Abstract
Background
Adapted ketogenic diet (AKD) and caloric restriction (CR) have been suggested as alternative therapeutic strategies for inflammatory, hyperproliferative and neurodegenerative diseases. Pro-inflammatory eicosanoids have been implicated in the pathogenesis of multiple sclerosis since they augment vascular permeability and induce leukocyte migration into the brain. We explored the impact of ketogenic diets on gene expression of biosynthetic enzymes for pro- (ALOX5, COX1, COX2) and anti-inflammatory (ALOX15) eicosanoids in patients with relapsing-remitting multiple sclerosis.
Methods
60 adults were prospectively recruited for this six months randomized controlled trial and the impact of dietary treatment on the Multiple Sclerosis Quality of Life-54 index (ClinicalTrials.gov (NCT01538355) has previously been published. Here we explored 24 patients (8 controls, 5 on CR and 11 on AKD). For statistical analysis we combined the two diet groups to a single pooled treatment group.
Findings
Inter-group comparison indicated that expression of the pro-inflammatory ALOX5 in the pooled treatment group was significantly (p < 0.05) reduced when compared with the control group. Moreover, intra-group comparison (same individuals before and after dietary treatment) suggested significantly impaired expression of other pro-inflammatory enzymes, such as COX1 (p < 0.001) and COX2 (p < 0.05). Finally, pretreatment cross-group analysis revealed a significant positive correlation between expression of pro-inflammatory ALOX5 and COX2 and an inverse correlation of ALOX5 and COX1 expression with the MSQoL-54 index.
Interpretation
Ketogenic diets can reduce the expression of enzymes involved in the biosynthesis of pro-inflammatory eicosanoids. Pharmacological interference with eicosanoid biosynthesis might constitute a strategy supplementing current therapeutic approaches for MS.
Keywords: Multiple Sclerosis, Adapted Ketogenic Diet, Caloric Restriction, Lipoxygenase, Cyclooxygenase, Multiple Sclerosis Quality of Life-54 Instrument (MSQOL-54), Eosinophils, Basophils, Inflammation, Neuroprotection, Eicosanoids
Research in context.
What is already known on this topic
Ketone bodies constitute a vital source of energy during fasting and low carbohydrate alimentation. They mediate neuroprotection in neurodegenerative diseases, stroke and traumatic brain injury. Oxidation of ketone bodies is compensated by an equal reduction in glucose oxidation. COX1, COX2 and ALOX5 are major pro-inflammatory enzymes, which have been implicated in demyelination and inflammation in multiple sclerosis and other chronic inflammatory diseases.
What this study adds
Ketogenic diets and caloric restriction inhibit systemic expression of key enzymes (COX1, COX2, ALOX5) involved in the biosynthesis of pro-inflammatory eicosanoids and multiple sclerosis patients benefit from such dietary treatment.
Alt-text: Unlabelled Box
1. Introduction
Multiple sclerosis is a chronic inflammatory demyelinating disease of the central nervous system (CNS), which is characterized by recurrent and progressive demyelination/remyelination cycles. Owing to the damage of white and gray matter, to axonal destruction and neuro-inflammation this disease leads to disability in young adults and finally to a loss of neuronal functionality [[1], [2], [3]]. Demyelination is accompanied by depletion of oligodendrocyte precursor cells, by loss of mature oligodendrocytes, astrogliosis, and infiltration of macrophages, microglia and T-lymphocytes [4]. Four patterns of morphological destruction have been described for multiple sclerosis lesions [5]. Pattern I and II lesions structurally resemble T-cell-mediated or T-cell plus antibody-mediated autoimmune encephalomyelitis, respectively. In contrast, pattern III and IV lesions are characterized by primary oligodendrocyte damage and degeneration, reminiscent of virus- or toxin-induced demyelination [6].
Fatty acids have been implicated in the pathogenesis of MS and the omega-3 fatty acid alpha-linolenic acid constitutes an inverse risk factor [7]. Whether linoleic acid, arachidonic acid (omega-6 fatty acid) and their metabolites also exhibit protective effects has controversially been discussed in the literature [8,9]. In the experimental autoimmune encephalomyelitis model (EAE) [8,[10], [11], [12]], which has frequently been employed as animal model for human MS, functional inactivation of the fatty acid metabolizing enzymes ALOX5 and ALOX15 leads to exacerbation of the clinical symptoms [13,14]. These data suggest that both enzymes exhibit protective activities. On the other hand, in human MS the arachidonic acid cascade appears to be activated during demyelination and increased expression of ALOX5 and cyclooxygenase-2 (COX2) in lesional areas has been reported [[15], [16], [17], [18]]. Moreover, increased levels of ALOX5 derived leukotrienes and COX1/COX2 derived prostaglandins have been detected in the cerebro-spinal fluid of multiple sclerosis patients [[19], [20], [21], [22], [23], [24]]. These data suggest a potential role for eicosanoid formation in the pathogenesis of MS and other cerebral inflammatory diseases. The molecular basis for the pro-inflammatory role of eicosanoids in these diseases is rather complex but disruption of the blood brain barrier, which leads to more severe infiltration of peripheral immune cells into the CNS, clearly contributes [[25], [26], [27]]. In human MS the ALOX5 pathway has been implicated in microglia activation and neuro-inflammation and thus, this enzyme also contributes to axonal damage and motor neuron dysfunction [20,22]. In early stages of multiple sclerosis inflammatory blood cells gain access into the brain taking advantage of the disruption of the blood–brain barrier [28]. These cells release pro-inflammatory mediators including prostaglandins and leukotrienes [29,30] and these lipid mediators impair inter-neuronal signal transduction, activate cerebral immune cells (microglia) and reduce the blood-brain barrier function of the vascular endothelium even further. Invading neutrophils and monocytes express key enzymes of eicosanoid biosynthesis (ALOX5, COX1, COX2), which leads to augmented formation of pro-inflammatory prostaglandins and leukotrienes in the brain [21,22,26].
In contrast to ALOX5, which constitutes a key enzyme in the biosynthesis of pro-inflammatory leukotrienes, ALOX15 exhibits anti-inflammatory properties [31,32]. In peripheral leukocytes this enzyme is mainly expressed in granulocytes (eosinophils, neutrophils) and has been implicated in the biosynthesis of anti-inflammatory and pro-resolving mediators such as lipoxins, resolvins and protectins [[33], [34], [35]]. In fact, experiments with ALOX15 knockout mice indicated that the clinical symptoms of EAE were significantly worsened when compared with corresponding control animals [13]. However, these findings were contradicted by pharmacological intervention studies, in which baicalein, an ALOX15 inhibitor with potent anti-oxidative properties, significantly attenuated the clinical severity of EAE [36]. The compound inhibited migration of autoimmune T cells into the central nervous system, reduced activation of microglia, decreased production of pro-inflammatory cytokines and induced expression of the peroxisome proliferator-activated receptor (PPAR)β/δ in microglia [36]. Although inhibition of the ALOX15 pathway has not directly been shown in this study the findings suggest an important regulatory role of ALOX15 in CNS inflammation.
For >100 years caloric restriction (CR) and ketogenic diet (KD), which both induce a metabolic switch from glucose to ketone bodies (fatty acid breakdown products) as primary brain energy source, are known for their beneficial effects on pharmaco-resistant seizures in children and on disorders of the energy metabolism of the brain [[37], [38], [39]]. In addition, corresponding human and mouse dietary studies point towards anti-inflammatory and anti-neurodegenerative effects of such diets as suggested by MRI studies and histology [[40], [41], [42], [43]]. The observed diet-induced improvement of neuronal resistance and axonal survival are clearly neuro-protective but the underlying mechanisms remain elusive [44]. To contribute to this discussion we recently initiated a randomized clinical trial (NCT01538355), in which we explored the impact of adapted ketogenic diet (AKD) and CR on quality of life parameters in MS patients. In 2014 the study was finalised and improvement of the health related quality of life index (MSQoL-54) and a decreased Expanded Disability Status Scale (EDSS) was reported [45]. We also observed a positive impact of AKD on blood lipids and on the intestinal microbiome [46,47].
In the present study we explored whether AKD and CR may impact the gene expression patterns of key enzymes of eicosanoid biosynthesis (ALOX5, COX1, COX2, ALOX15) in peripheral leukocyte. We found a significant diet-induced impairment in expression of COX1, COX2 and ALOX5, which are involved in the biosynthesis of pro-inflammatory eicosanoids.
2. Materials and methods
2.1. Clinical trial design
This study was a three-armed parallel grouped, single centered, controlled and randomized clinical pilot trial. The permuted-block randomization was generated online at http://randomization.com. An investigator blind to the randomization plan determined the patients' randomization number before they underwent randomization. This study was registered at http://www/clicaltrials.gov as NCT01538355.
2.2. Patients
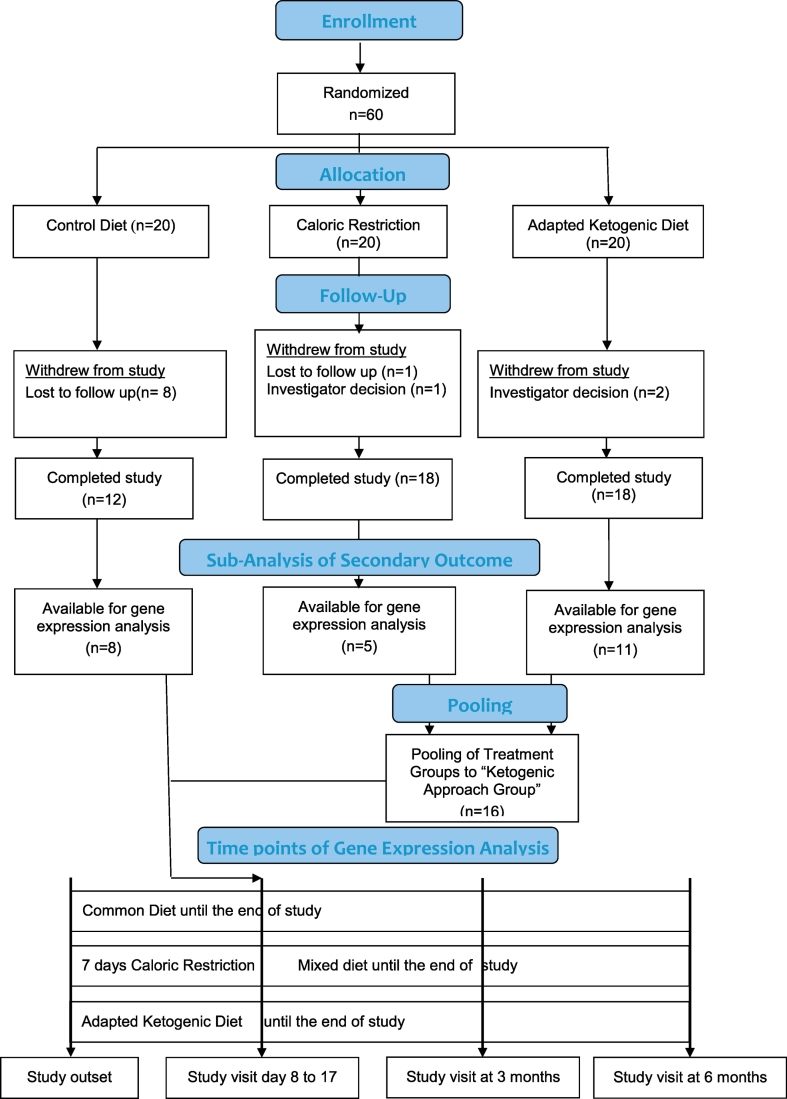
Patients were randomly allocated to three different experimental groups: i) AKD for 6 months or ii) CR for 7 days followed by common carbohydrate intake diet for 6 months or iii) CD (control diet) for 6 months. Before starting the dietary treatment blood was drawn from all participants for intra-group control purpose. The time interval of the initial blood removal did not exceed 2 months prior starting the dietary intervention and after initial blood removal the patients stayed on their regular diet. After the start of the dietary intervention blood was drawn again within the first month after changing the diet. At day 8 blood was taken from CR group patients and from the AKD and the CD patients blood was drawn between day 9 and day 17 from the start of the dietary intervention. Unfortunately, because of patients' dropouts and a loss of blood samples during the workup procedure expression profiles for only 24 patients (8 controls, 5 on CR and 11 on AKD) could be established (Scheme 1). The patients recruited originally met the following criteria: Age ≥ 18 ≤ 67 years, stable disease modifying therapy (DMT) for at least six months prior to inclusion, no DMT for at least six months or naive to therapy, expanded disability status scale (EDSS) ≤ 6.5 and BMI > 18 < 45. Exclusion criteria were primary or secondary progressive forms of multiple sclerosis, clinically relevant heart, lung, liver, and kidney diseases, pregnancy or breast-feeding, other neurologic disorders, cancer, weight loss therapy in the month prior to screening, relapse or steroid pulse therapy <30 days prior to screening, diabetes or other metabolic defects, bulimia, anorexia and drug abuse.
Scheme 1.
Flow chart of study design.
2.3. Study settings
A local ethics committee approved the study and all participants gave informed written consent according to the 1964 Declaration of Helsinki. Relapsing remitting multiple sclerosis patients fulfilling the current panel criteria were prospectively recruited from all over Germany and their health status was assessed at the ECRC [48].
2.4. Interventions
Multiple sclerosis patients who met the inclusion criteria (n = 60) were randomly assigned to three study dietary interventions (n = 20 per groups. To evaluate the food intake we used a 115 item dietary self-record with additional gaps for unlisted and individual foods or liquids and quantities prospectively over a period of 7 days before baseline and between all other visits (Optidiet software Version 5.1 GOE mbH, Büro Linden, Linden, Germany).
2.5. Caloric restriction diet (CR)
Single cycle of CR consist of Day 1 – pre-fasting followed by Day 2–8 – very low calorie diet. Day 1-prefasting consists of a 800 kcal (about 40% of normal caloric intake) monodiet (fruit, rice, or potatoes) by preference of individuals. CR consisted of 100 ml vegetable broth or vegetable juice with 1 tablespoon of linseed oil 3 times daily, plus additional calorie-free liquids. The daily calorie intake was predefined with 200–350 kcal (10–18% of normal caloric intake). We advised patients to drink 2–3 of unsweetened fluids each day (water, and herbal teas). After the 7-day fasting period solid foods were stepwise reintroduced after assessment on day eight.
2.6. Adapted ketogenic diet (AKD)
Patients received AKD for 6 months from study outset. The established therapeutic models of ketogenic diet in children were adapted by Dr. Bock in regard to increase feasibility of traditional, modified and Atkins ketogenic diet in adult patients. Thus AKD was adapted to achieve i) a modest ketosis (≥500 μmol/l ß-hydroxybutyrate) in blood, self-measure after dinner twice a week (FreeStyle Precision, Abbott Diabetes Care Ltd.), ii) a modest ketosis (≥500 μmol/l acetoacetate) in urine, self- measure after dinner once a week (Ketostix, Bayer Consumer Care AG) and iii) to maintain patient compliance. Patients received a booklet (Dr. Bock protocol) with meal suggestions over 28 balanced days and were encouraged not to limit fat and vegetables ingestion. We adapted the common ketogenic diets to an average daily intake of <50 g carbohydrates, >160 g fat (not exceeding omega 6 vs omega 3 ratio 2:1), average protein intake ≤100 g per day, high fibers intake. Patients received detailed information about nutritional facts, glycemic load and learned how to handle carbohydrates by an experienced nutritional coach.
2.7. Control diet (CD)
Patients on CD met the criteria of a regular diet in German population as described in the “National Nutrition Survey II” (https://www.mri.bund.de/de/institute/ernaehrungsverhalten/publikationen/forschungsprojekte/nvsii/). We advised patients to stay on their regular diet.
2.8. Primary clinical outcome measures
To judge the clinical outcome of this study we quantified the Multiple Sclerosis Quality of Life-54 questionnaire (MSQOL-54), which consists of 54 items. Two composite scores quantifying physical and mental health were calculated from the outcome of quantification of the following 12 independent scoring parameters: Physical Function, Health Perception, Energy/Fatigue, Role Limitations Physical, Pain, Sexual Function, Social Function, Health Distress, Overall Quality of Life, Emotional Well Being, Role Limitations Emotional, Cognitive Function. All patients including the control individuals completed the questionnaire before the beginning of dietary treatment as well as 3 and 6 months after the first blood removal. In addition, all members of the CR group were assessed before breaking the fasting at day 8. The diet-induced alterations in the MSQOL-54 index, which constitute the primary outcome measure of this study, have been reported before [45]. This paper only involves secondary outcome measures and these data have recently been generated and were not available when the original report was filed [45].
2.9. Secondary outcome measures
As secondary outcome measure the expression levels of selected biosynthetic enzymes of eicosanoid biosynthesis (ALOX5, ALOX15, COC1, COX2) in peripheral leukocytes were quantified. For this purpose RNA was extracted from peripheral blood. From the 48 patients who finished the study the target genes could only be determined for 24 since the blood samples of the remaining patients got lost during the workup procedure and direct pairing was not possible.
2.10. Gene expression analyses
We collected whole blood from all patients in PAXgene blood RNA tubes. The samples were stored at −80°. RNA isolation was performed using PAX gene RNA extraction kit. The extracted total RNA was quantified (absorbance at 260 nm) and a aliquot was then reversely transcribed into cDNA. The target cDNAs were subsequently quantified by qRT-PCR employing internal (GAPDH) and external (amplicons for the genes of interest and the internal standard) amplification standards. For this purpose the amplicons were prepared by RT-PCR, subcloned into a suitable cloning vector and amplified to obtain external standard stock solutions. The real-time PCRs were carried out on a DNA Engine Opticon® 2 (MJ Research, Inc, Biozym), using the QuantiTect SYBR Green PCR Kit from Qiagen, according to the recommendations of the vendors. For amplification of the target genes the primer combinations specified in Supplemental Table S1 were used and the following PCR protocol was applied: 15-min hot start at 95 °C, followed by 40 cycles of denaturation (30 s at 94 °C), annealing (30 s at 60 °C) and synthesis (30 s at 72 °C, total volume of 20 μl). For melting curve analysis, the temperature was elevated slowly from 60 °C to 95 °C. Data were acquired and analyzed with the Opticon Monitor software (version 2). The amplification kinetics were recorded as sigmoid progress curves, for which the fluorescence was plotted against the number of amplification cycles. The threshold cycle number (CT) was used to define the initial amounts for each template. CT was the first cycle, for which a detectable fluorescence signal was observed. Homogeneity of the amplified PCR products was tested by melting-curve analysis. The initial template concentration varied between 2 × 103 and 106 copy numbers and a reaction volume of 20 μl was adjusted.
To confirm the expression differences of our target genes on the protein level we carried out immunoblotting for ALOX15 and COX2. Unfortunately, owing to the large concentrations of hemoglobin in the protein extracts of the blood cells we were unable to detect specific immunoreactive bands in the expected molecular weight range. Our attempts to specifically remove the hemoglobin from the protein extracts were not successful and thus, these experiments were not very informative.
2.11. Serum biomarkers and anthropometrics
Prior to blood collection all patients were on an overnight fast and samples were always taken at the same time (±1 h). Blood count, and glucose were analyzed according to international standards at all visits. Serum and plasma were stored at −80 °C. For the quantitative determination of 3-hydroxybutyrate we used the cyclic enzymatic Wako Autokit 3-HB and Insulin was measured with PerkinElmer AutoDELFIA time-resolved fluoroimmunassay (B080-101). Body weight, fat and lean masses were specified by Air-Displacement Plethysmography (Bod Pod, Life Measurements, Inc. Concord, CA), BMI was calculated as weight (kg)/height2(m).
2.12. Statistical analysis
To test uniformity of the variables baseline characteristics of the three intervention groups (AKD, CR, CD) were first compared using the non-parametric Kruskal-Wallis test. Owing to the small n-number of the CR group (n = 5) reliable statistical analyses was not possible. To overcome this problem we combined the to two dietary intervention groups (CR and AKD) to a joint ketogenic approach (KA) group. Two sets of experimental data justified this data combination: i) The two intervention groups (CR and AKD) showed similar results in the gene expression experiments: ii) Caloric restriction and the employed adapted ketogenic diet induce similar metabolic alterations, which are indicated by an increase in peripheral keto bodies. Combination of the gene expression data from the two dietary intervention groups did increase the statistical power of our study. Under these conditions we observed statistically significant differences between the pooled treatment group and the control group. Such significant differences were not detected when the two intervention groups were separately evaluated. All differences between the control group and the ketogenic approach (KA) group were calculated using analysis of covariance (ANCOVA) to adjust for baseline dependencies. ANCOVA was reported as the most reliable method in trials with baseline and follow up measurements [49]. For intra-group comparison (comparison of a corresponding variable for an individual of an intervention group before and after dietary intervention) before and after dietary intervention) we used the paired Students t-test. Baseline associations between variables were assessed using Spearman's rank correlation coefficient (rs). All tests should be understood as constituting exploratory data analysis, as no previous power calculation and adjustments for multiple testing were made. All graphs are based on mean and standard error of the mean (SEM) data. The test level for statistical significance of differences between (inter-group comparisons) and within (intra-group comparison) the treatment groups was defined as p = 0.05 (two-sided) for all tests. For statistical analyses the following software was used: SPSS, version 21 (IBM, Armonk, New York, US) and Graph Pad Prism, Version 5.04 (GraphPad Software, CA, US).
3. Results
3.1. Situation prior to dietary intervention
3.1.1. Baseline inter-group comparison: no differences in readout parameters
Patients demographics did not differ significantly when the different intervention groups were compared (Table 1). The Disease Modifying Treatments (DTM) (Glatirameracetate and Interferone beta) were significantly different between the groups but this is not relevant because all participants were stable on their DMT at least six months prior to inclusion an did not change the DMT during the study. When we compared the expression levels of the genes of interest (ALOX5, ALOX15, COX1, COX2) we did not observe significant differences between the three groups prior to dietary interventions (Table 1). These data indicate that the different dietary habits of the patients involved in the study did not significantly impact the expression patterns of the genes of interest. Nevertheless, since even small differences in baseline expression levels might impact the basic conclusions, we used baseline expression as covariate for all inter-group evaluations. In order to control our data for plausibility we first analyzed (Table 2; Supplemental Fig. S1a–d; Supplemental Table 2) the cross-group correlation (involving all individuals of all experimental groups at baseline) with respect to the following readout parameters:
Table 1.
Baseline differences between the three dietary groups. Data are mean (SD), number (%) or median (inter quartile range). Baseline data were available for 24 patients (CD = control diet, CR = caloric restriction diet, AKD = adapted ketogenic diet).
| Baseline characteristics | CD (n = 8) | SD IQR | CR (n = 5) | SD IQR | AKD (n = 11) | SD IQR | ap-value |
|---|---|---|---|---|---|---|---|
| Age in years | 47.5 | 10.8 | 36.2 | 12.8 | 43.1 | 8.8 | 0.3618 |
| Gender F/M | 6/2 (75/25) | 4/1 (80/20) | 10/1 (91/9) | 0.6514 | |||
| Expanded disability status scale | 2.8 | 1,5–4 | 2 | 1.8–3.5 | 3.1 | 2.0–3.5 | 0.838 |
| Disease Duration in years | 7.6 | 2.9 | 3.5 | 3.1 | 6.1 | 4.5 | 0.1864 |
| Relapse rate 12 months prior study outset | 0.13 | 0.4 | 0.4 | 0.5 | 0.27 | 0.5 | 0.5372 |
| No immune modulating drugs | 2 (25) | 0 | 4 (36) | 0.313 | |||
| Glatirameracetate | 5 (63) | 1 (20) | 1 (9) | 0.0413 | |||
| Interferon Beta | 0 | 4 (80) | 3 (27) | 0.0102 | |||
| Fingolimod | 0 | 0 | 2 (18) | 0.2907 | |||
| Natalizumab | 1 (12.5) | 0 | 1 (9) | 0.7343 | |||
| BMI | 27.6 | 8.4 | 26.4 | 7.1 | 25.9 | 4.3 | 0.9824 |
| Percent Body Fat | 38.2 | 12.7 | 32.1 | 12.8 | 35.6 | 9.8 | 0.5518 |
| Fasting Blood Sugar mmol/l | 4.5 | 0.6 | 4.5 | 0.4 | 4.3 | 0.6 | 0.779 |
| Insulin mU/l | 6.8 | 3.6 | 9.4 | 2.7 | 7.8 | 3.0 | 0.1013 |
| ß-Hydroxy-butyrat mmol/l | 94.04 | 54.85 | 167.1 | 155.7 | 123.2 | 78.03 | 0.7468 |
| Neutrophils/nl | 3.2 | 0.7 | 3.9 | 2.5 | 3.7 | 1.6 | 0.8766 |
| COX1b | 19687 | 6577 | 17269 | 6776 | 21134 | 8610 | 0.7108 |
| ALOX5b | 213708 | 65541 | 228977 | 82679 | 279228 | 105975 | 0.4555 |
| COX2b | 13282 | 5978 | 18312 | 7435 | 20219 | 5913 | 0.0569 |
| ALOX15b | 74814 | 59677 | 63052 | 46475 | 52652 | 28327 | 0.8139 |
non parametric Kruskal Wallis Test to compare between all three groups.
Data are mean mRNA copies/106 GAPDH mRNA copies.
Table 2.
Baseline analysis of cross-group (n = 24) correlation between the target gene expression and disease related or subject reported parameters.
| Parameter | ALOX5 |
ALOX15 |
COX1 |
COX2 |
EDSS |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| COX2 | Neutrophils | Physical Function | Eosinophils | Basophils | Lympho-cytes | Physical Health | Physical Function | EDSS | Physical Function | |
| aCorrelation Coefficient | 0.618 | 0.599 | −0.412 | 0.891 | 0.504 | 0.441 | −0.563 | −0.255 | 0.286 | −0.734 |
| p (2-tailed) | .001 | .002 | .045 | <.000001 | .012 | .031 | .012 | .229 | .176 | <.0001 |
EDSS = Expanded Disability Status Scale.
Spearmans Rho.
3.1.2. The expanded disability status scale (EDSS) did inversely correlate with Multiple Sclerosis Quality of Life-54 (MSQOL-54) index
Initially we compared the two clinically most relevant readout parameters (MSQOL-54 and EDSS) quantified in our study. The MSQOL-54 score, which was determined by a questionnaire, mirrors the degree of subjective wellbeing of the patients. The EDSS score was calculated on the basis of the outcome of neurological examination and quantifies the degree of neurological disability. We found a highly significant inverse correlation (p = 0.00004, r = −0.734) between these two clinical scores (Table 2 + Supplemental Fig. S1a) confirming that patients reporting limitations in their quality of life suffer from more severe neurological dysfunctions [50].
3.1.3. Positive correlation between ALOX5 and COX2 gene expression
At baseline we observed a significant cross-group correlation (p = 0.001, r = 0.618) between the expression of COX2 and ALOX5 in peripheral leukocytes (Table 2 + Supplemental Fig. S1b). Both enzymes catalyze key reactions in the biosynthesis of pro-inflammatory mediators (ALOX5 is the key enzyme of leukotriene formation and COX2 catalyzes the initial reaction in the biosynthesis of pro-inflammatory prostaglandins) and thus, a parallel upregulation of their expression is biologically meaningful (see discussion).
3.1.4. Expression levels of ALOX5 and ALOX15 positively correlated with the counts of different white blood cells
Next, we carried out cross-group correlation analysis for the expression levels of pro- and anti-inflammatory target genes (ALOX15, ALOX5, COX1, COX2) and the counts of circulating blood cells (neutrophils, eosinophils and basophils). Here, we observed a statistically significant positive correlation between ALOX5 expression and the neutrophil count (p = 0.002, r = 0.599, Table 2 + Supplemental Fig. S1c). This correlation is not surprising since among peripheral blood cells neutrophils are a rich source of ALOX5 expression. Furthermore, we detected a significant positive correlation between ALOX15 expression and the counts of eosinophils (p = 5.3−9, r = 0.891, Supplemental Fig. S1d), basophils (p = 0.012, r = 0,504, Supplemental Fig. S1e) and lymphocytes (p = 0.031, r = 0.441 Supplemental Fig. S1f). The highly significant positive correlation of ALOX15 expression and the eosinophil count can be explained by the fact that among all peripheral leukocytes eosinophils are the richest source of ALOX15 expression [[51], [52], [53]].
3.1.5. The quality of life index MSQOL-54 did inversely correlate with the expression of pro-inflammatory genes
Finally, we correlated the MSQOL-54 index as clinically most relevant parameter of patients' personal wellbeing with the expression levels of key enzymes of eicosanoid biosynthesis in a cross-group approach. In theory, a high MSQOL-54 index, which indicates that the patients feel well, should inversely correlate with the expression levels of classical pro-inflammatory enzymes. We found that ALOX5 expression did inversely correlate with the MSQOL-54 index (p = 0.045, r = −0.412). Similarly, we observed a significant inverse correlation (p = 0.012, r = −0.563) between COX1 expression and the MSQOL-54 index (Table 2 + Supplemental: Fig. S1g, h and Table S2). A trend for an inverse correlation was also observed when COX2 expression was correlated with the MSQOL-54 index but owing to the relatively small n-numbers this correlation did not reach the level of statistical significance (Supplemental: Table S2 + Fig. S1i).
The EDSS index was expected to positively correlate with the expression levels of pro-inflammatory enzymes as rising EDSS scores reflect increasing neurological dysfunction. We observed for all target genes a statistical trend for positive correlation with the EDSS index but owing to the relatively small n-numbers these correlations did not reach the level of significance (Supplemental: Table S2 + Fig. S1j + k). The scattered plots visualizing the correlation of the different readout parameters are given in supplemental Fig. S1.
3.2. Expression alterations of target genes as consequence of dietary intervention
3.2.1. Intra-group comparison: ketogenic diets induced significant reductions in the expression of pro-inflammatory genes in peripheral blood leukocytes
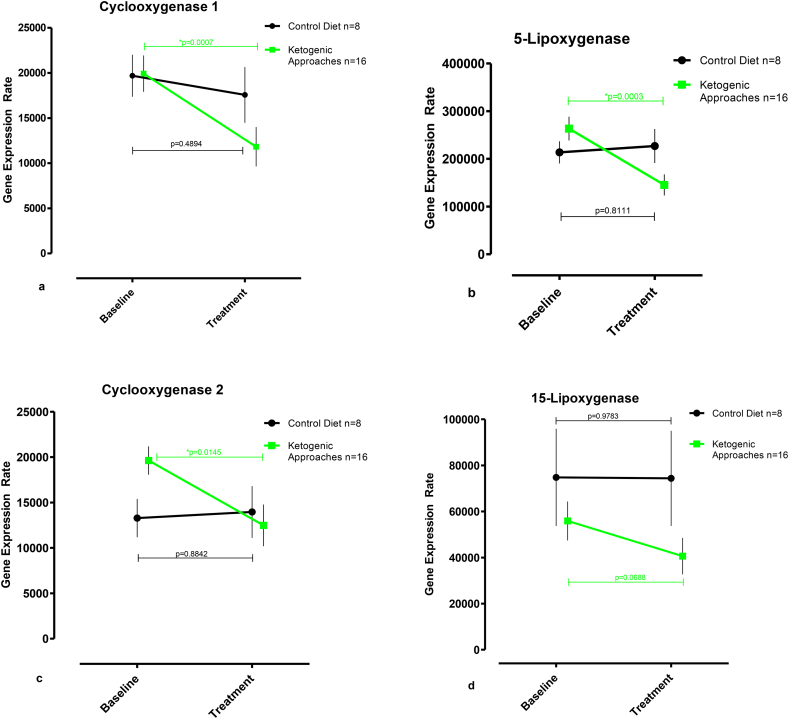
The data summarized in the supplemental Fig. S2a–d and in the supplemental Table S3 suggest that treatment of the patients with the two different types of ketogenic diets (CR and AKD) down-regulated the expression levels of all target enzymes. In fact, after dietary intervention the gene expression rates are always smaller than in patients with no dietary intervention. In contrast, in the control group the gene expression rates before study onset and during the study are always similar. To improve the statistical power of our study we pooled the CR + AKD groups to one “ketogenic approach” (KA) group. For intra-group comparison we quantified the readout parameters of all individuals in the treatment group before and after dietary intervention. Similarly, we quantified the corresponding readout parameters of the individuals in the control group. The members of the control group did not receive a special diet and thus, there should not be any differences in the gene expression levels before and after the intervention period (Fig. 1a–d). As expected there was no significant difference in the expression level of any gene product in the control group. This finding excludes unspecific dietary effects on the expression of the target genes. When we profiled the expression of Cyclooxygenase-1 (Fig. 1a) within the pooled ketogenic approach group (KA) we found a significant decrease in the expression levels from 19,927 COX1 mRNA copies per 106 GAPDH copies at baseline (no dietary intervention) to 11,824 COX1 mRNA copies per 106 GAPDH copies (p < 0.001; 95% CI -12,179 to −4026) after intervention. A similar decline was also observed for expression of pro-inflammatory 5-Lipoxygenase from 263,524 ALOX5 mRNA copies per 106 GAPDH copies at baseline to 145,441 ALOX5 mRNA copies per 106 GAPDH copies (p < 0.0005; 95% CI -172,249 to −63,918) after the treatment period (Fig. 1b). Moreover, we observed a significant drop in the expression level of pro-inflammatory Cyclooxygenase-2 (Fig. 1c) from 19,623 COX2 mRNA copies per 106 GAPDH copies at baseline to 12,484 COX2 mRNA copies per 106 GAPDH copies (p < 0.05; 95% CI 12,643 to 1634) on diet. In contrast, expression of 15-Lipoxygenase in peripheral leucocytes was not significantly altered by dietary intervention (Fig. 1d).
Fig. 1.
Intra-group comparison - Ketogenic approaches lower the gene expression of pro-inflammatory enzymes involved in eicosanoid biosynthesis. (a–d) Alterations of gene expression induced by caloric restriction/adapted ketogenic diet (ketogenic approaches) when compared with common western diet (control diet). The steady state concentrations (copy numbers of target gene mRNA per 106 copies of GAPDH mRNA) for the following transcripts were evaluated: a) COX1, b) ALOX5, c)COX2, and d) LOX15. Data represent mean ± standard error of the mean (SEM) and were measured at baseline and retested between day 8 and 17 after the onset of dietary intervention. Horizontal bars indicate paired t-Test analysis.
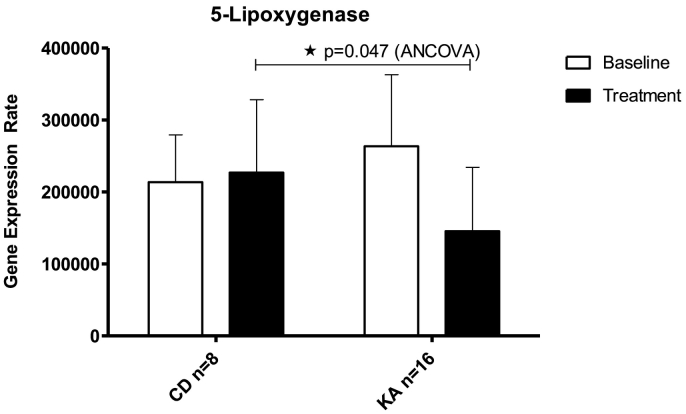
3.2.2. Inter-group comparison: ketogenic diets lower ALOX5 expression in comparison to normal diet
Next, we compared the expression levels of the target genes between the members of the control group and the pooled dietary intervention groups (KA). In this inter-group comparison we found that the difference in COX2 expression between the members of the control group and the members of the pooled dietary intervention groups did not reach the level of statistic significance (Table 3 + Supplemental Fig. S3a). Similar observations were made for ALOX15 (Table 3 + Supplemental Fig. S3b). However, we observed (Fig. 2 + Table 3) a significant decrease in the gene expression levels of ALOX5 when the members of the control group were compared with the members of the pooled KA group (adjusted difference 89,010 ± 42,201; 95% CI 1249 to 176,772 ALOX5 mRNA copies/106 GAPDH mRNA copies, p < 0.05). In this inter-group comparison we also observed a statistic trend for declined expression of COX1 (adjusted difference 5890 ± 3238; 95% CI 843 to 12,623 COX1 mRNA copies/106 GAPDH mRNA copies) but the p-value of 0.08 suggested only borderline significance (Table 3 + Supplemental Fig. S3c).
Table 3.
Impact of ketogenic approaches (KA) on expression of pro−/anti-inflammatory enzymes when compared with common western diet (CD). Adjusted comparison between pooled ketogenic approaches (caloric restriction + adapted ketogenic diet) and control group (common western diet). Data were available for 24 patients (CD = control diet, KA = ketogenic approaches) and were measured at baseline and retested between day 8 and 17 after the onset of dietary intervention. Mean mRNA copies/106 GAPDH mRNA copies (SD).
| Gene | Timepoint | CD (n = 8) | SD | KA (n = 16) | SD | ap-value |
|---|---|---|---|---|---|---|
| COX1 | Baseline | 19687 | 6577 | 19927 | 8068 | |
| Treatment | 17564 | 8748 | 11824 | 8717 | 0.083 | |
| ALOX5 | Baseline | 213708 | 65541 | 263524 | 99442 | |
| Treatment | 226943 | 101181 | 145441 | 88723 | 0.047 | |
| COX2 | Baseline | 13282 | 5978 | 19623 | 6235 | |
| Treatment | 13953 | 8060 | 12484 | 9181 | 0.842 | |
| ALOX15 | Baseline | 74814 | 59677 | 55902 | 33700 | |
| Treatment | 74392 | 58376 | 40614 | 31658 | 0.143 |
Analysis of covariance (ANCOVA) to adjust for baseline dependencies.
Fig. 2.
Ketogenic approaches (KA) lower ALOX5 expression in the inter-group comparison to common western diet (CD). Comparison of ALOX5 gene expression during caloric restriction and adapted ketogenic diet (pooled ketogenic approach group, n = 16) with common western diet (control diet). Data were measured at baseline and retested between day 8 and 17 after the onset of dietary intervention. See also supplemental Fig. S2a–c for COX1, COX2, and ALOX15 expression results. Data represent mean ± standard error of the mean (SEM). *p ≤ 0.05, analysis of covariance (ANCOVA) to adjust for baseline dependencies.
3.3. Changes in body composition and blood parameters change as consequence of ketogenic intervention
3.3.1. Inter-group comparison indicates no differences in baseline readout parameters
First, we compared BMI, fasting blood sugar, ketone bodies (ß-hydroxy-butyrate), insulin and neutrophil counts between the three experimental groups before the intervention. Here we did not observe any significant differences (Table 1 + Supplemental Fig. S4a–e).
3.3.2. Ketogenic approaches reduce BMI and increase ketone body levels in comparison to normal diet
Caloric restriction and an adapted ketogenic diet are known to induce weight loss, to increase the levels of ketone bodies in the blood, and to reduce the circulating insulin and glucose concentrations. These metabolic alterations are characteristic for a diet defective in carbohydrates. We compared BMI, blood glucose, ß-hydroxy-butyrate, insulin and neutrophil levels between control individuals and those patients receiving a ketogenic diet (KA) (Table 4 + Supplemental Fig. S4). Here we observed significantly elevated beta hydroxybutyrate blood levels (adjusted difference 1442.3 mmol/l ± 537.7; 95% CI for difference 324.1 to 2560.4, p < 0.05) and a significant loss in BMI (adjusted difference −0.78 ± 0.3; 95% CI for difference −1.3 to −0.2, p < 0.01).
Table 4.
Impact of ketogenic approaches (KA) on anthropometric, metabolic and immunologic outcome when compared with common western diet (CD). Adjusted comparison between pooled ketogenic approaches (caloric restriction + adapted ketogenic diet) and control group (common western diet). Data were available for 24 patients (CD = control diet, KA = ketogenic approaches).
| Parameter | Timepoint | CD (n = 8) | SD | KA (n = 16) | SD | ap-value |
|---|---|---|---|---|---|---|
| BMI | Baseline | 27.61 | 8.4 | 26.05 | 5.1 | |
| Treatment | 27.62 | 8.3 | 25.31 | 5.0 | 0.008 | |
| Fasting Blood Sugar mmol/l | Baseline | 4.52 | 0.6 | 4.40 | 0.5 | |
| Treatment | 4.52 | 0.5 | 3.99 | 1.1 | 0.241 | |
| Insulin mU/l | Baseline | 5.75 | 3.6 | 8.288 | 2.9 | |
| Treatment | 5.75 | 2.4 | 5.481 | 2.8 | 0.493 | |
| ß-Hydroxy-butyrat mmol/l | Baseline | 94.04 | 54.9 | 136.9 | 104.7 | |
| Treatment | 92.53 | 106.8 | 1439.2 | 1451.3 | 0.014 | |
| Neutrophils/nl | Baseline | 3.241 | 0.7 | 3.763 | 1.9 | |
| Treatment | 3.459 | 0.9 | 2.952 | 1.1 | 0.176 |
Analysis of covariance (ANCOVA) to adjust for baseline dependencies.
3.3.3. Distinct effects of CR and AKD on BMI, insulin and ketone body levels
Under our experimental conditions CR was more efficient in reducing BMI, inducing ketosis and lowering insulin levels when compared with AKD (Supplemental Table 4). In fact, an inter-group comparison of the two types of ketogenic diets revealed three interesting results: i) CR induced a significantly stronger reduction in BMI (adjusted difference −1.248 ± .184; 95% CI for difference −1.632 to −.865, p < 0.001). ii) CR induced significantly higher beta hydroxybutyrate concentration (adjusted difference 2131.1 mmol/l ± 486.6; 95% CI for difference 1116 to 3146.3, p < 0.001). iii) CR induced significantly lower blood insulin levels (adjusted difference −3.602 mU/l ± 1.222; 95% CI for difference −6.201 to −1.103, p < 0.01).
4. Discussion
This study is – to the best of our knowledge – the first study that investigated the impact of ketogenic diets (CR and AKD) on the gene expression profiles of key enzymes involved in eicosanoid biosynthesis (ALOX5, ALOX15, COX1 and COX2) in multiple sclerosis patients. Employing a strictly quantitative real-time PCR approach we were able to explore whether expression of selected target genes were up- or downregulated in peripheral blood leukocytes during dietary intervention in MS patients. One such gene with obvious implications for MS pathology is ALOX5, the expression of which was significantly downregulated in peripheral blood leukocytes during dietary intervention. The expression product of the ALOX5 gene (5-lipoxygenase) catalyzes the two initial steps in the conversion of arachidonic acid to biologically active leukotrienes. As classical pro-inflammatory mediators leukotrienes induce an increase in the vascular permeability during inflammation, trigger leukocyte migration and improve leukocyte chemotaxis. Moreover, leukotrienes induce tissue edema, play an important role in host defense mechanisms against pathogens and promote disruption of the blood brain barrier [27,[54], [55], [56], [57], [58]].
However, the precise role of ALOX5 in the pathogenesis of MS is far from clear. In fact, controversial data have been obtained in mouse experimental MS models (Emerson and LeVine, [13]) and in MS patients (Mirshafiey and Jadidi-Niaragh, [18]). But even in the mouse EAE model controversial roles of ALOX5 have been reported. Inactivation of the ALOX5 gene leads to exacerbation of the neurological symptoms (Emerson and LeVine, [13]) suggesting a protective role of ALOX5 in this animal disease model. On the other hand, pharmacological inhibition of the ALOX5 pathway delayed the onset of EAE and reduced disease severity (Marusic et al., [14]). These data suggest a promoting role of ALOX5 in EAE pathogenesis. This conclusion was supported by more recent finding that leukotriene B4 (LTB4), the biosynthesis of which requires a functional ALOX5, promotes Th17 lymphocyte differentiation and improves recruitment of these cells into the brain of EAE mice [59,60]. According to these data ALOX5 contributes to neuroinflammation in the EAE model. In summary, it may be concluded that the role of ALOX5 in the pathogenesis of MS is still a matter of discussion and that additional experimental data are needed to characterize the precise role of this enzyme in inflammatory cerebral diseases.
Cyclooxygenase-isoforms (COX1, COX2) are key enzymes in the biosynthesis of pro-inflammatory prostaglandins since these two enzymes catalyze the rate-limiting step in the formation of these lipid mediators [[61], [62], [63]]. COX1 is constitutively expressed in many cells and tissues whereas expression of COX2 is strongly upregulated in activated inflammatory cells [64]. In neurons, COX2 expression is induced by glutamate receptor agonists and inhibition of COX2 exhibits beneficial effects in the mouse EAE model [14,23,[65], [66], [67], [68], [69]]. In humans another group reported that Th17 cells from MS patients expressed higher levels of prostaglandin EP2 receptors and that overexpression of EP2 in Th17 cells from healthy adults induced a pathogenic Th17 gene expression profile [70]. Thus the development of pathogenic autoreactive Th17 cells seems to be directly linked to PGE2 signaling. Interestingly PGE2 exerts anti-inflammatory properties by EP4 receptor-mediated action [71]. Moreover, a Th17/Treg imbalance is characteristic of childhood intractable epilepsy and was corrected by ketogenic diet [72].
Inhibitors of COX-isoforms are frequently prescribed as non-steroidal anti-inflammatory drugs (NSAIDs) but they may also exhibit neuroprotective activity [66,67,73,74]. Because of their pharmacological profile NSAIDs have been employed for the management of side effects, when interferon beta is employed for multiple sclerosis treatment. Moreover, the classical COX1 inhibitor aspirin is frequently used to limit the severity of multiple sclerosis related fatigue and premenstrual associated pseudo-exacerbations [[75], [76], [77]]. Although COX1 in contrast to COX2 may not be considered a classical pro-inflammatory enzyme it contributes to the biosynthesis of pro-inflammatory prostaglandins [63]. Consequently, patients with high ALOX5 and COX1 expression levels are likely to develop more severe inflammatory symptoms.
The use of COX-2 inhibitors for treating multiple sclerosis has currently been challenged owing to cardiovascular side effects of some COX-2 inhibitors [78,79]. Our intra-group comparisons indicated that expression of COX1 and COX2 in human peripheral leukocytes is downregulated by ketogenic diets and these data may explain the beneficial effects of these types of diet for multiple sclerosis patients. Moreover, our data are consistent with previous findings of other groups reporting that AKD impaired systemic and local COX2 gene expression [[80], [81], [82]]. Interestingly, we found an association of COX- and LOX-isoforms expression with the MSQOL-54 index, which did robustly correlate with the neurological symptoms (EDSS index), which does not mean a causal connection between expression levels of our target genes and disease symptoms [50]. Furthermore, previous cross-sectional and longitudinal studies have indicated that the MSQOL-54 index did correlate with lesion burden and brain volume measures even in early stages of MS [83,84]. Although we did not directly measure such morphological readout parameters it might well be that the expression of COX/ALOX-isoforms in peripheral leukocytes may have predictive value for the extent of such morphological alterations. However, to definitely answer this question a more powerful case-control study must be carried out, in which these morphological characteristics are directly determined.
We recently reported that ketogenic diets in MS patients improved the MSQOL-54 index and lowered the peripheral lymphocyte count [45]. These data suggest that this therapeutic approach may silence the intensity of the immune response, which might slow down the progress of the disease. Although our intra-group comparisons suggest that expression of the classical pro-inflammatory enzymes COX2 and ALOX5 in peripheral leukocytes is silenced by ketogenic diets these data have to be interpreted with caution. In fact, our inter-group comparisons did not confirm these data for COX2 and thus, additional studies are required to prove or disprove this hypothesis. On the other hand, our observations are consistent with the outcome of previous studies [[80], [81], [82]], which indicate that ketogenic diet reduces the activity of pro-inflammatory enzymes. Dual inhibition of the COX/LOX pathways in neuroinflammatory diseases induced promising alterations [[85], [86], [87]] but the future will tell whether these promises can be confirmed in large clinical trials. The mechanisms on how ketogenic diets reduce COX/LOX gene expression remain still elusive but it is likely that during the initial mitochondrial and cellular adaptation to ketogenic diets ROS is involved in the regulation of gene expression of several target enzymes [80,88]. Thus a complex interaction of ROS as effector molecules to orchestrate gene networks may regulate major intracellular signaling cascades and cellular homeostatic mechanisms under ketogenic diets [80].
Our data have indicated a significant correlation between the expression level of ALOX5 and the blood counts of eosinophils, basophils, neutrophils and lymphocytes. These data are not surprising since all leukocytic cell types are rich sources for this classical pro-inflammatory enzymes [58,61,62,89]. The ketogenic diets did not suppress circulating neutrophils (Table 4 Supplemental Table S4), which could have been also an explanation of an ALOX5 decrease.
The steady state concentration of ALOX5 mRNA in peripheral leukocytes is higher than COX1 and COX2 mRNA levels. This is an interesting finding but it remains to be explored whether a similar ratio can be confirmed on the protein level. To address this question we performed immunoblot experiments for ALOX15 and COX2 but were not able to obtain specific signals in the expected molecular weight range. In these experiments we struggled with the large hemoglobin concentrations in the blood cell samples, which interfered with immunostaining. Our attempts to specifically remove the hemoglobin form the samples were not successful and thus, for the time being it remains unclear whether the mRNA levels quantified in this study mirror the in vivo activities of the corresponding enzymes. To answer this question quantitative metabolome studies should be carried out in the future.
We also observed that expression of ALOX5 and COX2 in peripheral leukocytes did correlate with each other. Although the molecular mechanisms of gene expression regulation of the two enzymes are very different both enzymes constitute classical pro-inflammatory gene products and thus, parallel expression regulation is biologically meaningful [58,61,62,89]. However, it remains unclear which molecular mechanisms are the basis for parallel expression regulation of these two enzymes. A compensatory shunting of arachidonic acid metabolism between the two pathways is possible and has been reported for other conditions [[90], [91], [92]]. We also noticed that the HOMA-IR (unpublished data) improved under both ketogenic treatments, which is clinically meaningful because insulin resistance seems to be associated with disability, chronic inflammatory processes and oxidative stress [[93], [94], [95]].
A major limitation of our study is its small sample size (n = 24 analyzed patients vs. n = 48 total patients who finished the study) and the limited selection of target genes. However, based on our results more comprehensive expression profiles (genome covering expression microarrays), which also quantify the expression other classical pro- and anti-inflammatory gene products, should be carried out during follow-up studies. Thus, there is urgent need for follow up and validation of our results with a larger cohort. However, despite this caveat our study suggests that ketogenic diets offer an elegant way to modulate leukocytic ALOX/COX expression, which seems to be a promising effect in neuro-inflammatory diseases [87,96]. Our findings are of immediate medical interest since such dietary therapeutic strategies have well been characterized in the past decades and exhibit little unwanted side effects [44,45,97].
Acknowledgments
Acknowledgment
The analysis of the presented gene products was funded by Genzyme GmbH – Siemensstr. 5b – 63263 Neu-Isenburg Sanofi Genzyme – Germany –, which was not involved in any decision-making processes relating the study or its participants.
The study was funded by Myelin Projekt e.V. and Familie Ernst Wendt Stiftung Stadt Koeln, which were not involved in any decision-making processes relating the study or its participants. All authors have completed the disclosure form and declare: no financial relationships with any organisations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
Declaration of interests
The authors declare no competing interests.
Author contributions
Conceptualization, M.B. and H.K.; Methodology, M.B., and H.K.; Investigation, M.B.,M.K. and H.K.; Writing – Original Draft, M.B. and H.K.; Writing – Review & Editing, M.B.,M.K. and H.K.; Funding Acquisition, M.B.; Resources, M.B. and H.K.; Supervision, H.K..
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.08.057.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Kidd D., Barkhof F., McConnell R., Algra P.R., Allen I.V., Revesz T. Cortical lesions in multiple sclerosis. Brain J Neurol. 1999;122:17–26. doi: 10.1093/brain/122.1.17. Pt 1. [DOI] [PubMed] [Google Scholar]
- 2.Lucchinetti C., Brück W., Noseworthy J. Multiple sclerosis: recent developments in neuropathology, pathogenesis, magnetic resonance imaging studies and treatment. Curr Opin Neurol. 2001;14:259–269. doi: 10.1097/00019052-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Noseworthy J.H., Lucchinetti C., Rodriguez M., Weinshenker B.G. Multiple sclerosis. N Engl J Med. 2000;343:938–952. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- 4.Barnett M.H., Prineas J.W. Relapsing and remitting multiple sclerosis: pathology of the newly forming lesion. Ann Neurol. 2004;55:458–468. doi: 10.1002/ana.20016. [DOI] [PubMed] [Google Scholar]
- 5.Lucchinetti C.F., Bruck W., Lassmann H. Evidence for pathogenic heterogeneity in multiple sclerosis. Ann Neurol. 2004;56:308. doi: 10.1002/ana.20182. [DOI] [PubMed] [Google Scholar]
- 6.Lucchinetti C., Brück W., Parisi J., Scheithauer B., Rodriguez M., Lassmann H. Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann Neurol. 2000;47:707–717. doi: 10.1002/1531-8249(200006)47:6<707::aid-ana3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 7.Bjørnevik K., Chitnis T., Ascherio A., Munger K.L. Polyunsaturated fatty acids and the risk of multiple sclerosis. Mult Scler Houndmills Basingstoke Engl. 2017 doi: 10.1177/1352458517691150. 1352458517691150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbige L.S., Sharief M.K. Polyunsaturated fatty acids in the pathogenesis and treatment of multiple sclerosis. Br J Nutr. 2007;98(Suppl. 1) doi: 10.1017/S0007114507833010. (S46-53) [DOI] [PubMed] [Google Scholar]
- 9.Farinotti M., Vacchi L., Simi S., Di Pietrantonj C., Brait L., Filippini G. Dietary interventions for multiple sclerosis. Cochrane Database Syst Rev. 2012;12 doi: 10.1002/14651858.CD004192.pub3. [DOI] [PubMed] [Google Scholar]
- 10.Dworkin R.H., Bates D., Millar J.H., Paty D.W. Linoleic acid and multiple sclerosis: a reanalysis of three double-blind trials. Neurology. 1984;34:1441–1445. doi: 10.1212/wnl.34.11.1441. [DOI] [PubMed] [Google Scholar]
- 11.Meade C.J., Mertin J., Sheena J., Hunt R. Reduction by linoleic acid of the severity of experimental allergic encephalomyelitis in the guinea pig. J Neurol Sci. 1978;35:291–308. doi: 10.1016/0022-510x(78)90010-2. [DOI] [PubMed] [Google Scholar]
- 12.Hughes D., Keith A.B., Mertin J., Caspary E.A. Linoleic acid therapy in severe experimental allergic encephalomyelitis in the guinea-pig: suppression by continuous treatment. Clin Exp Immunol. 1980;40:523–531. [PMC free article] [PubMed] [Google Scholar]
- 13.Emerson M.R., LeVine S.M. Experimental allergic encephalomyelitis is exacerbated in mice deficient for 12/15-lipoxygenase or 5-lipoxygenase. Brain Res. 2004;1021:140–145. doi: 10.1016/j.brainres.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 14.Marusic S., Thakker P., Pelker J.W., Stedman N.L., Lee K.L., McKew J.C. Blockade of cytosolic phospholipase A2 alpha prevents experimental autoimmune encephalomyelitis and diminishes development of Th1 and Th17 responses. J Neuroimmunol. 2008;204:29–37. doi: 10.1016/j.jneuroim.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Marusic S., Leach M.W., Pelker J.W., Azoitei M.L., Uozumi N., Cui J. Cytosolic phospholipase A2 alpha-deficient mice are resistant to experimental autoimmune encephalomyelitis. J Exp Med. 2005;202:841–851. doi: 10.1084/jem.20050665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalyvas A., Baskakis C., Magrioti V., Constantinou-Kokotou V., Stephens D., Lopez-Vales R. Differing roles for members of the phospholipase A2 superfamily in experimental autoimmune encephalomyelitis. Brain. 2009;132:1221–1235. doi: 10.1093/brain/awp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara Y., Matsushita T., Kita Y., Uematsu S., Akira S., Kira J. Targeted lipidomics reveals mPGES-1-PGE2 as a therapeutic target for multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106:21807–21812. doi: 10.1073/pnas.0906891106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirshafiey A., Jadidi-Niaragh F. Immunopharmacological role of the leukotriene receptor antagonists and inhibitors of leukotrienes generating enzymes in multiple sclerosis. Immunopharmacol Immunotoxicol. 2010;32:219–227. doi: 10.3109/08923970903283662. [DOI] [PubMed] [Google Scholar]
- 19.Neu I., Mallinger J., Wildfeuer A., Mehlber L. Leukotrienes in the cerebrospinal fluid of multiple sclerosis patients. Acta Neurol Scand. 1992;86:586–587. doi: 10.1111/j.1600-0404.1992.tb05491.x. [DOI] [PubMed] [Google Scholar]
- 20.Whitney L.W., Ludwin S.K., McFarland H.F., Biddison W.E. Microarray analysis of gene expression in multiple sclerosis and EAE identifies 5-lipoxygenase as a component of inflammatory lesions. J Neuroimmunol. 2001;121:40–48. doi: 10.1016/s0165-5728(01)00438-6. [DOI] [PubMed] [Google Scholar]
- 21.Minghetti L. Cyclooxygenase-2 (COX-2) in inflammatory and degenerative brain diseases. J Neuropathol Exp Neurol. 2004;63:901–910. doi: 10.1093/jnen/63.9.901. [DOI] [PubMed] [Google Scholar]
- 22.Arthur A.T., Armati P.J., Bye C., Consortium S.M.G., Heard R.N., Stewart G.J. Genes implicated in multiple sclerosis pathogenesis from consilience of genotyping and expression profiles in relapse and remission. BMC Med Genet. 2008;9:17. doi: 10.1186/1471-2350-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson N.G., Rojas M.A., Redd J.W., Tang P., Wood B., Hill K.E. Cyclooxygenase-2 expression in oligodendrocytes increases sensitivity to excitotoxic death. J Neuroinflammation. 2010;7:25. doi: 10.1186/1742-2094-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prüss H., Rosche B., Sullivan A.B., Brommer B., Wengert O., Gronert K. Proresolution lipid mediators in multiple sclerosis - differential, disease severity-dependent synthesis - a clinical pilot trial. PLoS One. 2013;8:e55859. doi: 10.1371/journal.pone.0055859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis E.A., Rhee S.G., Billah M.M., Hannun Y.A. Role of phospholipase in generating lipid second messengers in signal transduction. FASEB J Off Publ Fed Am Soc Exp Biol. 1991;5:2068–2077. doi: 10.1096/fasebj.5.7.1901288. [DOI] [PubMed] [Google Scholar]
- 26.Kalyvas A., David S. Cytosolic phospholipase A2 plays a key role in the pathogenesis of multiple sclerosis-like disease. Neuron. 2004;41:323–335. doi: 10.1016/s0896-6273(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L., Maruvada R., Sapirstein A., Peters-Golden M., Kim K.S. Cysteinyl leukotrienes as novel host factors facilitating Cryptococcus neoformans penetration into the brain. Cell Microbiol. 2017;19 doi: 10.1111/cmi.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich D.S., Lucchinetti C.F., Calabresi P.A. Multiple sclerosis. N Engl J Med. 2018;378:169–180. doi: 10.1056/NEJMra1401483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yiangou Y., Facer P., Durrenberger P., Chessell I.P., Naylor A., Bountra C. COX-2, CB2 and P2X7-immunoreactivities are increased in activated microglial cells/macrophages of multiple sclerosis and amyotrophic lateral sclerosis spinal cord. BMC Neurol. 2006;6:12. doi: 10.1186/1471-2377-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yagami T., Koma H., Yamamoto Y. Pathophysiological roles of cyclooxygenases and prostaglandins in the central nervous system. Mol Neurobiol. 2016;53:4754–4771. doi: 10.1007/s12035-015-9355-3. [DOI] [PubMed] [Google Scholar]
- 31.Ackermann J.A., Hofheinz K., Zaiss M.M., Krönke G. The double-edged role of 12/15-lipoxygenase during inflammation and immunity. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbalip.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Kühn H., O'Donnell V.B. Inflammation and immune regulation by 12/15-lipoxygenases. Prog Lipid Res. 2006;45:334–356. doi: 10.1016/j.plipres.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Chandrasekharan J.A., Sharma-Walia N. Lipoxins: nature's way to resolve inflammation. J Inflamm Res. 2015;8:181–192. doi: 10.2147/JIR.S90380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sansbury B.E., Spite M. Resolution of acute inflammation and the role of resolvins in immunity, thrombosis, and vascular biology. Circ Res. 2016;119:113–130. doi: 10.1161/CIRCRESAHA.116.307308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duvall M.G., Levy B.D. DHA- and EPA-derived resolvins, protectins, and maresins in airway inflammation. Eur J Pharmacol. 2016;785:144–155. doi: 10.1016/j.ejphar.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J., Zhang Y., Xiao Y., Ma S., Liu Q., Dang S. Inhibition of 12/15-lipoxygenase by baicalein induces microglia PPARβ/δ: a potential therapeutic role for CNS autoimmune disease. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kossoff E.H., Zupec-Kania B.A., Rho J.M. Ketogenic diets: an update for child neurologists. J Child Neurol. 2009;24:979–988. doi: 10.1177/0883073809337162. [DOI] [PubMed] [Google Scholar]
- 38.Barañano K.W., Hartman A.L. The ketogenic diet: uses in epilepsy and other neurologic illnesses. Curr Treat Options Neurol. 2008;10:410–419. doi: 10.1007/s11940-008-0043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neal E.G., Chaffe H., Schwartz R.H., Lawson M.S., Edwards N., Fitzsimmons G. The ketogenic diet for the treatment of childhood epilepsy: a randomised controlled trial. Lancet Neurol. 2008;7:500–506. doi: 10.1016/S1474-4422(08)70092-9. [DOI] [PubMed] [Google Scholar]
- 40.Piccio L., Stark J.L., Cross A.H. Chronic calorie restriction attenuates experimental autoimmune encephalomyelitis. J Leukoc Biol. 2008;84:940–948. doi: 10.1189/jlb.0208133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esquifino A.I., Cano P., Jimenez-Ortega V., Fernández-Mateos M.P., Cardinali D.P. Immune response after experimental allergic encephalomyelitis in rats subjected to calorie restriction. J Neuroinflammation. 2007;4:6. doi: 10.1186/1742-2094-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D.Y., Hao J., Liu R., Turner G., Shi F.-D., Rho J.M. Inflammation-mediated memory dysfunction and effects of a ketogenic diet in a murine model of multiple sclerosis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stafstrom C.E., Rho J.M. The ketogenic diet as a treatment paradigm for diverse neurological disorders. Front Pharmacol. 2012;3:59. doi: 10.3389/fphar.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masino S.A., Rho J.M. Mechanisms of ketogenic diet action. In: Noebels J.L., Avoli M., Rogawski M.A., Olsen R.W., Delgado-Escueta A.V., editors. Jaspers Basic Mech. Epilepsies. 4th ed. National Center for Biotechnology Information (US); Bethesda (MD): 2012. [PubMed] [Google Scholar]
- 45.Choi I.Y., Piccio L., Childress P., Bollman B., Ghosh A., Brandhorst S. A diet mimicking fasting promotes regeneration and reduces autoimmunity and multiple sclerosis symptoms. Cell Rep. 2016;15:2136–2146. doi: 10.1016/j.celrep.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock M., Michalsen A., Paul F. Ketogenic diet and prolonged fasting improve health related quality of life and blood lipid profile in multiple sclerosis - a randomized controlled trial. ECTRIMS Online Libr. 2015;116359:P1509. [Google Scholar]
- 47.Swidsinski A., Dörffel Y., Loening-Baucke V., Gille C., Göktas Ö., Reißhauer A. Reduced mass and diversity of the colonic microbiome in patients with multiple sclerosis and their improvement with ketogenic diet. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polman C.H., Reingold S.C., Edan G., Filippi M., Hartung H.-P., Kappos L. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 49.Vickers A.J., Altman D.G. Statistics notes: Analysing controlled trials with baseline and follow up measurements. BMJ. 2001;323:1123–1124. doi: 10.1136/bmj.323.7321.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vickrey B.G., Lee L., Moore F., Moriarty P. EDSS change relates to physical HRQoL while relapse occurrence relates to overall HRQoL in patients with multiple sclerosis receiving subcutaneous interferon β-1a. Mult Scler Int. 2015;2015 doi: 10.1155/2015/631989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sigal E., Grunberger D., Cashman J.R., Craik C.S., Caughey G.H., Nadel J.A. Arachidonate 15-lipoxygenase from human eosinophil-enriched leukocytes: partial purification and properties. Biochem Biophys Res Commun. 1988;150:376–383. doi: 10.1016/0006-291x(88)90531-1. [DOI] [PubMed] [Google Scholar]
- 52.Nadel J.A., Ueki I.F., Schuster A., Conrad D.J., Sigal E. Arachidonate 15-lipoxygenase: immunocytochemical localization in blood and airway cells. Trans Assoc Am Physicians. 1990;103:145–153. [PubMed] [Google Scholar]
- 53.Nadel J.A., Conrad D.J., Ueki I.F., Schuster A., Sigal E. Immunocytochemical localization of arachidonate 15-lipoxygenase in erythrocytes, leukocytes, and airway cells. J Clin Invest. 1991;87:1139–1145. doi: 10.1172/JCI115110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samuelsson B., Dahlén S.E., Lindgren J.A., Rouzer C.A., Serhan C.N. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 55.Lewis R.E., Granger H.J. Diapedesis and the permeability of venous microvessels to protein macromolecules: the impact of leukotriene B4 (LTB4) Microvasc Res. 1988;35:27–47. doi: 10.1016/0026-2862(88)90048-9. [DOI] [PubMed] [Google Scholar]
- 56.Ford-Hutchinson A.W., Gresser M., Young R.N. 5-Lipoxygenase. Annu Rev Biochem. 1994;63:383–417. doi: 10.1146/annurev.bi.63.070194.002123. [DOI] [PubMed] [Google Scholar]
- 57.Funk C.D., Chen X.S. 5-Lipoxygenase and leukotrienes. Transgenic mouse and nuclear targeting studies. Am J Respir Crit Care Med. 2000;161:S120–S124. doi: 10.1164/ajrccm.161.supplement_1.ltta-24. [DOI] [PubMed] [Google Scholar]
- 58.Haeggström J.Z., Funk C.D. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 59.Chen H., Qin J., Wei P., Zhang J., Li Q., Fu L. Effects of leukotriene B4 and prostaglandin E2 on the differentiation of murine Foxp3+ T regulatory cells and Th17 cells. Prostaglandins Leukot Essent Fatty Acids. 2009;80:195–200. doi: 10.1016/j.plefa.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Wonyong L., Hyeong S.K., Ryol L.G. Leukotrienes induce the migration of Th17 cells. Immunol Cell Biol. 2015;93:472–479. doi: 10.1038/icb.2014.104. [DOI] [PubMed] [Google Scholar]
- 61.Smith W.L., DeWitt D.L., Garavito R.M. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 62.Fitzpatrick F.A. Cyclooxygenase enzymes: regulation and function. Curr Pharm Des. 2004;10:577–588. doi: 10.2174/1381612043453144. [DOI] [PubMed] [Google Scholar]
- 63.Seo M.-J., Oh D.-K. Prostaglandin synthases: Molecular characterization and involvement in prostaglandin biosynthesis. Prog Lipid Res. 2017;66:50–68. doi: 10.1016/j.plipres.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Smith W.L., Dewitt D.L. Prostaglandin endoperoxide H synthases-1 and -2. Adv Immunol. 1996;62:167–215. doi: 10.1016/s0065-2776(08)60430-7. [DOI] [PubMed] [Google Scholar]
- 65.Reder A.T., Thapar M., Sapugay A.M., Jensen M.A. Prostaglandins and inhibitors of arachidonate metabolism suppress experimental allergic encephalomyelitis. J Neuroimmunol. 1994;54:117–127. doi: 10.1016/0165-5728(94)90238-0. [DOI] [PubMed] [Google Scholar]
- 66.Hewett S.J., Uliasz T.F., Vidwans A.S., Hewett J.A. Cyclooxygenase-2 contributes to N-methyl-D-aspartate-mediated neuronal cell death in primary cortical cell culture. J Pharmacol Exp Ther. 2000;293:417–425. [PubMed] [Google Scholar]
- 67.Carlson N.G. Neuroprotection of cultured cortical neurons mediated by the cyclooxygenase-2 inhibitor APHS can be reversed by a prostanoid. J Neurosci Res. 2003;71:79–88. doi: 10.1002/jnr.10465. [DOI] [PubMed] [Google Scholar]
- 68.Miyamoto K., Miyake S., Mizuno M., Oka N., Kusunoki S., Yamamura T. Selective COX-2 inhibitor celecoxib prevents experimental autoimmune encephalomyelitis through COX-2-independent pathway. Brain J Neurol. 2006;129:1984–1992. doi: 10.1093/brain/awl170. [DOI] [PubMed] [Google Scholar]
- 69.Ni J., Shu Y.-Y., Zhu Y.-N., Fu Y.-F., Tang W., Zhong X.-G. COX-2 inhibitors ameliorate experimental autoimmune encephalomyelitis through modulating IFN-gamma and IL-10 production by inhibiting T-bet expression. J Neuroimmunol. 2007;186:94–103. doi: 10.1016/j.jneuroim.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 70.Kofler D.M., Marson A., Dominguez-Villar M., Xiao S., Kuchroo V.K., Hafler D.A. Decreased RORC-dependent silencing of prostaglandin receptor EP2 induces autoimmune Th17 cells. J Clin Invest. 2014;124:2513–2522. doi: 10.1172/JCI72973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang E.H.C., Libby P., Vanhoutte P.M., Xu A. Anti-inflammation therapy by activation of prostaglandin EP4 receptor in cardiovascular and other inflammatory diseases. J Cardiovasc Pharmacol. 2012;59:116–123. doi: 10.1097/FJC.0b013e3182244a12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ni F.-F., Li C.-R., Liao J.-X., Wang G.-B., Lin S.-F., Xia Y. The effects of ketogenic diet on the Th17/Treg cells imbalance in patients with intractable childhood epilepsy. Seizure. 2016;38:17–22. doi: 10.1016/j.seizure.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 73.Nogawa S., Zhang F., Ross M.E., Iadecola C. Cyclo-oxygenase-2 gene expression in neurons contributes to ischemic brain damage. J Neurosci. 1997;17:2746–2755. doi: 10.1523/JNEUROSCI.17-08-02746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagayama M., Niwa K., Nagayama T., Ross M.E., Iadecola C. The cyclooxygenase-2 inhibitor NS-398 ameliorates ischemic brain injury in wild-type mice but not in mice with deletion of the inducible nitric oxide synthase gene. J Cereb Blood Flow Metab Off J Int Soc Cereb Blood Flow Metab. 1999;19:1213–1219. doi: 10.1097/00004647-199911000-00005. [DOI] [PubMed] [Google Scholar]
- 75.Leuschen M.P., Filipi M., Healey K. A randomized open label study of pain medications (naproxen, acetaminophen and ibuprofen) for controlling side effects during initiation of IFN beta-1a therapy and during its ongoing use for relapsing-remitting multiple sclerosis. Mult Scler Houndmills Basingstoke Engl. 2004;10:636–642. doi: 10.1191/1352458504ms1114oa. [DOI] [PubMed] [Google Scholar]
- 76.Wingerchuk D.M., Benarroch E.E., O'Brien P.C., Keegan B.M., Lucchinetti C.F., Noseworthy J.H. A randomized controlled crossover trial of aspirin for fatigue in multiple sclerosis. Neurology. 2005;64:1267–1269. doi: 10.1212/01.WNL.0000156803.23698.9A. [DOI] [PubMed] [Google Scholar]
- 77.Wingerchuk D.M., Rodriguez M. Premenstrual multiple sclerosis pseudoexacerbations: Role of body temperature and prevention with aspirin. Arch Neurol. 2006;63:1005–1008. doi: 10.1001/archneur.63.7.1005. [DOI] [PubMed] [Google Scholar]
- 78.Fitzgerald G.A. Coxibs and cardiovascular disease. N Engl J Med. 2004;351:1709–1711. doi: 10.1056/NEJMp048288. [DOI] [PubMed] [Google Scholar]
- 79.Topol E.J. Failing the public health—rofecoxib, Merck, and the FDA. N Engl J Med. 2004;351:1707–1709. doi: 10.1056/NEJMp048286. [DOI] [PubMed] [Google Scholar]
- 80.Stafford P., Abdelwahab M.G., Kim D.Y., Preul M.C., Rho J.M., Scheck A.C. The ketogenic diet reverses gene expression patterns and reduces reactive oxygen species levels when used as an adjuvant therapy for glioma. Nutr Metab. 2010;7:74. doi: 10.1186/1743-7075-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeong E.A., Jeon B.T., Shin H.J., Kim N., Lee D.H., Kim H.J. Ketogenic diet-induced peroxisome proliferator-activated receptor-γ activation decreases neuroinflammation in the mouse hippocampus after kainic acid-induced seizures. Exp Neurol. 2011;232:195–202. doi: 10.1016/j.expneurol.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Terrone G., Pauletti A., Salamone A., Rizzi M., Villa B.R., Porcu L. Inhibition of monoacylglycerol lipase terminates diazepam-resistant status epilepticus in mice and its effects are potentiated by a ketogenic diet. Epilepsia. 2018;59:79–91. doi: 10.1111/epi.13950. [DOI] [PubMed] [Google Scholar]
- 83.Mowry E.M., Beheshtian A., Waubant E., Goodin D.S., Cree B.A., Qualley P. Quality of life in multiple sclerosis is associated with lesion burden and brain volume measures. Neurology. 2009;72:1760–1765. doi: 10.1212/WNL.0b013e3181a609f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen M., Lebrun C., Aufauvre D., Chanalet S., Filleau-Bertogliatti C., Camu W. Longitudinal study of health related quality of life in multiple sclerosis: correlation with MRI parameters. Rev Neurol (Paris) 2010;166:894–900. doi: 10.1016/j.neurol.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 85.Simmons R.D., Hugh A.R., Willenborg D.O., Cowden W.B. Suppression of active but not passive autoimmune encephalomyelitis by dual cyclo-oxygenase and 5-lipoxygenase inhibition. Acta Neurol Scand. 1992;85:197–199. doi: 10.1111/j.1600-0404.1992.tb04027.x. [DOI] [PubMed] [Google Scholar]
- 86.Moon C., Ahn M., Wie M.-B., Kim H.-M., Koh C.-S., Hong S.-C. Phenidone, a dual inhibitor of cyclooxygenases and lipoxygenases, ameliorates rat paralysis in experimental autoimmune encephalomyelitis by suppressing its target enzymes. Brain Res. 2005;1035:206–210. doi: 10.1016/j.brainres.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 87.Kong W., Hooper K.M., Ganea D. The natural dual cyclooxygenase and 5-lipoxygenase inhibitor flavocoxid is protective in EAE through effects on Th1/Th17 differentiation and macrophage/microglia activation. Brain Behav Immun. 2016;53:59–71. doi: 10.1016/j.bbi.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 88.Noh H.S., Lee H.P., Kim D.W., Kang S.S., Cho G.J., Rho J.M. A cDNA microarray analysis of gene expression profiles in rat hippocampus following a ketogenic diet. Brain Res Mol Brain Res. 2004;129:80–87. doi: 10.1016/j.molbrainres.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 89.Rådmark O., Werz O., Steinhilber D., Samuelsson B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim Biophys Acta. 1851;2015:331–339. doi: 10.1016/j.bbalip.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 90.Sánchez-Borges M., Capriles-Hulett A., Caballero-Fonseca F. NSAID-induced urticaria and angioedema: a reappraisal of its clinical management. Am J Clin Dermatol. 2002;3:599–607. doi: 10.2165/00128071-200203090-00002. [DOI] [PubMed] [Google Scholar]
- 91.Ganesh R., Marks D.J.B., Sales K., Winslet M.C., Seifalian A.M. Cyclooxygenase/lipoxygenase shunting lowers the anti-cancer effect of cyclooxygenase-2 inhibition in colorectal cancer cells. World J Surg Oncol. 2012;10:200. doi: 10.1186/1477-7819-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park S.-W., Heo D.-S., Sung M.-W. The shunting of arachidonic acid metabolism to 5-lipoxygenase and cytochrome p450 epoxygenase antagonizes the anti-cancer effect of cyclooxygenase-2 inhibition in head and neck cancer cells. Cell Oncol (Dordr) 2012;35:1–8. doi: 10.1007/s13402-011-0051-7. [DOI] [PubMed] [Google Scholar]
- 93.Wens I., Dalgas U., Stenager E., Eijnde B.O. Risk factors related to cardiovascular diseases and the metabolic syndrome in multiple sclerosis - a systematic review. Mult Scler Houndmills Basingstoke Engl. 2013;19:1556–1564. doi: 10.1177/1352458513504252. [DOI] [PubMed] [Google Scholar]
- 94.Oliveira S.R., Simão A.N.C., Kallaur A.P., de Almeida E.R.D., Morimoto H.K., Lopes J. Disability in patients with multiple sclerosis: influence of insulin resistance, adiposity, and oxidative stress. Nutr Burbank Los Angel Cty Calif. 2014;30:268–273. doi: 10.1016/j.nut.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 95.Tettey P., Simpson S., Taylor B.V., van der Mei I.A.F. Vascular comorbidities in the onset and progression of multiple sclerosis. J Neurol Sci. 2014;347:23–33. doi: 10.1016/j.jns.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 96.Di Penta A., Chiba A., Alloza I., Wyssenbach A., Yamamura T., Villoslada P. A trifluoromethyl analogue of celecoxib exerts beneficial effects in neuroinflammation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pfeifer H.H., Lyczkowski D.A., Thiele E.A. Low glycemic index treatment: implementation and new insights into efficacy. Epilepsia. 2008;49(Suppl. 8):42–45. doi: 10.1111/j.1528-1167.2008.01832.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2