which focus on static and/or single modality brain connectivity, we also survey advanced approaches and their performances in building dynamic and multi-modal brain graphs. We show results from both simulated and real data from healthy controls and patients with mental illnesse. We outline the advantages and challenges of these various techniques. By summarizing and inspecting recent studies which analyzed brain imaging data based on graph theory, this article provides a guide for developing new powerful tools to explore complex brain networks.
Index Terms: brain graph, nodes, dynamic, multi-modal
I. Introduction
Brain performance has been characterized and quantitatively studied using state of the art noninvasive brain imaging techniques [1–3]. For example, magnetic resonance imaging (MRI) studies have shown that brain regions are not only structurally connected [4–6] but also functionally associated when performing cognitive tasks or even in resting state [7–9]. In the last decade, graph theory based analysis has become a powerful and popular approach for assessing brain networks [10–41], largely because of its potential to quantitatively illuminate the static architecture in structure and function [42–60], the organization of dynamic behavior over time in resting state or during different cognitive tasks [61–83], the development across the lifespan [84–103], and changes in mental disorders [104–161].
Technically, when building a brain graph using imaging data, the first step is typically to define the nodes, and then build edges between them [162]. The use of fixed spatial regions of interest (ROIs) assessed by anatomical atlases based on brain structure is one of the popular methods for defining brain nodes [163]. This approach is appropriate for structural brain imaging data. However, the selected ROIs do not necessarily respect the functional boundaries of the brain [164]. Thus, more approaches have been developed for defining the nodes when building brain graphs using functional magnetic resonance imaging (fMRI) data [165], such as independent component analysis (ICA) which is adopted to decompose the whole-brain fMRI into independent spatial components (called ICA nodes) [129, 166]. Briefly, both fixed (including ROIs, voxels) and data-driven (such as ICA) methods are widely used to define the nodes in brain graph studies using fMRI data [167].
While the dynamics of both structural and functional brain connectivity across different age stages have been investigated, most previous studies hypothesize that the functional brain connectivity is static over a scanning time. However, recent fMRI experiments have shown that functional brain connectivity is dynamic on the scale of tens of seconds even during the resting state [168, 169]. Currently, more studies are investigating time varying brain graph performance [82]. While structural brain graphs can be built across different ages, functional brain graphs are built not only across the life span, but also on short time scales such as across different cognitive tasks, or even across a few minutes of the resting state [170–172].
In addition to dynamic analysis, techniques to perform multimodal fusion analysis of brain imaging data are required, because collecting multiple types of brain data from the same individual using various non-invasive imaging techniques has become a common practice [173]. A key motivation for jointly analyzing multimodal data is to take advantage of the cross-information of the existing data, thereby potentially revealing important variations that may only be partially detected by a single modality [174–176]. Recent studies have developed powerful methods to build multimodal brain graphs.
In this article, we briefly review and compare the research findings in static and dynamic (across the life span, at different ages, during various cognitive tasks, or during the resting state), structural (gray matter and diffusion image data) and functional brain graphs with nodes defined by different methods in not only healthy controls, but also patients with mental disorder. We survey advanced techniques for defining the brain nodes, building dynamic and multimodal brain graphs, and point out potential directions for developing new tools to build and characterize brain graphs. The rest of this paper is organized as follows: In Section II, we review and compare research results from static structural and functional brain graphs. In Section III, we focus on dynamic brain graphs. In Section IV, we survey studies creating brain graphs with multimodal data. Finally, we discuss the limitations of existing methods and suggest possible directions for developing new approaches in investigating different brain graphs.
II. STATIC STRUCTURAL AND FUNCTIONAL BRAIN GRAPHS
In gray matter and diffusion image structural brain graph studies, nodes are typically defined using ROIs though at multiple spatial scales. Edges are usually defined using correlation of gray matter volume or thickness, or fiber tracks (connections) between pairs of ROIs in gray matter and diffusion magnetic resonance imaging (dMRI) data respectively.
When building brain graphs in fMRI data, spatial brain components evaluated by group ICA have been used to define data-driven nodes, as opposed to fixed atlas-based ROI nodes. Edges are defined using a variety of metrics including Pearson correlation, partial correlation, or wavelet-based frequency dependent correlation between the time courses of any pair of nodes.
A. Structural Brain Graphs Built with ROIs
A main finding of gray matter structural brain graph studies is that the topology of structural brain networks shows “small world” properties rather than random. Previous studies have defined the nodes in ROIs from different ways, such as using 3 × 3 × 3 voxel cubes (size corresponding to 6 × 6 × 6 mm3) as nodes [177], or using 104 ROIs defined by atlas images as nodes [115]. It has also been shown that different cortical scales lead to cortical networks with different values of small-worldness when building brain graphs at 23 cortical scales (number of nodes varied from 66 to 1494) based on the Desikan-Killiany atlas [178].
Consistent with gray matter structural brain graphs, diffusion brain image-based graphs are also not random. When using 78 cortical ROIs from the automated anatomical labeling (AAL) atlas [179] as nodes, the topology of the diffusion brain image-based graph resembled a small-world architecture characterized by an exponentially truncated power-law distribution. In addition, the diffusion brain image-based graph was characterized by major hub regions in association cortices that were connected by bridge connections following long-range white matter pathways [180]. Importantly, the observed inter-scan reproducibility of the graph measures was high [181].
Diffusion brain image-based graphs have also been built at multiple scales. In a diffusion spectrum imaging (DSI) and diffusion tensor imaging (DTI) study, 90, 180, 360, 720 ROIs defined by AAL atlas, 110, 220, 440, and 880 ROIs defined by the Harvard-Oxford Atlas (HO), and 54, 108, 216 and 432 ROIs defined by the LONI Probabilistic Brain Atlas (LPBA40) were used as nodes respectively. Basic connectivity properties and several graph metrics displayed high reproducibility and low variability in both DSI and DTI networks [182].
An impressive property of previous diffusion brain image-based graphs is the so-called “rich club” organization initially identified by a study using 82 ROIs defined by Freesurfer suite as nodes. A rich club organization is characterized by a tendency for high-degree nodes to be more densely connected among themselves than nodes of a lower degree in the graph [183]. A following study which used 1170 ROIs as nodes showed that the set of pathways linking rich club regions formed a central high-cost, high-capacity backbone for global brain communication [184].
A wide application of brain graph analysis is for the detection of potential biomarkers of mental illness such as schizophrenia. Altered brain graph properties have been revealed in schizophrenia using DTI data with different nodes. When using 82 ROIs [104] and 108 ROIs [185] as nodes to build the graphs respectively, it was shown that though small-world attributes were conserved in schizophrenia, the cortex was interconnected more sparsely and up to 20% less efficiently [104], and node specific path lengths were longer in patients [185]. Another DTI study which used 90 ROIs defined by AAL as nodes discovered decreased global efficiency in schizophrenia [186].
In summary, structural brain graphs have been extensively studied using multi scale ROIs as nodes. Structural brain graphs show small-world properties with hub nodes, modular, and rich club organization, and graph measures are robust across scans. Nodes with different cortical scales may lead to different levels of small-world organization. Graph metrics including path length and global efficiency have been shown to be disrupted in schizophrenia [185, 186].
B. Functional Brain Graphs Built with ROIs
In line with structural graphs, the functional brain graphs also show small world rather than random topology. This topological property has been consistently revealed by graphs with different nodes such as the 90 ROIs come from AAL atlas [47, 105, 179, 187], and single voxels [188, 189]. For a review of the small world topology of functional brain networks see [190]. In addition to small-world property, functional brain graph show scale-free topology when using single voxels as nodes [188, 189], and significantly non-random modular organization when using AAL-based 90 ROIs as nodes [84]. Modules have been found to be consistent with cognitive function when using five different node definitions, in which each module was associated with a discrete cognitive function [191]. Moreover, functional brain graph studies strongly suggested that brain hubs play important roles in information integration underpinning numerous aspects of complex cognitive function [192].
The reliability of graph measures in functional brain connectivity has also been studied. A test-retest study of graphs with 90 ROIs defined by AAL as nodes estimated the reliability of various graph measures including clustering coefficient, characteristic path length, local and global efficiency, assortativity, modularity, hierarchy and small-worldness. Results showed that the second order metrics (small-worldness, hierarchy, assortativity) tended to be more robust than first order metrics (clustering coefficient, characteristic path length, modularity, global and local efficiency) [193].
Similar to structural brain graphs, functional brain graphs have been built with ROIs in multiple spatial resolutions as well. It was found that the graphs with nodes of higher resolution exhibited the small-world properties more prominently. And region-based graphs fragmented more at high thresholds than voxel-based graphs, suggesting region-based graphs are less robust [194]. Although the degree distributions of all graphs followed an exponentially truncated power law rather than a true power law, the higher the resolution, the closer the distribution was to a power law. Furthermore, voxel-based analyses enhanced visualization of the results in the 3D brain space. These results demonstrated benefits of constructing the brain graph at the finest scale [194]. Another study built functional brain graphs with seven different parcellation resolutions (84, 91, 230, 438, 890, 1314, and 4320 regions) based on the AAL atlas. Results showed that gross inferences regarding graph topology, such as whether the brain was small-world or scale-free, were robust. But both absolute values of, and individual differences in, specific parameters such as path length, clustering, small-worldness, and degree distribution descriptors varied considerably across the resolutions [49]. Brain graphs with 90 (AAL) or 70 (Automated Nonlinear Imaging Matching and Anatomical Labelling [ANIMAL-atlas] [195]) ROIs as nodes were found to show robust small world properties and truncated power-law connectivity degree distribution. However, significant differences in multiple topological parameters (e.g., small-worldness and degree distribution) between the two graphs were revealed [196].
Functional brain graphs have been extensively investigated in brain disorders. Using AAL-based 90 ROIs as nodes, pairwise functional connectivity was found to be decreased and the variance was increased in schizophrenia [124]. Graph measures including degree, clustering coefficient, global efficiency, and local efficiency were decreased and path length was increased in patients with schizophrenia [122]. These findings were repeated in another fMRI study which used 72 ROIs defined by AAL as nodes [107]. Altered community structure was also reported in schizophrenia in graphs with 278 ROIs nodes defined with FSL’s cortical and subcortical Harvard-Oxford probabilistic atlas [197].
In addition to schizophrenia, disruptions of functional brain graphs have also been studied in other brain disorders including Alzheimer’s disease [198, 199], amnestic mild cognitive impairment (aMCI) – the prodromal stage of Alzheimer’s disease [200], major depressive disorder [201], children with frontal lobe epilepsy (FLE) [202], children with attention-deficit/hyperactivity disorder (ADHD) [203], and mesial temporal lobe epilepsy (mTLE) patients [204] using different ROI nodes.
In summary, functional brain graphs with high resolution voxel level rather than low resolution brain region level nodes showed less fragmentation. The higher resolution graphs exhibited more prominent small-world properties, and the higher the resolution, the closer the degree distribution was to a power law [194]. However, there were some disadvantages when defining the nodes at voxel level. For example, some voxels may be located completely in white matter, thus it was difficult to remove the effects of white matter signal fluctuations [49]. In addition, computational burden was high for calculating graph metrics in weighted voxel-based brain graphs. Optimization of these computational algorithms is an important goal [194]. Notably, graph measures were largely disrupted in brain disorders.
C. Functional Brain Graphs Built with ICA Nodes
There are some limitations when defining the ROI nodes in functional MRI data based on anatomical brain atlases. For example, The ROIs may not correspond well to real functional boundaries [167] in the brain or fully capture individual subject variability [179, 205–207]. Although functional connectivity-based data-driven methods for brain parcellation have been developed both at the group [208–212] and individual level [213, 214], no study to our knowledge has used these approaches to define brain nodes for building graphs. In contrast, ICA is a popular data-driven method for defining the nodes to mitigate these limitations of ROI-based nodes in building functional brain graphs using fMRI data. In this approach, the fMRI data of a group subjects are firstly decomposed into a number of spatial components. Then single subject components and their associated time courses are back-reconstructed [215–218]. The graph nodes of spatial brain components evaluated by ICA are functionally homogeneous and may capture individual differences better than anatomical atlas-based ROI nodes [166, 215, 216, 219]. Using this node definition approach, the topology of brain graphs has also been revealed to be small world and modular with hub nodes. Moreover, alterations of graph measures including connectivity strength, clustering coefficient, local efficiency, characteristic path length, global efficiency, modularity, and rich club parameters were discovered in schizophrenia [55, 127–129]. However, limitations of defining nodes using ICA include spatial overlap of some of the nodes (though this can potentially be an advantage as well [220]), and the number of nodes/components need to be selected [129, 220].
D. Comparing ROI and ICA Methods for Building Functional Brain Graphs using Simulations
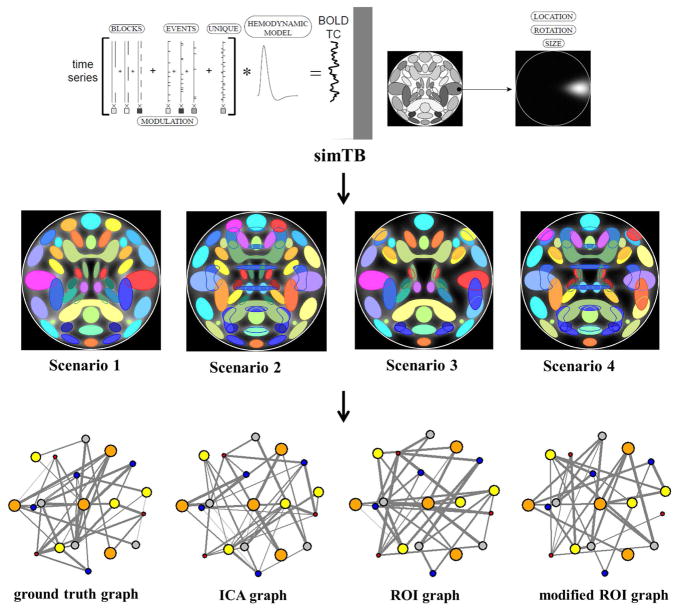
An important and open question is that which method (atlas-based ROI vs data-driven ICA) can better define the nodes of brain graph in fMRI data. Using simulated data to evaluate the node definition methods may be promising, because different scenarios can be estimated and graph measures computed with different nodes can be compared to a known ground truth. In one recent study [221], simulated data was generated using SimTB (http://mialab.mrn.org/software/simtb/), a MATLAB toolbox which implements a data generation model consistent with spatiotemporal separability, that is, data can be expressed as the product of time courses (TCs) and spatial maps (SMs) (for details of the data generation model in SimTB see Erhardt et al 2012 [222]). Four scenarios involving different SM configurations were analyzed. In scenario 1, spatial maps (SMs) were created with little overlap. In scenario 2, twenty-nine SMs were with little overlap plus three artificial SMs. In scenario 3, SMs were with large overlapping. In scenario 4, SMs were with large overlapping and included 3 “artefactual” SMs.
Four kinds of undirected weighted graphs were built based on the simulated data in each scenario. The number of nodes (N) was same in all graphs (N = 29). In scenarios 2 and 4, three artefactual SMs associated nodes were excluded when building the four types of graphs.
Ground truth graph. In this graph, the nodes were SMs (excluding artifacts if any) in the simulation. Edges were the Pearson correlation between each pair of simulated time courses of the SMs.
ICA graph. In this graph, the nodes were spatial independent components obtained by group ICA performed on the simulated data. Edges were the Pearson correlation between each pair of ICA time courses of the components.
ROI graph. In this graph, the nodes were ROIs which were defined base on the SMs of the simulation using a threshold of 0.8. Time series of voxels within each ROI were averaged into one representative time course. Edge weights of the graph were the Pearson correlation between each pair of representative time courses of the ROIs.
Modified ROI graph (MROI-graph). When using ROIs as graph nodes, the ROIs are usually defined base on a brain atlas which will not perfectly match the variation in an individual subject. To simulate the effects of individual subject variability in brain shape and functional domains, and compare with the ideal ROI scenario, a MROI-graph (in which nodes deviate from the ground truth ROIs to reflect subject variability) was built and examined. In this graph, the nodes were spatially moved up to 15% of the area of the ROI. See Yu et al. [221] for details of the method. See figure 1 for a pipeline of the analysis in that study.
Fig. 1.
An analysis pipeline of the simulation study which compared graphs with ROI nodes and ICA nodes [221]. Simulated fMRI data were generated in four scenarios using SimTB. Group ICA was performed on the simulated data. Graphs with different nodes (ground truth nodes, ICA nodes, ROI nodes, and modified ROI nodes) were then built. Graph metrics between the ground truth graph and any other graph were compared. (The figure was reproduced with permission from both author and journal of Yu et al. 2017 [221].)
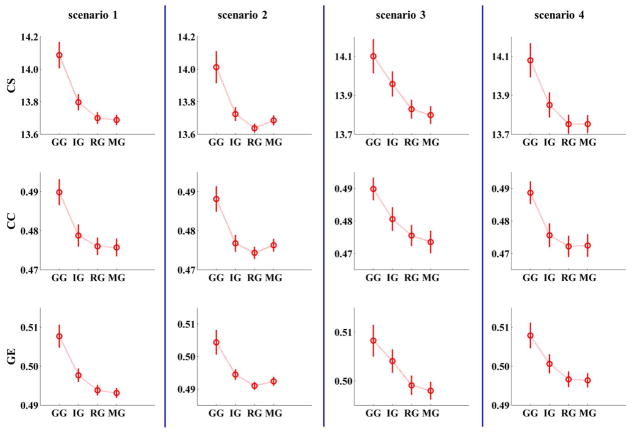
When comparing the measures including connectivity strength, clustering coefficient, and global efficiency of graphs with ROI nodes and ICA nodes with ground truth, results showed that in all scenarios, values of all graph metrics for the data-driven ICA graph (compared to the ROI and MROI graphs) were closer to the ground truth graph. In the cases with no artifacts (scenarios 1 and 3), graph measures in the MROI graph were far away from ground truth compared to the ROI graph. However, in cases with artifacts (scenarios 2 and 4), graph measures of the MROI graph were closer to ground truth than the ROI graph (see Figure 2). Since ROIs are typically defined using anatomical atlases which may not correspond well to real functional boundaries in the brain or to individual subject variability, these findings suggest that a data-driven ICA method is more accurate compared to a fixed ROI method for defining graph nodes in functional brain network studies. Similar conclusions have also been shown in another study [223].
Fig. 2.
Results of a simulation study which compared graphs with ROI nodes and ICA nodes (Yu et al. [221]). The measures of graphs with different nodes (GG: ground truth graph; IG: ICA graph; RG: ROI graph; MG: modified ROI graph) in all four scenarios. All measures of ICA graphs are closer to ground truth than ROI graphs and MROI graphs. (CS: connectivity strength; CC: clustering coefficient; GE: global efficiency). (The figure was reused with permission from both author and journal of Yu et al. 2017 [221].)
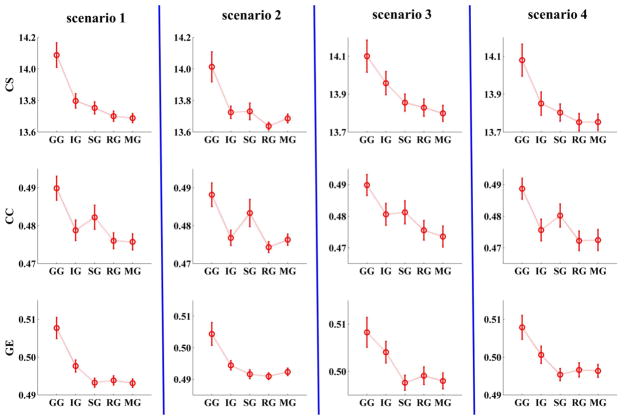
Since previous studies have shown some benefits of using single voxel than ROI with a group of voxels as graph node, a voxel level graph was also estimated in that simulation study. In this graph, each node was the voxel with the highest intensity value in each of the 29 SMs. Results were mixed. The clustering coefficient for the voxel level graph was closer to ground truth than the ICA graph, the ROI graph, and the MROI graph. Connectivity strength of voxel level graph was closer to ground truth then the ICA graph in scenario 2. Global efficiency of voxel level graph was further from the ground truth than the ICA graph (see Figure 3). These findings suggest that graph metrics of voxel level graphs do not consistently reflect ground truth better than ICA graph.
Fig. 3.
Measures of graphs built with different node definition methods in simulated data. Clustering coefficient of voxel level graph is closer to ground truth than ICA graph, ROI graph, and MROI graph. Connectivity strength of voxel level graph is closer to ground truth then ICA graph in scenario 2. Global efficiency of voxel level graph is further to ground truth than ICA graph. (GG: ground truth graph; IG: ICA graph; SG: single voxel level graph; RG: ROI graph; MG: modified ROI graph; CS: connectivity strength; CC: clustering coefficient; GE: global efficiency). (The figure was reproduced with permission from both author and journal of Yu et al. 2017 [221].).
However, there are limitations of the simulation findings. The simulated data were generated using a linear model (the product of time courses and spatial maps). While ICA, which is based on a linear model, has revealed robust brain networks in both resting state and task fMRI data, it is not clear whether the linear model is a perfect match for real fMRI data. In addition, the conclusions are limited to the four scenarios considered here. Future studies may simulate more scenarios for generating the ROIs such as functional connectivity based pacellation methods [208–214].
E. Comparing graphs with ROI nodes and ICA nodes to differentiate schizophrenia patients from controls
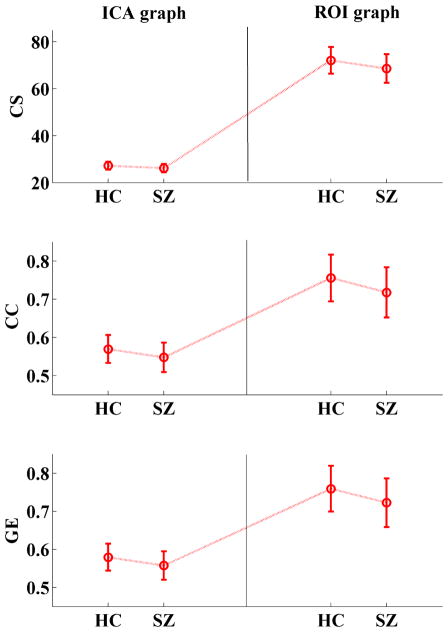
To evaluate the ability of differentiating patients from controls using graphs with different nodes, we build both an ROI graph (in which nodes are 96 ROIs selected from AAL such that each ROI has more than one gray matter voxel) and an ICA graph (in which nodes are 48 spatial brain components) in resting state fMRI data of 164 (82 controls vs 82 patients with schizophrenia) subjects. This work has not been reported before. However, for details on data collection and preprocessing see Yu et al. [172]. Results show that though two sample t-tests (controls vs patients) of graph measures (including connectivity strength, clustering coefficient, and global efficiency) are significant in both ICA and ROI graphs, p-values are lower and effect sizes (Hedges’ g) [224] are higher in the ICA graph compared to the ROI graph (see Table 1). Thus, the ICA nodes outperform the ROI nodes for differentiating patients from controls. Group mean values for the graph measures are reported in Figure 4. In order to provide a statistical view and to eliminate the possibility that the findings may due to the different number of nodes between the two types of graphs, we perform 1000 permutation analysis by randomly selecting 48 ROIs from the 96 ROIs to build ROI graphs and perform t-tests between HCs and SZs on graph measures. Results show that the p values and (Hedges’ g) effect sizes of the ICA graphs listed in Table 1 are located at about the 10th percentile of the corresponding 1000 permutated values.
TABLE I.
P values and effect sizes (Hedges’ g) of two sample t-tests (controls vs patients) on graph measures in ICA graph, ROI graph, and voxel level graph. (CS: connectivity strength; CC: clustering coefficient; GE: global efficiency)
| CS | CC | GE | ||
|---|---|---|---|---|
| P value | ICA graph | 7.73 × 10 (-5) | 1.13 × 10 (-4) | 7.12 × 10 (-5) |
| ROI graph | 1.89 × 10 (-4) | 1.91 × 10 (-4) | 1.91 × 10 (-4) | |
| Voxel level graph | 5.15 × 10 (-5) | 5.09 × 10 (-5) | 5.24 × 10 (-5) | |
| Effect size | ICA graph | 0.63 | 0.62 | 0.63 |
| ROI graph | 0.59 | 0.59 | 0.59 | |
| Voxel level graph | 0.65 | 0.65 | 0.65 |
Fig. 4.
Group mean of graph measures in controls and patients with schizophrenia in graph with ICA nodes and ROIs. Error bars correspond to standard deviation. Two sample t-tests show that graph measures are significantly (p < 0.001) lower in patients than controls in both ICA graph and ROI graph. However, p values are lower and effect sizes (Hedges’ g) are higher in ICA graph then ROI graph (see Table 1). (HC: healthy controls; SZ: patients with schizophrenia; CS: connectivity strength; CC: clustering coefficient; GE; global efficiency)
A voxel level graph (with 96 nodes) is also constructed to estimate the ability of single voxel graphs in differentiating the two groups. In this graph, each of 96 nodes is a gray matter voxel randomly selected in each of the 96 AAL ROIs. Two sample t-tests of graph measures in the two groups of subjects show that p values are a little lower than those of the ICA graph and the ROI graph, and effect sizes (Hedges’ g) are a little higher than the results of those two graphs (see Table 1). Again, we perform 1000 permutation analyses by randomly selecting 48 ROIs from the 96 ROIs and build voxel level graphs with 48 single voxels from these 48 ROIs. T-tests (HCs vs SZs) are performed in each permutation. This time, the p values and (Hedges’ g) effect sizes of the ICA graphs listed in Table 1 are located at about the 40th percentile of the corresponding 1000 permutated values. Due to computational load for calculating graph measures of large weighted graphs [194], we do not run the tests with all voxels in this study.
III. Dynamic BRAIN graphs
Recent brain imaging studies have shown that brain connectivity is dynamic rather than static over time [169]. The dynamics of brain connectivity have been studied at different age stages (life span), during different cognitive tasks, and during the resting state. Dynamic graph analysis is a powerful tool to quantitatively characterize the time evolving brain performance in a system level.
A. Development of Structural and Functional Brain Graphs with ROI Nodes
Using 82 ROIs as graph nodes, a DTI study investigated the development throughout childhood and adolescence (7 –23 years). Results showed a simultaneous age-related decrease in average path length and increase in node strength and network clustering, which may reflect fine-tuning of topological organization in brain development [95]. When assessing the developmental changes of diffusion brain image-based graph in an age range (19 – 85 years) using 78 cortical ROIs from AAL as graph nodes, a reduction in overall cortical connectivity with age, and decreased local efficiency in older brains were revealed [225]. Another DTI brain graph study which involved 882 participants (ages 8 – 22) showed that structural modules became more segregated with age. Evolving modular topology facilitated global efficiency and was driven by age-related strengthening of hub edges. In addition, both modular segregation and graph efficiency were associated with enhanced executive performance and mediated the improvement of executive functioning with age. These results delineated a process of structural network maturation that supports executive function in youth [75]. Another more recent brain graph study also used DTI data of 882 participants (ages 8 – 22) to investigate how structural connectivity facilitates changes and constrains patterns of dynamics in the developing brain. That work draw on the computational tools and conceptual frameworks of theoretical physics and engineering to study two complementary predictors of brain dynamics, controllability and synchronizability, which separately predict the brain’s ability to transition to nearby vs. distant states, or to maintain a single state characterized by a stable temporal dynamic. Results showed that white matter connectivity becomes increasingly optimized for a diverse range of predicted dynamics in development. Notably, stable controllers in subcortical areas were negatively related to cognitive performance. These findings suggested that the brain optimizes the controllability at the expense of the synchronizability. That optimization occurred during development in youth aged 8 – 22 years, and individual differences in control architecture of white matter were correlated with individual differences in cognitive performance. Impressively, that work used forward-modeling computational approaches to identify constrained evolutionary trajectories which provided evidence that network control is a key mechanism in development [226].
Functional brain graphs in resting state fMRI data were studied using 264 ROIs which were identified by meta-analyses in task fMRI data and connectivity mapping of resting state fMRI data [227–229] as nodes to quantify the formation of graph modules across age 8 – 22 years. It was shown that functional graph organization changes in youth through a process of modular evolution that was governed by the specific cognitive roles of each system, as defined by the balance of within- vs. between-module connectivity, which suggested that dynamic maturation of network modules in youth may be a critical driver for the development of cognition [68]. Another study used the same 264 ROIs as nodes and revealed that brain networks in the elderly (older: 59 – 74 years; younger: 18 –26 years) showed decreased modularity and decreased local efficiency [230].
Using task fMRI data, a study investigated age-related change of brain connectivity during memory encoding and recognition. Ninety ROIs from AAL atlas were used as nodes. It was found that age-related changes mainly occurred in long-range connections with widespread reductions. The older adults (75 – 87 years) had longer path lengths linking different regions in the functional brain networks as compared to the younger adults (20 – 27 years). The increases in shortest path length in the networks were combined with the loss of long-range connections. These findings suggest that normal aging was associated with disruption of large-scale brain systems during the performance of memory tasks [231].
Development of functional brain graphs have also been examined with nodes at multiple scales. One study in which subjects were 10 – 20 years old used two methods to define nodes: 1. voxel-wise approach in which each 4 mm3 gray matter voxel as a node; 2. 160 ROIs defined based on a meta-analysis of fMRI activation. Results showed that hub architecture was evident in late childhood and was stable from adolescence to early adulthood. Connectivity between hub and non-hub regions changed with development from childhood to adolescence [232].
When examining the developmental changes in graph organization, connectivity strength, and integration to inhibitory control development across four stages, childhood (10 – 12 y olds), early adolescence (13 –15 y olds), late adolescence (16 – 19 y olds), and adulthood (20 – 26 y olds), using 264 ROIs which were identified by meta-analyses in task fMRI data and connectivity mapping of resting state fMRI data [227–229] as nodes, results indicated that network organization was stable throughout adolescence. However, cross-network integration increased with age. Those findings provided compelling evidence that the transition to adult-level inhibitory control was dependent upon the refinement and strengthening of integration between specialized networks, and supported a novel, two-stage model of neural development, in which networks stabilize prior to adolescence and subsequently increase their integration to support the cross-domain incorporation of information processing critical for mature cognitive control [170].
In summary, brain graphs with ROI nodes initially change with age by decreasing path length, and increasing connectivity strength, clustering coefficient, local efficiency, and modularity for enhancing the integration of information processing from childhood to adolescence (young adult). The change of these graph measures was reversed from young adults to elder adults.
B. Functional Brain Graphs across Cognitive Tasks
When using single voxels as nodes to build brain graphs using fMRI data of different tasks or cognitive states including resting state, visual stimulation, and multisensory (auditory and visual stimulation) conditions, results showed that despite stability of graph measures at global level, brain graphs exhibited considerable task-induced changes in connectivity, efficiency, and community structure at the nodal level [171].
Another task fMRI study used 264 ROIs which were identified by meta-analyses in task fMRI data and connectivity mapping of resting state fMRI data [227–229] as nodes, and found that the frontoparietal brain network (FPN)’s brain-wide functional connectivity pattern shifted more than those of other networks across a variety of task states. These connectivity patterns could be used to identify the current task which supported a central role for frontoparietal flexible hubs in cognitive control and adaptive implementation of task demands [233].
C. Sliding Window Based Functional Dynamic Brain Graphs
Sliding window is a popular method for assessing dynamic functional connectivity in fMRI data across a short time (a few minutes to a few hours) [169, 234]. When using the sliding window method to build a dynamic resting state fMRI graphs with 264 ROIs, which were identified by meta-analyses in task fMRI data and connectivity mapping of resting state fMRI data [227–229] as nodes, it has been shown that brain regions spontaneously changed their module affiliations on a temporal scale of seconds. These dynamics were highly reproducible across repeated scanning sessions [82]. Another sliding window graph study which used ROIs with multiple scales as nodes discovered that the most dynamic connections were inter-module, and localize to known hubs of default mode and frontoparietal systems. In addition, spatially distributed regions spontaneously increased the efficiency with which they can transfer information, producing temporary, globally efficient network states, which suggested that brain dynamics give rise to variations in complex network properties over time, possibly achieving a balance between efficient information-processing and metabolic expenditure [83].
However, some properties have been found to be static across sliding window graphs. For example, when using 90 ROIs from AAL atlas and a random parcellation of 1024 ROIs as nodes, dynamic functional graphs exhibited evident small-world and assortativity architecture, with several regions (e.g., insula, sensorimotor cortex and medial prefrontal cortex) emerging as functionally persistent hubs though possessing large temporal variability in their degree centrality [235].
Another interesting dynamic graph called a hypergraph [236] can be built based on time-varying graphs. For example, a study firstly established sliding window graphs using 194 ROIs as nodes in fMRI data from a task-free resting state, an attention-demanding state, and two memory-demanding states. Then hypergraphs in which nodes were the edges in the graph of 194 ROIs were constructed. Results identified the presence of groups of functional interactions that fluctuated coherently in strength over time both within (task-specific) and across (task-general) brain states, demonstrating that brain adaptability can be described by common processes that drive the dynamic integration of cognitive systems [67]. Another study used a hypergraph approach in fMRI data of 780 participants (ages 8 – 22) to investigate the development of functional brain connectivity. Three distinct classes of subnetworks (hyperedges) including clusters, bridges, and stars were revealed. Cluster hyperedges showed a strong resemblance to previously described functional modules of the brain including somatomotor, visual, default mode, and salience systems. In contrast, star hyperedges represented highly localized subnetworks centered on a small set of regions, and were distributed across the entire cortex. Finally, bridge hyperedges linked clusters and stars in a core-periphery fashion, with the greatest developmental effects occurring in networked hyperedges within the functional core [89]. The hypergraph method has also been used to evaluate individual differences in dynamic functional brain connectivity across the human lifespan [88].
When assessing dynamic functional brain connectivity using the sliding window approach, brain states with different connectivity patterns can be detected by k-means clustering or decomposition methods [237]. Both k-means and decomposition methods require selecting the number of states [168]. One can also use time-varying graph measures to examine the brain states during the resting state using ICA nodes. This method was developed by a study which firstly constructed time-varying dynamic graphs with ICA nodes using sliding windows. Then connectivity states were detected based on the correlation of nodal connectivity strength between time-varying brain graphs. Results show that patients with schizophrenia exhibit decreased variance in the dynamic graph metrics [172]. For a pipeline of this approach see Figure 5.
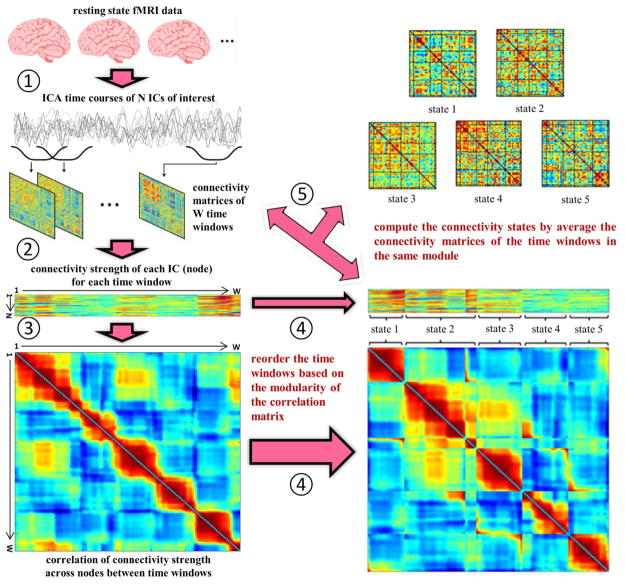
Fig. 5.
The flowchart of the algorithmic pipeline for a method of evaluating connectivity states base on graph analysis of ICA nodes developed in one of our studies [172]. Five steps were labeled as follows. ➀ do group ICA, segment ICA time courses, and calculate the correlation between any pair of (N = 48) independent components (ICs) for each time-window; ➁ compute nodal connectivity strength of the weighted brain graph for each time-window; ➂ calculate the correlation of nodal connectivity strength between any pair of time-windows (W = 131) across (N = 48) ICs; ➃ reorder the time-windows based on the modular organization of the correlation matrix; ➄ compute the brain connectivity states by averaging the connectivity matrices of the time windows belonging to the same module. (The figure was reused with permission from both author and journal of Yu et al. 2015 [172].)
In summary, functional brain graphs are indeed dynamic across different time scales. Sliding window is the most popular method to build dynamic brain graphs across a relatively short time (a few minutes to a few hours), though there are many other approaches which are adapted as well [238].
IV. Multimodal graphs
As there has been increasing interest in performing multimodal analysis of brain imaging data, more approaches are being developed to build multi-modal brain graphs which may be helpful to understand the physiological, electrophysiological, or even genetic basis of the topological properties in brain connectivity.
A. Associate Single Modal Brain Graph with Other Modalities
Combing functional and structural brain data may reveal the structural basis of the functional brain graphs. To this end, one study built functional brain graphs with nodes defined by ROIs of AAL atlas using fMRI data acquired during an episodic memory-for-context task in both healthy controls and patients with schizophrenia, and performed a morphometric analysis to investigate schizophrenia-related deficits. Functional graphs showed significant reductions in local efficiency in schizophrenia. Structural data showed several key network “hub” regions including bilateral dorsal anterior cingulate gyrus with reduced gray matter volume in schizophrenia patients. These findings suggest that loss of gray matter volume may contribute to local inefficiencies in the architecture of the functional graph underlying memory-for-context in schizophrenia [239]. Another study combined high-resolution diffusion weighted imaging (DWI) with resting-state fMRI. DWI data were used to build structural graph with 1170 ROIs segmented by Freesurfer as nodes, and eleven resting state brain networks were detected by ICA in fMRI data. By associating structural graphs with functional networks, new evidence suggesting that the brain’s rich club serves as a macroscopic anatomical substrate to cross-link functional networks was provided. Such links likely play an important role in the integration of information between segregated functional domains of the human cortex [240].
The relationship of functional hubs to measures of brain physiology (regional cerebral blood flow, rCBF) had been studied using fMRI data and arterial-spin-labeling (ASL) perfusion data during resting state and an N-back working-memory task. During the resting state, functional graphs were built at the voxel-level and hubs with higher functional connectivity strength (FCS) were identified. The FCS showed a striking spatial correlation with rCBF. During the working memory task, task-induced changes of FCS and rCBF in the lateral-parietal lobe positively correlated with behavioral performance. Together, these findings suggest a tight coupling between blood supply and brain functional topology during rest and its modulation in response to task demands [241].
B. Associate Graphs Built in Different Modalities
Building graphs with same nodes using different datasets is powerful, because the graphs may be directly compared. A life span study built fMRI and diffusion brain image-based graphs using 114 ROIs which demonstrated that both whole brain functional and structural connectivity exhibit reorganization with age. Components of the control, default mode, saliency/ventral attention, dorsal attention, and visual networks became less functionally cohesive, as evidenced by decreased component modularity. Paralleling this functional reorganization was a decrease in the density and weight of anatomical white-matter connections. Hub regions were affected by these changes, and the capacity of those regions to communicate with other regions exhibits a lifelong pattern of decline. Functional connectivity along multi-step structural paths tends to be stronger in older subjects than in younger subjects [85].
Associations between sMRI (gray matter) graphs and magnetoencephalography (MEG) graphs were also investigated. One study used 78 AAL cortical ROIs as nodes in both sMRI and MEG data to build structural and functional brain networks respectively in both healthy controls and multiple sclerosis (MS) patients. A beamformer approach was adopted to map the MEG data from sensor level to source space within the cortical ROIs. In MS patients, a more regular network organization for structural covariance graphs and for functional graphs in the theta band and a more random network organization for functional graphs in the alpha2 band were revealed. By computing the correlation coefficient between structural and functional connectivity measures across the nodes, a positive association between covariation in thickness and functional connectivity in especially the theta band in MS patients was revealed [242]. Another work studied the association among three graphs (fMRI, MEG, and structural MRI). FMRI and MEG graphs were built using the 78 AAL ROIs as well. By computing an overlap metric, a high overlap of nodes with high degree in these two resting-state functional graphs was found in fMRI and especially alpha band in MEG. This overlap was characterized by a strongly interconnected functional core network in temporo-posterior brain regions. After combining with structural data, by building a distance vs degree reconfiguration model, it was discovered that this functional core network could be explained by a trade-off between the product of the degrees of structurally-connected regions and the Euclidean distance between them. For both fMRI and MEG, the product of the degrees of connected regions was the most important predictor for functional network connectivity. Together, these results indicate that, irrespective of the modality, a functional core network in the human brain is especially shaped by communication between high degree nodes of the structural network [243].
C. Graphs with Multimodal Nodes
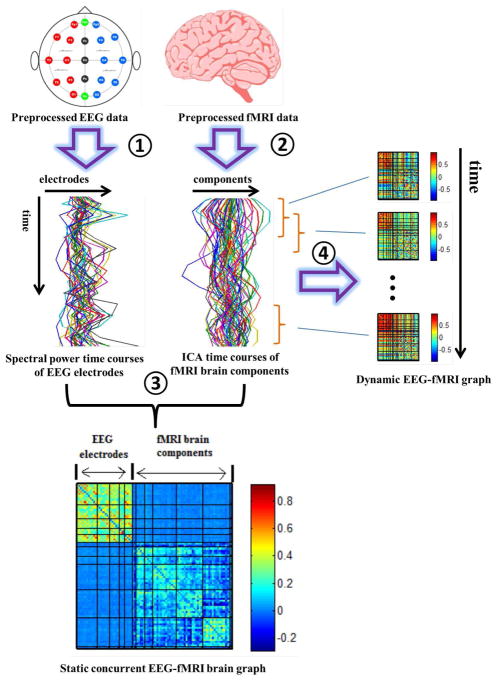
Another interesting approach for analyzing multimodal graph is to build a graph with nodes from multi-modalities. For example, an approach for building concurrent EEG-fMRI multi-modal brain graphs has been developed in which nodes are fMRI ICA spatial maps and EEG electrodes [244]. In that study, both static and dynamic EEG-fMRI graphs were estimated. Concurrent EEG-fMRI data were simultaneously collected during eyes open (EO) and eyes closed (EC) resting states. EEG time series were segmented into time windows with 2 seconds length (which was the TR of the fMRI data), and then spectral power of 5 frequency bands (delta; theta; alpha; beta; low gamma) of each time window was computed. Thus, the temporal resolution of EEG spectral power time courses was matched to that of the fMRI time series. At the global level, static graph measures and properties of dynamic graph measures were different across frequency bands and were mainly showing higher values in eyes closed than eyes open. Nodal level graph measures of a few brain components were also showing higher values during eyes closed in specific frequency bands. Overall, these findings incorporated fMRI spatial localization and EEG frequency information which could not be obtained by examining only one modality [244]. However, graph metrics were computed based on the formula defined for a single-modal (classical) graph. It is unclear how global level graph measures within a multi-modal graph are affected by the distribution of edges and nodes from different modalities. Thus, future studies should define new methods for computing topological measures in graphs with multimodal nodes. See Figure 6 for a pipe line of this method.
Fig. 6.
Pipeline for building concurrent EEG-fMRI multi-modal brain graphs (from Yu et al. [244]). ➀ Segment EEG signal into 2s time windows, and compute the average spectral power within a selected frequency window. ➁ Perform group ICA on fMRI data. ➂ Compute the correlation coefficient within and across the EEG spectral power’s and fMRI ICA’s full time courses, generating one EEG-fMRI static connectivity matrix for each frequency band. ➃ Segment EEG spectral power and fMRI ICA time courses into time windows, then compute the correlation between each pair of time windowed time courses to get dynamic EEG-fMRI brain graphs. (The figure was reused with permission from both author and journal of Yu et al. 2016 [244].)
Another work constructed graphs with multi-modality nodes and investigated how different brain areas were associated to genetic disorders and risk genes. In particular, a tripartite graph with genes, genetic diseases, and brain areas as nodes was constructed based on the associations among them reported in the literature through text mining. In the resulting graph, a disproportionately large number of gene-disease and disease-brain associations were attributed to a small subset of genes, disease, and brain areas. Furthermore, a small number of brain areas were found to be associated with a large number of the same genes and diseases. These core brain regions encompassed the areas identified by the previous genome-wide association studies, and suggest potential areas of focus in the future imaging genetics research [245].
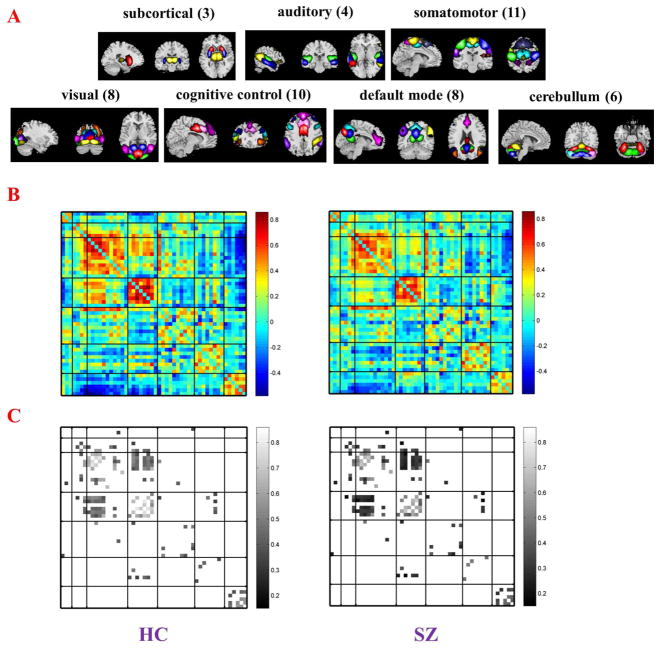
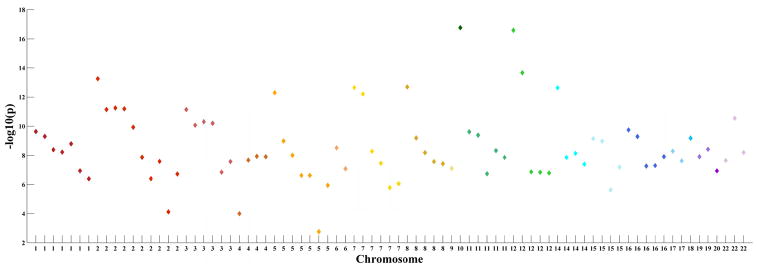
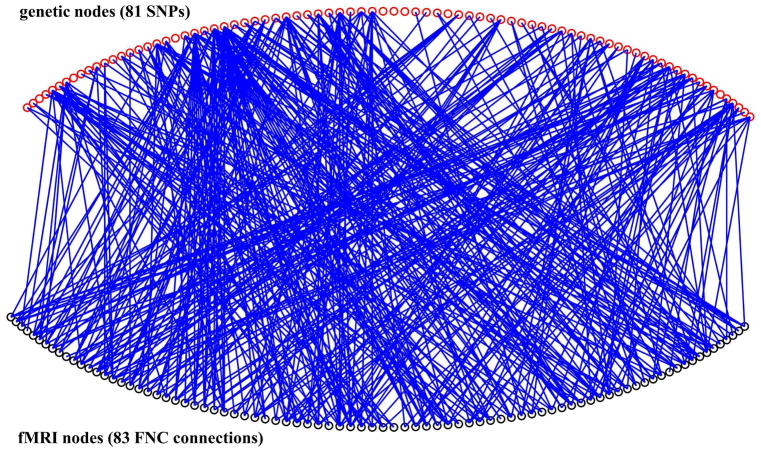
To explore genetic basis of functional brain connectivity, we build a bipartite graph which includes fMRI nodes and genetic nodes using a cohort consisted of 97 healthy controls and 70 patients with schizophrenia for which good-quality resting state fMRI and single nucleotide polymorphism (SNP) data have been collected. This analysis has not been reported before. However, for details on recruiting information and data processing see Damaraju et al. [246] and Chen et al. [247]. To define the brain nodes, fMRI are firstly decomposed into spatial components by group ICA. Then functional network connectivity (FNC) [248, 249] is estimated using the correlation between time courses of 50 brain components (Figure 7AB). In the bipartite graph which is built, fMRI nodes are not brain components. Instead, each fMRI node is a connection in the FNC. A total of 83 connections (Figure 7C) of the FNC show significant group difference (p < 0.05, FDR correction) are selected as the fMRI nodes. The genetic nodes in the bipartite graph are 81 schizophrenia associated SNPs selected base on the Psychiatric Genomics Consortium (PGC) study [250]. See Figure 8 for a Manhattan plot of these 81 SNPs. To define edges in the bipartite graph, the association between each pair of nodes (one from fMRI and the other from SNPs) is assessed across all 167 subjects using a multivariate regression model (http://mialab.mrn.org/software/mancovan) in which fMRI connectivity (correlation values of FNC connections) is the dependent variable, SNP allele (0, 1, or 2) is the independent variable, and data sites, diagnosis groups, and race are covariates. If the p value of any pair of fMRI node and SNP node is smaller than 0.05 (uncorrected), there is a link between them. Thus, the bipartite graph is a binary network. For the structure of this bipartite graph see Figure 9. Degree of each node is computed. SNPs with high degree in this SNP-FNC bipartite graph may modulate more functional brain connectivity. See Table 2 for the name of 81 SNP nodes and their degree. This analysis provides a new model to examine brain imaging and genetic associations. It can be adopted to investigate genetic basis of functional brain connectivity that is further related to mental illnesses.
Fig. 7.
Spatial maps (A) of 50 brain components which are selected to compute the functional network connectivity (FNC). Brain components are divided into groups and arranged based on their anatomical and functional properties. Structure of group mean FNC (B; HC: healthy controls; SZ: patients with schizophrenia) shows that brain connectivity is lower in SZ. Eighty three connections (C) in the FNC are selected to be nodes for constructing the SNP-FNC bipartite graph.
Fig. 8.
Scatter plots which showing schizophrenia associations of the selected 81 SNPs as genetic nodes and the distribution of them across chromosomes. The x axis is chromosomal position and the y axis is the significance (−log10(p)) of association as reported by the PGC study [250].
Fig. 9.
Structure of the genetic-fMRI bipartite graph. Some SNP nodes are showing high degree.
TABLE II.
Eighty one SNP nodes and degree of them in the fMRI-genetic bipartite graph.
| SNP | degree | SNP | degree | SNP | degree |
|---|---|---|---|---|---|
| ‘rs10503253’ | 31 | ‘rs778341’ | 5 | ‘rs9906500’ | 2 |
| ‘rs4674917’ | 24 | ‘rs2068012’ | 5 | ‘rs12903146’ | 2 |
| ‘rs7201930’ | 21 | ‘rs12129573’ | 4 | ‘rs4648845’ | 2 |
| ‘rs79235996’ | 15 | ‘rs9611520’ | 4 | ‘rs67401222’ | 2 |
| ‘rs604362’ | 14 | ‘rs4481150’ | 4 | ‘rs1209749’ | 2 |
| ‘rs3784399’ | 14 | ‘rs5995756’ | 4 | ‘rs12446487’ | 2 |
| ‘rs12623667’ | 14 | ‘rs1501362’ | 4 | ‘rs7722581’ | 2 |
| ‘rs7267348’ | 13 | ‘rs10206411’ | 4 | ‘rs10860955’ | 2 |
| ‘rs4908939’ | 12 | ‘rs7197756’ | 4 | ‘rs6867549’ | 2 |
| ‘rs11693094’ | 11 | ‘rs12939020’ | 4 | ‘rs2007044’ | 1 |
| ‘rs1009080’ | 9 | ‘rs2789605’ | 4 | ‘rs7108770’ | 1 |
| ‘rs34026011’ | 8 | ‘rs215411’ | 4 | ‘rs2973155’ | 1 |
| ‘rs11210892’ | 8 | ‘rs4129585’ | 3 | ‘rs34538000’ | 1 |
| ‘rs12132780’ | 8 | ‘rs11688767’ | 3 | ‘rs217326’ | 1 |
| ‘rs7896519’ | 7 | ‘rs16880919’ | 3 | ‘rs7438’ | 1 |
| ‘rs7240986’ | 7 | ‘rs2053079’ | 3 | ‘rs134900’ | 1 |
| ‘rs67733815’ | 7 | ‘rs11956240’ | 3 | ‘rs2693698’ | 1 |
| ‘rs4788190’ | 7 | ‘rs6846161’ | 3 | ‘rs17149781’ | 1 |
| ‘rs6670165’ | 7 | ‘rs7838490’ | 3 | ‘rs211792’ | 1 |
| ‘rs2851447’ | 6 | ‘rs1355585’ | 3 | ‘rs4546329’ | 0 |
| ‘rs9876421’ | 6 | ‘rs2300990’ | 3 | ‘rs12705304’ | 0 |
| ‘rs1076884’ | 6 | ‘rs10745572’ | 3 | ‘rs68002929’ | 0 |
| ‘rs4728408’ | 6 | ‘rs8039305’ | 2 | ‘rs11229116’ | 0 |
| ‘rs7018304’ | 6 | ‘rs13240464’ | 2 | ‘rs1470276’ | 0 |
| ‘rs7927176’ | 6 | ‘rs787983’ | 2 | ‘rs832190’ | 0 |
| ‘rs13227554’ | 5 | ‘rs2514218’ | 2 | ‘rs302321’ | 0 |
| ‘rs11693528’ | 5 | ‘rs66691851’ | 2 | ‘rs2909456’ | 0 |
In summary, there are three main ways to perform graph analysis in multimodal data. 1. Graphs built in one modality and then are associated with information from other modalities. 2. Graphs built in different modalities with the same nodes so that they are comparable. 3. A single multimodal graph in which nodes are formed using multi-modality data. Such approaches can expand our knowledge of the brain by linking together various complementary modalities.
V. Conclusion and perspective
In this article, we briefly review the findings of brain graphs in which nodes are defined using different methods. While structural (gray matter and diffusion image-based) brain graphs can be constructed using ROI nodes at different spatial resolutions, functional brain graphs may be built using either ROI or ICA nodes. Generally, both structural and functional brain graphs show features of complex networks, such as small world and scale free topology, rich club and modular organization with highly connected hubs. Moreover, those graph properties and some other measures such as clustering coefficient, local efficiency, global efficiency, path length show changes in patients with brain disorders in both graphs with ROI nodes and ICA nodes. These brain graph features have been studied at different time scales (across the life span, in different cognitive states, or moment to moment) as well as in multi-modality data.
When building brain graphs with ROI nodes at different spatial resolutions, it has been shown that the higher the resolution the more prominent is small-world properties. Region-based graphs fragmented more tremendously at high thresholds than voxel-based graphs, implying more robust graphs with high resolution nodes. ICA is a data-driven method for defining brain nodes in fMRI data that may mitigate some of the limitations of anatomical atlas-based nodes by providing regions that are temporally coherent and also adaptive to individual subjects. Graphs with different numbers of ICA spatial components have not been fully investigated. ROI and ICA-based methods for defining the graph nodes were compared in a simulation study, which revealed that graphs built with ICA nodes were closer to the ground truth than graphs built with ROI nodes, though this conclusion is limited to the scenarios that were simulated. More comparisons among graphs with nodes defined by different methods using real fMRI data are required in future studies. A potential direction is to perform data mining methods including deep learning, support vector machine classification, or clustering on topological measures of the brain graphs built with different nodes to evaluate which method performs better for classifying healthy controls and patients. Such approaches may have some clinical utility in the future; though it is also important to recognize that classification accuracy does not equate to advances in understanding the mechanisms of brain disease. For this, it is important to evaluate the predictive features in order to build new models of disease that can be tested in future work. Future studies may also try to define nodes using ICA or source based morphometry (SBM) [251] in structural MRI data.
In terms of dynamic brain graph, nodes with different resolutions within and/or between methods (ROI vs ICA) have rarely been compared. Future studies may build multi-layered brain graphs by incorporating both spatial-varying and time-varying information to investigate the development, growth, and state changes of the human brain [252].
Although multimodal graphs are helpful to better understand the topological properties of the brain by combining information from different modalities, and multi-modal parcellation of the human brain methods have also been developed [253], how to define the nodes is still a challenge when building a multimodal graph. Because different modalities provide different spatial or temporal information of the brain, it is hard to define comparable nodes across modalities. However, a promising way may be to build bipartite or tripartite graphs in which edges may only present within modality nodes not between modality nodes. In addition, network control theory may be adopted to investigate the relationship among modalities [254, 255]. Building multilayer brain graphs is another approach for performing multimodal data analysis [256–260].
Acknowledgments
“This work is supported by the National Institutes of Health (NIH) grants including a COBRE grant (P20GM103472/5P20RR021938), R01 grants (R01EB005846, 1R01EB006841, 1R01DA040487, R01REB020407, R01EB000840, and R37MH43775) and the National Science Foundation (NSF) grants #1539067, #1618551 and #1631838. This work is also partly supported by the “100 Talents Plan” of Chinese Academy of Sciences, the state high-tech development plan of China (863) 2015AA020513 (PI: JS), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB02060005), Chinese NSF (81471367, 61773380, PI: Sui J; 61703253, PI: YHD) and Natural Science Foundation of Shanxi (2016021077, PI: YHD).”
Biographies

Qingbao Yu Dr. Yu received his B.S. degree in physics and PhD. degree in biomedical engineering: neuroinformatics from Dalian University of Technology in 2003 and 2009 respectively. Then he worked at the Mind Research Network, NM, USA as a postdoctoral fellow and got promoted to Research Scientist in 2013. His research topics include ICA and graph theory analysis in human brain neuroimaging data (fMRI, EEG) and their applications in brain diseases. His ongoing work is trying to build multimodal brain graphs and to combine with deep learning methods to classify patients with mental illness from healthy controls. He has published more than 50 peer-reviewed journal papers. He also serves as reviewers for more than 20 peer-reviewed journals.

Yuhui Du Dr. Du is currently a Research Scientist at the Mind Research Network. She aims to develop new methods to accurately estimate brain functional networks and identify biomarkers from brain images. She also focuses on distinguishing different mental disorders using advanced machine learning methods, in order to achieve a biologically meaningful nosology and benefit individual diagnosis. She has published about 50 publications and won Trainee Abstract Travel Awards from the Organization for Human Brain Mapping in 2011 and 2016. She successfully obtained a patent and software for brain image analysis. As PI or co-investigator on several grants, she laid the groundwork by developing novel methods for brain network estimation and disease classification. She also serves as a reviewer for more than 20 journals in the neuroimaing field.

Jiayu Chen Dr. Chen received the Ph.D. degree in electrical engineering and is currently a research scientist at The Mind Research Network, Albuquerque, NM, USA. Her research is focused on developing computational algorithms for integrating multi-modal imaging, genetic and phenotypic data to better understand psychiatric disorders including but not limited to schizophrenia, bipolar disorder and attention deficit/hyperactivity disorder. She has published more than 40 peer-reviewed papers in this interdisciplinary field involving engineering, neuroscience, genetics and psychiatry.

Jing Sui (Senior Member, IEEE) received her B.S. and PhD. Degree in optical engineering with honors from Beijing Institute of Technology in 2002 and 2007 respectively. Then she worked at the Mind Research Network, NM, USA as a postdoctor fellow and got promoted to Research Scientist in 2010 and Assistant Professor of Translational Neuroscience in 2012. She is currently a full professor at the National Laboratory of Pattern Recognition & Brainnetome Center, Institute of Automation, Chinese Academy of Science (CAS). Her research interests include machine learning in neuroimaging, multi-modal brain imaging data fusion (fMRI, sMRI, dMRI, EEG, genetics), pattern recognition and their applications in mental illnesses. Her ongoing work in predictive data mining attempts to meet clinical challenges of making early and optimal intervention possible based on fundamental neuroimaging data. She is a recipient of One Hundred Talents plan of CAS in 2013. Now she has published 70 peer-reviewed journal articles and over 60 technical reports, abstracts and conference proceedings. She also serves as grant reviewer for China Natural Science Foundation, China Overseas Scholarship Council, Ministry of Science and Technology etc. and regular reviewers for more than 40 peer-reviewed journals.

Tülay Adali (S’89–M’92–SM’98–F’09) received the Ph.D. degree in Electrical Engineering from North Carolina State University, Raleigh, NC, USA, in 1992 and joined the faculty at the University of Maryland Baltimore County (UMBC), Baltimore, MD, the same year. She is currently a Distinguished University Professor in the Department of Computer Science and Electrical Engineering at UMBC. She has been active in conference and workshop organizations. She was the general or technical co-chair of the IEEE Machine Learning for Signal Processing (MLSP) and Neural Networks for Signal Processing Workshops 2001−2008, and helped organize a number of conferences including the IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP). She has served or currently serving on numerous editorial boards and technical committees of the IEEE Signal Processing Society. She was the chair of the technical committee on MLSP, 2003−2005 and 2011−2013, and the Technical Program Co-Chair for ICASSP 2017. She is the Special Sessions Chair for ICASSP 2018. Prof. Adali is a Fellow of the IEEE and the AIMBE, a Fulbright Scholar, and an IEEE Signal Processing Society Distinguished Lecturer. She was the recipient of a 2010 IEEE Signal Processing Society Best Paper Award, 2013 University System of Maryland Regents’ Award for Research, and an NSF CAREER Award. Her current research interests are in the areas of statistical signal processing, machine learning, and applications in medical image analysis and fusion.

Godfrey D. Pearlson Dr. Pearlson is currently Professor of Psychiatry & Neuroscience at Yale University Medical School, founding director of the Olin Neuropsychiatry Research Center (www.NRC-IOL.org) at the Institute of Living (IOL)/Hartford Hospital in Hartford CT and IOL Research Director. Dr. Pearlson’s research uses neuroimaging as a tool to address a broad array of questions regarding the neurobiology of major mental disorders, primarily psychosis (e.g. the NIMH-funded B-SNIP Consortium) and drug abuse (primarily alcohol & marijuana). He has received an NIMH MERIT award, a NARSAD Distinguished Investigator Award, the Ziskind-Sommerfeld Research Award from the Society of Biological Psychiatry, the Stanley Dean Award in Schizophrenia Research from the American College of Psychiatry, is a member of the Johns Hopkins Society of Scholars (for distinguished alumni) and was awarded the Albert M. Biele Lectureship from Jefferson Medical College, a Michael Visiting Professorship from the Weizmann Institute & is on the editorial board of several neuroimaging in psychiatric journals. He has published over 650 peer-reviewed research articles. He is also co-founder of the annual BrainDance Competition, which is open to high school and college students across New England. The BrainDance Awards encourage students to gain knowledge about psychiatric diseases and to develop a more tolerant and realistic perspective toward people with severe psychiatric problems.

Vince D. Calhoun, biography not available at the time of publication.
Contributor Information
Qingbao Yu, Mind Research Network, Albuquerque NM 87106 USA.
Yuhui Du, Mind Research Network, Albuquerque NM 87106 USA. And also with School of Computer & Information Technology, Shanxi University, Taiyuan, 030006, China.
Jiayu Chen, Mind Research Network, Albuquerque NM 87106 USA.
Jing Sui, University of Chinese Academy of Sciences, Beijing 100049 China. And also with CAS Center for Excellence in Brain Science and Intelligence Technology, Institute of Automation, Chinese Academy of Science (CAS), University of CAS, Beijing 100190 China.
Tulay Adali, Department of Computer Science and Electrical Engineering, University of Maryland, Baltimore County, Baltimore, MD 21250, USA.
Godfrey Pearlson, Olin Neuropsychiatry Research Center, Hartford, CT 06106, USA. And also with Departments of Psychiatry and Neurobiology, Yale University, New Haven, CT 06520, USA.
Vince D. Calhoun, Mind Research Network, Albuquerque NM 87106 USA. And also with Department of Electrical and Computer Engineering, University of New Mexico, Albuquerque, NM 87131, USA.
References
- 1.DeYoe EA, Bandettini P, Neitz J, Miller D, Winans P. Functional magnetic resonance imaging (FMRI) of the human brain. J Neurosci Methods. 1994 Oct;54:171–87. doi: 10.1016/0165-0270(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence SJD, Formisano E, Muckli L, de Lange FP. Laminar fMRI: Applications for cognitive neuroscience. Neuroimage. 2017 Jul 04; doi: 10.1016/j.neuroimage.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Heurling K, Leuzy A, Jonasson M, Frick A, Zimmer ER, Nordberg A, et al. Quantitative positron emission tomography in brain research. Brain Res. 2017 Jun 23; doi: 10.1016/j.brainres.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 4.Cabral J, Kringelbach ML, Deco G. Functional connectivity dynamically evolves on multiple time-scales over a static structural connectome: Models and mechanisms. Neuroimage. 2017 Mar 23; doi: 10.1016/j.neuroimage.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 5.Calamante F. Track-weighted imaging methods: extracting information from a streamlines tractogram. MAGMA. 2017 Feb 08; doi: 10.1007/s10334-017-0608-1. [DOI] [PubMed] [Google Scholar]
- 6.Castellanos FX, Cortese S, Proal E. Connectivity. Curr Top Behav Neurosci. 2014;16:49–77. doi: 10.1007/7854_2013_244. [DOI] [PubMed] [Google Scholar]
- 7.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echoplanar MRI. Magn Reson Med. 1995 Oct;34:537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 8.Friston KJ, Frith CD, Fletcher P, Liddle PF, Frackowiak RS. Functional topography: multidimensional scaling and functional connectivity in the brain. Cereb Cortex. 1996 Mar-Apr;6:156–64. doi: 10.1093/cercor/6.2.156. [DOI] [PubMed] [Google Scholar]
- 9.Friston KJ, Frith CD, Liddle PF, Frackowiak RS. Functional connectivity: the principal-component analysis of large (PET) data sets. J Cereb Blood Flow Metab. 1993 Jan;13:5–14. doi: 10.1038/jcbfm.1993.4. [DOI] [PubMed] [Google Scholar]
- 10.Vertes PE, Alexander-Bloch AF, Gogtay N, Giedd JN, Rapoport JL, Bullmore ET. Simple models of human brain functional networks. Proceedings of the National Academy of Sciences of the United States of America. 2012 Apr 10;109:5868–73. doi: 10.1073/pnas.1111738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zalesky A, Fornito A, Bullmore ET. Network-based statistic: Identifying differences in brain networks. Neuroimage. 2010 Dec;53:1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 12.Sporns O. From simple graphs to the connectome: Networks in neuroimaging. Neuroimage. 2011 Sep 10; doi: 10.1016/j.neuroimage.2011.08.085. [DOI] [PubMed] [Google Scholar]
- 13.Sporns O. The human connectome: a complex network. Annals of the New York Academy of Sciences. 2011 Apr;1224:109–25. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 14.Sporns O. Making sense of brain network data. Nat Methods. 2013 Jun;10:491–3. doi: 10.1038/nmeth.2485. [DOI] [PubMed] [Google Scholar]
- 15.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009 Mar;10:186–98. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 16.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010 Sep;52:1059–69. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, et al. Network Centrality in the Human Functional Connectome. Cerebral Cortex. 2011 Oct 12; doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]
- 18.He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007 Oct;17:2407–19. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, Zou Q, He Y, Yang Y. Topologically Reorganized Connectivity Architecture of Default-Mode, Executive-Control, and Salience Networks across Working Memory Task Loads. Cereb Cortex. 2016 Apr;26:1501–1511. doi: 10.1093/cercor/bhu316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Zuo X, He Y. Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci. 2010;4:16. doi: 10.3389/fnsys.2010.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassett DS, Gazzaniga MS. Understanding complexity in the human brain. Trends Cogn Sci. 2011 May;15:200–9. doi: 10.1016/j.tics.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muldoon SF, Bassett DS. Why network neuroscience? Compelling evidence and current frontiers. Comment on “Understanding brain networks and brain organization” by Luiz Pessoa. Phys Life Rev. 2014 Sep;11:455–7. doi: 10.1016/j.plrev.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Bassett DS, Sporns O. Network neuroscience. Nat Neurosci. 2017 Feb 23;20:353–364. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassett DS, Khambhati AN, Grafton ST. Emerging Frontiers of Neuroengineering: A Network Science of Brain Connectivity. Annu Rev Biomed Eng. 2017 Jun 21;19:327–352. doi: 10.1146/annurev-bioeng-071516-044511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stam CJ. Characterization of anatomical and functional connectivity in the brain: a complex networks perspective. Int J Psychophysiol. 2010 Sep;77:186–94. doi: 10.1016/j.ijpsycho.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 26.Goni J, van den Heuvel MP, Avena-Koenigsberger A, Velez de Mendizabal N, Betzel RF, Griffa A, et al. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014 Jan 14;111:833–8. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papo D, Buldu JM, Boccaletti S, Bullmore ET. Complex network theory and the brain. Philos Trans R Soc Lond B Biol Sci. 2014 Oct 05;369 doi: 10.1098/rstb.2013.0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friston KJ, Kahan J, Razi A, Stephan KE, Sporns O. On nodes and modes in resting state fMRI. Neuroimage. 2014 Oct 01;99:533–47. doi: 10.1016/j.neuroimage.2014.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sporns O. Cerebral cartography and connectomics. Philos Trans R Soc Lond B Biol Sci. 2015 May 19;370 doi: 10.1098/rstb.2014.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Heuvel MP, Bullmore ET, Sporns O. Comparative Connectomics. Trends Cogn Sci. 2016 May;20:345–61. doi: 10.1016/j.tics.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Rao H, Wang DJ, Yang Y, He Y. Neuroimaging of brain networks and function. Biomed Res Int. 2015;2015:509141. doi: 10.1155/2015/509141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fornito A, Zalesky A, Breakspear M. Graph analysis of the human connectome: promise, progress, and pitfalls. Neuroimage. 2013 Oct 15;80:426–44. doi: 10.1016/j.neuroimage.2013.04.087. [DOI] [PubMed] [Google Scholar]
- 33.Guye M, Bettus G, Bartolomei F, Cozzone PJ. Graph theoretical analysis of structural and functional connectivity MRI in normal and pathological brain networks. MAGMA. 2010 Dec;23:409–21. doi: 10.1007/s10334-010-0205-z. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser M. A tutorial in connectome analysis: Topological and spatial features of brain networks. Neuroimage. 2011 Aug 1;57:892–907. doi: 10.1016/j.neuroimage.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011 Jun 15;56:2068–79. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- 36.Telesford QK, Simpson SL, Burdette JH, Hayasaka S, Laurienti PJ. The brain as a complex system: using network science as a tool for understanding the brain. Brain Connect. 2011;1:295–308. doi: 10.1089/brain.2011.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sporns O. The non-random brain: efficiency, economy, and complex dynamics. Frontiers in Computational Neuroscience. 2011 Feb 8;5 doi: 10.3389/fncom.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Straaten ECW, Stam CJ. Structure out of chaos: Functional brain network analysis with EEG, MEG, and functional MRI. European Neuropsychopharmacology. 2013 Jan;23:7–18. doi: 10.1016/j.euroneuro.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Alexander-Bloch A, Giedd JN, Bullmore ET. Imaging structural co-variance between human brain regions. Nature Reviews Neuroscience. 2013 May;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sporns O. Structure and function of complex brain networks. Dialogues Clin Neurosci. 2013 Sep;15:247–62. doi: 10.31887/DCNS.2013.15.3/osporns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014 May;17:652–60. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 42.Bullmore E, Sporns O. The economy of brain network organization. Nat Rev Neurosci. 2012 May;13:336–49. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 43.Collin G, Sporns O, Mandl RC, van den Heuvel MP. Structural and Functional Aspects Relating to Cost and Benefit of Rich Club Organization in the Human Cerebral Cortex. Cereb Cortex. 2013 Apr 3; doi: 10.1093/cercor/bht064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis FC, Knodt AR, Sporns O, Lahey BB, Zald DH, Brigidi BD, et al. Impulsivity and the modular organization of resting-state neural networks. Cereb Cortex. 2013 Jun;23:1444–52. doi: 10.1093/cercor/bhs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goni J, Sporns O, Cheng H, Aznarez-Sanado M, Wang Y, Josa S, et al. Robust estimation of fractal measures for characterizing the structural complexity of the human brain: Optimization and reproducibility. Neuroimage. 2013 Jul 3;83C:646–657. doi: 10.1016/j.neuroimage.2013.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008 Jul 01;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Comput Biol. 2007 Feb 2;3:e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bassett DS, Bullmore ET, Meyer-Lindenberg A, Apud JA, Weinberger DR, Coppola R. Cognitive fitness of cost-efficient brain functional networks. Proc Natl Acad Sci U S A. 2009 Jul 14;106:11747–52. doi: 10.1073/pnas.0903641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fornito A, Zalesky A, Bullmore ET. Network scaling effects in graph analytic studies of human resting-state FMRI data. Front Syst Neurosci. 2010;4:22. doi: 10.3389/fnsys.2010.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Front Neurosci. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. Hierarchical modularity in human brain functional networks. Front Neuroinform. 2009;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen ZJ, He Y, Rosa-Neto P, Germann J, Evans AC. Revealing modular architecture of human brain structural networks by using cortical thickness from MRI. Cereb Cortex. 2008 Oct;18:2374–81. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, et al. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PLoS One. 2009;4:e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014 Jul 2;83:238–51. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu Q, Plis SM, Erhardt EB, Allen EA, Sui J, Kiehl KA, et al. Modular Organization of Functional Network Connectivity in Healthy Controls and Patients with Schizophrenia during the Resting State. Frontiers in systems neuroscience. 2011;5:103. doi: 10.3389/fnsys.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010 Aug;20:519–34. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Misic B, Betzel RF, de Reus MA, van den Heuvel MP, Berman MG, McIntosh AR, et al. Network-Level Structure-Function Relationships in Human Neocortex. Cereb Cortex. 2016 Jul;26:3285–96. doi: 10.1093/cercor/bhw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng G, Chen HC, Zhu Z, He Y, Wang S. Dynamic brain architectures in local brain activity and functional network efficiency associate with efficient reading in bilinguals. Neuroimage. 2015 Oct 01;119:103–18. doi: 10.1016/j.neuroimage.2015.05.100. [DOI] [PubMed] [Google Scholar]
- 59.Wang ZJ, Dai ZJ, Gong GL, Zhou CS, He Y. Understanding Structural-Functional Relationships in the Human Brain: A Large-Scale Network Perspective. Neuroscientist. 2015 Jun;21:290–305. doi: 10.1177/1073858414537560. [DOI] [PubMed] [Google Scholar]
- 60.Park HJ, Friston KJ. Structural and Functional Brain Networks: From Connections to Cognition. Science. 2013 Nov 1;342:579-+. doi: 10.1126/science.1238411. [DOI] [PubMed] [Google Scholar]
- 61.Karuza EA, Thompson-Schill SL, Bassett DS. Local Patterns to Global Architectures: Influences of Network Topology on Human Learning. Trends Cogn Sci. 2016 Aug;20:629–40. doi: 10.1016/j.tics.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A. 2011 May 3;108:7641–6. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bassett DS, Wymbs NF, Rombach MP, Porter MA, Mucha PJ, Grafton ST. Task-based core-periphery organization of human brain dynamics. PLoS Comput Biol. 2013;9:e1003171. doi: 10.1371/journal.pcbi.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bassett DS, Yang M, Wymbs NF, Grafton ST. Learning-induced autonomy of sensorimotor systems. Nat Neurosci. 2015 May;18:744–51. doi: 10.1038/nn.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braun U, Schafer A, Walter H, Erk S, Romanczuk-Seiferth N, Haddad L, et al. Dynamic reconfiguration of frontal brain networks during executive cognition in humans. Proc Natl Acad Sci U S A. 2015 Sep 15;112:11678–83. doi: 10.1073/pnas.1422487112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlson JM, Alderson DL, Stromberg SP, Bassett DS, Craparo EM, Guiterrez-Villarreal F, et al. Measuring and modeling behavioral decision dynamics in collective evacuation. PLoS One. 2014;9:e87380. doi: 10.1371/journal.pone.0087380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davison EN, Schlesinger KJ, Bassett DS, Lynall ME, Miller MB, Grafton ST, et al. Brain network adaptability across task states. PLoS Comput Biol. 2015 Jan;11:e1004029. doi: 10.1371/journal.pcbi.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu S, Satterthwaite TD, Medaglia JD, Yang M, Gur RE, Gur RC, et al. Emergence of system roles in normative neurodevelopment. Proc Natl Acad Sci U S A. 2015 Nov 03;112:13681–6. doi: 10.1073/pnas.1502829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Medaglia JD, Lynall ME, Bassett DS. Cognitive network neuroscience. J Cogn Neurosci. 2015 Aug;27:1471–91. doi: 10.1162/jocn_a_00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Telesford QK, Lynall ME, Vettel J, Miller MB, Grafton ST, Bassett DS. Detection of functional brain network reconfiguration during task-driven cognitive states. Neuroimage. 2016 Nov 15;142:198–210. doi: 10.1016/j.neuroimage.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soto FA, Bassett DS, Ashby FG. Dissociable changes in functional network topology underlie early category learning and development of automaticity. Neuroimage. 2016 Nov 01;141:220–41. doi: 10.1016/j.neuroimage.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chai LR, Mattar MG, Blank IA, Fedorenko E, Bassett DS. Functional Network Dynamics of the Language System. Cereb Cortex. 2016 Oct 17;26:4148–4159. doi: 10.1093/cercor/bhw238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bassett DS, Mattar MG. A Network Neuroscience of Human Learning: Potential to Inform Quantitative Theories of Brain and Behavior. Trends Cogn Sci. 2017 Apr;21:250–264. doi: 10.1016/j.tics.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmalzle R, Brook O’Donnell M, Garcia JO, Cascio CN, Bayer J, Bassett DS, et al. Brain connectivity dynamics during social interaction reflect social network structure. Proc Natl Acad Sci U S A. 2017 May 16;114:5153–5158. doi: 10.1073/pnas.1616130114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baum GL, Ciric R, Roalf DR, Betzel RF, Moore TM, Shinohara RT, et al. Modular Segregation of Structural Brain Networks Supports the Development of Executive Function in Youth. Curr Biol. 2017 Jun 05;27:1561–1572 e8. doi: 10.1016/j.cub.2017.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ashourvan A, Gu S, Mattar MG, Vettel JM, Bassett DS. The energy landscape underpinning module dynamics in the human brain connectome. Neuroimage. 2017 Jun 07;157:364–380. doi: 10.1016/j.neuroimage.2017.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Telesford QK, Ashourvan A, Wymbs NF, Grafton ST, Vettel JM, Bassett DS. Cohesive network reconfiguration accompanies extended training. Hum Brain Mapp. 2017 Jun 24; doi: 10.1002/hbm.23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bota M, Sporns O, Swanson LW. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci U S A. 2015 Apr 21;112:E2093–101. doi: 10.1073/pnas.1504394112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petersen SE, Sporns O. Brain Networks and Cognitive Architectures. Neuron. 2015 Oct 07;88:207–19. doi: 10.1016/j.neuron.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Betzel RF, Fukushima M, He Y, Zuo XN, Sporns O. Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage. 2016 Feb 15;127:287–97. doi: 10.1016/j.neuroimage.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohr H, Wolfensteller U, Betzel RF, Misic B, Sporns O, Richiardi J, et al. Integration and segregation of large-scale brain networks during short-term task automatization. Nature Communications. 2016 Nov 3;7 doi: 10.1038/ncomms13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liao X, Cao M, Xia M, He Y. Individual differences and time-varying features of modular brain architecture. Neuroimage. 2017 May 15;152:94–107. doi: 10.1016/j.neuroimage.2017.02.066. [DOI] [PubMed] [Google Scholar]
- 83.Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. Proc Natl Acad Sci U S A. 2014 Jul 15;111:10341–6. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009 Feb 1;44:715–23. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 85.Betzel RF, Byrge L, He Y, Goni J, Zuo XN, Sporns O. Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage. 2014 Nov 15;102(Pt 2):345–57. doi: 10.1016/j.neuroimage.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 86.Chen ZJ, He Y, Rosa-Neto P, Gong G, Evans AC. Age-related alterations in the modular organization of structural cortical network by using cortical thickness from MRI. Neuroimage. 2011 May 1;56:235–45. doi: 10.1016/j.neuroimage.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 87.Wu K, Taki Y, Sato K, Kinomura S, Goto R, Okada K, et al. Age-related changes in topological organization of structural brain networks in healthy individuals. Hum Brain Mapp. 2011 Mar 9; doi: 10.1002/hbm.21232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davison EN, Turner BO, Schlesinger KJ, Miller MB, Grafton ST, Bassett DS, et al. Individual Differences in Dynamic Functional Brain Connectivity across the Human Lifespan. PLoS Comput Biol. 2016 Nov;12:e1005178. doi: 10.1371/journal.pcbi.1005178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gu S, Yang M, Medaglia JD, Gur RC, Gur RE, Satterthwaite TD, et al. Functional hypergraph uncovers novel covariant structures over neurodevelopment. Hum Brain Mapp. 2017 Aug;38:3823–3835. doi: 10.1002/hbm.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Otte WM, van Diessen E, Paul S, Ramaswamy R, Subramanyam Rallabandi VP, Stam CJ, et al. Aging alterations in whole-brain networks during adulthood mapped with the minimum spanning tree indices: the interplay of density, connectivity cost and life-time trajectory. Neuroimage. 2015 Apr 01;109:171–89. doi: 10.1016/j.neuroimage.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 91.Smit DJ, de Geus EJ, Boersma M, Boomsma DI, Stam CJ. Life-Span Development of Brain Network Integration Assessed with Phase Lag Index Connectivity and Minimum Spanning Tree Graphs. Brain Connect. 2016 May;6:312–25. doi: 10.1089/brain.2015.0359. [DOI] [PubMed] [Google Scholar]
- 92.Sinke MR, Dijkhuizen RM, Caimo A, Stam CJ, Otte WM. Bayesian exponential random graph modeling of whole-brain structural networks across lifespan. Neuroimage. 2016 Jul 15;135:79–91. doi: 10.1016/j.neuroimage.2016.04.066. [DOI] [PubMed] [Google Scholar]
- 93.van den Heuvel MP, Kersbergen KJ, de Reus MA, Keunen K, Kahn RS, Groenendaal F, et al. The Neonatal Connectome During Preterm Brain Development. Cereb Cortex. 2015 Sep;25:3000–13. doi: 10.1093/cercor/bhu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koenis MMG, Brouwer RM, van den Heuvel MP, Mandl RCW, van Soelen ILC, Kahn RS, et al. Development of the Brain’s Structural Network Efficiency in Early Adolescence: A Longitudinal DTI Twin Study. Human Brain Mapping. 2015 Dec;36:4938–4953. doi: 10.1002/hbm.22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wierenga LM, van den Heuvel MP, van Dijk S, Rijks Y, de Reus MA, Durston S. The development of brain network architecture. Hum Brain Mapp. 2016 Feb;37:717–29. doi: 10.1002/hbm.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Betzel RF, Avena-Koenigsberger A, Goni J, He Y, de Reus MA, Griffa A, et al. Generative models of the human connectome. Neuroimage. 2016 Jan 01;124:1054–64. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zuo XN, He Y, Betzel RF, Colcombe S, Sporns O, Milham MP. Human Connectomics across the Life Span. Trends Cogn Sci. 2017 Jan;21:32–45. doi: 10.1016/j.tics.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 98.Zhao T, Cao M, Niu H, Zuo XN, Evans A, He Y, et al. Age-related changes in the topological organization of the white matter structural connectome across the human lifespan. Hum Brain Mapp. 2015 Oct;36:3777–92. doi: 10.1002/hbm.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cao M, Huang H, Peng Y, Dong Q, He Y. Toward Developmental Connectomics of the Human Brain. Front Neuroanat. 2016;10:25. doi: 10.3389/fnana.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhong S, He Y, Shu H, Gong GL. Developmental Changes in Topological Asymmetry Between Hemispheric Brain White Matter Networks from Adolescence to Young Adulthood. Cerebral Cortex. 2017 Apr;27:2560–2570. doi: 10.1093/cercor/bhw109. [DOI] [PubMed] [Google Scholar]
- 101.Cao M, Huang H, He Y. Developmental Connectomics from Infancy through Early Childhood. Trends Neurosci. 2017 Jul 03; doi: 10.1016/j.tins.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baker ST, Lubman DI, Yucel M, Allen NB, Whittle S, Fulcher BD, et al. Developmental Changes in Brain Network Hub Connectivity in Late Adolescence. J Neurosci. 2015 Jun 17;35:9078–87. doi: 10.1523/JNEUROSCI.5043-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hagmann P, Grant PE, Fair DA. MR connectomics: a conceptual framework for studying the developing brain. Front Syst Neurosci. 2012;6:43. doi: 10.3389/fnsys.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zalesky A, Fornito A, Seal ML, Cocchi L, Westin CF, Bullmore ET, et al. Disrupted axonal fiber connectivity in schizophrenia. Biol Psychiatry. 2011 Jan 1;69:80–9. doi: 10.1016/j.biopsych.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005 Sep;15:1332–42. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- 106.Olabi B, Ellison-Wright I, Bullmore E, Lawrie SM. Structural brain changes in first episode Schizophrenia compared with Fronto-Temporal Lobar Degeneration: a meta-analysis. BMC psychiatry. 2012 Aug 7;12:104. doi: 10.1186/1471-244X-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lynall ME, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, et al. Functional connectivity and brain networks in schizophrenia. J Neurosci. 2010 Jul 14;30:9477–87. doi: 10.1523/JNEUROSCI.0333-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lo CYZ, Su TW, Huang CC, Hung CC, Chen WL, Lan TH, et al. Randomization and resilience of brain functional networks as systems-level endophenotypes of schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2015 Jul 21;112:9123–9128. doi: 10.1073/pnas.1502052112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fornito A, Zalesky A, Pantelis C, Bullmore ET. Schizophrenia, neuroimaging and connectomics. Neuroimage. 2012 Oct 1;62:2296–314. doi: 10.1016/j.neuroimage.2011.12.090. [DOI] [PubMed] [Google Scholar]
- 110.Fornito A, Harrison BJ, Goodby E, Dean A, Ooi C, Nathan PJ, et al. Functional dysconnectivity of corticostriatal circuitry as a risk phenotype for psychosis. JAMA Psychiatry. 2013 Nov;70:1143–51. doi: 10.1001/jamapsychiatry.2013.1976. [DOI] [PubMed] [Google Scholar]
- 111.Fornito A, Bullmore ET. Reconciling abnormalities of brain network structure and function in schizophrenia. Curr Opin Neurobiol. 2015 Feb;30:44–50. doi: 10.1016/j.conb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 112.Fornito A, Bullmore ET. Connectomics: a new paradigm for understanding brain disease. Eur Neuropsychopharmacol. 2015 May;25:733–48. doi: 10.1016/j.euroneuro.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 113.Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014 Aug;137:2382–95. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bassett DS, Bullmore ET. Human brain networks in health and disease. Curr Opin Neurol. 2009 Aug;22:340–7. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 2008 Sep 10;28:9239–48. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alexander-Bloch AF, Vertes PE, Stidd R, Lalonde F, Clasen L, Rapoport J, et al. The anatomical distance of functional connections predicts brain network topology in health and schizophrenia. Cereb Cortex. 2013 Jan;23:127–38. doi: 10.1093/cercor/bhr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RC, Cahn W, et al. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry. 2013 Aug 1;70:783–92. doi: 10.1001/jamapsychiatry.2013.1328. [DOI] [PubMed] [Google Scholar]
- 118.Cao M, Wang Z, He Y. Connectomics in psychiatric research: advances and applications. Neuropsychiatr Dis Treat. 2015;11:2801–10. doi: 10.2147/NDT.S63470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gong Q, He Y. Depression, neuroimaging and connectomics: a selective overview. Biol Psychiatry. 2015 Feb 1;77:223–35. doi: 10.1016/j.biopsych.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 120.He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer’s disease. J Neurosci. 2008 Apr 30;28:4756–66. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Du Y, Li Z, Li L, Chen ZG, Sun SY, Chen P, et al. Distinct growth factor-induced dynamic mass redistribution (DMR) profiles for monitoring oncogenic signaling pathways in various cancer cells. J Recept Signal Transduct Res. 2009;29:182–94. doi: 10.1080/10799890902976933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Y, Liang M, Zhou Y, He Y, Hao Y, Song M, et al. Disrupted small-world networks in schizophrenia. Brain. 2008 Apr;131:945–61. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- 123.Xia M, He Y. Magnetic resonance imaging and graph theoretical analysis of complex brain networks in neuropsychiatric disorders. Brain Connect. 2011;1:349–65. doi: 10.1089/brain.2011.0062. [DOI] [PubMed] [Google Scholar]
- 124.Bassett DS, Nelson BG, Mueller BA, Camchong J, Lim KO. Altered resting state complexity in schizophrenia. Neuroimage. 2012 Feb 1;59:2196–207. doi: 10.1016/j.neuroimage.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Betzel RF, Bassett DS. Multi-scale brain networks. Neuroimage. 2016 Nov 11; doi: 10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu Q, Allen EA, Sui J, Arbabshirani MR, Pearlson G, Calhoun VD. Brain Connectivity Networks In Schizophrenia Underlying Resting State Functional Magnetic Resonance Imaging. Current topics in medicinal chemistry. 2012 Dec 21;12:2415–2425. doi: 10.2174/156802612805289890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu Q, Sui J, Kiehl KA, Pearlson G, Calhoun VD. State-related functional integration and functional segregation brain networks in schizophrenia. Schizophr Res. 2013 Nov;150:450–8. doi: 10.1016/j.schres.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu Q, Sui J, Liu J, Plis SM, Kiehl KA, Pearlson G, et al. Disrupted correlation between low frequency power and connectivity strength of resting state brain networks in schizophrenia. Schizophrenia research. 2013 Jan;143:165–71. doi: 10.1016/j.schres.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu Q, Sui J, Rachakonda S, He H, Gruner W, Pearlson G, et al. Altered topological properties of functional network connectivity in schizophrenia during resting state: a small-world brain network study. PLoS One. 2011;6:e25423. doi: 10.1371/journal.pone.0025423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu Q, Sui J, Rachakonda S, He H, Pearlson G, Calhoun VD. Altered small-world brain networks in temporal lobe in patients with schizophrenia performing an auditory oddball task. Front Syst Neurosci. 2011;5:7. doi: 10.3389/fnsys.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang J, Cheng W, Liu Z, Zhang K, Lei X, Yao Y, et al. Neural, electrophysiological and anatomical basis of brain-network variability and its characteristic changes in mental disorders. Brain. 2016 Aug;139:2307–21. doi: 10.1093/brain/aww143. [DOI] [PubMed] [Google Scholar]
- 132.Nelson BG, Bassett DS, Camchong J, Bullmore ET, Lim KO. Comparison of large-scale human brain functional and anatomical networks in schizophrenia. Neuroimage Clin. 2017;15:439–448. doi: 10.1016/j.nicl.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stam CJ. Modern network science of neurological disorders. Nat Rev Neurosci. 2014 Oct;15:683–95. doi: 10.1038/nrn3801. [DOI] [PubMed] [Google Scholar]
- 134.van Diessen E, Zweiphenning WJ, Jansen FE, Stam CJ, Braun KP, Otte WM. Brain Network Organization in Focal Epilepsy: A Systematic Review and Meta-Analysis. PLoS One. 2014;9:e114606. doi: 10.1371/journal.pone.0114606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Dellen E, Bohlken MM, Draaisma L, Tewarie PK, van Lutterveld R, Mandl R, et al. Structural Brain Network Disturbances in the Psychosis Spectrum. Schizophr Bull. 2016 May;42:782–9. doi: 10.1093/schbul/sbv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pievani M, Filippini N, van den Heuvel MP, Cappa SF, Frisoni GB. Brain connectivity in neurodegenerative diseases–from phenotype to proteinopathy. Nat Rev Neurol. 2014 Nov;10:620–33. doi: 10.1038/nrneurol.2014.178. [DOI] [PubMed] [Google Scholar]
- 137.van den Heuvel MP, Fornito A. Brain networks in schizophrenia. Neuropsychol Rev. 2014 Mar;24:32–48. doi: 10.1007/s11065-014-9248-7. [DOI] [PubMed] [Google Scholar]
- 138.Collin G, Kahn RS, de Reus MA, Cahn W, van den Heuvel MP. Impaired rich club connectivity in unaffected siblings of schizophrenia patients. Schizophr Bull. 2014 Mar;40:438–48. doi: 10.1093/schbul/sbt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Collin G, van den Heuvel MP, Abramovic L, Vreeker A, de Reus MA, van Haren NEM, et al. Brain Network Analysis Reveals Affected Connectome Structure in Bipolar I Disorder. Human Brain Mapping. 2016 Jan;37:122–134. doi: 10.1002/hbm.23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van den Heuvel MP, Scholtens LH, de Reus MA, Kahn RS. Associated Microscale Spine Density and Macroscale Connectivity Disruptions in Schizophrenia. Biological Psychiatry. 2016 Aug 15;80:293–301. doi: 10.1016/j.biopsych.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 141.Yeo RA, Ryman SG, van den Heuvel MP, de Reus MA, Jung RE, Pommy J, et al. Graph Metrics of Structural Brain Networks in Individuals with Schizophrenia and Healthy Controls: Group Differences, Relationships with Intelligence, and Genetics. Journal of the International Neuropsychological Society. 2016 Feb;22:240–249. doi: 10.1017/S1355617715000867. [DOI] [PubMed] [Google Scholar]
- 142.Kambeitz J, Kambeitz-Ilankovic L, Cabral C, Dwyer DB, Calhoun VD, van den Heuvel MP, et al. Aberrant Functional Whole-Brain Network Architecture in Patients With Schizophrenia: A Meta-analysis. Schizophrenia Bulletin. 2016 Jul;42:S13–S21. doi: 10.1093/schbul/sbv174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reess TJ, Rus OG, Schmidt R, de Reus MA, Zaudig M, Wagner G, et al. Connectomics-based structural network alterations in obsessive-compulsive disorder. Translational Psychiatry. 2016 Sep 6;6 doi: 10.1038/tp.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Romme IAC, de Reus MA, Ophoff RA, Kahn RS, van den Heuvel MP. Connectome Disconnectivity and Cortical Gene Expression in Patients With Schizophrenia. Biological Psychiatry. 2017 Mar 15;81:495–502. doi: 10.1016/j.biopsych.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 145.Galantucci S, Agosta F, Stefanova E, Basaia S, van den Heuvel MP, Stojkovic T, et al. Structural Brain Connectome and Cognitive Impairment in Parkinson Disease. Radiology. 2017 May;283:515–525. doi: 10.1148/radiol.2016160274. [DOI] [PubMed] [Google Scholar]
- 146.Bos DJ, Oranje B, Achterberg M, Vlaskamp C, Ambrosino S, de Reus MA, et al. Structural and functional connectivity in children and adolescents with and without attention deficit/hyperactivity disorder. Journal of Child Psychology and Psychiatry. 2017 Jul;58:810–818. doi: 10.1111/jcpp.12712. [DOI] [PubMed] [Google Scholar]
- 147.Crossley NA, Mechelli A, Ginestet C, Rubinov M, Bullmore ET, McGuire P. Altered Hub Functioning and Compensatory Activations in the Connectome: A Meta-Analysis of Functional Neuroimaging Studies in Schizophrenia. Schizophr Bull. 2016 Mar;42:434–42. doi: 10.1093/schbul/sbv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hulshoff Pol H, Bullmore E. Neural networks in psychiatry. Eur Neuropsychopharmacol. 2013 Jan;23:1–6. doi: 10.1016/j.euroneuro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 149.Cary RP, Ray S, Grayson DS, Painter J, Carpenter S, Maron L, et al. Network Structure among Brain Systems in Adult ADHD is Uniquely Modified by Stimulant Administration. Cereb Cortex. 2016 Jul 14; doi: 10.1093/cercor/bhw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bi Y, He Y. Connectomics reveals faulty wiring patterns for depressed brain. Biol Psychiatry. 2014 Oct 01;76:515–6. doi: 10.1016/j.biopsych.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 151.Dai Z, Yan C, Li K, Wang Z, Wang J, Cao M, et al. Identifying and Mapping Connectivity Patterns of Brain Network Hubs in Alzheimer’s Disease. Cereb Cortex. 2015 Oct;25:3723–42. doi: 10.1093/cercor/bhu246. [DOI] [PubMed] [Google Scholar]
- 152.He Y, Evans A. Magnetic resonance imaging of healthy and diseased brain networks. Front Hum Neurosci. 2014;8:890. doi: 10.3389/fnhum.2014.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.He Y. Imaging brain networks in neurodegenerative diseases. CNS Neurosci Ther. 2015 Oct;21:751–3. doi: 10.1111/cns.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cao M, Wang ZJ, He Y. Connectomics in psychiatric research: advances and applications. Neuropsychiatric Disease and Treatment. 2015;11:2801–2810. doi: 10.2147/NDT.S63470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015 Mar;16:159–72. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- 156.Klauser P, Baker ST, Cropley VL, Bousman C, Fornito A, Cocchi L, et al. White Matter Disruptions in Schizophrenia Are Spatially Widespread and Topologically Converge on Brain Network Hubs. Schizophr Bull. 2017 Mar 01;43:425–435. doi: 10.1093/schbul/sbw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Micheloyannis S. Graph-based network analysis in schizophrenia. World J Psychiatry. 2012 Feb 22;2:1–12. doi: 10.5498/wjp.v2.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rubinov M, Bullmore E. Schizophrenia and abnormal brain network hubs. Dialogues Clin Neurosci. 2013 Sep;15:339–49. doi: 10.31887/DCNS.2013.15.3/mrubinov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rubinov M, Bullmore ET. Fledgling pathoconnectomics of psychiatric disorders. Trends in Cognitive Sciences. 2013 Dec;17:641–647. doi: 10.1016/j.tics.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 160.Deco G, Kringelbach ML. Great expectations: using whole-brain computational connectomics for understanding neuropsychiatric disorders. Neuron. 2014 Dec 03;84:892–905. doi: 10.1016/j.neuron.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 161.Bernhardt BC, Bonilha L, Gross DW. Network analysis for a network disorder: The emerging role of graph theory in the study of epilepsy. Epilepsy & Behavior. 2015 Sep;50:162–170. doi: 10.1016/j.yebeh.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 162.Butts CT. Revisiting the foundations of network analysis. Science. 2009 Jul 24;325:414–6. doi: 10.1126/science.1171022. [DOI] [PubMed] [Google Scholar]
- 163.Smith SM. The future of FMRI connectivity. Neuroimage. 2012 Aug 15;62:1257–66. doi: 10.1016/j.neuroimage.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 164.Khullar S, Michael AM, Cahill ND, Kiehl KA, Pearlson G, Baum SA, et al. ICA-fNORM: Spatial Normalization of fMRI Data Using Intrinsic Group-ICA Networks. Frontiers in systems neuroscience. 2011;5:93. doi: 10.3389/fnsys.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Du YH, Li HM, Wu H, Fan Y. Identification of subject specific and functional consistent ROIs using semi-supervised learning. Medical Imaging 2012: Image Processing. 2012;8314 [Google Scholar]
- 166.Calhoun VD, Eichele T, Adali T, Allen EA. Decomposing the brain: components and modes, networks and nodes. Trends Cogn Sci. 2012 May;16:255–6. doi: 10.1016/j.tics.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stanley ML, Moussa MN, Paolini BM, Lyday RG, Burdette JH, Laurienti PJ. Defining nodes in complex brain networks. Front Comput Neurosci. 2013 Nov 22;7:169. doi: 10.3389/fncom.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2014 Mar;24:663–76. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Calhoun VD, Miller R, Pearlson G, Adali T. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron. 2014 Oct 22;84:262–74. doi: 10.1016/j.neuron.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Marek S, Hwang K, Foran W, Hallquist MN, Luna B. The Contribution of Network Organization and Integration to the Development of Cognitive Control. Plos Biology. 2015 Dec;13 doi: 10.1371/journal.pbio.1002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moussa MN, Vechlekar CD, Burdette JH, Steen MR, Hugenschmidt CE, Laurienti PJ. Changes in cognitive state alter human functional brain networks. Front Hum Neurosci. 2011;5:83. doi: 10.3389/fnhum.2011.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yu Q, Erhardt EB, Sui J, Du Y, He H, Hjelm D, et al. Assessing dynamic brain graphs of time-varying connectivity in fMRI data: Application to healthy controls and patients with schizophrenia. Neuroimage. 2015 Feb 15;107:345–55. doi: 10.1016/j.neuroimage.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sui J, Adali T, Yu Q, Chen J, Calhoun VD. A review of multivariate methods for multimodal fusion of brain imaging data. Journal of neuroscience methods. 2012 Feb 15;204:68–81. doi: 10.1016/j.jneumeth.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Friston KJ. Modalities, modes, and models in functional neuroimaging. Science. 2009 Oct 16;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- 175.Sui J, Yu Q, He H, Pearlson GD, Calhoun VD. A selective review of multimodal fusion methods in schizophrenia. Frontiers in human neuroscience. 2012;6:27. doi: 10.3389/fnhum.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sui J, Huster R, Yu Q, Segall JM, Calhoun VD. Function-structure associations of the brain: evidence from multimodal connectivity and covariance studies. Neuroimage. 2014 Nov 15;102(Pt 1):11–23. doi: 10.1016/j.neuroimage.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tijms BM, Series P, Willshaw DJ, Lawrie SM. Similarity-based extraction of individual networks from gray matter MRI scans. Cereb Cortex. 2012 Jul;22:1530–41. doi: 10.1093/cercor/bhr221. [DOI] [PubMed] [Google Scholar]
- 178.Romero-Garcia R, Atienza M, Clemmensen LH, Cantero JL. Effects of network resolution on topological properties of human neocortex. Neuroimage. 2012 Feb 15;59:3522–3532. doi: 10.1016/j.neuroimage.2011.10.086. [DOI] [PubMed] [Google Scholar]
- 179.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002 Jan;15:273–89. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 180.Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, et al. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex. 2009 Mar;19:524–36. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Vaessen MJ, Hofman PAM, Tijssen HN, Aldenkamp AP, Jansen JFA, Backes WH. The effect and reproducibility of different clinical DTI gradient sets on small world brain connectivity measures. Neuroimage. 2010 Jul 1;51:1106–1116. doi: 10.1016/j.neuroimage.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 182.Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011 Jan 15;54:1262–79. doi: 10.1016/j.neuroimage.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 183.van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011 Nov 2;31:15775–86. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.van den Heuvel MP, Kahn RS, Goni J, Sporns O. High-cost, high-capacity backbone for global brain communication. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jul 10;109:11372–11377. doi: 10.1073/pnas.1203593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.van den Heuvel MP, Mandl RCW, Stam CJ, Kahn RS, Pol HEH. Aberrant Frontal and Temporal Complex Network Structure in Schizophrenia: A Graph Theoretical Analysis. Journal of Neuroscience. 2010 Nov 24;30:15915–15926. doi: 10.1523/JNEUROSCI.2874-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang QF, Su TP, Zhou Y, Chou KH, Chen IY, Jiang TZ, et al. Anatomical insights into disrupted small-world networks in schizophrenia. Neuroimage. 2012 Jan 16;59:1085–1093. doi: 10.1016/j.neuroimage.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 187.Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J Neurosci. 2006 Jan 4;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Eguiluz VM, Chialvo DR, Cecchi GA, Baliki M, Apkarian AV. Scale-free brain functional networks. Phys Rev Lett. 2005 Jan 14;94:018102. doi: 10.1103/PhysRevLett.94.018102. [DOI] [PubMed] [Google Scholar]
- 189.van den Heuvel MP, Stam CJ, Boersma M, Hulshoff Pol HE. Small-world and scale-free organization of voxel-based resting-state functional connectivity in the human brain. Neuroimage. 2008 Nov 15;43:528–39. doi: 10.1016/j.neuroimage.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 190.Bassett DS, Bullmore E. Small-world brain networks. Neuroscientist. 2006 Dec;12:512–23. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 191.Bertolero MA, Yeo BTT, D’Esposito M. The modular and integrative functional architecture of the human brain. Proceedings of the National Academy of Sciences of the United States of America. 2015 Dec 8;112:E6798–E6807. doi: 10.1073/pnas.1510619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013 Dec;17:683–96. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 193.Braun U, Plichta MM, Esslinger C, Sauer C, Haddad L, Grimm O, et al. Test-retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage. 2012 Jan 16;59:1404–1412. doi: 10.1016/j.neuroimage.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 194.Hayasaka S, Laurienti PJ. Comparison of characteristics between region-and voxel-based network analyses in resting-state fMRI data. Neuroimage. 2010 Apr 1;50:499–508. doi: 10.1016/j.neuroimage.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Human Brain Mapping. 1995;3:190–208. [Google Scholar]
- 196.Wang J, Wang L, Zang Y, Yang H, Tang H, Gong Q, et al. Parcellation-dependent small-world brain functional networks: a resting-state fMRI study. Hum Brain Mapp. 2009 May;30:1511–23. doi: 10.1002/hbm.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Alexander-Bloch A, Lambiotte R, Roberts B, Giedd J, Gogtay N, Bullmore E. The discovery of population differences in network community structure: new methods and applications to brain functional networks in schizophrenia. Neuroimage. 2012 Feb 15;59:3889–900. doi: 10.1016/j.neuroimage.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sanz-Arigita EJ, Schoonheim MM, Damoiseaux JS, Rombouts SARB, Maris E, Barkhof F, et al. Loss of ‘Small-World’ Networks in Alzheimer’s Disease: Graph Analysis of fMRI Resting-State Functional Connectivity. Plos One. 2010 Nov 1;5 doi: 10.1371/journal.pone.0013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhao XH, Liu Y, Wang XB, Liu B, Xi Q, Guo QH, et al. Disrupted Small-World Brain Networks in Moderate Alzheimer’s Disease: A Resting-State fMRI Study. Plos One. 2012 Mar 23;7 doi: 10.1371/journal.pone.0033540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang JH, Zuo XN, Dai ZJ, Xia MR, Zhao ZL, Zhao XL, et al. Disrupted Functional Brain Connectome in Individuals at Risk for Alzheimer’s Disease. Biological Psychiatry. 2013 Mar 1;73:472–481. doi: 10.1016/j.biopsych.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 201.Zhang JR, Wang JH, Wu QZ, Kuang WH, Huang XQ, He Y, et al. Disrupted Brain Connectivity Networks in Drug-Naive, First-Episode Major Depressive Disorder. Biological Psychiatry. 2011 Aug 15;70:334–342. doi: 10.1016/j.biopsych.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 202.Vaessen MJ, Braakman HMH, Heerink JS, Jansen JFA, Debeijvan Hall MHJA, Hofman PAM, et al. Abnormal Modular Organization of Functional Networks in Cognitively Impaired Children with Frontal Lobe Epilepsy. Cerebral Cortex. 2013 Aug;23:1997–2006. doi: 10.1093/cercor/bhs186. [DOI] [PubMed] [Google Scholar]
- 203.Wang L, Zhu C, He Y, Zang Y, Cao Q, Zhang H, et al. Altered small-world brain functional networks in children with attention-deficit/hyperactivity disorder. Hum Brain Mapp. 2009 Feb;30:638–49. doi: 10.1002/hbm.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liao W, Zhang Z, Pan Z, Mantini D, Ding J, Duan X, et al. Altered functional connectivity and small-world in mesial temporal lobe epilepsy. PLoS One. 2010;5:e8525. doi: 10.1371/journal.pone.0008525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Van Horn JD, Grafton ST, Miller MB. Individual Variability in Brain Activity: A Nuisance or an Opportunity? Brain Imaging Behav. 2008 Dec 1;2:327–334. doi: 10.1007/s11682-008-9049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Barnes KA, Cohen AL, Power JD, Nelson SM, Dosenbach YB, Miezin FM, et al. Identifying Basal Ganglia divisions in individuals using resting-state functional connectivity MRI. Front Syst Neurosci. 2010;4:18. doi: 10.3389/fnsys.2010.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Finn ES, Scheinost D, Finn DM, Shen X, Papademetris X, Constable RT. Can brain state be manipulated to emphasize individual differences in functional connectivity? Neuroimage. 2017 Oct 15;160:140–151. doi: 10.1016/j.neuroimage.2017.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Craddock RC, James GA, Holtzheimer PE, 3rd, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012 Aug;33:1914–28. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Eickhoff SB, Thirion B, Varoquaux G, Bzdok D. Connectivity-based parcellation: Critique and implications. Hum Brain Mapp. 2015 Dec;36:4771–92. doi: 10.1002/hbm.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011 Sep;106:1125–65. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Shen X, Papademetris X, Constable RT. Graph-theory based parcellation of functional subunits in the brain from resting-state fMRI data. Neuroimage. 2010 Apr 15;50:1027–35. doi: 10.1016/j.neuroimage.2009.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wig GS, Laumann TO, Petersen SE. An approach for parcellating human cortical areas using resting-state correlations. Neuroimage. 2014 Jun;93(Pt 2):276–91. doi: 10.1016/j.neuroimage.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, et al. Parcellating cortical functional networks in individuals. Nat Neurosci. 2015 Dec;18:1853–60. doi: 10.1038/nn.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chong M, Bhushan C, Joshi AA, Choi S, Haldar JP, Shattuck DW, et al. Individual parcellation of resting fMRI with a group functional connectivity prior. Neuroimage. 2017 Aug 1;156:87–100. doi: 10.1016/j.neuroimage.2017.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001 Nov;14:140–51. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD. Comparison of multi-subject ICA methods for analysis of fMRI data. Hum Brain Mapp. 2011 Dec;32:2075–95. doi: 10.1002/hbm.21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Du YH, Allen EA, He H, Sui J, Wu L, Calhoun VD. Artifact removal in the context of group ICA: A comparison of single-subject and group approaches. Hum Brain Mapp. 2016 Mar;37:1005–25. doi: 10.1002/hbm.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Du YH, Fan Y. Group information guided ICA for fMRI data analysis. Neuroimage. 2013 Apr 1;69:157–97. doi: 10.1016/j.neuroimage.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 219.Calhoun VD, Kiehl KA, Pearlson GD. Modulation of temporally coherent brain networks estimated using ICA at rest and during cognitive tasks. Hum Brain Mapp. 2008 Jul;29:828–38. doi: 10.1002/hbm.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Xu J, Potenza MN, Calhoun VD, Zhang R, Yip SW, Wall JT, et al. Large-scale functional network overlap is a general property of brain functional organization: Reconciling inconsistent fMRI findings from general-linear-model-based analyses. Neurosci Biobehav Rev. 2016 Dec;71:83–100. doi: 10.1016/j.neubiorev.2016.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yu Q, Du Y, Chen J, He H, Sui J, Pearlson G, et al. Comparing brain graphs in which nodes are regions of interest or independent components: a simulation study. J Neurosci Methods. 2017 Aug 11; doi: 10.1016/j.jneumeth.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Erhardt EB, Allen EA, Wei Y, Eichele T, Calhoun VD. SimTB, a simulation toolbox for fMRI data under a model of spatiotemporal separability. Neuroimage. 2012 Feb 15;59:4160–7. doi: 10.1016/j.neuroimage.2011.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, et al. Network modelling methods for FMRI. Neuroimage. 2011 Jan 15;54:875–91. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 224.Hedges LV. Distribution theory for Glass’ estimator of effect size and related estimators. Journal of Educational Statistics. 1981;6:107–128. [Google Scholar]
- 225.Gong G, Rosa-Neto P, Carbonell F, Chen ZJ, He Y, Evans AC. Age- and gender-related differences in the cortical anatomical network. J Neurosci. 2009 Dec 16;29:15684–93. doi: 10.1523/JNEUROSCI.2308-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tang E, Giusti C, Baum GL, Gu S, Pollock E, Kahn AE, et al. Developmental increases in white matter network controllability support a growing diversity of brain dynamics. Nat Commun. 2017 Nov 1;8:1252. doi: 10.1038/s41467-017-01254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional Network Organization of the Human Brain. Neuron. 2011 Nov 17;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, et al. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010 Jul 15;67:156–70. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, et al. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008 May 15;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Geerligs L, Renken RJ, Saliasi E, Maurits NM, Lorist MM. A Brain-Wide Study of Age-Related Changes in Functional Connectivity. Cerebral Cortex. 2015 Jul;25:1987–1999. doi: 10.1093/cercor/bhu012. [DOI] [PubMed] [Google Scholar]
- 231.Wang L, Li Y, Metzak P, He Y, Woodward TS. Age-related changes in topological patterns of large-scale brain functional networks during memory encoding and recognition. Neuroimage. 2010 Apr 15;50:862–72. doi: 10.1016/j.neuroimage.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 232.Hwang K, Hallquist MN, Luna B. The Development of Hub Architecture in the Human Functional Brain Network. Cerebral Cortex. 2013 Oct;23:2380–2393. doi: 10.1093/cercor/bhs227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. Multi-task connectivity reveals flexible hubs for adaptive task control. Nature Neuroscience. 2013 Sep;16:1348-U247. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Vidaurre D, Abeysuriya R, Becker R, Quinn AJ, Alfaro-Almagro F, Smith SM, et al. Discovering dynamic brain networks from big data in rest and task. Neuroimage. 2017 Jun 29; doi: 10.1016/j.neuroimage.2017.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liao X, Yuan L, Zhao T, Dai Z, Shu N, Xia M, et al. Spontaneous functional network dynamics and associated structural substrates in the human brain. Front Hum Neurosci. 2015;9:478. doi: 10.3389/fnhum.2015.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Grafton ST. Cross-linked structure of network evolution. Chaos. 2014 Mar;24:013112. doi: 10.1063/1.4858457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Du YH, Pearlson GD, He H, Wu L, CJY, Calhoun VD. Identifying brain dynamic network states via GIG-ICA: Application to schizophrenia, bipolar and schizoaffective disorders. IEEE 12th International Symposium on Biomedical Imaging (ISBI); 2015; 2015. pp. 478–481. [Google Scholar]
- 238.Calhoun VD, Adali T. Time-Varying Brain Connectivity in fMRI Data Whole-brain data-driven approaches for capturing and characterizing dynamic states. Ieee Signal Processing Magazine. 2016 May;33:52–66. [Google Scholar]
- 239.Wang LA, Metzak PD, Honer WG, Woodward TS. Impaired Efficiency of Functional Networks Underlying Episodic Memory-for-Context in Schizophrenia. Journal of Neuroscience. 2010 Sep 29;30:13171–13179. doi: 10.1523/JNEUROSCI.3514-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.van den Heuvel MP, Sporns O. An Anatomical Substrate for Integration among Functional Networks in Human Cortex. J Neurosci. 2013 Sep 4;33:14489–500. doi: 10.1523/JNEUROSCI.2128-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Liang X, Zou Q, He Y, Yang Y. Coupling of functional connectivity and regional cerebral blood flow reveals a physiological basis for network hubs of the human brain. Proc Natl Acad Sci U S A. 2013 Jan 29;110:1929–34. doi: 10.1073/pnas.1214900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tewarie P, Steenwijk MD, Tijms BM, Daams M, Balk LJ, Stam CJ, et al. Disruption of structural and functional networks in longstanding multiple sclerosis. Hum Brain Mapp. 2014 Dec;35:5946–61. doi: 10.1002/hbm.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tewarie P, Hillebrand A, van Dellen E, Schoonheim MM, Barkhof F, Polman CH, et al. Structural degree predicts functional network connectivity: a multimodal resting-state fMRI and MEG study. Neuroimage. 2014 Aug 15;97:296–307. doi: 10.1016/j.neuroimage.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 244.Yu Q, Wu L, Bridwell DA, Erhardt EB, Du Y, He H, et al. Building an EEG-fMRI Multi-Modal Brain Graph: A Concurrent EEG-fMRI Study. Front Hum Neurosci. 2016;10:476. doi: 10.3389/fnhum.2016.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hayasaka S, Hugenschmidt CE, Laurienti PJ. A network of genes, genetic disorders, and brain areas. PLoS One. 2011;6:e20907. doi: 10.1371/journal.pone.0020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Damaraju E, Allen EA, Belger A, Ford JM, McEwen S, Mathalon DH, et al. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage Clin. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen J, Calhoun VD, Lin D, Perrone-Bizzozero NI, Bustillo JR, Pearlson GD, et al. Shared Genetic Risk of Schizophrenia and Gray Matter Reduction in 6p22. 1. Schizophrenia bulletin. 2018 doi: 10.1093/schbul/sby010. vol. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jafri MJ, Pearlson GD, Stevens M, Calhoun VD. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage. 2008 Feb 15;39:1666–81. doi: 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Calhoun VD, de Lacy N. Ten Key Observations on the Analysis of Resting-state Functional MR Imaging Data Using Independent Component Analysis. Neuroimaging Clin N Am. 2017 Nov;27:561–579. doi: 10.1016/j.nic.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014 Jul 24;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD. Source-Based Morphometry: The Use of Independent Component Analysis to Identify Gray Matter Differences With Application to Schizophrenia. Human Brain Mapping. 2009 Mar;30:711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Khambhati AN, Sizemore AE, Betzel RF, Bassett DS. Modeling and interpreting mesoscale network dynamics. Neuroimage. 2017 Jun 20; doi: 10.1016/j.neuroimage.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016 Aug 11;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gu S, Pasqualetti F, Cieslak M, Telesford QK, Yu AB, Kahn AE, et al. Controllability of structural brain networks. Nat Commun. 2015 Oct 01;6:8414. doi: 10.1038/ncomms9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gu S, Betzel RF, Mattar MG, Cieslak M, Delio PR, Grafton ST, et al. Optimal trajectories of brain state transitions. Neuroimage. 2017 Mar 01;148:305–317. doi: 10.1016/j.neuroimage.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Vaiana M, Muldoon SF. Multilayer Brain Networks. Journal of Nonlinear Science. 2018 Jan 03; [Google Scholar]
- 257.Battiston F, Nicosia V, Chavez M, Latora V. Multilayer motif analysis of brain networks. Chaos. 2017 Apr;27:047404. doi: 10.1063/1.4979282. [DOI] [PubMed] [Google Scholar]
- 258.Muldoon SF, Bassett DS. Network and Multilayer Network Approaches to Understanding Human Brain Dynamics. Philosophy of Science. 2016 Dec;83:710–720. [Google Scholar]
- 259.Halu A, De Domenico M, Arenas A, Sharma A. The multiplex network of human diseases. bioRxiv. 2017 doi: 10.1038/s41540-019-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.De Domenico M. Multilayer modeling and analysis of human brain networks. Gigascience. 2017 May 1;6:1–8. doi: 10.1093/gigascience/gix004. [DOI] [PMC free article] [PubMed] [Google Scholar]