Abstract
Epilepsy includes a group of disorders of the brain characterized by an enduring predisposition to generate epileptic seizures. Although familial epilepsy has a genetic component and heritability, the etiology of the majority of non-familial epilepsies has no known associated genetic mutations. In epilepsy, recent epigenetic profiles have highlighted a possible role of microRNAs in its pathophysiology. In particular, molecular profiling identifies a significant number of microRNAs (miRNAs) altered in epileptic hippocampus of both animal models and human tissues. In this review, analyzing molecular profiles of different animal models of epilepsy, we identified a group of 20 miRNAs commonly altered in different epilepsy-animal models. As emerging evidences highlighted the poor overlap between signatures of animal model tissues and human samples, we focused our analysis on miRNAs, circulating in human biofluids, with a principal role in epilepsy hallmarks, and we identified a group of 8 diagnostic circulating miRNAs. We discussed the functional role of these 8 miRNAs in the epilepsy hallmarks. A few of them have also been proposed as therapeutic molecules for epilepsy treatment, revealing a great potential for miRNAs as theranostic molecules in epilepsy.
Keywords: microRNA, miRNA, epilepsy, biomarker, diagnosis, theranostic molecules
Graphical Abstract
Main Text
Epilepsy is a global health care issue affecting 50–70 million people worldwide with 2.4 million people being diagnosed with epilepsy each year.1 It affects ∼1% of the population worldwide at the age of 20 years and 3% at the age of 75 years;2 ∼80% of epilepsy cases occur in the developing countries, and three-fourths of the affected individuals do not get appropriate treatment.1
Epilepsy is a highly heterogeneous condition that encompasses a spectrum of clinical subtypes sharing the common feature of persistently increased neuronal excitability and spontaneous seizure generation. Although clinical classifications are constantly evolving, in 2017, the International League Against Epilepsy (ILAE) announced new classification schemes for both seizures and epilepsies, and it proposed to classify epilepsies on seizure types, epilepsy types, and epileptic syndromes, dividing seizures into focal onset and generalized onset.3, 4, 5, 6
Traditional estimates of heritability for epilepsy varied greatly, and growing evidence has illustrated that genetic factors may play a role in many epilepsy syndromes.6, 7, 8 A recent work reported heritability as 32% for all epilepsy classes, 23% for focal epilepsy, and 36% for nonfocal epilepsy.7
Currently available interventions and antiepileptic drugs (AEDs) are not effective in over 30% of the patients.8 Most of these AEDs are primarily targeted against neuronal ion channels or neuronal receptors, such as gamma-aminobutyric acid (GABA) and glutamate receptors.9 Thus, there is a need to better understand the underlying disease mechanisms in order to identify novel drug targets or treatment strategies.10
The pathogenesis of epilepsy is supposed to involve large-scale alterations in the expression of genes controlling neurotransmitter signaling, ion channels, synaptic structure, neuronal death, gliosis, and inflammation.11
Numerous gene defects underlying different forms of epilepsy have been identified, with most of these genes encoding ion channel proteins.12 However, the etiology of the majority of non-familial epilepsies has no known associated genetic mutations, even though genetic linkage analyses have identified several loci that may contain mutation-susceptible sequences in epilepsy.13 Copy number variants associated with epilepsies have been observed through genome-wide analysis, and single-point mutations have been detected through exome sequencing or whole-genome sequencing.14
Epigenetic mechanisms unrelated to altered DNA sequences that may affect epileptogenesis include transcriptional or post-transcriptional regulation. The pathogenesis of different types of epilepsy involves many important biological pathways, some of which have been shown to be regulated by microRNAs (miRNAs), small non-coding RNA of about 22 nt in length, which are able to regulate the stability of several mRNAs by binding to their complementary sequence at the 3′ UTR of a coding mRNA.15 miRNA works in a particular way for activity-dependent regulation of mRNA stability and translation.11, 16 Previous studies have focused on the regulations of miRNA on its target genes, and some studies analyzed the effects of the altered expression of single miRNA, increasing or decreasing miRNA levels in mouse and rat models, and observing the amelioration of the pathological features of epilepsy.17, 18, 19, 20
The SNPs in miRNA genes (miRNA-SNPs) are one example of point mutation that could affect miRNA function in one of the following three possible ways: altering transcription of the primary miRNA transcript, processing primary miRNA (pri-miRNA) and precursor miRNA (pre-miRNA), and through their effects on the modulation of miRNA-mRNA interactions.21, 22 As a result, miRNA-SNPs have been associated with the pathogenesis of human disease, including epilepsy.23
The alteration in the expression levels of specific miRNAs has been described as a possible cause in the onset of different pathologies, from cancer24 to brain pathologies, such as Parkinson’s disease,25 Alzheimer’s disease (AD),26 or other neurodegenerative diseases.27 In epilepsy, several studies showed an alteration of global miRNA expression profiles in a variety of animal models and humans.19 Specific miRNAs in brain tissue have been linked to seizure-induced neuronal death or neuroprotection.28 Components of the miRNA biogenesis pathway that have been found to be altered in brain tissue from epilepsy patients result in select changes to miRNAs regulating neuronal microstructure, cell death, inflammation, and apoptosis. Over the past 5 years, several target studies and genome-wide miRNA expression-profiling studies18, 29, 30, 31, 32 have identified over 100 different miRNAs in epilepsy patients or animal models, and they have provided compelling evidence that epilepsy is associated with widespread changes of miRNA expression. The first study on miRNA in human epilepsy was published in 2010 and reported an increase in hippocampal levels of miR-146a, an miRNA linked to the control of inflammatory responses.33
More recently, a potential use of miRNA in clinical application has been proposed, using miRNA mimic or inhibitor sequences as possible therapeutic molecules in epilepsy, although several limits are emerging (i.e., targeted delivery and multi-targeting effect).11, 16
miRNAs can be detected in blood serum, making them suitable candidates for potential biomarkers to assess disease risk and treatment responses. miRNAs have been found stable in serum, and the test of miRNAs in blood is broadly accessible, rapid, noninvasive, and economically sustainable.34 Previous studies have documented that circulating miRNAs have been proposed as biomarkers, with potential application in aging and neurological diseases such as AD and multiple sclerosis (MS).35, 36 Altered miRNA profiles in biofluids may be potentially useful biomarkers of epileptogenesis. In addition to their potential use as therapeutic tools, epilepsy-associated miRNAs offer prospects as biomarkers for identifying patients at risk.37
Given that one miRNA is able to regulate several target genes belonging to different functional pathways, in this review, we highlight the role of miRNAs in controlling epilepsy functional hallmarks starting from the analysis of miRNA profiles of focal epilepsy animal models. We then tried to assess those miRNAs found commonly altered in tissues from animal models and in human focal epilepsy tissues, and we discussed their role in functional pathways altered in temporal lobe epilepsy (TLE) (Figure 1). As circulating miRNAs could become the targets of therapeutic tools, we then focused on those human circulating miRNAs that, having a main role in the control of specific functional hallmarks associated with focal epilepsy, could become potential pharmacological targets for focal epilepsy (Figure 1).
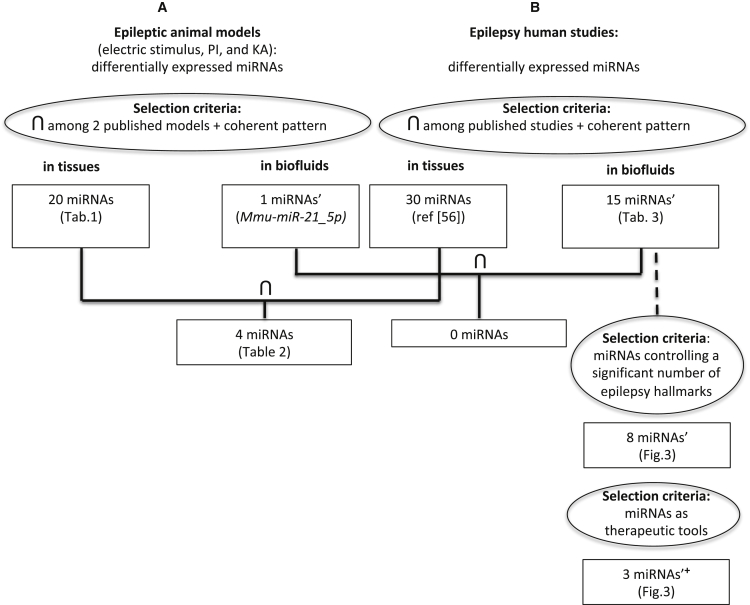
Figure 1.
Flowchart
Flowchart representing the logic flux of the analysis for the identification of animal-derived (A) and human (B) miRNAs with a main role in epilepsy. The selection criteria in the (A) and (B) approaches are indicated in the ovals. The horseshoe symbol indicates selecting miRNAs in common among. Coherent pattern means selecting an miRNA only if the miRNA has constantly been found with an upregulated or downregulated pattern. Circulating miRNAs previously found with therapeutic effect are indicated (’+) and the other circulating miRNAs are also indicated (’).
miRNA Production
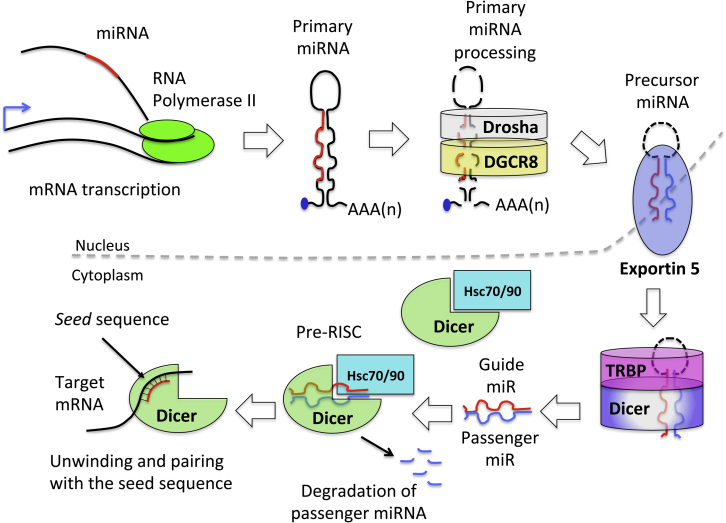
miRNA sequence is transcribed by RNA polymerase II (Pol ll) as a long primary transcript (pri-miRNA). The majority of miRNAs are located in intronic regions of protein-coding mRNA.38 As soon as the mRNA is spliced, pri-miRNAs emerge as a long hairpin structure. It undergoes several steps of processing, becoming pre-miRNA and mature miRNA to form miRNA duplex.38 During the maturation, miRNA loses part of its sequence, being cut by Drosha in the nucleus and DICER complex in the cytoplasm.38 One strand of the duplex miRNA, the guide strand, is recognized and loaded into the RNA-induced silencing complex (RISC) to elicit its silencing effect. The other strand, the passenger strand (indicated as miRNA *), is usually degraded.38 Once miRNA recognizes the complementary sequence on the target mRNA (called the seed sequence), it binds to it, leading to mRNA degradation or repression, depending on the level of pairing with the seed sequence38 (Figure 2).
Figure 2.
miRNA Biogenesis
The life of an miRNA begins in the nucleus, where RNA polymerase II (Pol II) transcribes the whole RNA along with miRNA. The long primary miRNA (pri-miRNA) (approximately 1 kb) with its hairpin structure is processed by the Drosha-DCGR8 complex. The microprocessor complex, formed by Drosha and its cofactor DCGR8, lead to the maturation of the pri-miR into precursor miRNA (pre-miRNA). The RNase activity of Drosha removes the stem loop structure, releasing a small hairpin RNA of approximately 65 bp. Several other proteins collaborate with the microprocessor to guide the correct maturation of the pri-miRNA (p53, SMAD1-3, p68, and TDP43). After Drosha processing, pre-miRNAs are exported into the cytoplasm by the transport complex formed by Exportin 5 and RAN-GTP. Once the complex is translocated into the cytoplasm, GTP is hydrolyzed to GDP, leading to complex disassembly. In the cytoplasm, another RNase guides the maturation of pre-miRNA into the duplex form,.i.e., Dicer. Dicer interacts with the double-strand hairpin structure of the cytosolic pre-miRNA and, in collaboration with TAR RNA-binding protein, cleaves the RNA duplex; further, the mature miRNA is loaded directly into the RNA-induced silencing complex (RISC), whereas the passenger miRNA is usually degraded. The RISC complex needs the help of two heat shock cognate proteins (HSC70 and 90) that, using ATP, mediate the opening of the Argonaute (AGO) protein to receive the miRNA duplex. The RISC receives the duplex, unwinds it, and degrades the passenger miRNA. The pairing between the RISC-mature miRNA and the seed sequence on the target mRNA determines mRNA degradation or mRNA repression.38
In mammals, 60% of the mRNAs have a known seed sequence for miRNA binding; thus, miRNAs play an essential role in controlling all the cellular processes.39 In the brain, miRNAs are particularly abundant and control neurogenesis.40 In Dicer−/− mouse model, the biogenesis of miRNA is blocked, leading to neuronal loss and premature animal death.41
Tissue-Associated and Circulating miRNAs in Focal TLE Animal Models
Some epileptic models have been developed to understand the onset and development of the pathology. Initially, to study epilepsy, simple acute seizure models were developed, as those obtained by pentylenetetrazol or the volatile chemoconvulsant flurothyl treatment42 to induce seizures, or maximal electroshock seizure models, but these models failed to mimic the spontaneous onset of seizures, typical of epilepsy.43
Epilepsy has been studied in several organisms, starting from the simpler (i.e., Drosophila melanogaster, Caenorhabditis elegans, and Danio rerio) to non-human primates.44 Nowadays, the most commonly used rodent models to study epilepsy are Rattus norvegicus and Mus musculus. The use of these animals has been helpful to generate genetic models of epilepsy. These models have been developed from the observation that single-gene mutations could be the cause of epileptic disease, such as mutation in ion (voltage-gated sodium channels, potassium channels, or calcium channels) and neurotransmitter receptor.44 In the last case, epilepsy is a comorbid phenotype caused by other pathologies, such as AD, autism, or other neurological disorders.44
More recently, other rodent models have been developed to study the status epilepticus (SE), which is an episode of prolonged seizures generated by an initial precipitant injury, affecting the hippocampus and/or the temporal lobe. These animal models were generated to mimic TLE, the most common form of epilepsy that displays a clinical history including an onset injury, a pre-epileptic latent period before the manifestation of spontaneous seizures, a chronic clinical manifestation, and the typical histopathological neuronal changes of human TLE. No animal model has been proposed until now to describe the development of non-focal epilepsy, possibly because the origin of this form of epilepsy is not completely understood. To generate TLE-associated status, the main models of acquired epilepsy are due to intracerebral or systemic injection of drugs (such as pilocarpine, kainic acid [KA], or bicuculine) or of toxins (tetanus toxins) or mimicking the etiology due to febrile seizures by hyperthermia treatment, sustained electrical stimulation, or ischemia.44 Hyperthermia is a good model to mimic TLE, while the treatment with KA (an agonist of ionotropic glutamate receptor) or pilocarpine (an agonist of cholinergic muscarinic receptor) is the most commonly used model, presenting a histopathological profile similar to that observed in mesial TLE patients.45
The research of miRNAs in biofluids of animal models of epilepsy could be helpful in order to identify new, specific, sensitive, reproducible, predictive, and accurate biomarkers and to develop new, non-invasive, standardized, cost-affordable, and easy-to-perform detection assay for the differential diagnosis of TLE.46 We have found a few publications that propose a diagnostic signature of circulating miRNAs obtained by the study of animal models (i.e., Gorter et al.32 and Hu et al.47), while several single miRNAs have been shown to possess diagnostic properties (i.e., Liu et al.42). Some papers discussed the miRNA profile of brain tissues of focal TLE animal models (i.e., Hu et al.47 and Li et al.48) and also presented data regarding circulating miRNAs (i.e., Gorter et al.32). These miRNAs are summarized in Table 1, grouped for each animal model. Some papers propose a single miRNA as a possible epilepsy biomarker, but without presenting an miRNA profile able to describe the pathology.49
Table 1.
Summary of miRNAs Involved in Focal TLE, as Obtained by the Study of the miRNA Profiles in Tissues and/or Biofluids of Three Different Animal Models
| Epilepsy Model | Tissue | Up miRNA | Down miRNA | Differential Diagnosis | Reference |
|---|---|---|---|---|---|
| Amygdala electric stimulation | H | miR-54, miR-296-5p, miR-345-3p, miR-365-5p, miR-423-3p, miR-455-3p | miR-296-5p | focal TLE | 48 |
| H | acute + latent miR-21; latent + chronic miR-23a, -27a, -143 | – | focal TLE | 32 | |
| DG | acute + latent + chronic miR-21, -132, -132*, -212; acute + latent miR-142-5p; chronic miR-23a, -146a, -212* acute + chronic miR-223 |
acute + latent miR-485 chronic miR-138 |
focal TLE | 32 | |
| plasma | miR-21-5p, -142-5p, -146-5p | focal TLE | 32 | ||
| H | miR-20a-5p, -345-3p, -365-5p, -764-3p, -874-3p | miR-99b-3p | focal TLE | 50 | |
| DG | miR-21-5p, -132-3p, -212-3p | miR-7a-5p, -187-3p, -551b-3p | focal TLE | 31 | |
| PI rat model | H | miR-22, -24, -26a, -29b, -30c, -34a, -99a, -101-1, -124a, -125a/b, -132, -145, -150, -151, -196b, -199a, -213, -375 | miR-10b, -21, -25, -29a, -181b/c, -215 | focal TLE | 47 |
| blood | miR-22, miR-34a, miR-125a | miR-21 | focal TLE | 47 | |
| H | miR-23a, -27a, -31, -34a, -146a, -152, -203, -210, -211 | miR-19a, -33, -135b, -136, -138*, -144, -153, -190, -296*, -301a, -325-5p, -380, -542-3p, -542-5p, -543 | focal TLE | 18 | |
| H | miR-22-5p, -23a-3p, -27a-3p, -129-5p, -129-2-3p, -132-3p, -135b-5p, -199a-5p, -203-3p, -212-3p, -221-3p, -222-3p, -297a-5p, -297c-5p, -455-3p, -466f-3p, -467c-3p/ -467e-3p, -467e-5p, -467 g, -494-3p, -669c-5p, and -669f-3p | let-7d-3p, let-7f-1-3p, miR-29b-1-5p, -34b-3p, -92b-3p, -130a-3p, -138-1-3p, -140-3p, -181a-5p, -187-3p, -191-5p, -194-5p, -210-3p, -298-5p, -324-5p, -325-3p, -330-3p, -331-3p, -338-5p, -345-5p, -350-3p, -409-5p, -431-3p, -551b-3p, -674-5p, -676-3p, -767, -875-3p, -1949 | focal TLE | 51 | |
| KA-induced rat model | H | let-7a, miR-99a, -125b-5p, -193, 202-3p, -219a-3p, -292-3p, -294, -302b, -346, -383, -490, -666-5p, -672, -770-3p | – | focal TLE | 53 |
| H | let-7f, miR-7, -9, -28, -29a, -29c, -30a-3p, -31, -34b, -34c, -100, -129, -130b, -132, -140, -148b, -184, -204, -299-5p, miR-335, -369-3p, -375, -376a, -409-5p, -448 | – | focal TLE | 54 | |
| H | – | miR-9, -125a-5p, -128, -138, -145, -150 | focal TLE | 55 | |
| H | miR-10b, -21, -27a, -29a, -30e, -107, -132, -134, -139, -145, -146b, -148b, -153, -181c, -199a, -200a, -219, -323, -326, -328, -375, -425, -451, -487b, -507, -509, -518d, -532 | miR-101, -103, -125a, -127, -133b, -330, -374, -381, -422b, -497, -520b, -657 | focal TLE | 56 |
H, hippocampus; DG, dentate gyrus; up miRNAs, upregulated miRNAs; down miRNAs, downregulated miRNAs; PI, pilocarpine-induced; KA, kainic acid.
In 2014, three papers analyzed the miRNA profile of brain tissue extracted from two different rat epileptic models. High-throughput sequencing analysis of rat hippocampus generated by amygdala stimulation revealed that 7 miRNAs were differentially expressed in TLE models with respect to a non-stimulated animal model48 (Table 1). In particular, all these miRNAs were upregulated in TLE, except miR-296-5p, which was downregulated. These miRNAs were validated by RT-PCR. The analysis of the functional role of these miRNAs in epileptic hippocampus revealed that, in chronic TLE, the main function controlled by these miRNAs is apoptosis, which is observed as a neuronal loss induced by TLE.
The second paper32 presented the analysis of rat brain hippocampal and dentate gyrus (DG) tissues from electrically induced SE compared to control tissues. This work revealed that several miRNAs were altered at different time points (day 1 and day 7 to simulate acute phase and 3 months after seizures at latent stage). In particular, miR-21 was upregulated in both acute and latent stages; miR-143, miR-23a, and miR-27a were upregulated in both latent and chronic stages in the CA1 region of the hippocampus (Table 1). In DG, 4 miRNAs were upregulated at all time points: miR-132, -132*, -21, and -212. miR-142-5p was upregulated in acute and latent stages, but not in chronic phase, while three miRNAs were upregulated only in chronic and latent phases: miR-23a, -146a, and -212*. miR-223 was upregulated only in acute and chronic phases. In the same paper, plasma miRNA analysis was performed. It confirmed that miR-21-5p, -146-5p, and miR-142-5p were upregulated; in particular, miR-21-5p increased significantly at 1 week post-SE, while miR-146a-5p showed an increased expression 3–4 months after SE and not 1 week after SE, as observed in the tissues. miR-142-5p was increased in plasma 24 hr and not 1 week after SE. Regarding the role controlled by these miRNAs in the epileptic models after 1 week from SE, the main functions are those related to the inflammatory process (transforming growth factor β [TGF-β] signaling and Toll-like and interleukin-1 receptor signaling). The inflammatory pathway is controlled by miR-19b, miR-20b, miR-21, and miR-146a, which are all upregulated, and miR-33, which is downregulated. In the tissues, miR-21 and miR-21* are the most upregulated miRNAs after 1 week of SE in all the rat brain tissues. Also, these miRNAs seem to be involved in the regulation of inflammatory process, associated with Toll-like receptor signaling.
The third paper47 presented the results of the analysis of rat hippocampus from a pilocarpine-induced epileptic model; miRNA array profiling revealed 19 upregulated and 7 downregulated miRNAs (Table 1). The comparative analysis of these miRNAs in the hippocampus from this epilepsy model showed that miR-21 was decreased in hippocampal tissue, while miR-22, miR-34a, and miR-125a-5p were increased. This paper presented also the results of miRNA profiling in peripheral blood samples of the same epileptic model. Some miRNAs were investigated; miR-21-5p was decreased, while miR-22, miR-34a, and miR-125a were confirmed to be increased. The deregulated miRNAs were predicted to control the mitogen-activated protein kinase (MAPK)-signaling pathway, TGF-β-signaling pathway, long-term potentiation pathway, tight junction pathway, glycan structure-biosynthesis 1, and axon guidance.47
In another paper, high-throughput miRNA profiling was used to analyze differentially expressed miRNAs in rat hippocampus 24 hr after amygdala electric stimulation50 (Table 1). Five miRNAs were increased in 1 week, while only miR-99b-3p was decreased 3 weeks after SE. The expression of miR-365-5p and miR-99b-3p correlated with neuronal apoptosis observed in SE tissues.50
The analysis of the DG of the amygdala in electrically stimulated rats revealed that 9 miRNAs were upregulated, while 57 miRNAs were downregulated.31 The miRNAs with the highest downregulation were miR-7a-5p, miR-187-3p, and miR-551b-3p, while those with the highest increase were miR-21-5p, miR-212-3p, and miR-132-3p. The analysis of potential miRNA targets revealed that several biological functions could be affected, such as transcription, response to wounding, apoptosis, cell proliferation, and immune response.31
The miRNA expression profile analysis of the rat models of SE and TLE induced by pilocarpine treatment revealed that 9 miRNAs were upregulated and 15 were downregulated18 (Table 1). Among the others, miR-34a was significantly upregulated.18
In another pilocarpine-induced epilepsy mouse model on hippocampus, miRNA profile revealed that, at a later time (28 days after the induction of SE with pilocarpine treatment), 51 miRNAs were significantly deregulated. In particular, 22 miRNAs were upregulated while 29 were downregulated51 (Table 1).
DG and hippocampus tissues isolated from a KA mouse model or saline-injected mouse were subjected to multi-omics profile.52, 53 Among the 189 dysregulated miRNAs,53 the authors highlighted the upregulation of 15 hippocampal miRNAs, with an enrichment of brain-specific miRNAs (Table 1). In another publication,54 CA3 regions from control or KA-treated animals were isolated and subjected to miRNA profile. The analysis revealed an increase of 25 miRNAs in epileptic models versus control (Table 1). In Pichardo-Casas et al.,55 KA-treated hippocampus was analyzed for specific miRNAs, and the authors identified a general reduction of all considered miRNAs, with a significant reduction in miR-145 expression. In Jimenez-Mateos et al.,56 the miRNA profile was extracted from the ipsilateral CA3 subfield 24 hr after focal-onset SE in KA animal models, and 45 miRNAs were found altered in SE versus controls (Table 1).
Regarding the miRNA profile of the KA mouse or rat model of epilepsy, no paper actually exists with an miRNA profiling of epilepsy-associated plasma or serum. Only one paper presented the miRNA profile for brain ischemia, hemorrhage, and KA-induced seizure tissues and blood;57 but, in this paper, the list of all the miRNA specifically altered in the blood of SE models versus control animals is not specifically reported.
All the results highlight the importance of miRNA in the epilepsy phenotype, but a general agreement on a few common groups of miRNAs as possible diagnostic or prognostic biomarkers of epilepsy is still lacking. By comparing the lists of altered miRNAs in all the analyzed papers, we sum up with the tissue-associated miRNAs with a coherent pattern of expression in two models of epilepsy (with coherent we mean those miRNAs that are increased or decreased in the same way in two different epilepsy models), representing a core list of 20 possible epilepsy-associated miRNAs altered in at least 2 of 3 animal models of chronic epilepsy: miR-23a, miR-27a, miR-31, miR-99a, miR-125b-5p, miR-129-5p, miR-132-3p, miR-142-5p, miR-146a, miR-212-3p, and miR-375 all upregulated; miR-10b, miR-21, miR-29a, miR-138, miR-153, miR-181c, miR-187-3p, miR-330-3p, and miR-551b-3p all downregulated.
Translation of miRNAs Identified in TLE Animal Models to Human TLE
Analyzing the literature to identify all the publications on whole miRNA profiling on human epileptic tissues (hippocampus or DG), we identified only one new recent paper that proposes the whole miRNA profile of human hippocampus of TLE subjects.58 Considering the 20 tissue-isolated miRNAs in common between at least two TLE animal models obtained by an analysis of Table 1 (namely, miR-23a, miR-27a, miR-31, miR-99a, miR-125b-5p, miR-129-5p, miR-132-5p, miR-142-5p, miR-146a, miR-212-3p, and miR-375 all upregulated; miR-10b, miR-21, miR-29a, miR-138, miR-153, miR-181c, miR-187-3p, miR-330-3p, and miR-551b-3p all downregulated), we found some present in the profiles of human epileptic tissue. In particular, we found 4 miRNAs that are in common with the miRNA hippocampus profile of human TLE subjects.58 Following the new annotation suggested by the database http://mirgenedb.org/,59 the four miRNAs are Hsa-miR-132-P1, Hsa-miR-142-P1-v1_5p, Hsa-miR-146-P1_5p, and Hsa-miR-138-P1_5p.
A recent publication by Korotkov60 proposed a meta-analytic approach to identify unique miRNA profiles by comparison of three post-epileptic seizure models. In our approach, Korotkov’s analysis includes electrical stimulation, pilocarpine, and KA models. The authors compared profiles obtained by their analyses with the miRNA profile of human TLE with hippocampal sclerosis, which instead was not included in our study. The Korotkov analyses also considered different stages of epileptogenesis in animal models, while we focused on miRNA profiles of the latent and chronic epileptic statuses of animal models, which, in our opinion, are more similar to human disease when it is diagnosed. Some miRNAs of latent and chronic stages described by Korotkov have also been found in our analysis (i.e., miR-23a and miR-146a-5p among the upregulated miRNAs and miR-551b-3p among the downregulated miRNAs).
This limited overlap between miRNA profiling in human and animal tissues may possibly be because of the different techniques for miRNA isolation and profiling to the different brain regions analyzed or to the low abundance of altered miRNAs in epilepsy.
To understand the role of the 4 overlapping miRNAs between human and animal models in epilepsy development, we evaluated their functions through a pathway enrichment analysis, identifying a group of pathways significantly enriched with miRNA targets. We used 1,077 pathways derived from the Reactome,61 BioCarta,62 and Kyoto Encyclopedia of Genes and Genomes (KEGG)63 databases. The enrichment for each miRNA was evaluated using the Fisher’s exact test between miRNA targets and the selected 1,077 pathways. We considered a pathway to be enriched if the p value was <0.01; p values were adjusted using the Benjamini-Hochberg procedure for multiple testing correction.64 The association between miRNA and its target gene was performed using experimentally validated databases, such as miRTAR,65 miRWalk,66 and miRTarBase.67 An R package, SpidermiR, was used to integrate the data.68, 69, 70 The only miRNAs that control functional pathways, in a statistically significant way, are Hsa-miR-132-P1, Hsa-miR-138-P1_5p, Hsa-miR-142-P1-v1_5p, and Hsa-miR-146-P1_5p (Table 2).
Table 2.
Summary of Genes and Functional Pathways that Are Targets of Epilepsy-Associated miRNA in Common among at Least Two Temporal Lobe Epilepsy Animal Models and TLE Human Tissues, with Coherent Behavior
| miRNA | Functional Pathway | Target Genes in the Functional Pathway |
|---|---|---|
| Hsa-miR-132-P1 | cell cycle, proliferation, apoptosis | CDKN1A, PSMA2, PSMD12, RB1, CCNA2, CCNB1, MAPK1, CDKN1A, CRK, HBEGF, FOXO1 |
| neuroinflammation | NGF signaling: CDKN1A, CRK, ECT2, FOXO1, IRAK1, MEF2A, MAPK1, RTN4, MAP3K3, BDNF, IRAK4 | |
| PDGF signaling: CDKN1A, CRK, FOXO1, MAPK1, RASA1, THBS1 | ||
| ILS signaling: CRK, IRAK1, MAP3K3, IRAK4, MAPK1 | ||
| gene expression | MAPK signaling: CDKN1A, CRK, FOXO1, MAPK1, RASA1, IRAK1, MEF2A, IRAK4 | |
| tissue remodeling | extracellular signaling: SEC61A1, RPSA, RPL7, RPS5, SSR3 | |
| Hsa-miR-138-P1_5p | cell cycle, proliferation, apoptosis | HIF1A, NFKB1, RARA, CASP3, CCND1, CDH1, CEBPA, PPARG |
| focal adhesion | IGF1R, PTK2, AKT1, CCND1 | |
| Hsa-miR-142-P1-v1_5p | cell cycle, proliferation, apoptosis | HIF1A, TGFB2, TGFBR2, SMAD3, PTEN, RAC1 |
| neuroinflammation | RARA, CEBPA, EIF4EBP1, AKT1, NFKB1, CCND1 | |
| focal adhesion | IGF1R, PTK2, AKT1, RELN, ROCK2, CCND1, CCND3 | |
| tissue remodeling | CDH2, AKT1, FABP4, RHOC, LPL, PPARG, PTK2, CCND3, ROCK2 | |
| Hsa-miR-146-P1_5p | cell cycle, proliferation, apoptosis | CD40LG, CFH, FADD, ICAM1, IFI27, IRF7, ISG15, KIF22, MX2, OASL, OSBPL1A, RAC1, SIKE1 |
| neuroinflammation | NGF signaling: CDKN1A, IRAK1, IRAK2, | |
| ILS signaling: IRAK1, IRAK2, STAT1 | ||
| immune response: TLR2, TLR4, TRAF6 | ||
| tissue remodeling | extracellular signaling: IFIT1, IFIT3, IFITM1, IFITM3, ITGB2 |
Among the pathways controlled by these four miRNAs, there are (1) cell cycle, proliferation, and apoptosis; (2) neuroinflammation; (3) gene expression; and (4) tissue remodeling (Table 2).
-
(1)
Cell cycle, proliferation, and apoptosis. Hsa-miR-132-P1 has been already studied in cancer, playing a role in the control of proliferation, apoptosis,71 and epilepsy-associated inflammation.72
In TLE, miR-146 upregulation is usually associated with not only neuroinflammation73 but also the suppression of proliferation74 and decrease of vascular endothelial growth factor (VEGF) expression and vasculogenesis.75 It has been already reported that, in epilepsy, a loss of neurons and an increase in inflammation are among the main histopathological features.76
-
(2)
Neuroinflammation. Several pro-inflammatory cytokines increase during epilepsy or after an epileptic event, and, as a consequence, their related miRNAs are also affected. Regarding miR-146a-5p, previous publications have demonstrated it to play a role in the increase of IL-1beta and tumor necrosis factor alpha (TNF-α),77 being upregulated in hippocampal astrocytes of epileptic rat models and in human brain tissues.33 It has been suggested that the upregulation of miR-146a-5p could represent an attempt of the astrocytes to modulate and reduce inflammatory response due to IL-1b, perhaps by modulating IL-1R1-associated protein kinase-1 and -2 (IRAK1 and IRAK2) and TNF receptor-associated factor 6 (TRAF6).78, 79 In rodents, pharmacological treatment with a blockage of IL-1b synthesis reduced seizure, decreasing the inflammation associated with SE.80 Thus, miR-146a-5p could be proposed as a possible target to inhibit epilepsy-associated neuroinflammation.
-
(3 and 4)
Gene expression and tissue remodeling. The control of miRNAs on gene expression could lead to different functional outcomes. A clear role of miR138-P1_5p in homeostasis and tissue remodeling already has been demonstrated,81 possibly due to its ability also to control sirtuin genes (i.e., SIRT-1). This miRNA also seems to play a role in angiogenesis and hypoxia response.82, 83
Neuronal miR-132-3p, necessary for neuronal development, plays a role in brain morphogenesis, controlling neurotrophin, the MAPK-signaling pathway, and apoptosis.56 Thus, this observation highlights the ability of this miRNA to control the gene expression, which determines its influence on several functional processes.84 On miR-146, several papers demonstrate its involvement in remodeling by modulating axonal growth85, as well as in heart tissue remodeling.86
Circulating miRNA in Human Epilepsy
One interesting feature of miRNAs is that they can be secreted and found in several human biofluids (lacrimae, milk, serum, plasma, saliva, and urine) under both physiological and pathological conditions. This feature makes miRNAs a promising molecule for diagnosis, prognosis, or the development of new therapeutic tools. Although the miRNA expression level in biofluids is not abundant, several commercial kits allow their isolation, identification, and amplification by real-time qPCR or next-generation sequencing (NGS) approaches on a low-cost basis. The main problem is the method for the selection of the group of epilepsy-associated miRNAs.
In animal models, the main miRNA isolated in the blood of at the least two different TLE models is miR-21-5p.32, 55
Looking for “circulating diagnostic miRNA in human epilepsy,” we found only 6 papers in PubMed (November 2017). To identify a reliable diagnostic biomarker able to identify the pathology, we analyzed those 6 publications87, 88, 89, 90, 91, 92 and the results are presented here (Table 3).
Table 3.
Summary of Circulating Human miRNAs Involved in Temporal Lobe Epilepsy Development, with Diagnostic Properties
| Human Biofluid | Circulating miRNA | Aim | Reference |
|---|---|---|---|
| CSF | miR-19b-3p (down); miR-451a (up) | TLE diagnosis (AUC = 0.82) | 87 |
| Serum | let-7d-5p, miR-106b-5p, -130a-3p, -146a-5p (up); miR-15a-5p, -194a-5p (down) | TLE diagnosis (AUC all = 0.88) (AUC miR-106b-5p = 0.882) | 88 |
| Plasma | miR-34a (down) | mesial TLE diagnosis (AUC = 0.671) | 89 |
| Serum | miR-106b-5p, -146a (up) | TLE diagnosis (AUC = 0.887) | 90 |
| Plasma exosomes | miR-3613-5p (up); miR-197-5p, miR-4322, miR-4668-5p, miR-6781-5p, miR-8071 (down) | TLE diagnosis (AUC for miR-3613-5p = 0.8444) | 91 |
One paper was excluded because it describes the procedures for the standardization of biomarker analysis in epilepsy without proposing any miRNA profile related to the disease.46
In the second paper, the circulation of the miRNA profile in humans was analyzed in cerebrospinal fluid (CSF), this fluid being in close contact with the brain tissue involved in the pathology.87 CSFs from 15 TLE patients were compared to those of 15 healthy subjects. The authors identified miR-19b-3p as significantly downregulated in epilepsy versus control, while miR-451a was significantly higher in TLE versus control samples. Receiver operator characteristic (ROC) analysis revealed that miR-19b-3p shows a good performance in the diagnosis of epilepsy (area under the curve [AUC] 0.73) and that the combination of miR-19a-3p and miR-451a has the best AUC value (0.82).87
In the third paper, serum profiles of the miRNA of 30 epileptic patients versus 30 healthy control were analyzed.88 Among 2,578 serum miRNAs analyzed with the Illumina platform, the authors found that 4 miRNAs were significantly upregulated (let-7d-5p, miR-106b-5p, miR-130a-3p, and miR-146a-5p), whereas 6 miRNAs (miR-15a-5p, -144-5p, -181c-5p, -194-5p, -889-3p, and novel-mir-96) were downregulated. The RT-PCR validation phase on 30 epileptic versus 30 healthy subject serum profiles revealed that miR-15a-5p, miR-194-5p, and miR-96 were downregulated, while let-7d-5p, miR-106b-5p, -130a-3p, and -146a-5p were increased in epileptic versus normal subjects. The ROC analysis on 112 patients versus 112 healthy controls revealed that 6 miRNAs could be proposed as diagnostic biomarkers of epilepsy (let-7d-5p, miR-15a-5p, miR-106b-5p, miR-130a-3p, miR-146a-5p, and miR-194-5p) and that serum miR-106b-5p is the best diagnostic circulating miRNA for epilepsy, with 80.3% sensitivity and 81.2% specificity.88 The authors proposed miR-106b-5p as the best diagnostic circulating miRNA for epilepsy diagnosis.
In the fourth paper, plasma miRNAs were analyzed, and decreased expression of miR-134a was proposed as a circulating biomarker of mesial TLE (MTLE) validated on a cohort of 65 MTLE and 83 controls.89
In serum, four epilepsy-associated miRNAs (miR-106b, miR-146b, miR-194-5p, and miR-301a) have been validated by RT-PCR in 90 epileptic subjects against 90 control subjects in the fifth paper.90 The authors found that miR-106b, miR-146b, and miR-301a significantly increased while miR-194-5p decreased in TLE samples. Moreover, the use of miR-106b and miR-146b has been proposed as a two-miRNA signature with diagnostic ability.90
Plasma miRNAs have been isolated from exosomes as well.91 Among the 6 differentially expressed miRNAs in human plasmatic exosomes among epileptic and healthy subjects, miR-3613 emerges as the best diagnostic molecule (AUC = 0.8444).91
In the sixth paper,92 a comparative analysis of circulating miRNAs in epilepsy has been proposed. In recent years, several miRNAs have been proposed as circulating biomarkers, showing dynamic changes of miRNA expression in relation to the history of the pathology. Some miRNAs could be expressed soon after the onset of seizures (i.e., miR-9a-3p), whereas others were expressed only after the chronicization of the disease (i.e., miR-21-5p).92
Considering both animal models and human samples, several inconsistencies emerge among the identified epilepsy-associated circulating miRNAs, and a unique miRNA signature associated with epilepsy is not detectable. In particular, when a comparison between miRNA profiles obtained from biofluids in an animal model and the circulating miRNA profile of human epilepsy is performed, only Hsa-miR-146-P1_5p and Hsa-miR-34-P1 are commonly present in the plasma of epilepsy animal models32, 55 and in human serum samples.88, 90 Circulating Hsa-miR-146-P1_5p has already been associated with human TLE by several papers (i.e., Korotkov), and Hsa-miR-34-P1 could potentially become a new diagnostic circulating biomarker of epilepsy. The limited overlap among human and mouse model circulating miRNA profiles could be due to the differences of the species used for animal models (rat versus mouse) and human pathology, different time points during pathology development in animal models, the different techniques used for epilepsy generation (electric stimulus versus pilocarpine injection versus KA injection), or the different methods used for miRNA analysis (NGS or RT-PCR). With regard to human biofluid samples, it is difficult to identify a main miRNA that plays a key role in the pathology as the analysis is performed on different tissues, such as CSF, serum, and plasma. The analysis of a recent database,93 containing “up-to-date information on all publications related to microRNA and epilepsy,” revealed that some miRNAs were reportedly upregulated in human blood and downregulated in human brain tissues (i.e., Hsa-miR-146-P1_5p) or vice versa. Other miRNAs are only reported to be upregulated in brain tissues and no confirmation highlighted their presence in human blood (i.e., Hsa-miR-34-P1).
To understand which miRNAs have a key role to play in epilepsy development, we considered single circulating miRNA or small-profile miRNA, described in Table 3, with an in silico approach in relation to their role in epileptic hallmarks, with epileptic hallmarks being the main biological pathways affected in epilepsy.94 We consider epilepsy hallmarks all the pathophysiological processes that are altered in epilepsy, giving rise to spontaneous seizures, such as those reported in Brennan and Henshall16 and Shao and Chen:92 neuroinflammation, neurodegeneration, neurogenesis, gene expression, oxidative response, neuronal plasticity, tissue remodeling, circadian rhythm, and drug resistance.16
TLE-Associated Circulating miRNAs Control Cell Proliferation and Apoptosis, Brain Tissue Remodeling, and Neuroinflammation
Some papers have described the role of miRNAs on epileptic hallmarks (i.e., Brennan and Henshall16) but without analyzing in detail the molecular pathway and the genes within those pathways, which are targets of the identified miRNAs by an in silico approach. In other recent reviews (i.e., Henshall), a detailed description of the use of miRNAs as potential therapeutic tools is offered. In our approach, we used bioinformatics tools to identify miRNAs with a major role in the control of functions altered in epilepsy while also considering miRNAs as potential therapeutic molecules. In particular, we focused mainly on the use of those miRNAs that we selected with our approach as potential therapeutic tools.
To identify a small number of miRNAs with a key role in epilepsy development, we evaluated the role of all the 15 circulating miRNAs in Table 3 (let-7d-5p, miR-15a-5p, -19b-3p, -34a, -106b-5p, -130a-3p, -146a-5p, -194a-5p, -197-5p, -451a, -3613-5p, -4322, -4668-5p, -6781-5p, and -8071) through a Pathway Enrichment Analysis, as described previously, involving the identification of a group of pathways significantly enriched with miRNA targets.
Figure 3 shows pathways enriched with miRNA-validated targets for 8 of the 15 miRNAs of Table 3, namely, let-7d-5p, miR-15a-5p, miR-19b-3p, miR-34a, miR-106b-5p, miR-130a-3p, miR-146a-5p, and miR-451. Indeed, no validated miRNA targets were found for miR-194a-5p, -451a, -3613-5p, -4322, -4668-5p, -6781-5p, and -8071.
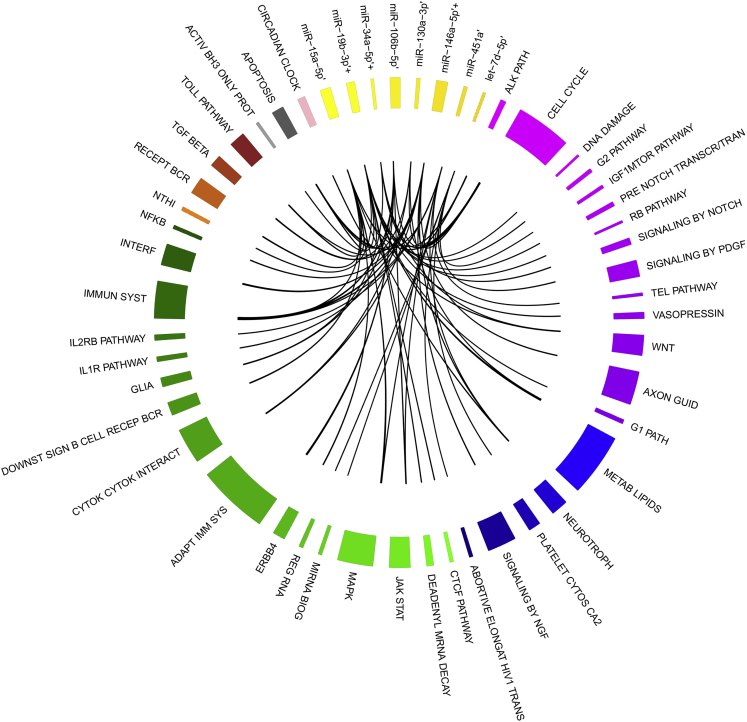
Figure 3.
Circulating miRNAs with a Main Role in Epilepsy Hallmarks
Circos plot showing the relationships among eight circulating miRNAs in epilepsy (yellow gradation, ’) and their target pathways. The pathways are grouped into six main sets: purple gradation, cell cycle control; blue gradation, tissue remodeling ion channel modification and neuronal plasticity; green gradation, transcription regulation and gene expression; brown gradation, inflammation pathway; black gradation, apoptosis; and pink, stemness. The width of each pathway slice is proportional to the number of genes of each pathway, and that of each miRNA slice is proportional to the number of gene targets of those miRNAs of all pathways. The circos plot was constructed using R-package circlize.125 The miRNAs that have been proposed as therapeutic molecules are indicated with a plus sign.
Among the most enriched pathways identified, there were the following: immune system, cell cycle, apoptosis, and neurotrophin-signaling pathway (Figure 3).
Focusing on the 8 human circulating epilepsy-associated miRNAs, we analyzed their possible role in epilepsy-associated cellular functions.16 From our analysis, the known, major physiological processes altered in TLE include the following (Figure 3):
-
(1)
Neuroinflammation with the involvement of microglia, which was called “immune system” in in silico analysis (Figure 3 in brown gradation). This pathway includes several genes regulated by miR-15a-5p (downregulated), miR-106b-5p, miR-146, and miR-451 (see Table S1). As already seen, several pro-inflammatory cytokines are increased during epilepsy or after an epileptic event. miR-15b-5p has been found to be specifically upregulated in the CSF of subjects who were Alzheimer’s patients95 correlated to plaque scores. On the contrary, in another pathology, i.e., that of diabetic retinopathy, miR-15a-5p has been proposed as a key regulator of both pro-inflammatory and proangiogenic pathways via the regulation of acid sphingomyelinase (ASM) and VEGF-A.96 In epilepsy, this miRNA has been found to be downregulated, suggesting a possible loss of control over the enzyme ASM, leading to the conversion of sphingomyelin into pro-inflammatory and pro-apoptotic ceramide.97
miR-106b, found upregulated in epilepsy, was described to be involved in AD-associated inflammation.98 As simvastatin has been proposed as a possible anti-inflammatory and anti-apoptotic drug in AD,98 recently statins also have been been tried as potential anti-seizure drugs, highlighting a possible protective, anti-inflammatory mechanism.99 Also miR-451, involved in the maintenance of the inflammatory status in several brain pathologies (i.e., Churov et al.100), has been proposed as a potential target of several statins.101
Regarding miR-146, we have already described its possible role in neuroinflammation.
Some papers described the upregulation of miR-451 in TLE54 and associated it with inflammation in the brain (i.e., Cogswell et al.102).
-
(2)
Neurogenesis, cell cycle control, and cell proliferation, called “cell cycle” in in silico analysis (Figure 3 in purple gradation). This pathway includes all those genes involved in neuronal cell proliferation and differentiation, which are controlled by miR-15a-5p, miR-34a, miR-106b-5p, and miR-146 (see Table S2).
In epilepsy, the observed downregulation of miR-15a-5p could lead to the reducing of its control activity on its targets, including ubiquitin ligase FBXW7 that destabilizes Cyclin E, leading to the block of the cell cycle in the S phase.103 This could reflect partially the inhibition of neurogenesis, which, with the parallel increase of neuronal apoptosis, leads to the neuronal loss observed in epileptic subjects.104
Regarding a possible role of miR-34a in neurogenesis, it has been recently demonstrated that the miR-34/449 family is a key regulator of mitotic spindle orientation during cortex development.105 Moreover, miR-34 family members are the main upregulated miRNAs in differentiated neurons, and they have a role in the control of cell cycle and the blocking of apoptosis,106 suggesting that the observed downregulation of miR-34a in epilepsy could lead to cell cycle block, apoptosis activation, and neuronal loss.106, 107
Also in the rat model of epilepsy induced by electric stimulation, miR-106b-5p has been reported to be upregulated in the early phase, suggesting a potential role of this miRNA in the induction of neuronal cell cycle block and neuronal apoptosis.31
Regarding miR-146, we have already described its possible role in the control of cell proliferation.
-
(3)
Apoptosis (Figure 3 in black gradation). This pathway includes all genes involved in pro- or anti-apoptotic signaling, which are validated targets of miR-15a-5p (downregulated), miR-106b-5p, miR-146, and miR-451 (see Table S3).
The main role of miR-15a-5p in the brain has been described as an ischemic modulator.108 After ischemic brain injury, miR-15a/16 cluster level is usually increased. It has been observed that the treatment with antagomiR or the genetic loss of this miRNA cluster is able to induce the upregulation of anti-apoptotic proteins (such as Bcl2 and Bcl-w) and suppress pro-inflammatory molecules.108 It is possible that the observed miR-15a-5p downregulation in epilepsy is mainly caused by the effect of miR-15a-5p decrease on the modulation of neuroinflammatory cytokines.
miR-106b-5p has been predicted to regulate caspase 6 (CASP6) and MAPK-binding protein 1 (MAPKBP1) (inflammation and neuronal apoptosis).88 SE induces CASP6 expression and activation in rat hippocampus, leading to neuronal apoptosis in different epilepsy models.109, 110
The upregulation of miR-146 has been found in several epilepsy models,18 and it could play a role also in the regulation of neuronal apoptosis.111, 112, 113
The upregulation of miR-451 decreases proliferation and promotes apoptosis in other pathologies as well.114, 115 In CSF, the upregulation of miR-451a has been associated with several CNS pathologies.116 Regarding the role of miR-451, it is known that this miRNA is able to control the AMPK-mTOR pathway.117 miR-451 increase, observed in epileptic patients, could modulate autophagy and neuronal loss, as observed in brains after cerebral ischemia.118
miRNA-Based Treatment in Epilepsy
miRNA modulation in epileptic animal models could be proposed as a therapeutic tool for modulating the epilepsy phenotype and could be obtained by the administration of synthetic oligonucleotides. These oligonucleotides could encode for the same sequence of the miRNA (miRNA mimic) if it is necessary to increase miRNA sequence as it is downregulated in epilepsy, or the reverse sequence (miRNA inhibitor) if it is necessary to decrease miRNA sequence as it is upregulated in epilepsy. Considering the role of miRNAs in different epilepsy hallmarks, some of the mimics or inhibitor molecules designed on miRNA sequence have been tried as potential therapeutic tools (Figure 3, indicated with a plus sign).
Regarding miRNA mimic or inhibitory synthetic oligonucleotides with potential therapeutic application, several miRNAs considered for the synthetic modulation belong to the class of circulating human TLE miRNAs, as if the modulation of a circulating molecule, used for the communication between cells of the same tissue, could be easier with respect to the modulation of intracellular molecules. In particular, we focused on those 8 of the 15 human circulating epileptic miRNAs that, playing a role in controlling epilepsy hallmark functions, could become the target of new therapies (let-7d-5p, miR-15a-5p, -19b-3p, -34a, -106b-5p, -130a-3p, -146a-5p, -451).
Among these, anti-miR-146a has been administered via intranasal delivery in a pilocarpine-induced TLE mouse model.113 The percentage of animals with seizure onset was reduced to 6.7%, with a parallel increase in seizure latency and severity. This was possibly caused by the observed decrease in inflammatory modulators, such as nuclear factor κB (NF-κB), TNF-α, IL-1b, and IL-6, as well as IRAK1 and TRAF6.113 With the same aim, in 2017, Iori et al.119 utilized the mimic miR-146a injection to block IL-1 receptor/Toll-like receptor (IL-1R1/TLR4) molecular pathway, obtaining a reduction of chronic seizure recurrence and preventing the progression of the disease obtained by KA injection. On the contrary, He et al.120 first demonstrated a significant increase of miR-146a 1 week after the induction of SE in rat by electrical stimulation. They analyzed the effect of antagomiR-146 on both human astrocytoma cell lines and rat epileptic models by injection in the lateral ventricle of the brain. The treatment increased the latency of the first seizures, decreasing the duration and the frequency of the phenomenon.120
The differences in the results described in these two publications could be because of different models of epilepsy used. Iori et al.119 analyzed the modulation of miR-146a in a mouse model obtained by intra-amygdala KA injection. The authors analyzed the effect of mimic miR-146 pretreatment before the onset of epilepsy or in a mouse model of acquired epilepsy. The authors show that the mimic miR-146 pretreatment decreases hippocampal excitability and acute seizure manifestation, also after KA injection. They hypothesized this occurs by reducing TRAF6 and IRAK2, key proteins for IL-1R1/TLR signaling. Next, the authors studied the effect of miR-146 increase on the acquired epileptic mouse model, and they showed a transient reduction in the number and frequency of seizures. He et al.120 analyzed the effect of antagomiR-146 on the hippocampus of an electric rat model of epilepsy. In their paper, the authors demonstrated that Complement factor H (CFH) is a direct target that possibly mediates the observed effect of antagomiR-146a. This target gene is described as a critical repressor of the complement and innate immune system, being a specific inhibitor of the C3-to-C3b transition in the complement pathway. It can thus be speculated that, in the initial phases of the disease, the acute phase response signaling is activated (the activation of IL-1R1/TLR signaling), causing the release of numerous proteins in the blood that serve as indicators of an inflammatory state. These indicators activate the complement pathway (second phase) that is required to resolve the cause of inflammation. This two-step process leads to the acquired epilepsy manifestation. We could speculate that in the initial phase, mimic miR-146 is able to silence key transducers of the acute phase response, causing a transient reduction of clinical manifestation of epilepsy, while in the second phase, when the complement system is activated, antagomir-146 is effective in activating a repressor of the complement system, thus reducing the neuroinflammation associated with the pathology. Recently, a model of activity has been proposed in which the upregulation of miR-146a causes an increase in IL-1b and a parallel decrease of CFH. The increase in IL-1b generates a feedback loop that upregulates miR-146a, which generates the chronicization of the inflammatory state observed in epileptic subjects.121
In addition, miR-34a has been proposed as a potential therapeutic target because the administration via the injection of miR-34a antagomiR, reducing its hippocampal levels, has a small modulatory effect on apoptosis, with the exception of severe or moderate SE.107 Nanoparticles containing miR-34a plasmid have been successfully developed for the modulation of miR-34 within breast cancer treatment.122 The use of antagomiR of miR-34a has been proposed as a therapeutic approach in a pilocarpine-induced epilepsy rat model, reducing the seizure-induced neuronal cell death at day 7 post-SE.18 The use of antagomiR-34a in combination with nanoparticles could be proposed as an opportunity for treating epilepsy.
Another research proposed to use mimic miR-19b for the modulation of this miRNA.19 In particular, the authors incorporated derived microparticles into endothelial cells to modulate the atherosclerosis phenotype in a mouse model. The intravenous injection of the microparticles promoted inflammatory cytokine secretion and macrophage infiltration in perivascular adipose tissues.19 As miR-19b-3p is downregulated in epileptic subjects, these miR-19b-filled microparticles could be proposed for the treatment of epileptic animal models to decrease the inflammation of epileptic subjects, thus translating this miRNA-based therapy to epileptic patients.
No experiment has been proposed to analyze the in vivo effect of antagomiR-451, let-7d inhibitor, antagomiR-106b, antagomiR-130a, or mimic miR-15a-5p in SE models. These miRNAs could be proposed as possible target molecules for thedevelopment of new oligonucleotide-based therapeutic options. No publication is currently available on the role of miR-197-5p, -3613-5p, -4322, -4668-5p, -6781-5p, and -8071 in epilepsy.
Among other antagomiRs analyzed for therapeutic application in epilepsy, antagomiR-134 is one of the most promising miRNAs; a single intracerebroventricular injection of locked nucleic acid (LNA)-antagomiR-134 reduced kainate-induced seizures by 50% and reduced hippocampal cell damage, suggesting the use of this approach in epilepsy.17 A similar result was obtained by the use of antagomiR-132 in pilocarpine-induced epileptic rats. This treatment reduced SE-associated degeneration of neurons and spontaneous seizures.123
Further, miR-184 has been modulated in in vitro experiments, and its overexpression significantly decreased secretion of the pro-inflammatory cytokines IL-6 and IL-1b without affecting mouse neuronal viability.124
Until now, no clinical trial has been proposed using miRNA-based treatment as a possible therapeutic option for epilepsy, but some interest is emerging on the role of these molecules during the onset, development, and healing of the disease (ClinicalTrials.gov: NCT02359188).
Conclusions
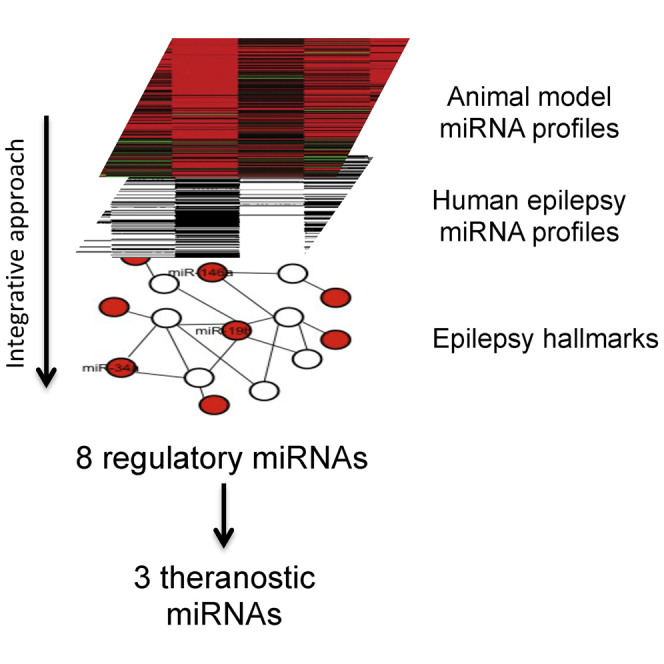
In this review, we analyzed miRNAs related to epilepsy with diagnostic properties. In particular, we considered all the TLE animal models, and we identified a group of 20 miRNAs (upregulated: miR-23a, -27a, -31, -99a, -125b-5p, -129-5p, -132-3p, -142-p, -146a, -212-3p, and -375; downregulated: miR-10b, -21, -29a, -138, -153, -181c, -187-3p, -330-3p, and -551b-3p) that are commonly altered in at least two different focal TLE animal models. An emerging problem is related to the translation of some of these miRNAs to the human pathology, because there is a poor overlap in the miRNA profiles of human tissues and animal model tissues. Indeed, we found four miRNAs that are common between animal model and human tissue profiles and could be proposed as diagnostic tissue-associated miRNAs (Hsa-miR-132-P1_5p, -138-P1_5p, -142-P1-v1_5p, and -146-P1_5p).
Looking for a small group of TLE-associated circulating miRNAs, we analyzed circulating miRNA profiles of different human body fluids, identifying a group of 15 circulating miRNAs (let-7d-5p, miR-15a-5p, -19b-3p, -34a, -106b-5p, -130a-3p, -146a-5p, -194a-5p, -197-5p, -451a, -3613-5p, -4322, -4668-5p, -6781-5p, and -8071) with potential diagnostic properties in humans. Of these, only miR-146-P1_5p and miR-34-P1 have been found in biofluids of animal models of TLE, and 8 of the 15 human circulating miRNAs seem to control neuroinflammation, cell proliferation, apoptosis, and brain tissue remodeling, all histopathological features affected in TLE. Until now, miR-194a-5p, -197-5p, -451a, -3613-5p, -4322, -4668-5p, -6781-5p, and -8071 have an unknown role in epilepsy development.
For the development of new therapeutic molecules, theranostic research has selected some miRNAs belonging to the group of TLE-altered tissue miRNAs (i.e., miR-132), although it seems easier for the successful modulation of a circulating molecule (i.e., miR-34 family members or miR-146) with respect to one intracellular miRNA. From our analysis, other miRNAs, especially those circulating (i.e., miR-19b-3p, -34a, and -146a-5p), could emerge as innovative therapeutic molecules for the development of theranostic miRNA-based approaches.
Author Contributions
C.C. and I.M. made substantial contributions to the conception and design of the work and to data analysis. A.G., G.B., and I.C. made substantial contributions to guiding and revising the work critically for important intellectual content, and they were involved in drafting the manuscript, answering the reviewers’ comments, and giving final approval of the version to be published. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
The authors would like to thank the following for financial support: INTEROMICS flagship project (Interomics PB05), National Research Council CUP Grant B91JI2000190001, and the project grant SysBioNet, Italian Roadmap Research Infrastructures 2012.
Footnotes
Supplemental Information includes three tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.008.
Contributor Information
Antonio Gambardella, Email: a.gambardella@unicz.it.
Gloria Bertoli, Email: gloria.bertoli@ibfm.cnr.it.
Supplemental Information
References
- 1.Megiddo I., Colson A., Chisholm D., Dua T., Nandi A., Laxminarayan R. Health and economic benefits of public financing of epilepsy treatment in India: An agent-based simulation model. Epilepsia. 2016;57:464–474. doi: 10.1111/epi.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browne T., Holmes G.L. Fourth Edition. Lippincott Williams & Wilkins; Philadelphia, PA: 2008. Handbook of Epilepsy. [Google Scholar]
- 3.Fisher R.S., Acevedo C., Arzimanoglou A., Bogacz A., Cross J.H., Elger C.E., Engel J., Jr., Forsgren L., French J.A., Glynn M. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. 2014;55:475–482. doi: 10.1111/epi.12550. [DOI] [PubMed] [Google Scholar]
- 4.Fisher R.S., Cross J.H., French J.A., Higurashi N., Hirsch E., Jansen F.E., Lagae L., Moshé S.L., Peltola J., Roulet Perez E. Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:522–530. doi: 10.1111/epi.13670. [DOI] [PubMed] [Google Scholar]
- 5.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher R.S., Cross J.H., D’Souza C., French J.A., Haut S.R., Higurashi N., Hirsch E., Jansen F.E., Lagae L., Moshé S.L. Instruction manual for the ILAE 2017 operational classification of seizure types. Epilepsia. 2017;58:531–542. doi: 10.1111/epi.13671. [DOI] [PubMed] [Google Scholar]
- 7.Speed D., O’Brien T.J., Palotie A., Shkura K., Marson A.G., Balding D.J., Johnson M.R. Describing the genetic architecture of epilepsy through heritability analysis. Brain. 2014;137:2680–2689. doi: 10.1093/brain/awu206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Striano P., Vari M.S., Mazzocchetti C., Verrotti A., Zara F. Management of genetic epilepsies: From empirical treatment to precision medicine. Pharmacol. Res. 2016;107:426–429. doi: 10.1016/j.phrs.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Meldrum B.S., Rogawski M.A. Molecular targets for antiepileptic drug development. Neurotherapeutics. 2007;4:18–61. doi: 10.1016/j.nurt.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staley K. Molecular mechanisms of epilepsy. Nat. Neurosci. 2015;18:367–372. doi: 10.1038/nn.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henshall D.C., Hamer H.M., Pasterkamp R.J., Goldstein D.B., Kjems J., Prehn J.H.M., Schorge S., Lamottke K., Rosenow F. MicroRNAs in epilepsy: pathophysiology and clinical utility. Lancet Neurol. 2016;15:1368–1376. doi: 10.1016/S1474-4422(16)30246-0. [DOI] [PubMed] [Google Scholar]
- 12.Lerche H., Shah M., Beck H., Noebels J., Johnston D., Vincent A. Ion channels in genetic and acquired forms of epilepsy. J. Physiol. 2013;591:753–764. doi: 10.1113/jphysiol.2012.240606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-Palma E., Helbig I., Klein K.M., Anttila V., Horn H., Reinthaler E.M., Gormley P., Ganna A., Byrnes A., Pernhorst K. Heterogeneous contribution of microdeletions in the development of common generalised and focal epilepsies. J. Med. Genet. 2017;54:598–606. doi: 10.1136/jmedgenet-2016-104495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T., Giri M., Xia Z., Subedi Y.N., Li Y. Genetic and epigenetic mechanisms of epilepsy: a review. Neuropsychiatr. Dis. Treat. 2017;13:1841–1859. doi: 10.2147/NDT.S142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Bartel D.P. Widespread Influence of 3′-End Structures on Mammalian mRNA Processing and Stability. Cell. 2017;169:905–917.e11. doi: 10.1016/j.cell.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan G.P., Henshall D.C. microRNAs in the pathophysiology of epilepsy. Neurosci. Lett. 2018;667:47–52. doi: 10.1016/j.neulet.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez-Mateos E.M., Engel T., Merino-Serrais P., McKiernan R.C., Tanaka K., Mouri G., Sano T., O’Tuathaigh C., Waddington J.L., Prenter S. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu K., Xie Y.Y., Zhang C., Ouyang D.S., Long H.Y., Sun D.N., Long L.L., Feng L., Li Y., Xiao B. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012;13:115. doi: 10.1186/1471-2202-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Li C., Li S., Zhang F., Wu M., Liang H., Song J., Lee C., Chen H. Endothelial microparticles-mediated transfer of microRNA-19b promotes atherosclerosis via activating perivascular adipose tissue inflammation in apoE−/− mice. Biochem. Biophys. Res. Commun. 2018;495:1922–1929. doi: 10.1016/j.bbrc.2017.11.195. [DOI] [PubMed] [Google Scholar]
- 20.Reschke C.R., Henshall D.C. microRNA and Epilepsy. Adv. Exp. Med. Biol. 2015;888:41–70. doi: 10.1007/978-3-319-22671-2_4. [DOI] [PubMed] [Google Scholar]
- 21.Saunders M.A., Liang H., Li W.H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan R., Pak C., Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 23.Manna I., Labate A., Mumoli L., Pantusa M., Ferlazzo E., Aguglia U., Quattrone A., Gambardella A. Relationship between genetic variant in pre-microRNA-146a and genetic predisposition to temporal lobe epilepsy: a case-control study. Gene. 2013;516:181–183. doi: 10.1016/j.gene.2012.09.137. [DOI] [PubMed] [Google Scholar]
- 24.Markopoulos G.S., Roupakia E., Tokamani M., Chavdoula E., Hatziapostolou M., Polytarchou C., Marcu K.B., Papavassiliou A.G., Sandaltzopoulos R., Kolettas E. A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol. (Dordr.) 2017;40:303–339. doi: 10.1007/s13402-017-0341-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Yang Z., Le W. Tiny But Mighty: Promising Roles of MicroRNAs in the Diagnosis and Treatment of Parkinson’s Disease. Neurosci. Bull. 2017;33:543–551. doi: 10.1007/s12264-017-0160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dehghani R., Rahmani F., Rezaei N. MicroRNA in Alzheimer’s disease revisited: implications for major neuropathological mechanisms. Rev. Neurosci. 2018;29:161–182. doi: 10.1515/revneuro-2017-0042. [DOI] [PubMed] [Google Scholar]
- 27.Quinlan S., Kenny A., Medina M., Engel T., Jimenez-Mateos E.M. MicroRNAs in Neurodegenerative Diseases. Int. Rev. Cell Mol. Biol. 2017;334:309–343. doi: 10.1016/bs.ircmb.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Henshall D.C. MicroRNAs in the pathophysiology and treatment of status epilepticus. Front. Mol. Neurosci. 2013;6:37. doi: 10.3389/fnmol.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y.J., Tian X.B., Zhang S., Zhang Y.X., Li X., Li D., Cheng Y., Zhang J.N., Kang C.S., Zhao W. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res. 2011;1387:134–140. doi: 10.1016/j.brainres.2011.02.073. [DOI] [PubMed] [Google Scholar]
- 30.Kan A.A., van Erp S., Derijck A.A., de Wit M., Hessel E.V., O’Duibhir E., de Jager W., Van Rijen P.C., Gosselaar P.H., de Graan P.N., Pasterkamp R.J. Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell. Mol. Life Sci. 2012;69:3127–3145. doi: 10.1007/s00018-012-0992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bot A.M., Dębski K.J., Lukasiuk K. Alterations in miRNA levels in the dentate gyrus in epileptic rats. PLoS ONE. 2013;8:e76051. doi: 10.1371/journal.pone.0076051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorter J.A., Iyer A., White I., Colzi A., van Vliet E.A., Sisodiya S., Aronica E. Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol. Dis. 2014;62:508–520. doi: 10.1016/j.nbd.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Aronica E., Fluiter K., Iyer A., Zurolo E., Vreijling J., van Vliet E.A., Baayen J.C., Gorter J.A. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 34.Jackson D.B. Serum-based microRNAs: are we blinded by potential? Proc. Natl. Acad. Sci. USA. 2009;106:E5. doi: 10.1073/pnas.0809999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiko T., Nakagawa K., Tsuduki T., Furukawa K., Arai H., Miyazawa T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J. Alzheimers Dis. 2014;39:253–259. doi: 10.3233/JAD-130932. [DOI] [PubMed] [Google Scholar]
- 36.Gandhi R., Healy B., Gholipour T., Egorova S., Musallam A., Hussain M.S., Nejad P., Patel B., Hei H., Khoury S. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann. Neurol. 2013;73:729–740. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 37.Ma Y. The Challenge of microRNA as a Biomarker of Epilepsy. Curr. Neuropharmacol. 2018;16:37–42. doi: 10.2174/1570159X15666170703102410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 39.Li M., Marin-Muller C., Bharadwaj U., Chow K.H., Yao Q., Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J. Surg. 2009;33:667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosik K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer A., O’Carroll D., Tan C.L., Hillman D., Sugimori M., Llinas R., Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L., Liu L., Shi J., Tan M., Xiong J., Li X., Hu Q., Yi Z., Mao D. MicroRNA-34b mediates hippocampal astrocyte apoptosis in a rat model of recurrent seizures. BMC Neurosci. 2016;17:56. doi: 10.1186/s12868-016-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Löscher W. Critical review of current animal models of seizures and epilepsy used in the discovery and development of new antiepileptic drugs. Seizure. 2011;20:359–368. doi: 10.1016/j.seizure.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Gu B., Dalton K.A. Models and detection of spontaneous recurrent seizures in laboratory rodents. Zool. Res. 2017;38:171–179. doi: 10.24272/j.issn.2095-8137.2017.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thom M. Review: Hippocampal sclerosis in epilepsy: a neuropathology review. Neuropathol. Appl. Neurobiol. 2014;40:520–543. doi: 10.1111/nan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Vliet E.A., Puhakka N., Mills J.D., Srivastava P.K., Johnson M.R., Roncon P., Das Gupta S., Karttunen J., Simonato M., Lukasiuk K. Standardization procedure for plasma biomarker analysis in rat models of epileptogenesis: Focus on circulating microRNAs. Epilepsia. 2017;58:2013–2024. doi: 10.1111/epi.13915. [DOI] [PubMed] [Google Scholar]
- 47.Hu K., Zhang C., Long L., Long X., Feng L., Li Y., Xiao B. Expression profile of microRNAs in rat hippocampus following lithium-pilocarpine-induced status epilepticus. Neurosci. Lett. 2011;488:252–257. doi: 10.1016/j.neulet.2010.11.040. [DOI] [PubMed] [Google Scholar]
- 48.Li M.M., Jiang T., Sun Z., Zhang Q., Tan C.C., Yu J.T., Tan L. Genome-wide microRNA expression profiles in hippocampus of rats with chronic temporal lobe epilepsy. Sci. Rep. 2014;4:4734. doi: 10.1038/srep04734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gross C., Yao X., Engel T., Tiwari D., Xing L., Rowley S., Danielson S.W., Thomas K.T., Jimenez-Mateos E.M., Schroeder L.M. MicroRNA-Mediated Downregulation of the Potassium Channel Kv4.2 Contributes to Seizure Onset. Cell Rep. 2016;17:37–45. doi: 10.1016/j.celrep.2016.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z., Yu J.T., Jiang T., Li M.M., Tan L., Zhang Q., Tan L. Genome-wide microRNA profiling of rat hippocampus after status epilepticus induced by amygdala stimulation identifies modulators of neuronal apoptosis. PLoS ONE. 2013;8:e78375. doi: 10.1371/journal.pone.0078375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kretschmann A., Danis B., Andonovic L., Abnaof K., van Rikxoort M., Siegel F., Mazzuferi M., Godard P., Hanon E., Fröhlich H. Different microRNA profiles in chronic epilepsy versus acute seizure mouse models. J. Mol. Neurosci. 2015;55:466–479. doi: 10.1007/s12031-014-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schouten M., Bielefeld P., Fratantoni S.A., Hubens C.J., Piersma S.R., Pham T.V., Voskuyl R.A., Lucassen P.J., Jimenez C.R., Fitzsimons C.P. Multi-omics profile of the mouse dentate gyrus after kainic acid-induced status epilepticus. Sci. Data. 2016;3:160068. doi: 10.1038/sdata.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schouten M., Fratantoni S.A., Hubens C.J., Piersma S.R., Pham T.V., Bielefeld P., Voskuyl R.A., Lucassen P.J., Jimenez C.R., Fitzsimons C.P. MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 2015;5:12448. doi: 10.1038/srep12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKiernan R.C., Jimenez-Mateos E.M., Sano T., Bray I., Stallings R.L., Simon R.P., Henshall D.C. Expression profiling the microRNA response to epileptic preconditioning identifies miR-184 as a modulator of seizure-induced neuronal death. Exp. Neurol. 2012;237:346–354. doi: 10.1016/j.expneurol.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pichardo-Casas I., Goff L.A., Swerdel M.R., Athie A., Davila J., Ramos-Brossier M., Lapid-Volosin M., Friedman W.J., Hart R.P., Vaca L. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Res. 2012;1436:20–33. doi: 10.1016/j.brainres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jimenez-Mateos E.M., Bray I., Sanz-Rodriguez A., Engel T., McKiernan R.C., Mouri G., Tanaka K., Sano T., Saugstad J.A., Simon R.P. miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 2011;179:2519–2532. doi: 10.1016/j.ajpath.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu D.Z., Tian Y., Ander B.P., Xu H., Stamova B.S., Zhan X., Turner R.J., Jickling G., Sharp F.R. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bencurova P., Baloun J., Musilova K., Radova L., Tichy B., Pail M., Zeman M., Brichtova E., Hermanova M., Pospisilova S. MicroRNA and mesial temporal lobe epilepsy with hippocampal sclerosis: Whole miRNome profiling of human hippocampus. Epilepsia. 2017;58:1782–1793. doi: 10.1111/epi.13870. [DOI] [PubMed] [Google Scholar]
- 59.Fromm B., Billipp T., Peck L.E., Johansen M., Tarver J.E., King B.L., Newcomb J.M., Sempere L.F., Flatmark K., Hovig E., Peterson K.J. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korotkov A., Mills J.D., Gorter J.A., van Vliet E.A., Aronica E. Systematic review and meta-analysis of differentially expressed miRNAs in experimental and human temporal lobe epilepsy. Sci. Rep. 2017;7:11592. doi: 10.1038/s41598-017-11510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishimura D. BioCarta. Biotech Softw. Internet Rep. 2004;2:117–120. [Google Scholar]
- 63.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 65.Hsu J.B., Chiu C.M., Hsu S.D., Huang W.Y., Chien C.H., Lee T.Y., Huang H.D. miRTar: an integrated system for identifying miRNA-target interactions in human. BMC Bioinformatics. 2011;12:300. doi: 10.1186/1471-2105-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dweep H., Sticht C., Pandey P., Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 67.Chou C.H., Chang N.W., Shrestha S., Hsu S.D., Lin Y.L., Lee W.H., Yang C.D., Hong H.C., Wei T.Y., Tu S.J. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cava C., Colaprico A., Bertoli G., Graudenzi A., Silva T.C., Olsen C., Noushmehr H., Bontempi G., Mauri G., Castiglioni I. SpidermiR: An R/Bioconductor Package for Integrative Analysis with miRNA Data. Int. J. Mol. Sci. 2017;18:E274. doi: 10.3390/ijms18020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cava C., Colaprico A., Bertoli G., Bontempi G., Mauri G., Castiglioni I. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinformatics. 2016;17(Suppl 12):348. doi: 10.1186/s12859-016-1196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Colaprico A., Cava C., Bertoli G., Bontempi G., Castiglioni I. Integrative Analysis with Monte Carlo Cross-Validation Reveals miRNAs Regulating Pathways Cross-Talk in Aggressive Breast Cancer. BioMed Res. Int. 2015;2015:831314. doi: 10.1155/2015/831314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z., Wang H., Cai H., Hong Y., Li Y., Su D., Fan Z. Long non-coding RNA MIAT promotes growth and metastasis of colorectal cancer cells through regulation of miR-132/Derlin-1 pathway. Cancer Cell Int. 2018;18:59. doi: 10.1186/s12935-017-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji Y.F., Wang D., Liu Y.R., Ma X.R., Lu H., Zhang B.A. MicroRNA-132 attenuates LPS-induced inflammatory injury by targeting TRAF6 in neuronal cell line HT-22. J. Cell. Biochem. 2018;119:5528–5537. doi: 10.1002/jcb.26720. [DOI] [PubMed] [Google Scholar]
- 73.Williams A.E., Perry M.M., Moschos S.A., Larner-Svensson H.M., Lindsay M.A. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem. Soc. Trans. 2008;36:1211–1215. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- 74.Hurst D.R., Edmonds M.D., Scott G.K., Benz C.C., Vaidya K.S., Welch D.R. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seo H.H., Lee S.Y., Lee C.Y., Kim R., Kim P., Oh S., Lee H., Lee M.Y., Kim J., Kim L.K. Exogenous miRNA-146a Enhances the Therapeutic Efficacy of Human Mesenchymal Stem Cells by Increasing Vascular Endothelial Growth Factor Secretion in the Ischemia/Reperfusion-Injured Heart. J. Vasc. Res. 2017;54:100–108. doi: 10.1159/000461596. [DOI] [PubMed] [Google Scholar]
- 76.Klein P., Dingledine R., Aronica E., Bernard C., Blümcke I., Boison D., Brodie M.J., Brooks-Kayal A.R., Engel J., Jr., Forcelli P.A. Commonalities in epileptogenic processes from different acute brain insults: Do they translate? Epilepsia. 2018;59:37–66. doi: 10.1111/epi.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balosso S., Ravizza T., Aronica E., Vezzani A. The dual role of TNF-α and its receptors in seizures. Exp. Neurol. 2013;247:267–271. doi: 10.1016/j.expneurol.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roseti C., van Vliet E.A., Cifelli P., Ruffolo G., Baayen J.C., Di Castro M.A., Bertollini C., Limatola C., Aronica E., Vezzani A., Palma E. GABAA currents are decreased by IL-1β in epileptogenic tissue of patients with temporal lobe epilepsy: implications for ictogenesis. Neurobiol. Dis. 2015;82:311–320. doi: 10.1016/j.nbd.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 80.Maroso M., Balosso S., Ravizza T., Iori V., Wright C.I., French J., Vezzani A. Interleukin-1β biosynthesis inhibition reduces acute seizures and drug resistant chronic epileptic activity in mice. Neurotherapeutics. 2011;8:304–315. doi: 10.1007/s13311-011-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tian F., Yuan C., Yue H. MiR-138/SIRT1 axis is implicated in impaired learning and memory abilities of cerebral ischemia/reperfusion injured rats. Exp. Cell Res. 2018;367:232–240. doi: 10.1016/j.yexcr.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 82.Zheng Z., Zhao B. Astragalus polysaccharide protects hypoxia-induced injury by up-regulation of miR-138 in rat neural stem cells. Biomed. Pharmacother. 2018;102:295–301. doi: 10.1016/j.biopha.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y., Du X., Li W., Sang H., Qian A., Sun L., Li X., Li C. Resveratrol Improves Endothelial Progenitor Cell Function through miR-138 by Targeting Focal Adhesion Kinase (FAK) and Promotes Thrombus Resolution In Vivo. Med. Sci. Monit. 2018;24:951–960. doi: 10.12659/MSM.906116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xia L., Li D., Lin C., Ou S., Li X., Pan S. Comparative study of joint bioinformatics analysis of underlying potential of ‘neurimmiR’, miR-212-3P/miR-132-3P, being involved in epilepsy and its emerging role in human cancer. Oncotarget. 2017;8:40668–40682. doi: 10.18632/oncotarget.16541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Spronsen M., van Battum E.Y., Kuijpers M., Vangoor V.R., Rietman M.L., Pothof J., Gumy L.F., van Ijcken W.F., Akhmanova A., Pasterkamp R.J., Hoogenraad C.C. Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS ONE. 2013;8:e74907. doi: 10.1371/journal.pone.0074907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu X., Dong Y., Chen S., Zhang G., Zhang M., Gong Y., Li X. Circulating MicroRNA-146a and MicroRNA-21 Predict Left Ventricular Remodeling after ST-Elevation Myocardial Infarction. Cardiology. 2015;132:233–241. doi: 10.1159/000437090. [DOI] [PubMed] [Google Scholar]
- 87.Raoof R., Jimenez-Mateos E.M., Bauer S., Tackenberg B., Rosenow F., Lang J., Onugoren M.D., Hamer H., Huchtemann T., Körtvélyessy P. Cerebrospinal fluid microRNAs are potential biomarkers of temporal lobe epilepsy and status epilepticus. Sci. Rep. 2017;7:3328. doi: 10.1038/s41598-017-02969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J., Yu J.T., Tan L., Tian Y., Ma J., Tan C.C., Wang H.F., Liu Y., Tan M.S., Jiang T., Tan L. Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci. Rep. 2015;5:9522. doi: 10.1038/srep09522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Avansini S.H., de Sousa Lima B.P., Secolin R., Santos M.L., Coan A.C., Vieira A.S., Torres F.R., Carvalho B.S., Alvim M.K., Morita M.E. MicroRNA hsa-miR-134 is a circulating biomarker for mesial temporal lobe epilepsy. PLoS ONE. 2017;12:e0173060. doi: 10.1371/journal.pone.0173060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.An N., Zhao W., Liu Y., Yang X., Chen P. Elevated serum miR-106b and miR-146a in patients with focal and generalized epilepsy. Epilepsy Res. 2016;127:311–316. doi: 10.1016/j.eplepsyres.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 91.Yan S., Zhang H., Xie W., Meng F., Zhang K., Jiang Y., Zhang X., Zhang J. Altered microRNA profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget. 2017;8:4136–4146. doi: 10.18632/oncotarget.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shao Y., Chen Y. Pathophysiology and Clinical Utility of Non-coding RNAs in Epilepsy. Front. Mol. Neurosci. 2017;10:249. doi: 10.3389/fnmol.2017.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mooney C., Becker B.A., Raoof R., Henshall D.C. EpimiRBase: a comprehensive database of microRNA-epilepsy associations. Bioinformatics. 2016;32:1436–1438. doi: 10.1093/bioinformatics/btw008. [DOI] [PubMed] [Google Scholar]
- 94.Jimenez-Mateos E.M., Henshall D.C. Epilepsy and microRNA. Neuroscience. 2013;238:218–229. doi: 10.1016/j.neuroscience.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 95.Sørensen S.S., Nygaard A.B., Christensen T. miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia - an exploratory study. Transl. Neurodegener. 2016;5:6. doi: 10.1186/s40035-016-0053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Q., Navitskaya S., Chakravarthy H., Huang C., Kady N., Lydic T.A., Chen Y.E., Yin K.J., Powell F.L., Martin P.M. Dual Anti-Inflammatory and Anti-Angiogenic Action of miR-15a in Diabetic Retinopathy. EBioMedicine. 2016;11:138–150. doi: 10.1016/j.ebiom.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Balosso S., Maroso M., Sanchez-Alavez M., Ravizza T., Frasca A., Bartfai T., Vezzani A. A novel non-transcriptional pathway mediates the proconvulsive effects of interleukin-1beta. Brain. 2008;131:3256–3265. doi: 10.1093/brain/awn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang W., Li Z., Zhao L., Zhao W. Simvastatin ameliorate memory deficits and inflammation in clinical and mouse model of Alzheimer’s disease via modulating the expression of miR-106b. Biomed. Pharmacother. 2017;92:46–57. doi: 10.1016/j.biopha.2017.05.060. [DOI] [PubMed] [Google Scholar]
- 99.Scicchitano F., Constanti A., Citraro R., De Sarro G., Russo E. Statins and epilepsy: preclinical studies, clinical trials and statin-anticonvulsant drug interactions. Curr. Drug Targets. 2015;16:747–756. doi: 10.2174/1389450116666150330114850. [DOI] [PubMed] [Google Scholar]
- 100.Churov A.V., Oleinik E.K., Knip M. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun. Rev. 2015;14:1029–1037. doi: 10.1016/j.autrev.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 101.Li J., Chen H., Ren J., Song J., Zhang F., Zhang J., Lee C., Li S., Geng Q., Cao C., Xu N. Effects of statin on circulating microRNAome and predicted function regulatory network in patients with unstable angina. BMC Med. Genomics. 2015;8:12. doi: 10.1186/s12920-015-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cogswell J.P., Ward J., Taylor I.A., Waters M., Shi Y., Cannon B., Kelnar K., Kemppainen J., Brown D., Chen C. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 103.Teplyuk N.M., Uhlmann E.J., Wong A.H., Karmali P., Basu M., Gabriely G., Jain A., Wang Y., Chiocca E.A., Stephens R. MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget. 2015;6:3770–3783. doi: 10.18632/oncotarget.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kapur J. Role of Neuronal Loss in the Pathogenesis of Recurrent Spontaneous Seizures. Epilepsy Curr. 2003;3:166–167. doi: 10.1046/j.1535-7597.2003.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fededa J.P., Esk C., Mierzwa B., Stanyte R., Yuan S., Zheng H., Ebnet K., Yan W., Knoblich J.A., Gerlich D.W. MicroRNA-34/449 controls mitotic spindle orientation during mammalian cortex development. EMBO J. 2016;35:2386–2398. doi: 10.15252/embj.201694056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen F., Hu S.J. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J. Biochem. Mol. Toxicol. 2012;26:79–86. doi: 10.1002/jbt.20412. [DOI] [PubMed] [Google Scholar]
- 107.Sano T., Reynolds J.P., Jimenez-Mateos E.M., Matsushima S., Taki W., Henshall D.C. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 2012;3:e287. doi: 10.1038/cddis.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang X., Tang X., Sun P., Shi Y., Liu K., Hassan S.H., Stetler R.A., Chen J., Yin K.J. MicroRNA-15a/16-1 Antagomir Ameliorates Ischemic Brain Injury in Experimental Stroke. Stroke. 2017;48:1941–1947. doi: 10.1161/STROKEAHA.117.017284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Narkilahti S., Pitkänen A. Caspase 6 expression in the rat hippocampus during epileptogenesis and epilepsy. Neuroscience. 2005;131:887–897. doi: 10.1016/j.neuroscience.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 110.Friedman L.K., Mancuso J., Patel A., Kudur V., Leheste J.R., Iacobas S., Botta J., Iacobas D.A., Spray D.C. Transcriptome profiling of hippocampal CA1 after early-life seizure-induced preconditioning may elucidate new genetic therapies for epilepsy. Eur. J. Neurosci. 2013;38:2139–2152. doi: 10.1111/ejn.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu R., Liu C., Chen D., Yang W.H., Liu X., Liu C.G., Dugas C.M., Tang F., Zheng P., Liu Y., Wang L. FOXP3 Controls an miR-146/NF-κB Negative Feedback Loop That Inhibits Apoptosis in Breast Cancer Cells. Cancer Res. 2015;75:1703–1713. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Park H., Huang X., Lu C., Cairo M.S., Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J. Biol. Chem. 2015;290:2831–2841. doi: 10.1074/jbc.M114.591420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tao H., Zhao J., Liu T., Cai Y., Zhou X., Xing H., Wang Y., Yin M., Zhong W., Liu Z. Intranasal Delivery of miR-146a Mimics Delayed Seizure Onset in the Lithium-Pilocarpine Mouse Model. Mediators Inflamm. 2017;2017:6512620. doi: 10.1155/2017/6512620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu W., Liu S.Y., He Y.B., Huang R.L., Deng S.Y., Ni G.X., Yu B. MiR-451 suppresses proliferation, migration and promotes apoptosis of the human osteosarcoma by targeting macrophage migration inhibitory factor. Biomed. Pharmacother. 2017;87:621–627. doi: 10.1016/j.biopha.2016.12.121. [DOI] [PubMed] [Google Scholar]
- 115.Peng Z., Zhang Y. Propofol inhibits proliferation and accelerates apoptosis of human gastric cancer cells by regulation of microRNA-451 and MMP-2 expression. Genet. Mol. Res. 2016;15:gmr7078. doi: 10.4238/gmr.15027078. [DOI] [PubMed] [Google Scholar]
- 116.Drusco A., Bottoni A., Laganà A., Acunzo M., Fassan M., Cascione L., Antenucci A., Kumchala P., Vicentini C., Gardiman M.P. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget. 2015;6:20829–20839. doi: 10.18632/oncotarget.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim Y., Powathil G., Kang H., Trucu D., Kim H., Lawler S., Chaplain M. Strategies of eradicating glioma cells: a multi-scale mathematical model with MiR-451-AMPK-mTOR control. PLoS ONE. 2015;10:e0114370. doi: 10.1371/journal.pone.0114370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xue T.F., Ding X., Ji J., Yan H., Huang J.Y., Guo X.D., Yang J., Sun X.L. PD149163 induces hypothermia to protect against brain injury in acute cerebral ischemic rats. J. Pharmacol. Sci. 2017;135:105–113. doi: 10.1016/j.jphs.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 119.Iori V., Iyer A.M., Ravizza T., Beltrame L., Paracchini L., Marchini S., Cerovic M., Hill C., Ferrari M., Zucchetti M. Blockade of the IL-1R1/TLR4 pathway mediates disease-modification therapeutic effects in a model of acquired epilepsy. Neurobiol. Dis. 2017;99:12–23. doi: 10.1016/j.nbd.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 120.He F., Liu B., Meng Q., Sun Y., Wang W., Wang C. Modulation of miR-146a/complement factor H-mediated inflammatory responses in a rat model of temporal lobe epilepsy. Biosci. Rep. 2016;36:e00433. doi: 10.1042/BSR20160290. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Li T.R., Jia Y.J., Ma C., Qiu W.Y., Wang Q., Shao X.Q., Lv R.J. The role of the microRNA-146a/complement factor H/interleukin-1β-mediated inflammatory loop circuit in the perpetuate inflammation of chronic temporal lobe epilepsy. Dis. Model. Mech. 2018;11:dmm031708. doi: 10.1242/dmm.031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin X., Chen W., Wei F., Zhou B.P., Hung M.C., Xie X. Nanoparticle Delivery of miR-34a Eradicates Long-term-cultured Breast Cancer Stem Cells via Targeting C22ORF28 Directly. Theranostics. 2017;7:4805–4824. doi: 10.7150/thno.20771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang Y., Guo J., Wang Q., Chen Y. MicroRNA-132 silencing decreases the spontaneous recurrent seizures. Int. J. Clin. Exp. Med. 2014;7:1639–1649. [PMC free article] [PubMed] [Google Scholar]
- 124.Danis B., van Rikxoort M., Kretschmann A., Zhang J., Godard P., Andonovic L., Siegel F., Niehusmann P., Hanon E., Delev D. Differential expression of miR-184 in temporal lobe epilepsy patients with and without hippocampal sclerosis - Influence on microglial function. Sci. Rep. 2016;6:33943. doi: 10.1038/srep33943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.