Abstract
Background
KRAS-mutant lung adenocarcinomas (LUADs) are heterogeneous and frequently occur in smokers. The heterogeneity of KRAS-mutant LUAD has been an obstacle for the drug discovery.
Methods
We integrated multiplatform datatypes and identified two corresponding subtypes in the patients and cell lines. We further characterized the features of these two subtypes and performed drug screening to identify subtype-specific drugs. Finally, we used the defining features of the KRAS subtypes for drug sensitivity prediction.
Findings
Patient-Subtype 1 (PS1) was characterized by increased smoking-related mutational signature activity, a low tumor-infiltrating lymphocyte (TIL)-associating score and STK11/KEAP1 co-mutations. Patient-Subtype 2 (PS2) was characterized by an increased smoking-related methylation signature activity, a high TIL-associating score and increased KRAS dependency. The cell line subtypes faithfully recapitulated all the patients' features. Drug screening of the two cell line subtypes yielded several potential candidates, such as cytarabine and enzastaurin for Cell-line-Subtype 1 (CS1) and a BTK inhibitor QL-XII-61 for Cell-line-Subtype 2 (CS2). The defining features, such as smoking-related methylation signature, were significantly associated with the sensitivity to several drugs.
Interpretation
The heterogeneity of KRAS-mutant LUAD is associated with smoking-related genomic and epigenomic aberration along with other features such as immunogenicity, KRAS dependency and STK11/KEAP1 co-mutations. These features might be used as biomarkers for drug sensitivity prediction.
Fund
This research was funded by the Young Scientists Fund of the National Natural Science Foundation of China, the Natural Science Foundation of Fujian Province, China and the Education and Research Foundation for Young Scholars of Education Department of Fujian Province, China.
Keywords: KRAS-mutant lung adenocarcinoma, KRAS subtypes, TIL-associating score, Smoking-related methylation signature, Smoking-related mutational signature, Drug sensitivity prediction
Research in context.
Evidence before this study
The heterogeneity of KRAS-mutant lung adenocarcinoma has been an obstacle for clinical treatment. However, the underlying mechanism has remained unclear. Therefore, only a subset of patients can benefit from either KRAS downstream targeted therapy or immune checkpoint blockades. We searched primary research studies in PubMed by using the terms ((((KRAS[Title]) AND (Heterogeneity[Title] OR subtype[Title] OR subtypes[Title] OR subset[Title] OR subsets[Title])) AND Lung[Title])). We identified 14 studies in which KRAS mutation (codon) subtypes are the major subject in these studies and only 1 study published in Cancer Discov. 2015 by Skoulidis F. et al. discussed the major subsets of KRAS-mutant lung adenocarcinoma and observed no difference of the KRAS mutation types among subsets. These authors performed an elegant study using gene expression data to group KRAS-mutant lung adenocarcinomas into three subtypes and found that HSP90 inhibitors selectively killed KRAS and STK11 co-mutant cancer cells. However, we hypothesized that the heterogeneity of KRAS-mutant lung adenocarcinoma may be the consequence of different features, such as smoking-induced genomic and epigenomic changes, the identification of which might be helpful for drug sensitivity prediction.
Added value of this study
KRAS-mutant lung cancer is associated with smoking activity and smoking can cause genomic and epigenomic changes. However, it is not clear that whether the smoking-related genomic and epigenomic changes contribute to the heterogeneity of KRAS-mutant lung adenocarcinomas. In this study, we found that the heterogeneity of KRAS-mutant lung adenocarcinomas was resulted from contributions by smoking-related DNA methylation, somatic mutational changes and the tumor-infiltrating leukocyte fraction. We were able to identify several promising drugs for each KRAS subtype and use the defining features of the subtypes for drug sensitivity prediction.
Implications of all the available evidence
Our findings indicate that the heterogeneity of KRAS-mutant lung adenocarcinomas is associated with smoking and can be interpreted at many levels, from genomic and epigenomic to transcriptomic and immunogenic levels. In addition, the multilevel features might be used as biomarkers for drug sensitivity prediction. However, additional studies that independently validate the clustering results are required. Moreover, it is necessary to use cell-line derived xenografts and/or patient-derived xenograft mouse models to further validate the potential drugs.
Alt-text: Unlabelled Box
1. Introduction
KRAS mutations occur in ~30% of lung adenocarcinomas [1,2]. Oncogenic KRAS is known to be “undruggable”; therefore, major efforts to combat KRAS-driven cancer focus on either inhibiting KRAS downstream targets [3,4] or screening for molecules that exhibit synthetic lethal interactions with oncogenic KRAS [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14]]. However, rare common hits have been identified among these screens. Fruitful results obtained from synthetical lethal screening suffered from poor reproducibility [9,11]. Moreover, only approximately 20% of KRAS-mutant lung cancer patients respond to MEK inhibitors, in contrast to a 60% response rate to EGFR inhibitors [4,15]. This lack of reproducibility is likely due to the heterogeneous background of both cell lines and patients harboring KRAS mutations. Therefore, it is critical to identify KRAS subtypes, characterize their features and further explore these features to predict of potential effective drugs.
How does one comprehensively evaluate KRAS heterogeneity? With the completion of The Cancer Genome Atlas (TCGA) project in April 2018, various datatypes and analyses including the immune landscape [[16], [17], [18]], oncogenic processes [17], cell of origin [19], and mutational signatures of cancer [20,21] are all now publicly available. Multiplatform analysis has become feasible and essential for elucidating the heterogeneity of cancer [[22], [23], [24], [25], [26]]. Moreover, several large-scale cell line drug sensitivity projects, including the Cancer Cell Line Encyclopedia (CCLE) [27], Cancer Therapeutics Response Portal (CTRP) [28,29], and The Genomics of Drug Sensitivity in Cancer Project (GDSC) [[30], [31], [32]], are all now publicly available for researchers' to explore.
Skoulidis et al. performed an elegant study using gene expression data to group KRAS-mutant lung adenocarcinoma into three subtypes and found that HSP90 inhibitors selectively killed KRAS and STK11 co-mutant cancer cells [33]. However, it is not clear whether the smoking-related genomic aberrations, epigenomic changes and tumor immunogenicity all contribute to the heterogeneity of KRAS-mutant lung adenocarcinoma. Given that KRAS-mutant lung cancer is associated with smoking activity [[34], [35], [36]] and smoking can cause genomic and epigenomic changes [37], it is very likely that smoking-induced genomic and epigenomic alterations may also contribute to the KRAS heterogeneity. Interestingly, Vaz et al. also found that chronic cigarette smoke condensate (CSC)-induced methylation changes are associated with KRAS-mutant lung cancer [38]. These authors hypothesized that oncogenic KRAS may contribute to the maintenance of smoking-induced DNA methylation. Therefore, in our study, we used a set of chronic CSC-induced DNA methylation according to Vaz et al. as metric for smoking-related DNA methylation changes and we calculated smoking-related mutational signature activity characterized by C > A transversions [20,39] to reflect smoking-induced genomic changes.
In this study, we took advantage of the aforementioned databases and identified two major subtypes of KRAS-mutant lung adenocarcinoma in patients by integrating multiplatform datatypes. We further validated the results obtained for patients using cell lines and found consistent features in cell line subtypes. Therefore, the cell line subtypes are useful surrogates for patient drug screening. We reanalyzed the publicly available drug sensitivity data using the cell line subtypes and found several promising drugs for each subtype. Interestingly, drug sensitivity was significantly associated with one or more oncogenic features of the KRAS subtypes.
2. Materials and methods
2.1. Data source
2.1.1. Patients
RNAseq (polyA+ IlluminaHiSeq), DNA methylation (Methylation450k), miRNA gene expression RNAseq (IlluminaHiSeq), somatic copy number variation (SCNA) (gene level, gistic2), somatic mutations (base substitution) (hg19, IlluminaGA) and clinical information for the patients (version: 2016-08-16) were downloaded from the UCSC Xena website: http://xena.ucsc.edu, dataset: TCGA-LUAD. There are 128 KRAS-mutant LUAD patients with all five data types, namely, gene expression RNAseq, DNA methylation, miRNA expression, SCNAs and somatic mutation (base substitution), available.
2.1.2. Cell lines
Gene expression array (Affymetrix Human Genome U219 array data at ArrayExpress (E-MTAB-3610)), DNA methylation (Methylation450k) and drug sensitivity data for the cell lines were downloaded from GDSC (https://www.cancerrxgene.org/). Somatic mutations of lung cancer cell lines were obtained from the COSMIC database [40]. Drug sensitivity data were also downloaded from CTRP (https://portals.broadinstitute.org/ctrp) and CCLE (https://portals.broadinstitute.org/ccle). Cell line histology information was obtained from GDSC, COSMIC or the previous literature. Of 35 KRAS-mutant lung cancer cell lines, we removed 3 small cell lung cancer (SCLC), 1 lung squamous cancer (LUSC) (according to GDSC records) and 3 LUAD cell lines with missing data types, resulting in the inclusion in the study of a total 28 KRAS-mutant cell lines with all three datatypes, namely, mRNA expression, DNA methylation and base substitution, available (Supplementary Table 1).
2.2. Somatic mutational signature analysis
The somatic mutational signatures of 543 LUAD patients and 178 lung cancer cell lines were decomposed and visualized using “SignatureAnalyzer” [41]. The results were compared with 30 reported mutational signatures from COSMIC to identify related etiologies. The similarity measures were based on the “cosine similarity”.
2.3. Data preprocessing
For the DNA methylation data, we first filtered out the following probes as previously reported [42]: 1. probes on chromosomes X and Y; 2. probes targeting multiple genes; and 3. probes containing an SNP, including the targeted CpG-site [43].
We then preprocessed the RNAseq, DNA methylation, miRNA expression and SCNA data in three steps as previously reported [23]. In brief, 1. Outlier removal: we deleted the features and samples that had >20% NAs using the R package DMwR [44]. 2. Missing data imputation: We imputed the missing data via the K nearest neighbor (KNN) imputation using the R package impute [45]. 3. Data normalization (Z-score).
In terms of preprocessing of the somatic base substitution data type, we calculated the frequency of six base substitutions (C > A, C > G, C > T, T > A, T > C and T > G).
The same preprocessing procedures were performed for the mRNA expression, DNA methylation and base substitution data using the lung cancer cell lines.
2.4. Similar network fusion and clustering
Similar network fusion (SNF) [23] was applied to integrate the above five preprocessed data types for patients, namely, RNAseq (16,661 identifiers × 128 samples), DNA methylation (331,515 identifiers × 128 samples), miRNA gene expression (554 identifiers × 128 samples), copy number (24,776 identifiers × 128 samples) and base substitutions (6 identifiers × 128 samples). For the cell lines, there are three data types available, namely, gene expression array data (17,484 identifiers × 28cell lines), DNA methylation (379,745 identifiers × 28 cell lines) and base substitution (6 identifiers × 28 cell lines) data. SNF created a similarity matrix for each data type and fused them into one similarity matrix. The network fusion step uses a nonlinear method based on the message-passing theory that iteratively updates every network and converges the data to a single network [23]. The fused SNF network was then subjected to consensus clustering (SNF-CC) by the function “ExecuteSNF.CC” from the R package “CancerSubtypes” [46]. The optimal parameters were tested and set as follows: 1. Patients clustering: K = 20, alpha = 0.5, t = 20, maxK = 10, pItem = 0.8, reps = 500. 2. Cell line clustering: K = 10, alpha = 0.5, t = 20, maxK = 10, pItem = 0.8, reps = 500.
Normalized mutual information (NMI) was calculated to assess the contributions to the network and compatibility of the data sources as described in ref. [23]. We calculated NMI values for the five data types using the function “rankFeaturesByNMI” in the R package “SNFtool” [23]. The percentages of the top-ranking features were used to select datatypes in cell lines for similar network fusion and clustering.
2.5. DNA methylation analysis
We used the R package “IlluminaHumanMethylation450kanno.ilmn12.hg19” [47] to analyze and annotate DNA methylation data. We also obtained a list of 847 unique smoking-related DNA methylation probes from the two repeats in the experiments conducted by Vaz et al. In their study, there were 633 CSC-induced probes in the first experiment and 242 CSC-induced probes in the repeated experiment. We considered the union of these two sets of experiments and there were a total of 847 unique probes from the two experiments. The smoking-related methylation signature was composed by calculating the mean of the β values of the 847 smoking-related DNA methylation probes for each patient and cell line. We then compared the difference in the smoking-related methylation signatures between the two subtypes and KRAS wild-type group (Kruskal-Wallis test and Wilcoxon test, the Q value is FDR-adjusted P value) in patients and cell lines.
2.6. Analysis of differentially expressed genes (DEGs)
To define genes that were differentially expressed between the two subtypes of patients and cell lines, we used the function “lmFit” in the R package “limma”.
For patients, we first selected genes at Q < 0.25. Then, we ordered these genes according to their log fold-change (logFC). We chose the genes with Q < 0.25, |logFC| > 0.5 as PS1-DEGs (n = 729) and PS2-DEGs (n = 2963).
Similarly, for cell lines, we also first selected genes at Q < 0.25. Then, we ordered these genes according to their logFC. We chose the genes with Q < 0.25, |logFC| > 0.5 as CS1-DEGs (n = 444) and CS2-DEGs (n = 356). The above DEGs were then subjected to gene set enrichment analysis (GSEA) using gene sets including Hallmark (MSigDB v6.1), KEGG (MSigDB v6.1) and Reactome (MSigDB v6.1).
2.7. Immune feature analysis
We used recently published TIL fraction data for TCGA LUAD patients according to Saltz et al. [18], who used deep learning methods (convolutional neural networks) to estimate TILs on hematoxylin and eosin stained (H&E-stained) slides. In our study, we took advantage of their data and built a linear model, Expi = β0 + β1 × TIL fractioni + εi to identify gene expression that was significantly associated with the TIL fraction of patients. We termed these genes “TIL-associating genes”. Q values are FDR adjusted P values. Genes with Q < 0.05 were considered TIL-associating genes. There were 214 positive TIL-associating genes and 3 negative TIL-associating genes (Supplementary Table 2). To explore whether TIL-associating genes were expressed in the cell lines, we plotted expression density curves of TIL-associating genes as well as 100 random background genes. After ensuring the expression of TIL-associating genes in both patients and cell lines, we calculated a composite score as the TIL-associating score according to the expression of TIL-associating genes. First, we median-centered each TIL-associating gene across the patients or cell lines. Then, we calculated the TIL-associating score by subtracting the mean of 3 negative TIL-associating genes from the mean of 214 positive TIL-associating genes. We then compared the TIL-associating score among the KRAS-mutant subtypes and KRAS wild-type group of LUAD patients and cell lines.
2.8. KRAS dependency score calculation
Two independent RAS gene expression signatures were used to calculate the KRAS dependency score. First, we calculated the “Singh Score” according to Singh et al. [48]. There were 262 KRAS-upregulated genes and 88 KRAS-downregulated genes in their study. We first median-centered these gene expression levels across all the samples. Next, we calculated the “Singh Score” by subtracting the mean of KRAS-downregulated genes from the mean of KRAS-upregulated genes.
Similarly, we calculated the “Loboda Score” according to Loboda et al [49]. Briefly, we calculated a composite score as described in [49,50]. There were 99 RAS-upregulated genes and 37 RAS-downregulated genes according to Loboda et al. [49]. We first median-centered these genes across the samples. Then we calculated the “Loboda Score” by subtracting the mean of KRAS-downregulated genes from the mean of KRAS-upregulated genes.
2.9. Drug screening data analysis
We compared the LN(IC50) of 265 drugs among the two CS and KRAS wild-type cell lines (Kruskal-Wallis test and Wilcoxon test). Drugs that were specifically sensitive to CS1 and CS2 were selected for further analysis. 1. CS1-specific drugs (n = 12), criteria: LN(IC50)CS1 < LN(IC50)CS2, P < 0.05; LN(IC50)CS1 < LN(IC50)WT, P < 0.05; and LN(IC50)CS2 ≈ LN(IC50)WT, P > 0.05. Wilcoxon test was used for pairwise comparison. 2. CS2-specific drugs: no drug met the following criteria: LN(IC50)CS2 < LN(IC50)CS1, P < 0.05; LN(IC50)CS2 < LN(IC50)WT, P < 0.05; LN(IC50)CS1 ≈ LN(IC50)WT, P > 0.05. There was only 1 drug that met the following criteria: LN(IC50)CS2 < LN(IC50)CS1, P < 0.05; LN(IC50)CS2 < LN(IC50)WT, P < 0.15; LN(IC50)CS1 ≈ LN(IC50)WT, P > 0.05. We listed and plotted all these subtype-specific drugs in Fig. 6, Supplementary Fig. 9 and Supplementary Table 3.
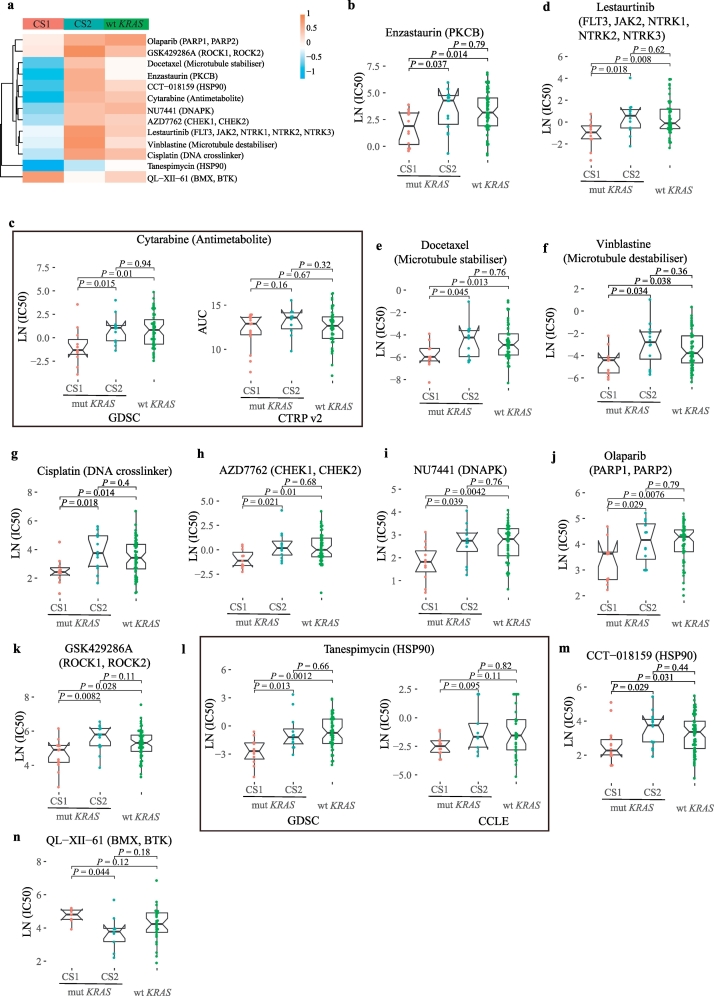
Fig. 6.
Screened drugs with selective sensitivity toward the KRAS subtypes.
(a) Drugs that selectively killed CS1 or CS2.
(b–m) CS1-specific drugs that are ordered by logFC: enzastaurin, lestaurtinib, cytarabine, docetaxel, vinblastine, cisplatin, AZD7762, NU7441, olaparib, GSK429286A, tanespimycin and CCT-018159. The Wilcoxon test was used for the pairwise comparison.
(n) CS2-specific drug: QL-XII-61. The Wilcoxon test was used for the pairwise comparison.
2.10. Univariate and multivariate analysis of drug prediction
We used the data of 28 KRAS-mutant lung cancer cell lines and built the following linear models for drug sensitivity prediction.
Univariate regression model:
Multivariate regression model:
3. Results
3.1. Identification of two major subtypes of KRAS-mutant lung adenocarcinoma
3.1.1. Patient subtypes
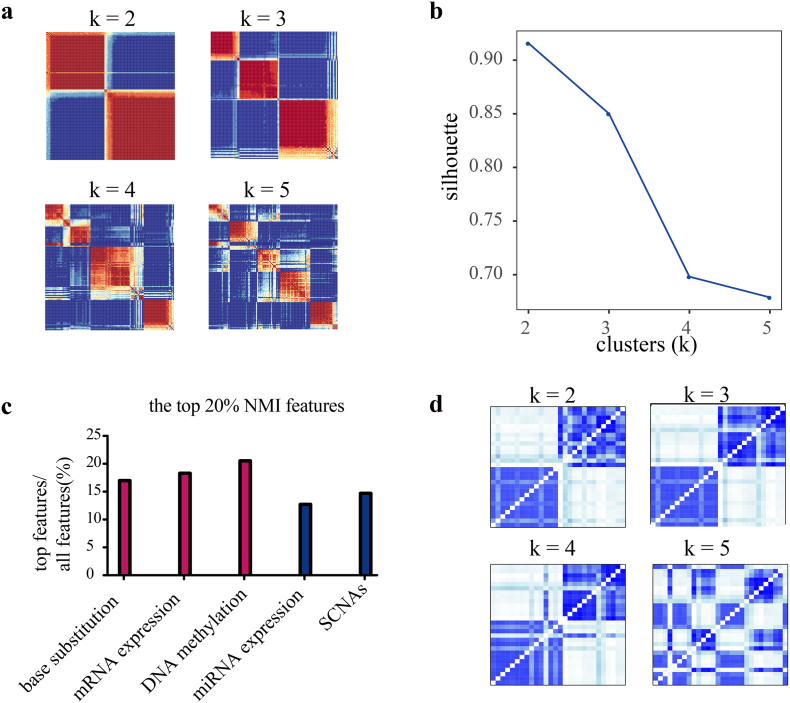
We integrated DNA methylation, mRNA expression, miRNA expression, SCNAs and base substitution (C > A, C > G, C > T, T > A, T > C and T > G), for a total of 5 data types, and we used SNF-CC [23,51] to cluster 128 KRAS-mutant lung adenocarcinoma patients (TCGA) into two subtypes (see 2.4 for detailed methods). The use of one data type yielded different classification results (Supplementary Fig. 1). By contrast, the SNF network captured both shared and complementary information from the above 5 data types and identified two subtypes in KRAS-mutant lung adenocarcinomas, PS1 and PS2 (silhouette = 0.92) (Fig. 1a and b). We next sought to evaluate the contribution of each data type to the fused network by NMI value [23]. The percentages of important features from each datatype were calculated based on the NMI values. We found that the fused network was mainly driven by three data types, mRNA expression (18.3% contribution), DNA methylation (20.5%) and base substitution (16.7%), according to the top 20% NMI (Fig. 1c) (Supplementary Table 4).
Fig. 1.
SNF-CC identifies two robust subsets of KRAS-mutant lung adenocarcinomas in patients and cell lines.
(a) SNF-CC integrated 5 data types of patients and similarity matrices for each class.
(b) Silhouette values for the k = 2 to k = 5 classes.
(c) The percentages of important features (top 20% NMI) from each data type that contribute to the fused network.
(d) SNF-CC integrated 3 data types of cell lines (N = 28) and similarity matrices for each class.
3.1.2. Cell lines' subtypes
To explore potential treatments for each subtype, we next sought to identify the subtypes in 28 LUAD cell lines (GDSC) harboring KRAS mutations. We used the three most important datatypes that contribute to the fused network from the patient clustering, namely, base substitution, DNA methylation and gene expression. Similarly, we identified two subtypes of mutant KRAS cell lines (N = 28), CS1 (N = 14) and CS2 (N = 14) (Fig. 1d).
3.2. Biological features of the KRAS subtypes
Given that DNA methylation, base substitution (somatic mutations), and gene expression were the 3 major datatypes contributing to the heterogeneity of KRAS-mutant LUAD, we extracted and characterized the biological features of the KRAS subtypes from these datatypes.
3.2.1. Smoking-related methylation signature
Since DNA methylation was the top 1 datatype contributing to the heterogeneity and smoking can cause epigenomic perturbations in lung tissues, we assessed both the global and smoking-induced DNA methylation patterns of the two subtypes.
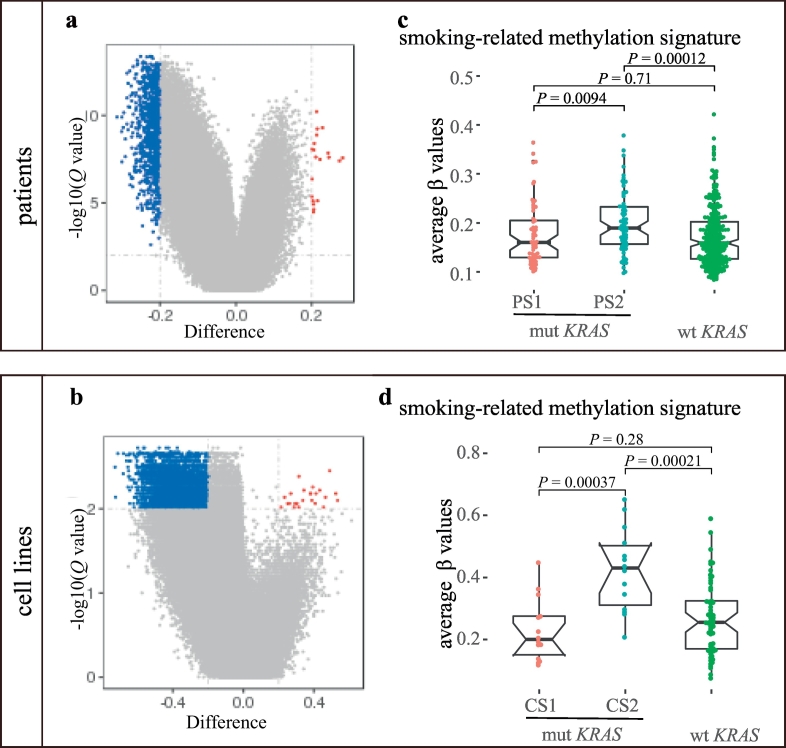
Both PS2 and CS2 displayed global hypermethylation compared with PS1 and CS1, respectively (995 differentially methylated probes (DMPs) in PS2 vs. 20 DMPs in PS1 (Fig. 2a); 4626 DMPs in CS2 vs. 624 DMPs in CS1 (Fig. 2b)). Next, we composed the smoking-related methylation signature activity using the average β values of 847 unique smoking-related probes from two repeats of the experiment according to Vaz et al. [38] and compared the signature activity between the two subtypes. We found that the activity of smoking-related methylation signature was significantly higher in PS2 than in PS1 (P = 0.0094, Wilcoxon test) and KRAS wild-type patients (P = 0.00012, Wilcoxon test) (Fig. 2c), whereas there was no difference in the smoking-related methylation signature activity between PS1 and KRAS wild-type patients (P = 0.71, Wilcoxon test) (Fig. 2c). The results suggested that PS2 exhibited more epigenomic alterations related to smoking compared with PS1 and KRAS wild-type patients. Similarly, CS2 displayed the highest smoking-related methylation signature activity compared with CS1 (P = 0.00037) and KRAS wild-type cell lines (P = 0.00021). Additionally, CS1 and KRAS wild-type cell lines showed similar lower smoking-related methylation signature activity (P = 0.28) (Fig. 2d).
Fig. 2.
Epigenomic features of the KRAS subtypes.
(a) and (b) Volcano plot of the global DNA methylation difference (difference in β values) between the subtypes of patients (a) and cell lines (b). Probes hypermethylated in PS1 or CS1 are labeled in dark red (difference > 0.2, Q < 0.1), whereas probes hypermethylated in PS2 or CS2 are labeled in dark blue (difference < −0.2, Q < 0.1). Wilcoxon test, Q values are FDR-adjusted P values.
(c) and (d) Comparison of the smoking-related methylation signature among the three groups in patients (c) and cell lines (d) Wilcoxon test was used for the comparison.
3.2.2. TIL-associating score
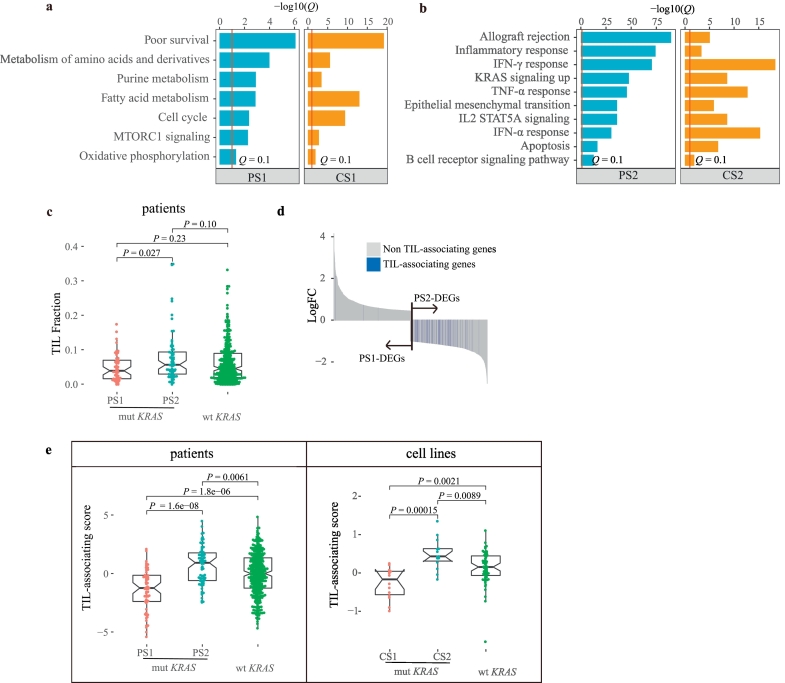
The second important datatype contributing to fused network for clustering was gene expression. We next analyzed the enriched pathways of the DEGs (Q < 0.25, |logFC| > 0.5) in each subtype (PS1/CS1-DEGs and PS2/CS2-DEGs). The enriched pathways were very similar between the corresponding subtypes of patients and cell lines (Fig. 3a and b). For example, PS1 and CS1 were both enriched for lung cancer poor survival signatures, cell cycle and metabolic pathways (Q < 0.1) (Fig. 3a); By contrast, PS2 and CS2 were both enriched for pathways such as allograft rejection, inflammatory response, interferon-gamma (IFN-γ) response, and KRAS signaling up, among others (Q < 0.1) (Fig. 3b).
Fig. 3.
The transcriptomic and immunological features of the KRAS subtypes.
(a) The enriched pathways of PS1 and CS1 according to GSEA, Q < 0.1 (Q is FDR adjusted P value).
(b) The enriched pathways of PS2 and CS2 by GSEA, Q < 0.1 (Q is FDR adjusted P value).
(c) Comparison of TIL fractions estimated from H&E-stained slides among the three groups. Wilcoxon test was used for the comparison.
(d) The distribution of TIL-associating genes in PS1-DEGs or PS2-DEGs (Hypergeometric test).
(e) Comparison of TIL-associating scores among the three groups of patients and cell lines. Wilcoxon test was used for the comparison.
Given that both PS2 and CS2 displayed active immune pathways, we decided to further characterize the immunological features of the KRAS subtypes. We first compared the tumor-infiltrating lymphocyte (TIL) fractions estimated from H&E-stained slides according to Saltz et al. [18] among the three groups. We found that PS2 exhibited the highest TIL fraction (median 0.056) compared with PS1 (median 0.030, P = 0.027) and wild-type (median 0.045, P = 0.10) (Fig. 3c). However, we could not compare the TIL fractions in cell lines due to the lacking of a tumor microenvironment.
To quantify the immunogenicity in the cell line subtypes, we derived a TIL-associating score using 217 TIL-associating genes (Supplementary Table 2) that were significantly associated with the TIL fraction (Q < 0.05) (see 2.7 for detailed methods). These genes were enriched in pathways such as allograft rejection, IFN-γ, and inflammatory response, among others (Supplementary Fig. 2a). Given that the tumor samples from the patients were infiltrated by immune cells and other cell types, we were concerned that part of TIL-associating genes might be contributed by infiltrated immune cells but not cancer cells. If this scenario was true, then a portion of TIL-associating genes would exhibit very low expression in cancer cell lines. Therefore, we checked the expression pattern of all 217 TIL-associating gene in the KRAS mutant cell lines. We split these 217 genes into 47 reported immune genes [52] and 170 non-immune genes. Importantly, we did not observe any differential expression patterns among the 47 immune genes, the other 170 genes and randomly sampled 100 background genes in lung cancer cell lines (Supplementary Fig. 2b), suggesting that the TIL-associating genes were indeed expressed by the tumor cells themselves. Interestingly, these genes were significantly enriched in PS2-DEGs, which indicated that PS2 tumors were more immunogenic than PS1 tumors (P = 3.25e-71, Hypergeometric test) (Fig. 3d). Thus, we composed a TIL-associating score according to the expression of the TIL-associating genes (see 2.7 for detailed methods). We then could compare the TIL-associating score not only in the patients but also in the cell lines. Indeed, we found that the TIL-associating score displayed the same trend between patients and cell lines. PS2/CS2 (median: PS2 = 0.92, CS2 = 0.43) had the highest TIL-associating score, followed by wild-type KRAS patients/cell lines (wtKRAS_P/wtKRAS_C) (median: wtKRAS_P = −0.0031, P = 0.0061 and wtKRAS_C = 0.16, P = 0.0089); PS1/CS1 had the lowest TIL-associating score (Fig. 3e) (median: PS1 = −1.24, P = 1.6e-08 and CS1 = −0.17, P = 0.00015).
In addition, we compared the spatial structural pattern of TIL according to Saltz et al. [18]. Interestingly, PS2 had the highest proportion of the “Brisk, diffuse” category (48.6%, 34/70) compared with PS1 (34.5%, 20/58) and KRAS wild-type group (32.2%, 122/379) (Supplementary Fig. 3). In contrast, PS1 had the highest proportion of “Non-Brisk, focal” category (13.8%, 8/58) compared with PS2 (8.6%, 6/70) and KRAS wild-type group (8.2%, 31/379) (Supplementary Fig. 3). All these findings indicated that PS2/CS2 were the most immunogenic, whereas PS1/CS1 were the least immunogenic and were even worse than wild-type KRAS patients/cell lines.
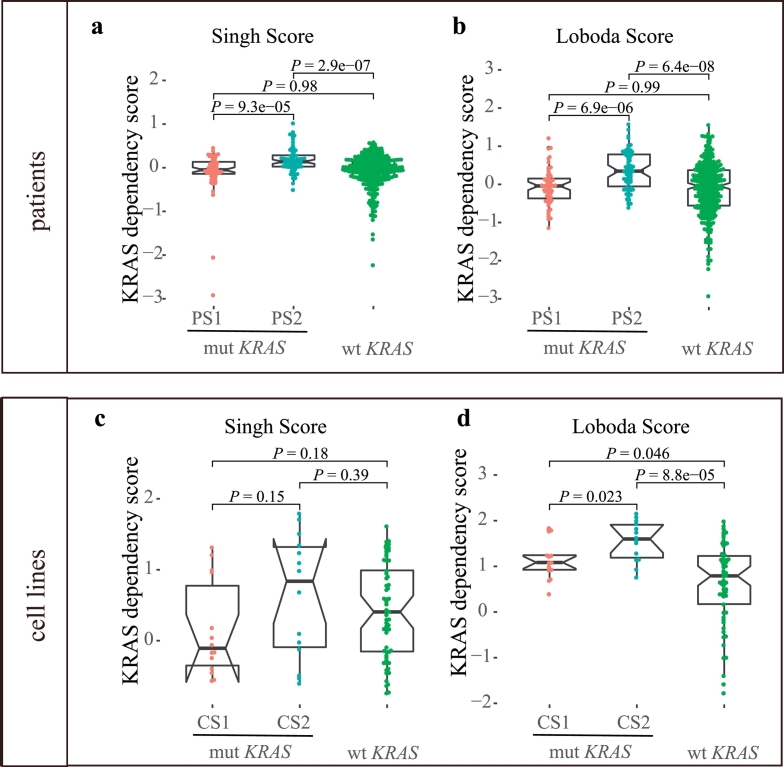
3.2.3. KRAS dependency score
Through pathway enrichment analysis, we found that KRAS signaling was significantly enriched in PS2 and CS2, although KRAS mutations existed in both subtypes. Additionally, there are reports suggesting that a gene expression signature-based pathway readout might be more appropriate than relying on a single indicator (KRAS mutation status) of pathway activity [48,49]. To measure Ras pathway activation, we calculated the KRAS dependency score (Singh Score and Loboda Score) according to two previous studies by Singh et al. [48] and Loboda et al. [49]. In both studies, they developed a method for the quantification of Ras-dependent gene expression that provides a better measure of Ras activity in cancer cells than KRAS mutation type analysis. Importantly, we found that PS2 had a significantly increased KRAS dependency score compared with PS1 (Singh Score: P = 9.3e-05 and Loboda Score: P = 6.9e-06, Wilcoxon test) and KRAS wild-type group (Singh Score: P = 2.9e-07 and Loboda Score: P = 6.4e-08) (Fig. 4a and b), supporting the presence of a hyperactive Ras pathway in PS2. PS1 and KRAS wild-type patients had similar KRAS dependency scores despite their different KRAS mutation statuses (Singh Score: P = 0.98 and Loboda Score: P = 0.99). The KRAS dependency score displayed a similar trend in cell lines, but with less significance. CS2 had the highest KRAS dependency score compared with CS1 (Loboda Score: P = 0.023 and Singh Score: P = 0.15). CS1 and KRAS wild-type cell lines had similar KRAS dependency scores (Singh Score: P = 0.18) (Fig. 4c and d).
Fig. 4.
Comparison of KRAS dependency scores among the KRAS subtypes.
(a, b) Comparison of KRAS dependency scores among PS1, PS2 and KRAS wild-type patients according to two independent studies.Wilcoxon test was used for the comparison.
(c, d) Comparison of KRAS dependency scores among CS1, CS2 and KRAS wild-type cell lines according to two independent studies. Wilcoxon test was used for the comparison.
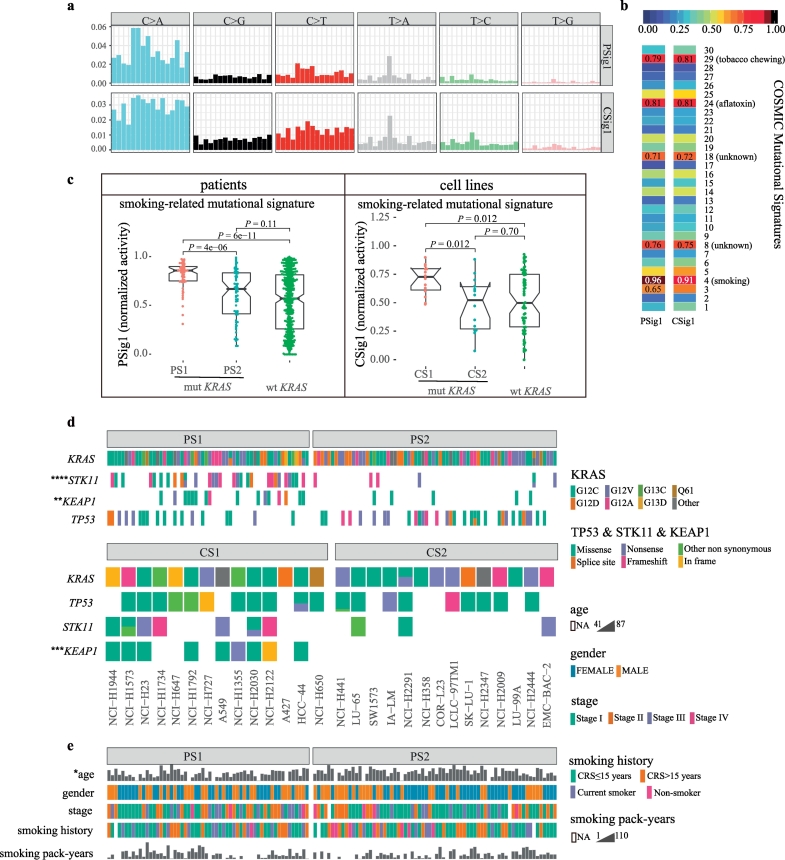
3.2.4. Smoking-related mutational signature
Finally, base substitution was the third important datatype contributing to the fused network for clustering. We assessed the somatic mutational pattern derived from base substitution and extracted 4 somatic mutational signatures from 543 lung adenocarcinoma patients (Supplementary Fig. 4a). Similarly, we extracted 5 somatic mutational signatures from 178 lung cancer cell lines (Supplementary Fig. 5a). Then, we compared these mutational signatures to 30 known somatic mutational signatures in the COSMIC database (cancer.sanger.ac.uk) using cosine similarity (CS) (Supplementary Fig.4b and 5b) [40,53]. Among them, signature 1 of patients (PSig1) and signature 1 of cell lines (CSig1) were both characterized primarily by C > A mutations and were highly similar to Signature 4 in COSMIC (smoking, CS = 0.96 and CS = 0.91, respectively) (Fig. 5a and b). Moreover, PSig1 accounted for 66% of the total somatic mutational signature activity in the patients, and CSig1 accounted for 50% of the total mutational signature activity in the cell lines. The remaining somatic signatures represented a relatively small fraction of the total normalized mutational signature activity (Supplementary Figs. 6 and 7). Therefore, the smoking-related mutational signature was the most important somatic mutational signature.
Fig. 5.
Genomic and clinical features of the KRAS subtypes.
(a) Smoking-related mutational signatures were retrieved from the mutational profiles of LUAD patients or lung cancer cell lines. The mutation types are displayed on the horizontal axis, whereas the vertical axis depicts the percentage of mutations attributed to a specific mutation type.
(b) Comparison of the smoking-related mutational signatures with the reported mutational signatures in COSMIC. The similarity measures are based on the cosine similarity.
(c) Differential activities of the smoking-related mutational signature among the subtypes of patients and cell lines. The Kruskal-Wallis test and Wilcoxon test were used for the comparison.
(d) Comparison of KRAS, STK11, KEAP1 and TP53 mutation types between the two subtypes of patients and cell lines. Fisher's exact test, ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
(e) The clinical features between the two subtypes of patients. Fisher's exact test or Wilcoxon test, *P < 0.05.
Importantly, we found that PSig1 was significantly increased in PS1 (median 0.86) compared with PS2 (median 0.67, P = 4.0e-06) and wild-type KRAS patients (median 0.57, P = 6.0e-11). Furthermore, smoking-related mutational signature activity was not significantly different between PS2 and KRAS wild-type patients (P = 0.11) (Fig. 5c). Very similarly, the smoking-related mutational signature activity was the highest in CS1 (median 0.73) compared with CS2 (median 0.52, P = 0.012) and wild-type KRAS cancer cell lines (median 0.50, P = 0.012). Additionally, there was no difference in the smoking-related mutational signature activity between CS2 and the wild-type KRAS cell lines (P = 0.70) (Fig. 5c).
3.2.5. STK11 and KEAP1 mutation status
Finally, we also assessed 54 significantly mutated genes in lung cancer from the Firehose Broad website (http://gdac.broadinstitute.org/; Jan 2016). We found that STK11 mutations (P = 5.22e-07, Fisher's exact test) and KEAP1 mutations (P = 0.0085) were significantly enriched within PS1, whereas the distribution of TP53 mutations was less significant between the two subtypes (P = 0.10) (Fig. 5d). Moreover, the types of KRAS mutation types were not significantly different between the two subtypes (patients: P = 0.43, cell lines: P = 0.34) (Fig. 5d and Supplementary Table 5). A similar enrichment pattern was also observed in the cell lines although the enrichment of STK11 mutations in the cell lines was not as significant as in the patients. STK11 mutations (P = 0.23) and KEAP1 mutations (P = 0.0007) occurred more frequently in CS1 than CS2, whereas the distribution of TP53 mutations was relatively even between the cell line subtypes (P = 0.43) (Fig. 5d).
3.2.6. Other features
Although the smoking-related mutational signature was significantly higher in PS1/CS1 while the smoking-related methylation signature was significantly higher in PS2/CS2, the smoking pack-years and smoking history of the patients did not show any difference between two the subtypes (Fig. 5e), suggesting that the molecular smoking signature is a more accurate molecular measurement of smoking-induced genomic damage and epigenomic alterations. Another interesting feature is that PS1 patients were significantly younger than PS2 patients (median age 63 vs. 69 years, P < 0.05) (Fig. 5e), providing further evidence supporting a poor prognosis for PS1.
3.3. Screening for compounds with selective sensitivity to the KRAS subtypes
After characterizing the key features of the subtypes, we next sought to explore potential drugs that were selective for each subtype using cell line models. We reanalyzed 265 drugs that tested on KRAS-mutant and KRAS wild-type lung adenocarcinoma cell lines from GDSC [[30], [31], [32]] and validated some of our results using two other large-scale cell line drug sensitivity projects, CCLE and CTRPv2 [28,29]. We were particularly interested in drugs with specific sensitivity to CS1 or CS2 (Fig. 6a). Twelve CS1-specific drugs and 1 CS2-specific drug were discovered according to the pairwise comparison of CS1, CS2 and the KRAS wild-type group.
CS1-specific drugs can roughly be classified into the following categories. First, a protein kinase C beta (PKCβ) inhibitor, enzastaurin, was found to preferentially kill CS1 (LN (IC50) = 1.89) compared with CS2 (LN (IC50) = 4.90, P = 0.044) and wild-type KRAS cell lines (LN (IC50) = 3.16, P = 0.013) (Fig. 6b). Second, a pyrimidine nucleoside analog, cytarabine, was applied. CS1 was sensitive to cytarabine treatment (LN (IC50) = −1.32) compared with CS2 (LN (IC50) = 1.05, P = 0.013, Wilcoxon test) and wild-type KRAS cell lines (LN (IC50) = 0.83, P = 0.010, Wilcoxon test). CS2 and wild-type KRAS cell lines were equally resistant to cytarabine (P = 0.94). Importantly, cytarabine showed similar specific toxicity with less significance to CS1 (AUC = 12.90) compared with CS2 (AUC = 13.59, P = 0.15) using drug sensitivity data from CTRP v2 (Fig. 6c). The structure of this drug mimics pyrimidine, and it can inhibit S phase of the cell cycle. Importantly, the cell cycle pathway was both enriched in both CS1 and PS1 (Fig. 3a). Therefore, cytarabine is a very promising drug for KRAS tailored therapy.
In addition, lestaurtinib, a multiple tyrosine kinase inhibitor (FLT3, JAK2), also showed specific killing effect for CS1 (LN (IC50) = −0.94) compared with CS2 (LN (IC50) = 0.58, P = 0.018) and wild-type KRAS cell lines (LN (IC50) = −011, P = 0.008) (Fig. 6d). It has been reported that FLT3 promotes the activation of RAS signaling and phosphorylation of downstream kinases in leukemia [54]. However, the role of this inhibitor in KRAS-mutant LUAD has been less explored.
Moreover, chemotherapy drugs, such as two microtubule-targeted drugs, docetaxel and vinblastine, and one DNA crosslinker, cisplatin, showed greater toxicity to CS1 than to CS2 (docetaxel: CS1_ LN (IC50) = −5.97, P = 0.045, vinblastine: CS1_ LN (IC50) = −4.41, P = 0.034, cisplatin: CS1_ LN (IC50) = 2.43, P = 0.018) and KRAS wild-type group (Fig. 6e–g). in addition, drugs involved in DNA damage response, such as the Chk1 inhibitor AZD7762, DNA-dependent protein kinase (DNA-PK) inhibitor, NU7441, and PARP1 inhibitor, olaparib, all showed better efficacy in CS1 than CS2 (AZD7762: CS1_LN (IC50) = −1.12, CS2_LN (IC50) = 0.17, P = 0.021; NU7441: CS1_LN (IC50) = 1.82, CS2_LN (IC50) = 2.74, P = 0.039; olaparib: CS1_LN (IC50) = 3.63, CS2_LN (IC50) = 4.16, P = 0.029) (Fig. 6h–j). Interestingly, DNA-PK gene expression was significantly increased in PS1 compared to PS2 (P = 0.0076) and a similar trend was observed in the cell lines, but with less significance (Supplementary Fig. 8a and b). Considering that the cell growth pathway (MTORC1 signaling) and the cell cycle pathway were activated in PS1/CS1 (Fig. 3a), it is reasonable that these chemotherapy drugs were more effective against the faster growing cells (CS1). In addition, given that smoking-induced genomic damage was more severe in PS1/CS1, it is plausible that that drugs involved in DNA damage repair were also more toxic to CS1.
Finally, previously reported compounds or targets [7,33], ROCK1, 2 inhibitor GSK429286A and HSP90 inhibitors tanespimycin (17-AAG) and CCT-018159, were also discovered in our study that were more toxic to CS1 compared with CS2 (GSK429286A: CS1_LN (IC50) = 4.91, CS2_LN (IC50) = 5.80, P = 0.0082; tanespimycin: CS1_LN (IC50) = −2.69, CS2_LN (IC50) = −1.21, P = 0.013; CCT-018159: CS1_LN (IC50) = 2.27, CS2_LN (IC50) = 3.75, P = 0.029) (Fig. 6k–m). A similar result, but with less significance was obtained for tanespimycin using data from CCLE (P = 0.095) (Fig. 6l).
The CS2-specific drug, QL-XII-61 (BMX, BTK inhibitor), which is related to the immune pathway, showed a selective killing effect against CS2 (CS1_LN (IC50) = 4.81, CS2_LN (IC50) = 3.78, P = 0.044) (Fig. 6n). Interestingly, both PS2 and CS2 were enriched for active B cell receptor (BCR) signaling (Fig. 3b). Moreover, BTK expression was significantly increased in PS2 than PS1 (P = 6.6e-10), and a similar trend, but with less significance was observed in the cell lines (Supplementary Fig. 8c and d). All evidences implied that BTK inhibitors might be potential candidates for PS2 patients.
In addition, as a positive control, we found that MEK1/2 and BRAF inhibitors killed both subtypes of KRAS-mutant cell lines but spared KRAS wild-type cell lines (Supplementary Fig. 9).
3.4. Drug response prediction by the subtype features
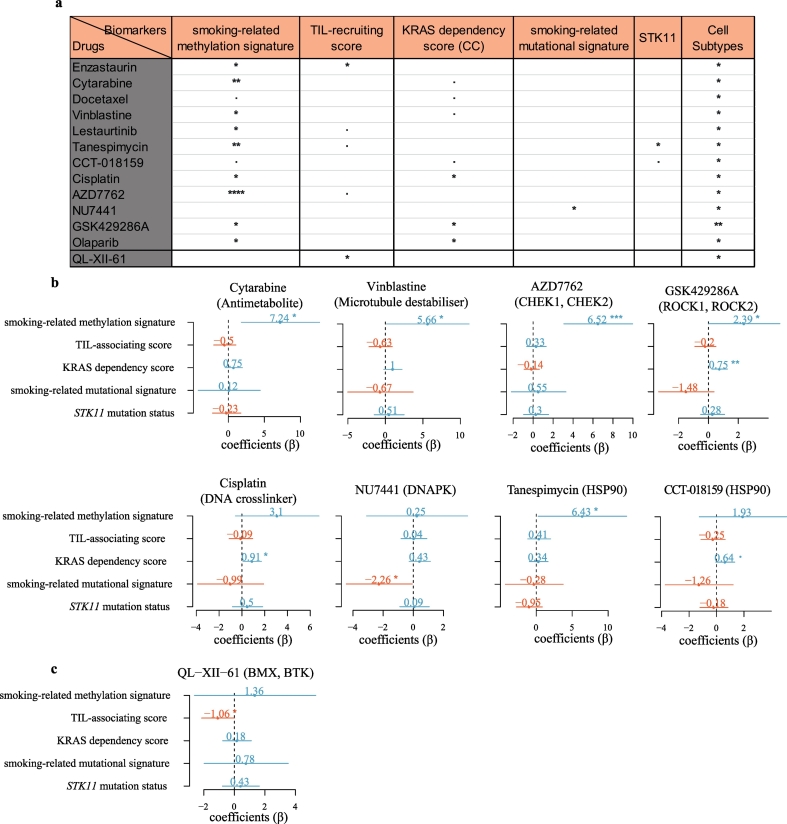
Given that we found several promising drugs that were synthetical lethal to the KRAS subtypes, we were interested in investigating whether the drug response could be predicted by the biological features of the KRAS subtypes using both univariate and multivariate regressions.
Importantly, all the candidate drugs could be predicted by one or more features in the univariate analysis and some of them could be predicted by the defining features in the multivariate analysis. For example, cytarabine could be predicted by the smoking-related methylation signature through univariate (coefficient (β) = 7.31, 95% CI: 2.83 to 11.79, P = 0.0027) or multivariate (β = 7.24, 95% CI: 1.81 to 12.67, P = 0.012) analysis (Fig. 7a and b). The positive β value for the smoking-related methylation signature suggested that IC50 increased with increases in smoking-related methylation signature activity.
Fig. 7.
Drug sensitivity can be predicted by the KRAS subtype features.
(a) The drug sensitivity was predicted by the KRAS subtype features in the univariate analysis. The P value is the significance of the coefficients (β) in the univariate regression model. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ·P < 0.1.
(b) Forest plots of coefficients (β) with 95% confidence intervals for features predicting the drug sensitivity in the multivariate analysis. the P value is the significance of the coefficients (β) in the multivariate regression model. **P < 0.01; *P < 0.05; ·P < 0.1.
In addition, the IC50 values of vinblastine (β = 5.66, 95% CI: 0.17 to 11.16, P = 0.044), AZD7762 (β = 6.52, 95% CI: 3.08 to 9.97, P = 0.00094) and GSK429286A (β = 2.39, 95% CI: −0.0065 to 4.78, P = 0.051) (Fig. 7b) were all positively associated with the smoking-related methylation signature in the multivariate analysis, indicating that the smoking-related methylation signature was a very strong predictor for multiple drugs.
Moreover, NU7441 could be predicted by the smoking-related mutational signature using either univariate (β = −1.85, 95% CI: −3.58 to −0.11, P = 0.038) or multivariate (β = −2.26, 95% CI: −4.45 to −0.070, P = 0.044) regression (Fig. 7a and b). The results indicated that NU7441 (DNA-PK inhibitor) would be more effective against tumor cells with higher smoking-related mutational signature activity.
Although CCT-018159 and tanespimycin are both HSP90 inhibitors, their predictors were quite different. CCT-018159 sensitivity could be predicted by the KRAS dependency score in both univariate (β = 0.53, 95% CI: −0.017 to 1.09, P = 0.057) and multivariate (β = 0.64, 95% CI: −0.056 to 1.33, P = 0.070) regressions (Fig. 7a and b). In contrast, tanespimycin could be predicted by either the smoking-related methylation signature (β = 8.40, 95% CI: 3.47 to 13.36, P = 0.0019, univariate analysis) (β = 6.43, 95% CI: 0.27 to 12.59, P = 0.042, multivariate analysis) or STK11 mutation status (β = −1.69, 95% CI: −3.35 to −0.039, P = 0.045, univariate analysis) in agreement with a previous report showing that co-mutant KRAS/STK11 lung cancer cells were sensitive to HSP90 inhibitors [33]. However, the STK11 mutation status was not any more significant when the smoking-related methylation signature was added as a predictor, suggesting that the smoking-related methylation signature is a stronger predictor than STK11 mutation status for tanespimycin.
Finally, the CS2-specific drug, QL-XII-61, was significantly predicted by the TIL-associating score both in univariate (β = −0.86, 95% CI: −1.69 to −0.033, P = 0.043) and multivariate (β = −1.06, 95% CI: −2.14 to 0.020, P = 0.054) analyses (Fig. 7a and b), suggesting that it is more effective against tumor cells with greater immunogenicity (a higher TIL-associating score).
4. Discussion
KRAS is one of the most frequently mutated genes in human cancers and related to smoking activity [34,35]. Gain-of-function mutations in KRAS are thought to be involved in tumor initiation, invasion and metastasis [55]. The design of therapeutics toward KRAS mutations has proven extremely challenging, although recent studies suggest that targeting “undruggable” oncogenic KRAS may be an attainable goal [[56], [57], [58]]. However, only recently has the heterogeneity of tumors harboring KRAS mutations been recognized [33], which may provide another layer of complexity to the treatment of this malignant tumor type.
In this study, we identified five important features of the heterogeneity of KRAS-mutant tumors and cell lines, including smoking-induced two processes. PS1 had an enhanced smoking-related mutational signature, while PS2 had increased smoking-related methylation signature although the reported smoking history were not different between the subtypes. We suspect that the brand of cigarettes and dosage and length of smoking are probably the underlying cause of the smoking-related features in the two subtypes. PS1 possibly consumed a higher dosage in a shorter period, causing more severe DNA damage, including the active smoking mutational signature and copy number variations, and the patients were younger in this category. Moreover, PS2 probably suffered from a longer exposure to a lower dosage of smoking, which was not enough to cause much genomic damage but led to the accumulation of smoking-specific DNA methylation. Furthermore, oncogenic KRAS contributed to both the accumulation of the smoking signature and the maintenance of smoking-induced methylation in these two subtypes. The smoking-history and smoking-pack years were not significant different between the two subtypes. This result was like due to an insufficient or incorrect patient-reported smoking history, suggesting that smoking molecular signatures were more accurate indicators for predicting KRAS subtypes and drug sensitivity than patient-reported smoking history which has also been suggested elsewhere [59].
Given the key role of immunotherapy treatments in contemporary cancer care, tumor-associated lymphocyte analysis is becoming increasingly important. Studies suggest that high densities of TILs correlate with favorable clinical outcomes, such as longer disease-free survival or improved overall survival (OS) in multiple cancer types [[60], [61], [62], [63]]. To compare the TIL-associated features both in the patients and cell lines, we derived a set of TIL-associating genes and composed a TIL-associating score so that the TIL feature could also be measured in the cell lines. By quantifying the TIL-associating score in the cell lines, we were able to use this new metric for drug prediction.
Finally, we reanalyzed three publicly available cell line drug sensitivity datasets and discovered several promising drugs along with previously reported HSP90 inhibitors. Among them, the RAS mutation was reported to disappeared after low-dose cytarabine treatment of in a 52-year-old women with myelodysplastic syndromes [64]. In addition, patients with acute myeloid leukemia (AML) carrying mutant RAS experienced a greater benefit from higher cytarabine doses than patients with wild-type RAS [65]. These studies together with our results suggest that cytarabine is a very promising drug for CS1/PS1. Another drug, NU7441, which is a DNA-PK inhibitor, is also toxic to CS1/PS1. DNA-PK plays a key role in the repair of DNA double-stranded breaks (DSBs) in cancer cells [[66], [67], [68]]. CS1 shows increased smoking-related mutational signature activity which in turn might activate DNA-PK. Moreover, the sensitivity to NU7441 can be predicted by the smoking-related mutational signature but by no other biological features, suggesting that NU7441 may be used for mutant KRAS tumors with higher smoking-related mutational signature activity. It is also worth noting that the CS2-specific drug, the BTK inhibitor QL-XII-61, can be predicted by the TIL-associating score, which corresponds to the observation that PS2/CS2 are highly immunogenic. It is known that BTK kinase is a key element of BCR signaling and plays important roles in the regulation of B-cell activation, proliferation and differentiation [69,70]. Therefore, our results might suggest a novel role of BTK inhibitors in the immunogenic KRAS subtype.
In summary, we identified two major subtypes of KRAS-mutant lung adenocarcinoma patients, PS1 is characterized by increased activity of the smoking-related mutational signature, a low TIL-associating score and STK11/KEAP1 co-mutations. PS2 is characterized by increased activity of the smoking-related methylation signature, an increased KRAS dependency and a high TIL-associating score. Importantly, the cell line subtypes faithfully recapitulated all the biological features in the patients. We also identified several KRAS subtype-specific drugs in the cell lines, and these drugs could be predicted by one or more biological features of the KRAS subtypes. Our results shed light on the understanding of the heterogeneity of KRAS-mutant lung adenocarcinomas and the discovery of associated targeted drug. However, since our research focus was a multiplatform analysis of KRAS-mutant lung adenocarcinomas, a relatively small sample size of patients and cell lines are currently available. Therefore, it is necessary to validate our results in new datasets and use cell-line derived xenografts and/or patient-derived xenograft mouse models to confirm these potential drugs in the future.
The following are the supplementary data related to this article.
Information of 35 KRAS-mutant lung cancer cells.
GDSC drugs sensitivity among CS1, CS2 and KRAS wild-type group.
Supplementary material
Acknowledgments
Acknowledgments
We would like to thank Xin Wang, Taoling Zhong, Liying Yu and Yudi Hu for constructive discussions.
Sources of funding
This research is supported by the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 81802823 to Ying Zhou), the Natural Science Foundation of Fujian Province, China (Grant No. 2018 J01054 to Ying Zhou) and the Education and Research Foundation for Young Scholars of Education Department of Fujian Province, China (Grant No. JAT170004 to Ying Zhou).
Declaration of interests
None.
Author contributions
The study was conceived and designed by Ying Zhou and Qiyuan Li. The data collection was done by Ke Liu and Ying Zhou. The data analysis was performed by Ke Liu, Jintao Guo, Kuai Liu, Peiyang Fan, Yuanyuan Zeng, Chaoqun Xu and Jiaxin Zhong. The data interpretation was done by Ke Liu, Jintao Guo and Ying Zhou. The manuscript was written by Ying Zhou, Ke Liu and Qiyuan Li.
Contributor Information
Qiyuan Li, Email: qiyuanli@xmu.edu.cn.
Ying Zhou, Email: yingzhou@xmu.edu.cn.
References
- 1.Heist R.S., Engelman J.A. SnapShot: Non-small cell lung cancer. Cancer Cell. 2012;21(3):448 e2. doi: 10.1016/j.ccr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Cox A.D. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinehart J. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22(22):4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 4.Stinchcombe T.E., Johnson G.L. MEK inhibition in non-small cell lung cancer. Lung Cancer. 2014;86(2):121–125. doi: 10.1016/j.lungcan.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y. miR-1298 inhibits mutant KRAS-driven tumor growth by repressing FAK and LAMB3. Cancer Res. 2016;76(19):5777–5787. doi: 10.1158/0008-5472.CAN-15-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538(7623):114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J. Suppression of KRas-mutant cancer through the combined inhibition of KRAS with PLK1 and ROCK. Nat Commun. 2016;7:11363. doi: 10.1038/ncomms11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corcoran R.B. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23(1):121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babij C. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71(17):5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- 10.Barbie D.A. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholl C. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Kim H.S. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155(3):552–566. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullis J. The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell. 2014;25(2):181–195. doi: 10.1016/j.ccr.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Luo J. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood K. Prognostic and predictive value in KRAS in non-small-cell lung cancer: A review. JAMA Oncol. 2016;2(6):805–812. doi: 10.1001/jamaoncol.2016.0405. [DOI] [PubMed] [Google Scholar]
- 16.Thorsson V. The immune landscape of cancer. Immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. [e14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding L. Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell. 2018;173(2):305–320. doi: 10.1016/j.cell.2018.03.033. [e10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saltz J. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(1):181–193. doi: 10.1016/j.celrep.2018.03.086. [e7] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoadley K.A. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(2):291–304. doi: 10.1016/j.cell.2018.03.022. [e6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov L.B. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov L.B. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354(6312):618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoadley K.A. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11(3):333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 24.Verhaak R.G. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N., Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network Electronic address, a.a.d.h.e. and N. cancer genome atlas research, integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203. doi: 10.1016/j.ccell.2017.07.007. [e13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seashore-Ludlow B. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5(11):1210–1223. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rees M.G. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12(2):109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iorio F. A Landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnett M.J. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skoulidis F. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dogan S. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riely G.J. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14(18):5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 37.Talikka M. Genomic impact of cigarette smoke, with application to three smoking-related diseases. Crit Rev Toxicol. 2012;42(10):877–889. doi: 10.3109/10408444.2012.725244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaz M. Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell. 2017;32(3):360–376. doi: 10.1016/j.ccell.2017.08.006. [e6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenthal R. DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forbes S.A. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48(6):600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturm D. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 43.Fortin J.P. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torgo L. Data mining with R, learning with case studies. 2010. http://www.dcc.fc.up.pt/~ltorgo/DataMiningWithR Available from:
- 45.Hastie T., T R., Narasimhan B., Chu G. 2017. Impute: Impute: Imputation for microarray data. [Google Scholar]
- 46.Xu T. CancerSubtypes: An R/Bioconductor package for molecular cancer subtype identification, validation and visualization. Bioinformatics. 2017;33(19):3131–3133. doi: 10.1093/bioinformatics/btx378. [DOI] [PubMed] [Google Scholar]
- 47.KD H. 2016. Illumina human methylation 450kanno.ilmn 12.hg19: Annotation for illumina's 450k methylation arrays. [Google Scholar]
- 48.Singh A. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loboda A. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loboda A. Biomarker discovery: Identification of a growth factor gene signature. Clin Pharmacol Ther. 2009;86(1):92–96. doi: 10.1038/clpt.2009.48. [DOI] [PubMed] [Google Scholar]
- 51.Wikerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyons Y.A. Immune cell profiling in cancer: Molecular approaches to cell-specific identification. NPJ Precis Oncol. 2017;1(1):26. doi: 10.1038/s41698-017-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov L.B. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3(1):246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizuki M. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96(12):3907–3914. [PubMed] [Google Scholar]
- 55.Boutin A.T. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 2017;31(4):370–382. doi: 10.1101/gad.293449.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ostrem J.M., Shokat K.M. Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat Rev Drug Discov. 2016;15(11):771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 57.Lito P. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351(6273):604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patricelli M.P. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6(3):316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 59.Rizvi N.A. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mlecnik B. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29(6):610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 61.Tosolini M. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 62.Lechner M.G. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J Immunother. 2013;36(9):477–489. doi: 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bindea G. The prognostic impact of anti-cancer immune response: a novel classification of cancer patients. Semin Immunopathol. 2011;33(4):335–340. doi: 10.1007/s00281-011-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Layton D.M. Loss of ras oncogene mutation in a myelodysplastic syndrome after low-dose cytarabine therapy. N Engl J Med. 1988;318(22):1468–1469. doi: 10.1056/NEJM198806023182215. [DOI] [PubMed] [Google Scholar]
- 65.Neubauer A. Patients with acute myeloid leukemia and RAS mutations benefit most from postremission high-dose cytarabine: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26(28):4603–4609. doi: 10.1200/JCO.2007.14.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66(10):5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 67.Ciszewski W.M. DNA-PK inhibition by NU7441 sensitizes breast cancer cells to ionizing radiation and doxorubicin. Breast Cancer Res Treat. 2014;143(1):47–55. doi: 10.1007/s10549-013-2785-6. [DOI] [PubMed] [Google Scholar]
- 68.Sunada S. Nontoxic concentration of DNA-PK inhibitor NU7441 radio-sensitizes lung tumor cells with little effect on double strand break repair. Cancer Sci. 2016;107(9):1250–1255. doi: 10.1111/cas.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aoki Y., Isselbacher K.J., Pillai S. Bruton tyrosine kinase is tyrosine phosphorylated and activated in pre-B lymphocytes and receptor-ligated B cells. Proc Natl Acad Sci U S A. 1994;91(22):10606–10609. doi: 10.1073/pnas.91.22.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H. Discovery of a BTK/MNK dual inhibitor for lymphoma and leukemia. Leukemia. 2016;30(1):173–181. doi: 10.1038/leu.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information of 35 KRAS-mutant lung cancer cells.
GDSC drugs sensitivity among CS1, CS2 and KRAS wild-type group.
Supplementary material