Abstract
Background
MicroRNA-122 (miR-122), a pivotal liver-specific miRNA, is frequently repressed in hepatocellular carcinoma (HCC) and associated with poor prognosis. Long non-coding RNA (lncRNA) HOTAIR has been proved to function as an oncogene in multiple cancers including HCC. However, the relationship between HOTAIR and miR-122 in HCC remains largely unknown.
Methods
We investigated the function of HOTAIR and miR-122 in HCC cell models and a xenograft mouse model. The regulatory network between HOTAIR and miR-122 was further detected following overexpression or knockdown of HOTAIR. DNA methylation status of miR-122 promoter region, as well as expression levels of DNMTs, EZH2 and Cyclin G1 were analyzed.
Findings
In this study, we found that HOTAIR was highly expressed whereas miR-122 was suppressed in HCC, and HOTAIR negatively regulated miR-122 expression in HCC cells. Furthermore, knockdown of HOTAIR dramatically inhibited HCC cell proliferation and induced cell cycle arrest in vitro and suppressed tumorigenicity in vivo by upregulating miR-122 expression. Mechanistically, a CpG island was located in the miR-122 promoter region. HOTAIR epigenetically suppressed miR-122 expression via DNMTs-mediated DNA methylation. Moreover, HOTAIR upregulated DNMTs expression via EZH2. In addition, suppression of miR-122 induced by HOTAIR directly reactivated oncogene Cyclin G1 expression. Collectively, our results suggest that HOTAIR epigenetically suppresses miR-122 expression via DNA methylation, leading to activation of Cyclin G1 and promotion of tumorigenicity in HCC, which provide new insight into the mechanism of HOTAIR-mediated hepatocarcinogenesis via suppressing miR-122.
Keywords: Hepatocellular carcinoma, LncRNA HOTAIR, microRNA-122, DNA methylation, Epigenetics
Abbreviations: HCC, hepatocellular carcinoma; lncRNA, long non-coding RNA; HOTAIR, Homeobox transcript antisense intergenic RNA; miR-122, microRNA-122; EZH2, Enhancer of zeste homolog 2; DNMT, DNA methyltransferase; CCNG1, Cyclin G1
Highlights
-
•
HOTAIR is highly expressed in HCC, and negatively regulates miR-122 expression in HCC cells.
-
•
HOTAIR increased HCC cell proliferation and tumor growth through downregulating miR-122 expression.
-
•
HOTAIR epigenetically suppressed miR-122 expression via DNMTs-mediated DNA methylation.
-
•
HOTAIR upregulated DNMTs expression via EZH2.
-
•
HOTAIR increased cyclin G1 expression through repressing miR-122.
Research in context.
Evidence before this study
MicroRNA-122 (miR-122) is a dominant liver-specific miRNA, playing a pivotal role in liver development, hepatocyte differentiation, as well as in hepatocarcinogenesis. MiR-122 has been reported to be frequently repressed in HCC and associated with poor prognosis. LncRNA HOTAIR has been proved to function as an oncogene in multiple cancers including HCC, by inhibiting several genes and microRNAs expression. However, the relationship between HOTAIR and miR-122 in HCC is still ill-defined.
Added value of this study
HOTAIR was overexpressed while miR-122 was suppressed in HCC, and HOTAIR negatively regulated miR-122 expression in HCC cells. Knockdown of HOTAIR was sufficient to inhibit tumorigenicity both in vitro and in vivo by upregulating miR-122 expression Mechanistically, HOTAIR epigenetically suppressed miR-122 expression via DNMTs-mediated DNA methylation, leading to activation of Cyclin G1 and promotion of tumorigenicity in HCC.
Implications of all the available evidence
Both HOTAIR and miR-122 play crucial roles in the development of HCC. Our study provides new insight into the novel mechanism of HOTAIR-mediated hepatocarcinogenesis via epigenetically suppressing miR-122, and propose the HOTAIR/miR-122 negative regulatory axis as a promising molecular target for HCC intervention.
Alt-text: Unlabelled Box
1. Introduction
Hepatocellular carcinoma (HCC) is the fifth most prevalent human malignancy and the third leading cause of cancer-related mortality worldwide [1,2]. In patients with HCC, the best treatment is surgical resection. However, only a little proportion of patients with HCC undergoes a radical operation, and even in patients who are suitable for radical surgery, the risk of recurrence is high. Despite the recent progress in HCC prevention, diagnosis and intervention, treatment for HCC still remains unsatisfactory [2]. Thus, elucidating the underlying mechanism of HCC progression and identifying novel potential targets for HCC therapies are urgently needed. Recently, accumulating studies demonstrated that non-coding RNAs play a part in regulating various biological processes in cancers including hepatocarcinogenesis [3,4], suggesting the potential role of non-coding RNAs for HCC diagnosis and intervention.
MicroRNA-122 (miR-122) is a dominant liver-specific miRNA, accounting for 70% and 52% of liver's total miRNAs in adult mouse and human, respectively [5,6]. Consequently, miR-122 plays a pivotal role in liver development, hepatocyte differentiation, lipid metabolism, hepatitis C virus (HCV) replication and hepatocarcinogenesis [[6], [7], [8], [9]]. Accumulating studies demonstrated that miR-122 was significantly suppressed in HCC tissues and cell lines. Decreased expression level of miR-122 was correlated with hepatocarcinogenesis, metastasis and poor prognosis of HCC [9,10]. Furthermore, deletion of miR-122 in miR-122-knockout (KO) mice developed hepatosteatosis, fibrosis and ultimately HCC, and restoration of miR-122 in HCC cells strongly reversed the tumorigenic properties of these cells and further prevent HCC development in miR-122-KO mice [9]. Moreover, restoration of miR-122 sensitized HCC cells to chemotherapeutic agents, as well as sorafenib [11,12]. Additionally, a variety of validated miR-122 targets including cyclin G1 (CCNG1), insulin-like growth factor 1 receptor (IGF1R), A disintegrin and metalloprotease 10 (ADAM10), Wnt family member 1 (WNT1) and pyruvate kinase M2 (PKM2) have been implicated in hepatocarcinogenesis, epithelial-mesenchymal transition and angiogenesis of HCC [[12], [13], [14], [15]]. These findings support that miR-122 functions as a tumor suppressor against HCC. Although an increasing number of studies have demonstrated that Peroxisome proliferator-activated receptor gamma (PPARγ), hepatitis B virus X protein (HBx), and REV-ERBα may be involved in the modulation of miR-122 downregulation [16,17], the precise mechanism underlying its suppression in HCC has not yet been fully elucidated.
Recently, long non-coding RNAs (lncRNAs) have attacked particular attentions in cancer research [3,18]. Accumulating evidences have revealed that several lncRNAs are frequently dysregulated in a variety of human cancers including HCC, and involved in pathogenesis and progression of cancers [18,19]. HOTAIR (Homeobox transcript antisense intergenic RNA) is a known lncRNA located in the Homeobox C (HOXC) gene cluster, and has been shown to regulate gene expression via epigenetic modifications, such as histone methylation and DNA methylation [20,21]. Numerous studies have revealed that HOTAIR was highly expressed in multiple cancers, including breast cancer, pancreatic cancer, colorectal cancer, and HCC [[20], [21], [22], [23]]. Overexpression of HOTAIR in HCC was positively associated with poor prognosis, tumor progression, and recurrence [22,23]. Knockdown of HOTAIR inhibited cell proliferation, migration, invasiveness and autophagy in HCC cells [[24], [25], [26]], implicating the important oncogenic role of HOTAIR in HCC development. A recent study has demonstrated that HOTAIR mediates tumorigenesis of HCC through inhibiting miR-218 expression [27], establishing a direct link between HOTAIR and miRNA dysregulation in HCC development. Moreover, HOTAIR has also been documented to negatively regulate the expression of miR-1 and miR-23b-3p in HCC, contributing to the malignancy of HCC [28,29]. However, there is no study investigating whether HOTAIR can regulate miR-122 expression, the most bountiful and frequently repressed miRNA in HCC.
In the present study, we showed that HOTAIR was upregulated whereas miR-122 was downregulated in HCC specimens, and HOTAIR negatively regulated miR-122 expression in HCC cells. Knockdown of HOTAIR was sufficient to inhibit tumorigenicity both in vitro and in vivo by upregulating miR-122 expression. Mechanistically, HOTAIR may epigenetically suppress miR-122 expression via DNA methylation, mediated by DNMTs. Moreover, downregulation of miR-122 induced by HOTAIR may directly reactivated Cyclin G1 expression. Collectively, our findings provide new insight into the mechanism of HOTAIR-mediated hepatocarcinogenesis via suppressing miR-122, and it would be helpful to develop a promising therapeutic strategy for HCC intervention.
2. Materials and methods
2.1. Cell culture and tissue specimens
A set of HCC cell lines including HepG2, Huh7, Hep3B, SMMC7721, MHCC97H, and immortalized non-tumorigenic hepatocyte MIHA cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% Penicillin-Streptomycin. 30 paired fresh clinic specimens including primary HCC specimens and their peri-tumor counterparts were obtained from surgical tumor resections of patients suffering HCC at the Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University (SYSU). The clinical data for these above patients was described in the Supplementary Table SI. The use of human tissues in this study was approved by the Ethics Committee of Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University.
2.2. RNA oligoribonucleotides and cell transfections
The RNA oligoribonucleotides targeting human HOTAIR, EZH2 and miR-122 were synthesized and purchased from Genepharma (Shanghai, China), and the sequences were listed in Supplementary Table SII. Human DNMT1-siRNA, DNMT3A-siRNA and DNMT3B-siRNA were purchased from Origene.
Lipofectamine 2000 (Invitrogen) was used for transfection of RNA oligoribonucleotides [30], and X-tremeGENE (Roche) was used for transfection of plasmid DNA, according to their manufacturer's instructions, respectively.
2.3. HOTAIR shRNA and overexpression plasmids
Lv-shHOTAIR and Lv-shNC (a small hairpin RNA which was nonhomologous to any human genome sequences, acted as negative control) plasmids were purchased from Genepharma (Shanghai, China). The HOTAIR overexpression plasmid (pHOTAIR) was purchased from Addgene.
2.4. Lentiviral miR-122 expression plasmid construction and lentiviruses production
A 160 bp sequence of pre-miR-122 containing the stem-loop was amplified and cloned into a lentiviral vector (designated as Lv-miR-122). Then lentiviruses were produced and purified. Briefly, Lv-miR-122 vector and three packaging vectors (pRRE, pRSV-REV and pCMV-VSVG) were co-transfected into 293 T cells to generate the pseudo-typed lentivirus. A lentiviral vector containing a non-targeting scramble RNA was used as negative control (Lv-NC).
2.5. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
The total RNA was extracted from HCC specimens or cells by using Trizol reagent (Invitrogen), and the reverse transcription was processed as described previously [30]. The specific Primers used were listed in the Supplementary Table SIII. GAPDH or U6 were used as endogenous controls.
2.6. Western blotting and immunocytochemistry
Protein lysates from HCC cells were subjected to Western blotting analysis by using anti-EZH2 (Abcam), anti-DNMT1 (Abcam), anti-DNMT3A (Abcam), anti-DNMT3B (Abcam), and anti-CCNG1 (Abcam) according to standard protocols as previously described [24,30]. Formaldehyde-fixed, paraffin-embedded sections were subjected to H&E staining and immunohistochemistry by using anti-Ki67 (Abcam) following the routine protocols.
2.7. Bisulfite sequencing analysis
The methylation status of miR-122 promoter was determined by Bisulfite sequencing PCR (BSP). Primers were listed in the Supplementary Table SIV. miR-122 DNA was extracted using a DNA kit (Qiagen), and the miR-122 DNA was subjected to bisulfite conversion using an EpiTect Bisulfite Kit (Qiagen). The transformed DNA was then PCR-amplified using the TaKaRa rTaq Kit (TaKaRa). The PCR amplification products were sequenced by Invitrogen Corporation, Shanghai, China.
2.8. Chromatin immunoprecipitation (ChIP)
ChIP assay was performed by using ChIP Kit (Millipore) according to the manufacturer's instructions as previously described [30]. Briefly, Chromatin was extracted, and DNA was sheared to 0.2- to 1-kb fragments. The sheared chromatin was immunoprecipitated with anti-DNMT1 (Abcam), anti-DNMT3A (Abcam), anti-DNMT3B (Abcam). DNA released from precipitated complexes was amplified by qRT-PCR.
2.9. Cell viability assay
Cell viability was analyzed by the Cell Counting Kit-8 system (Dojindo Laboratory, Kumamoto). Briefly, cells were seeded at 5 × 103 per well into 96-well plates and incubated under the indicated conditions. After 72 h, cell viabilities were detected by using a Benchmark microplate spectrometer (Bio-Rad).
2.10. Cell cycle analyses
For cell cycle distribution analysis, cells were seeded at 2 × 105 per well into 6-well plates. After cultured for 72 h, cells were then labeled with propidium iodide (PI) and analyzed by flow cytometry, as described previously [30].
2.11. Xenograft mouse model
The xenograft mouse models were carried out using female BALB/c nude mice at 4 weeks of age. HepG2 cells were infected with Lv-shHOTAIR or Lv-shNC or Lv-miR-122 or Lv-NC, respectively. Mice were injected subcutaneously with 1 × 106 infected cells into the dorsal flank. Tumor volumes (V) were surveyed using calipers to measure tumor length (L) and width (W) twice a week (V = 0.5 × L × W2). Tumor weight was measured at the endpoint of the study.
2.12. Statistical analysis
Data from three independent experiments were presented as the mean ± SD. Statistical analyses were performed by using a two-tailed Student's t-test or the Pearson's correlation in GraphPad Prism 7.0 (GraphPad Software, USA). P < 0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. HOTAIR negatively regulated miR-122 expression in HCC cells
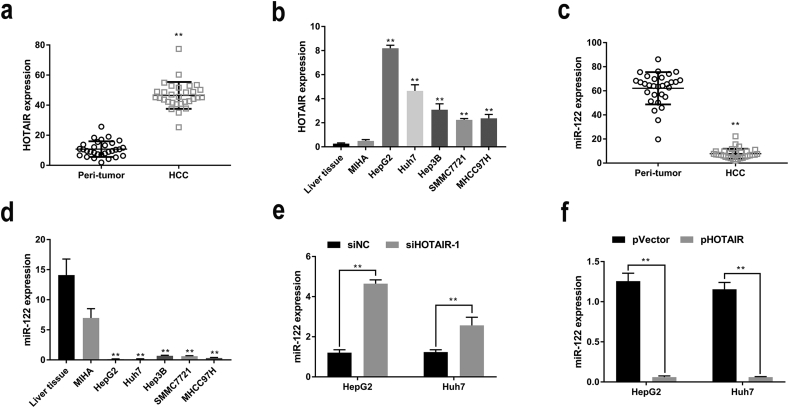
Previous studies have documented that lncRNA HOTAIR could negatively regulate a variety of miRNAs in several human cancers including HCC [27,29]. MiR-122 is frequently suppressed in HCC. To investigate whether HOTAIR plays a part in suppression of miR-122 in HCC development, we first test the expression levels of HOTAIR and miR-122 in HCC tissues and cell lines. Our results showed that HOTAIR was highly expressed in HCC tissues, as compared with the peri-tumor tissues (Fig. 1a). We also found that HOTAIR expression was markedly increased in HCC cell lines (Fig. 1b). Additionally, miR-122 expression was decreased in HCC tissues (Fig. 1c), as well as in HCC cell lines (Fig. 1d), which were consistent with previous studies [9]. Therefore, HOTAIR upregulation and miR-122 downregulation would be frequent events in HCC development, and an inverse relationship between HOTAIR and miR-122 in HCC would be observed. To further investigate the negative regulatory role of HOTAIR on miR-122 expression, we used the specific siRNAs (siHOTAIR) to silence HOTAIR. We found that HOTAIR expression was effectively knocked down by siHOTAIR-1 and siHOTAIR-2 (Fig. S1). Then siHOTAIR-1 was selected for silencing HOTAIR in the following experiments. Our results showed that knockdown of HOTAIR by siHOTAIR-1 dramatically increased the expression of miR-122 in HepG2 and Huh7 cells (Fig. 1e). On the other hand, overexpression of HOTAIR by pHOTAIR decreased miR-122 expression in HepG2 and Huh7 cells (Fig. 1f). These data indicated that HOTAIR negatively regulated miR-122 expression in HCC cells.
Fig. 1.
HOTAIR negatively regulated miR-122 expression in HCC cells.
(a–b) HOTAIR was upregulated in HCC specimens (a) and cell lines (b). (c–d) miR-122 was downregulated in HCC specimens (c) and cell lines (d). (e–f) miR-122 was upregulated by siHOTAIR-1 (e) whereas downregulated by pHOTAIR (f) in HepG2 and Huh7 cells. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
3.2. HOTAIR promoted cell growth through downregulating miR-122 expression in vitro
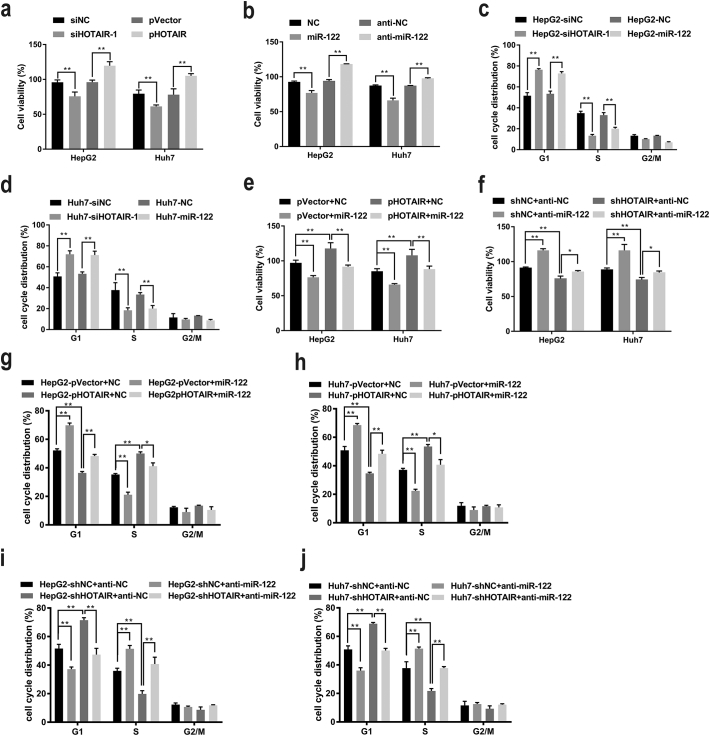
Previous studies have revealed that lncRNA HOTAIR promotes tumorigenesis and metastasis in several human cancers [20,21,27]. To investigate whether HOTAIR promotes hepatocarcinogenesis via suppressing miR-122 in HCC, we first test the proliferative function of HOTAIR in HCC. The HCC cell lines HepG2 and Huh7 were transfected with siHOTAIR or pHOTAIR, respectively, and cell proliferation were tested. Our results showed that knockdown of HOTAIR by siHOTAIR-1 suppressed cell proliferation while overexpression of HOTAIR by pHOTAIR promoted cell viability in HCC cells (Fig. 2a). Next, we studied the role of miR-122 on HCC cell proliferation and our results showed that miR-122 suppressed cell proliferation whereas anti-miR-122 promoted cell viability in HCC cells (Fig. 2b). Furthermore, the cell cycle distribution revealed that knockdown of HOTAIR by siHOTAIR-1 or ectopic miR-122 overexpression induced an enlarged percentage of HCC cells in G1-phase and fewer cells in S-phase (Fig. 2c–d), suggesting that siHOTAIR or miR-122 would inhibit HCC cell proliferation through inducing the cell cycle arrest. To further investigate the antagonistic effects of HOTAIR and miR-122 on HCC cell proliferation and cell cycle progression, miR-122 was transfected into the pHOTAIR-stably-infected HCC cells and cell viabilities was tested. Our results showed that overexpression of miR-122 reversed the cell proliferation induced by pHOTAIR (Fig. 2e). On the other hand, anti-miR-122 was transfected into the Lv-shHOTAIR-infected HCC cells and our result showed that inhibition of miR-122 by anti-miR-122 significantly abrogated cell growth inhibition induced by shHOTAIR (Fig. 2f). Moreover, the cell cycle distribution revealed that miR-122 reversed the cell cycle progression induced by pHOTAIR (Fig. 2g–h), whiles, anti-miR-122 abrogated the cell cycle arrest induced by shHOTAIR (Fig. 2i–j). These data suggested that HOTAIR promoted cell growth through downregulating miR-122 expression in HCC cells.
Fig. 2.
HOTAIR mediated cell growth through downregulating miR-122 expression in vitro.
(a) knockdown of HOTAIR suppressed whereas HOTAIR overexpression promoted cell viabilities in HCC cells. (b) miR-122 suppressed whereas anti-miR-122 promoted cell proliferation in HCC cells. (c–d) siHOTAIR-1 or miR-122 induced the G1-phase arrest in HCC cells. (e–f) miR-122 reversed the enhanced proliferative effect of HOTAIR overexpression (e) whereas anti-miR-122 rescued the suppressive effect of shHOTAIR (f) on HCC cell growth. (g–j) miR-122 reversed the cell cycle progression induced by HOTAIR overexpression (g–h) whereas anti-miR-122 abrogated the cell cycle arrest induced by shHOTAIR (i-j) in HCC cells. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
3.3. HOTAIR mediated tumorigenicity through miR-122 downregulation in vivo
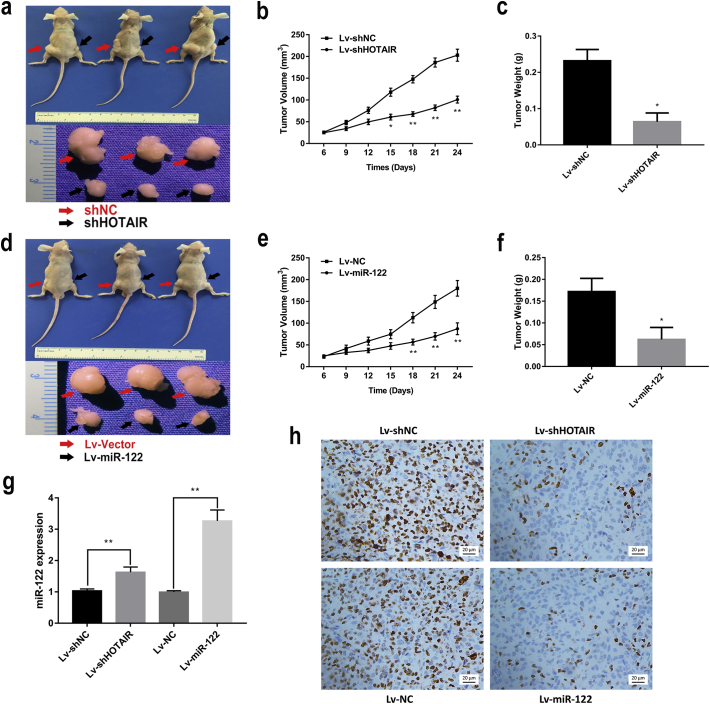
To further confirm these above findings in vitro, we performed an in vivo xenograft model that HepG2 cells transfected with Lv-shHOTAIR or Lv-miR-122 were injected subcutaneously into the dorsal flank of nude BALB/c mice. The in vivo results showed that the Lv-shHOTAIR group displayed a significant reduction in tumor volume and weight, as compared with the Lv-shNC group (Fig. 3a–c), and a similar anti-tumor effect of miR-122 on HCC cells in vivo was also observed, as compared with the Lv-NC group (Fig. 3d–f). Next, the xenograft tumors were removed and the expression level of HOTAIR and miR-122 were detected. Our results showed that expression level of miR-122 was increased in the Lv-shHOTAIR group, as compared with the Lv-shNC group (Fig. 3g). In addition, the cell proliferation marker Ki-67 was tested by immunohistochemistry (IHC) assay, and the results showed that Ki-67 expression was decreased in xenograft tumors treated with Lv-shHOTAIR or Lv-miR-122 (Fig. 3h). These data suggested that HOTAIR may mediate tumorigenicity in HCC, at least in part, by suppressing miR-122 expression.
Fig. 3.
HOTAIR mediated tumorigenicity through miR-122 downregulation in vivo.
HepG2 cells were infected with Lv-shHOTAIR or Lv-miR-122 injected subcutaneously into nude mice.
(a–d) Lv-shHOTAIR (a) and Lv-miR-122 (d) infected cells generated smaller tumors than their control cells. (b–e) the growth curves of tumor volumes were measured in the Lv-shHOTAIR (b) and Lv-miR-122 (e) group. (c–f) the tumor weights were measured in the Lv-shHOTAIR (c) and Lv-miR-122 (f) group. (g) miR-122 expression was increased in the xenograft tumors treated with Lv-shHOTAIR or Lv-miR-122. (h) the immunohistochemistry of Ki-67 stained sections followed by counterstaining with DAPI. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
3.4. HOTAIR regulated DNA methylation of miR-122 promoter region
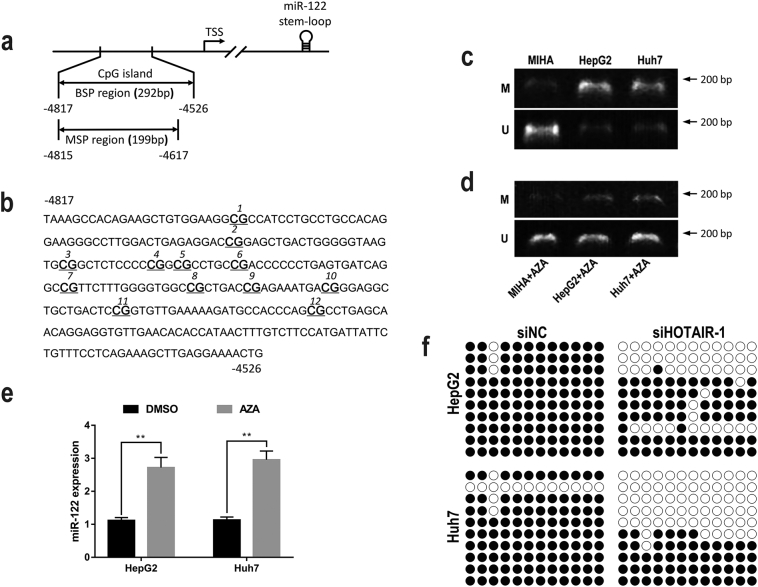
We next sought to elucidate the molecular mechanism by which HOTAIR mediates the downregulation of miR-122 in HCC. Previous study has reported that cotreatment of DNA methylation inhibitor (5-Aza-dC, AZA) and histone deacetylase inhibitor (PBA) restored miR-122 expression in HCC cells [16]. Our preliminary experiment found that AZA treatment without PBA also upregulated miR-122 expression in HCC cells (data unshown). Therefore, we focus on the potential role of DNA methylation in regulation of miR-122 expression. We examined the 5 kb genomic region up-stream of miR-122 stem-loop, where the putative promoter sequence of this miRNA is likely to be found [31]. Indeed, a CpG island was located in the miR-122 promoter region (Fig. 4a–b), suggesting a possible involvement of DNA methylation in the regulation of miR-122 expression. Next, we performed Methylation specific PCR (MSP) to evaluate the DNA methylation status of miR-122 promoter region in HCC cells, we found that the promoter region of miR-122 was hypermethylated in HCC cells, but hypomethylated in the hepatocyte MIHA cells (Fig. 4c). Then HCC cells were treated with 5-Aza-dC (AZA), a DNA methylation inhibitor, and the results showed that AZA treatment significantly decreased the DNA methylation level of miR-122 promoter region (Fig. 4d), and increased the expression of miR-122 in HCC cells (Fig. 4e), suggesting that suppression of miR-122 in HCC cells may be associated with DNA methylation status at the promoter region of this gene. To validate the regulatory effect of HOTAIR on DNA methylation level of miR-122 promoter region, Bisulfate sequencing PCR (BSP) was performed. Our results showed that knockdown of HOTAIR by siHOTAIR-1 in HepG2 and Huh7 cells significantly decreased DNA methylation level at the CpG island of miR-122 promoter region (Fig. 4f). These results suggested that HOTAIR may epigenetically suppress miR-122 expression via DNA methyaltion.
Fig. 4.
HOTAIR regulated DNA methylation of miR-122 promoter region in HCC cells.
(a) a CpG island was located in the miR-122 promoter region of −5000 bp upstream of the miR-122 stem-loop. (b) the sequence level detail of the miR-122 promoter region (−4817 to −4526 bp upstream of the miR-122 stem-loop). (c) miR-122 promoter region was hypermethylated in HCC cells, but unmethylated in MIHA cells analyzed by MSP. (d) 5-Aza-dC treatment decreased the DNA methylation level of miR-122 promoter region in HCC cells analyzed by MSP. (e) 5-Aza-dC treatment increased the expression level of miR-122 in HCC cells. (f) knockdown of HOTAIR decreased DNA methylation level of miR-122 promoter region in HCC cells analyzed by BSP. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
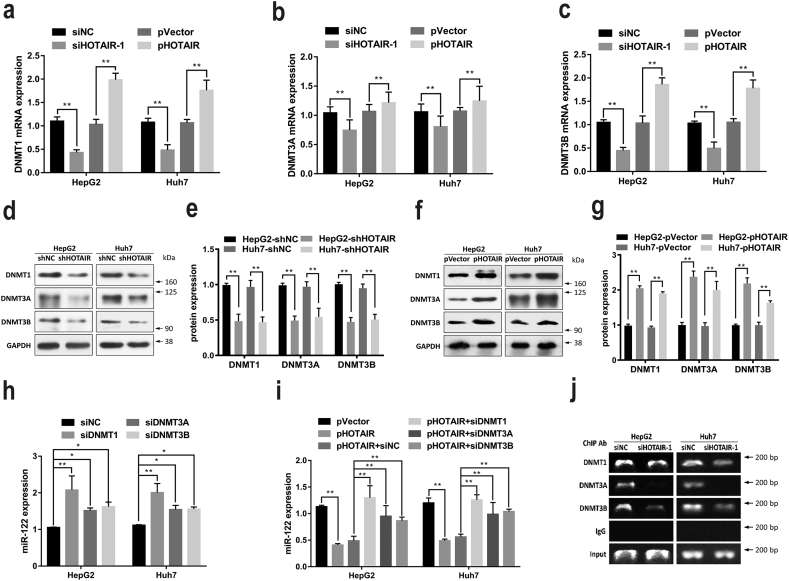
3.5. HOTAIR epigenetically suppressed miR-122 through DNMTs
Generally, DNMTs, including DNMT1, DNMT3A, and DNMT3B, are responsible for the regulation of DNA methylation at CpG islands [32]. To better understanding whether DNMTs were involved in the regulation of miR-122 methylation by HOTAIR, expression levels of DNMTs were detected. Our results showed that DNMT1, DNMT3A, and DNMT3B were upregualted in HCC cells (Fig. S2). Furthermore, DNMT1, DNMT3A, and DNMT3B were suppressed by siHOTAIR-1 and promoted by pHOTAIR at both mRNA and protein levels (Fig. 5a–g), indicating that HOTAIR are capable of inducing the upregulation of DNMTs in HCC cells, contributing to regulation of DNA methylation. To further determine whether DNMTs regulated miR-122 expression, the specific siRNAs against DNMT1, DNMT3A and DNMT3B were used respectively. Our results showed that either siDNMT1, or siDNMT3A, or siDNMT3B increased the expression level of miR-122 (Fig. 5h). Furthermore, either siDNMT1, or siDNMT3A, or siDNMT3B rescued the HOTAIR-induced miR-122 suppression (Fig. 5i). In addition, to assess whether HOTAIR is required for DNMTs binding to the miR-122 promoter region, ChIP was performed and our results showed that knockdown of HOTAIR caused a significant reduction of DNMTs binding to the promoter region of miR-122 (Fig. 5j). These data suggested that HOTAIR may epigenetically suppress miR-122 through DNMTs-mediated DNA methylation.
Fig. 5.
HOTAIR epigenetically suppressed miR-122 through DNMTs in HCC cells.
(a–g) DNMT1, DNMT3A, and DNMT3B expression were suppressed by siHOTAIR-1 and promoted by pHOTAIR at both mRNA (a–c) and protein (d–g) levels in HCC cells. (h) either siDNMT1, or siDNMT3A, or siDNMT3B increased miR-122 expression in HCC cells. (i) either siDNMT1, or siDNMT3A, or siDNMT3B rescued the HOTAIR-induced miR-122 suppression in HCC cells. (j) knockdown of HOTAIR caused a significant reduction of DNMTs binding to the miR-122 promoter region in HCC cells analyzed by ChIP assay. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
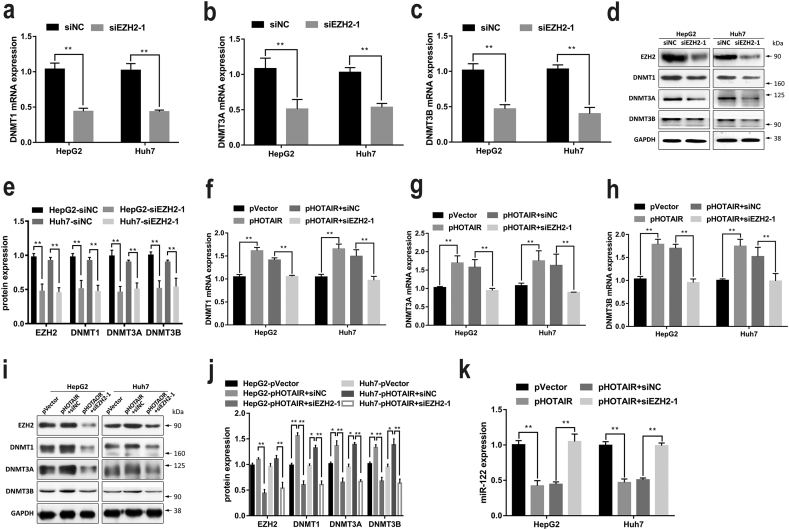
3.6. EZH2 mediated HOTAIR-induced DNMTs expression
Previous study have reported that HOTAIR promoted invasiveness and metastasis by recruiting polycomb repressive complex 2 (PRC2), thus leading to alteration of gene expression [20]. EZH2, an important component of PRC2, is frequently overexpressed in multiple cancers, acting as an oncogene [33]. Our results showed that EZH2 was elevated in HCC cells (Fig. S3). To further test whether HOTAIR could affect DNMTs expression in HCC cells by recruiting EZH2, the specific siRNAs against EZH2 (siEZH2) were used to silence EZH2 and its expression was downregulated by siEZH2-1 and siEZH2-2 (Fig. S4). Then siEZH2-1 was selected for silencing EZH2 in the following experiment. Our results showed that the expression levels of DNMT1, DNMT3A, and DNMT3B were significantly decreased by siEZH2-1 (Fig. 6a–e). Furthermore, siEZH2-1 reversed the upregulation of DNMTs expression induced by HOTAIR overexpression (Fig. 6f–j), indicating HOTAIR may regulate DNMTs expression via EZH2. Additionally, knockdown of EZH2 by siEZH2-1 not only increased the expression level of miR-122, but also abrogated the suppression of miR-122 induced by HOTAIR overexpression (Fig. 6k, Fig. S5). Collectively, these data suggested that HOTAIR might upregulate DNMTs expression by EZH2, leading to alterative expression of miR-122 via DNMTs-mediated DNA methylation in HCC cells.
Fig. 6.
EZH2 mediated HOTAIR-induced DNMTs expression in HCC cells.
(a–e) knockdown of EZH2 by siEZH2-1 downregulated the expression levels of DNMT1, DNMT3A, and DNMT3B mRNA (a–c) and protein (d–e) in HCC cells. (f–j) siEZH2-1 reversed the upregulation of DNMTs mRNA (f–h) and protein (i–j) expression induced by HOTAIR in HCC cells. (k) siEZH2-1 abrogated the suppression of miR-122 induced by HOTAIR in HCC cells. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
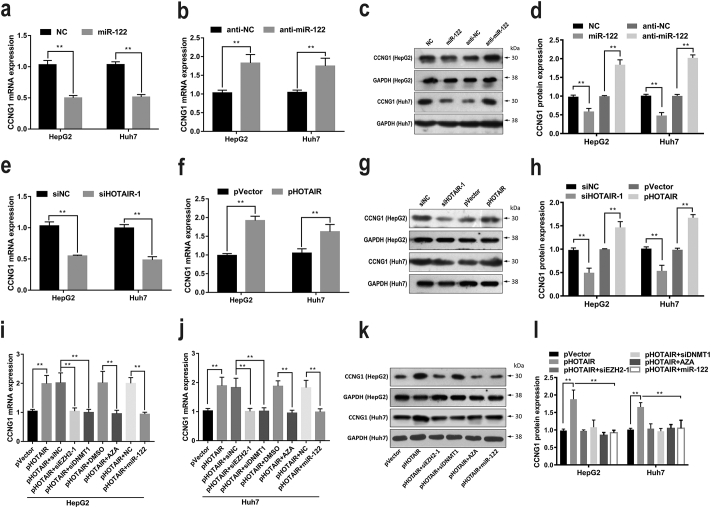
3.7. HOTAIR upregulated Cyclin G1 via repressing miR-122
Our previous results have demonstrated that knockdown of HOTAIR by siHOTAIR-1 or excess miR-122 induced cell cycle arrest (Fig. 2c–d). Moreover, it has been proved that Cyclin G1 (CCNG1) is a functional target of miR-122, which contributes to cell proliferation and cell cycle progression [[12], [13], [14], [15]]. Thus we suspected whether HOTAIR regulated cell cycle progression through CCNG1. Our results showed that miR-122 dramatically suppressed the endogenous expression of CCNG1 at both mRNA and protein levels, meanwhile, anti-miR-122 increased CCNG1 expression in HCC cells (Fig. 7a–d). As an oncogene, CCNG1 was frequently upregulated in HCC cells. Furthermore, our results showed that CCNG1 expression was suppressed by siHOTAIR-1 and increased by pHOTAIR at both mRNA and protein levels (Fig. 7e–h). In addition, either siEZH2-1, or siDNMT1, or AZA treatment, or miR-122 treatment reversed the enhanced expression of CCNG1 induced by HOTAIR overexpression (Fig. 7i–l). Collectively, these date suggested that HOTAIR may modulate carcinogenesis in HCC, at least in part, through suppressing miR-122 and activating CCNG1.
Fig. 7.
HOTAIR upregulated CCNG1 via repressing miR-122 in HCC cells.
(a–d) miR-122 suppressed whereas anti-miR-122 increased the expression levels of CCNG1 mRNA (a-b) and protein (c–d) in HCC cells. (e–h) CCNG1 expression was suppressed by siHOTAIR-1 whereas increased by pHOTAIR at both mRNA (e–f) and protein (g–h) levels in HCC cells. (i–l) either siEZH2-1, or siDNMT1, or AZA treatment, or miR-122 treatment reversed the enhanced expression of CCNG1 induced by HOTAIR at both mRNA (i–j) and protein (k–l) levels in HCC cells. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
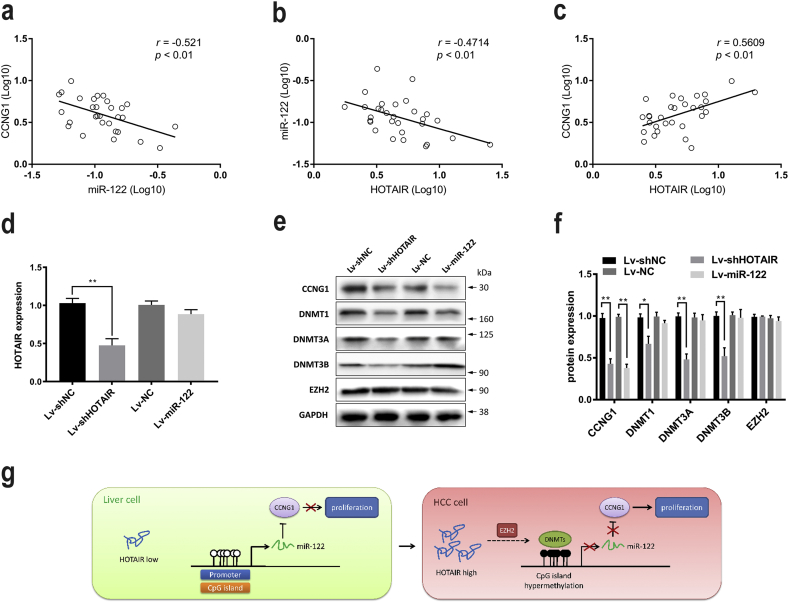
3.8. HOTAIR expression negatively correlated with miR-122 and positively correlated with Cyclin G1 in HCC
MiR-122 has been reported to be frequently repressed in HCC, whereas HOTAIR and Cyclin G1 (CCNG1) were upregulated in HCC, respectively [9,12,27]. To further study the correlation between HOTAIR, miR-122 and CCNG1, the association analyses were performed and our results showed that a significant negative relationship between HOTAIR and miR-122 expression was found in these HCC specimens (Fig. 8a). Moreover, a negative association between miR-122 and CCNG1 (Fig. 8b) and a positive correlation between HOTAIR and CCNG1 (Fig. 8c) were confirmed in these specimens. In addition, our in vivo results showed that knockdown of HOTAIR caused the increased expression of miR-122 and decreased expression of CCNG1, as well as decreased DNMTs (Fig. 8d–f). Taken together, our data suggested that lncRNA HOTAIR, upregulating DNMTs by EZH2, may epigenetically suppress miR-122 expression via DNA methylation, therefore leading to activation of CCNG1 and promotion of tumorigenicity in HCC (Fig. 8g).
Fig. 8.
HOTAIR expression negatively correlated with miR-122 and positively correlated with CCNG1 in HCC.
(a–c) the statistically significant association between HOTAIR, miR-122 and CCNG1 expression in HCC specimens. (d–f) the expression of HOTAIR, CCNG1, DNMT1, DNMT3A, DNMT3B, and EZH2 in xenograft tumors were analyzed by qRT-PCR and WB. (g) the schematic overview of HOTAIR/miR-122 mediated tumorigenesis in HCC. Data are representative of three independent experiments and are presented as mean ± SD. *P < 0.05, **P < 0.01 (Student's t-test).
4. Discussion
Accumulating studies have demonstrated that abnormal expression of non-coding RNAs including miRNAs and lncRNAs were associated with carcinogenesis and poor prognosis in several human cancers, functioning as oncogenes or tumor suppressors [3,4,15]. In this study, we confirmed that HOTAIR was overexpressed whereas miR-122 was suppressed in HCC specimens and cell lines, which were consistent with previous studies [9,27], and for the first time, we identified that HOTAIR negatively regulated miR-122 expression in HCC, which promoted cell growth and hepatocarcinogenesis.
Emerging studies have documented that HOTAIR has a crucial role in human cancers by inhibiting a list of miRNAs, including miR-130a in gallbladder cancer [34], miR-331-3p in gastric cancer [35], miR-34a and miR-663b in pancreatic cancer [36,37], miR-148a in cervical cancer [38], as well as miR-218 in HCC [27]. Considering miR-122 is a dominant liver-specific miRNA and its expression is decreased during hepatocarcinogenesis [9,15], whether HOTAIR is involved in the suppression of miR-122 in HCC remains unclear. In this study, we identified that HOTAIR negatively regulated miR-122 expression in which HOTAIR overexpression decreased miR-122 expression meanwhile knockdown of HOTAIR increased miR-122 expression. Moreover, this negative correlation between HOTAIR and miR-122 was confirmed in HCC specimens. It has been showed that HOTAIR not only promoted cancer metastasis, but also played key roles in cell proliferation and tumorigenicity of cancer cells [20,27,37]. Our results showed that knockdown of HOTAIR inhibited HCC cell proliferation and induced cell cycle arrest and effectively suppressed HCC tumor progression, which were mediated by negative regulation of miR-122 expression, indicating that HOTAIR might promote hepatocarcinogenesis through suppressing miR-122 expression. Nevertheless, the regulatory role of HOTAIR in the expression of miR-122 in HCC remains elusive.
DNA methylation has been recognized as one of the causes of tumor initiation and development. A number of coding genes such as RASSF1A, as well as non-coding miRNAs such as miR-124, containing CpG islands at their promoter regions, were suppressed in human cancers by DNA methylation [39,40]. We found that a CpG island was located in the miR-122 promoter region, and treatment of HCC cells with DNA methylation inhibitor AZA dramatically increased the miR-122 expression level, suggesting that miR-122 was epigenetically suppressed via DNA methylation in HCC cells. Numerous studies demonstrated that a list of lncRNAs could mediate the maintenance of DNA methylation status and consequently repress their target genes in several cancers [41,42]. Our study showed that knockdown of HOTAIR in HCC cells decreased DNA methylation level of miR-122 promoter region, resulting in elevated expression of miR-122. These data suggested that HOTAIR could modulate miR-122 expression via DNA methylation, thus contributing to HCC tumorigenesis.
DNA methylation has been reported to be catalyzed by a family of DNA methyltransferases (DNMTs). DNMT1 has an ability to induce both de novo and maintenance of methylation. DNMT3A and DNMT3B are mainly involved in the de novo methyltransferase [32]. Previous studies showed that HOTAIR affected HOXA1 methylation through regulating DNMT1 and DNMT3B in small cell lung cancers [43]. Thus, we hypothesized that HOTAIR-induced epigenetic suppression of miR-122 via DNA methylation was mediated by DNMTs. Indeed, we found that HOTAIR-induced miR-122 suppression was associated with enhanced expression of DNMTs, and abolished by inhibition of DNMTs, indicating that HOTAIR was capable of regulating DNMTs expression, leading to alteration of DNA methylation status. It has been proved that HOTAIR altered gene expression by recruiting EZH2 [20], and EZH2 could facilitate DNA methylation through DNMTs [44]. Besides, knockdown of EZH2 not only inhibited DNMT1 expression but also reduce DNMT1 presence at the target promoter region [45]. To validate the potential role of EZH2 between HOTAIR and DNMTs expression, we founded that knockdown of EZH2 decreased the expression of DNMTs, and further reversed the HOTAIR-induced DNMTs upregulation, suggesting HOTAIR regulated DNMTs via EZH2. Additionally, inhibition of either EZH2, or HOTAIR, or DNMTs reactivated the expression of miR-122 in HCC cells. Altogether, our data suggested that HOTAIR could upregulate DNMTs expression via EZH2, contributing to modulation of DNA methylation status at the miR-122 promoter region and consequently suppression of miR-122 expression. These finding might enrich our understanding about the regulatory mechanism of HOTAIR and DNA methylation in HCC.
As a well-known tumor suppressor against HCC, miR-122 has been reported to inhibit cell growth and suppress tumorigenesis by negatively regulating its targets [13,15]. Cyclin G1, a key cell cycle protein, plays an essential role in tumor growth, has been shown to be a target of miR-122, and highly expressed in HCC cells [13]. Our study confirmed that miR-122 negatively regulated Cyclin G1 expression in HCC cells. Furthermore, we identified a novel relationship between HOTAIR and Cyclin G1 in HCC that HOTAIR upregulated Cyclin G1 expression via suppression of miR-122, implicating that HOTAIR would play an important role in cell cycle regulation through the miR-122/Cyclin G1 pathway. Thus, our finding that overexpression of HOTAIR in HCC cells induced rapid cell proliferation and tumor growth, might at least in part be explained by miR-122-mediated upregulation of Cyclin G1 expression and further alteration of cell cycle progression.
In summary, our study showed for the first time that HOTAIR played a critical role in hepatocarcinogenesis through downregulation of miR-122. HOTAIR epigenetically suppressed miR-122 expression through DNMTs-mediated DNA methylation, leading to abnormal expression of Cyclin G1 in HCC cells. Therefore, our results provide a novel glimpse of the mechanistic link between two essential non-coding RNAs in HCC: HOTAIR and miR-122, and propose the HOTAIR/miR-122 negative regulatory axis as a promising molecular target for HCC intervention.
Acknowledgements and funding sources
This work was supported by grants from National Natural Science Foundation of China (No. 81000889), Natural Science Foundation of Guangdong Province (No. 2014A030311047; 2016A030313218; 2018A030313624), Science and Technology Planning Project of Guangdong Province (No. 2014A020212094), and Project of Sun Yat-Sen Memorial Hospital (No. YXQH201704).
Declaration of interest
The authors declared that they have no conflict of interest.
Authors' contributions
Study concept and design: ZJ, CR. Investigation and data acquisition: CD, DJ, ZB, HX, LG, MZ, YH, ZS, WL. Analysis and interpretation of data: CD, DJ, ZB, HX. Drafting of the manuscript: CD, DJ, ZB, HX. Critical revision of the manuscript: ZJ, CR, DX. Study supervision: ZJ, CR. Funding acquisition: ZJ, CR.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.08.055.
Contributor Information
Rufu Chen, Email: sums1866@126.com.
Jiajia Zhou, Email: jiajiazh2004@126.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Saran U., Humar B., Kolly P. Hepatocellular carcinoma and lifestyles. J. Hepatol. 2016;64(1):203–214. doi: 10.1016/j.jhep.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Wallace M.C., Preen D., Jeffrey G.P. The evolving epidemiology of hepatocellular carcinoma: a global perspective. Expert. Rev. Gastroenterol. Hepatol. 2015;9(6):765–779. doi: 10.1586/17474124.2015.1028363. [DOI] [PubMed] [Google Scholar]
- 3.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 4.Klingenberg M., Matsuda A., Diederichs S. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017;67(3):603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M., Rauhut R., Yalcin A. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 6.Girard M., Jacquemin E., Munnich A. MiR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008;48(4):648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Esau C., Davis S., Murray S.F. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Lanford R.E., Hildebrandt-Eriksen E.S., Petri A. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai W.C., Hsu S.D., Hsu C.S. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 2012;122(8):2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulouarn C., Factor V.M., Andersen J.B. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y., Xia F., Ma L. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310(2):160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Bai S., Nasser M.W., Wang B. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornari F., Gramantieri L., Giovannini C. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69(14):5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- 14.Jung C.J., Iyengar S., Blahnik K.R. Epigenetic modulation of miR-122 facilitates human embryonic stem cell self-renewal and hepatocellular carcinoma proliferation. PLoS ONE. 2011;6(11):27740–27753. doi: 10.1371/journal.pone.0027740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakao K., Miyaaki H., Ichikawa T. Antitumor function of microRNA-122 against hepatocellular carcinoma. J. Gastroenterol. 2014;49(4):589–593. doi: 10.1007/s00535-014-0932-4. [DOI] [PubMed] [Google Scholar]
- 16.Gatfield D., Le Martelot G., Vejnar C.E. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23(11):1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song K., Han C., Zhang J. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology. 2013;58(5):1681–1692. doi: 10.1002/hep.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y., Meng X.M., Huang C. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Rinn J.L., Kertesz M., Wang J.K. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta R.A., Shah N., Wang K.C. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu L., Zhu G., Zhang C. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res. Treat. 2012;136(3):875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 22.Gao J.Z., Li J., Du J.L. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol. Lett. 2016;11(3):1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z., Zhou L., Wu L.M. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J.J., Cheng D., He X.Y. Knockdown of Hotair suppresses proliferation and cell cycle progression in hepatocellular carcinoma cell by downregulating CCND1 expression. Mol. Med. Rep. 2017;6(4):4980–4986. doi: 10.3892/mmr.2017.7162. [DOI] [PubMed] [Google Scholar]
- 25.Ding C., Cheng S., Yang Z. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int. J. Mol. Sci. 2014;15(3):4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L., Zhang X., Li H. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol. BioSyst. 2016;12(8):2605–2612. doi: 10.1039/c6mb00114a. [DOI] [PubMed] [Google Scholar]
- 27.Fu W.M., Zhu X., Wang W.M. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J. Hepatol. 2015;63(4):886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Su D.N., Wu S.P., Chen H.T. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol. Lett. 2016;12(5):4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T., He X., Chen A. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670(1):114–122. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J.J., Chen R.F., Deng X.G. Hepatitis C virus core protein regulates NANOG expression via the stat3 pathway. FEBS Lett. 2014;588(4):566–573. doi: 10.1016/j.febslet.2013.11.041. [DOI] [PubMed] [Google Scholar]
- 31.Li Z.Y., Xi Y., Zhu W.N. Positive regulation of hepatic miR-122 expression by HNF4α. J. Hepatol. 2011;55(3):602–611. doi: 10.1016/j.jhep.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Cedar H., Bergman Y. Programming of DNA methylation patterns. Annu. Rev. Biochem. 2012;81(1):97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 33.Kim E., Kim M., Woo D.H. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma M.Z., Li C.X., Zhang Y. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer. 2014;13(1):156–169. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X.H., Sun M., Nie F.Q. LncRNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer. 2014;13(1):92–105. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C.H., Xiao Z., Tong J.H. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int. J. Cancer. 2017;140(1):120–129. doi: 10.1002/ijc.30414. [DOI] [PubMed] [Google Scholar]
- 37.Cai H., An Y., Chen X. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7(52):86857–86870. doi: 10.18632/oncotarget.13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J., Chu H., Ji J. Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int. J. Oncol. 2016;49(3):943–952. doi: 10.3892/ijo.2016.3589. [DOI] [PubMed] [Google Scholar]
- 39.Lambert M.P., Paliwal A., Vaissière T. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J. Hepatol. 2011;54(4):705–715. doi: 10.1016/j.jhep.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Murray-Stewart T., Sierra J.C., Piazuelo M.B. Epigenetic silencing of miR-124 prevents spermine oxidase regulation: implications for Helicobacter pylori-induced gastric cancer. Oncogene. 2016;35(42):5480–5488. doi: 10.1038/onc.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W., Zheng J., Deng J. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146(7):1714–1726. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Teschendorff A.E., Lee S.H., Jones A. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 2015;7(1):108–119. doi: 10.1186/s13073-015-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang S., Gao H., Tong Y. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab. Investig. 2016;96(1):60–68. doi: 10.1038/labinvest.2015.123. [DOI] [PubMed] [Google Scholar]
- 44.Viré E., Brenner C., Deplus R. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 45.Ning X., Shi Z., Liu X. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015;359(2):198–205. doi: 10.1016/j.canlet.2015.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material