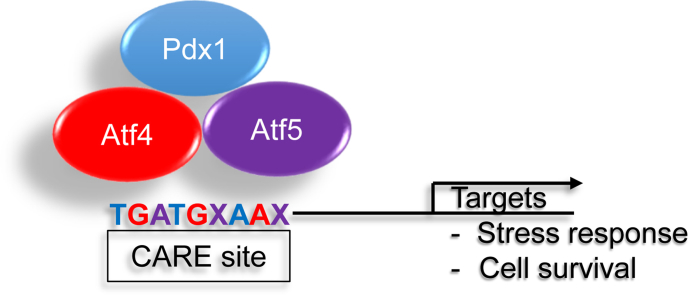
Abstract
Objective
Loss of insulin secretion due to failure or death of the insulin secreting β cells is the central cause of diabetes. The cellular response to stress (endoplasmic reticulum (ER), oxidative, inflammatory) is essential to sustain normal β cell function and survival. Pancreatic and duodenal homeobox 1 (PDX1), Activating transcription factor 4 (ATF4), and Activating transcription factor 5 (ATF5) are transcription factors implicated in β cell survival and susceptibility to stress. Our goal was to determine if a PDX1-ATF transcriptional complex or complexes regulate β cell survival in response to stress and to identify direct transcriptional targets.
Methods
Pdx1, Atf4 and Atf5 were silenced by viral delivery of gRNAs or shRNAs to Min6 insulinoma cells or primary murine islets. Gene expression was assessed by qPCR, RNAseq analysis, and Western blot analysis. Chromatin enrichment was measured in the Min6 β cell line and primary isolated mouse islets by ChIPseq and ChIP PCR. Immunoprecipitation was used to assess interactions among transcription factors in Min6 cells and isolated mouse islets. Activation of caspase 3 by immunoblotting or by irreversible binding to a fluorescent inhibitor was taken as an indication of commitment to an apoptotic fate.
Results
RNASeq identified a set of PDX1, ATF4 and ATF5 co-regulated genes enriched in stress and apoptosis functions. We further identified stress induced interactions among PDX1, ATF4, and ATF5. PDX1 chromatin occupancy peaks were identified over composite C/EBP-ATF (CARE) motifs of 26 genes; assessment of a subset of these genes revealed co-enrichment for ATF4 and ATF5. PDX1 occupancy over CARE motifs was conserved in the human orthologs of 9 of these genes. Of these, Glutamate Pyruvate Transaminase 2 (Gpt2), Cation transport regulator 1 (Chac1), and Solute Carrier Family 7 Member 1 (Slc7a1) induction by stress was conserved in human islets and abrogated by deficiency of Pdx1, Atf4, and Atf5 in Min6 cells. Deficiency of Gpt2 reduced β cell susceptibility to stress induced apoptosis in both Min6 cells and primary islets.
Conclusions
Our results identify a novel PDX1 stress inducible complex (es) that regulates expression of stress and apoptosis genes to govern β cell survival.
Keywords: Transcriptional regulation, Stress, Apoptosis, β cell, Pancreas
Graphical abstract
Highlights
-
•
PDX1 binds to composite CEBP/ATF (CARE) sites of stress and apoptosis genes.
-
•
A novel stress inducible transcriptional complex involving PDX1, ATF4, and ATF5 is discovered.
-
•
Novel stress induced targets of the complex involved in fate decisions are identified.
-
•
Silencing of one of these targets, Gpt2, protects β cells from apoptosis due to stress.
Abbreviations
- ATF4
Activating transcription factor 4
- ATF5
Activating Transcription Factor 5
- BZIP
basic leucine zipper domain
- CARE
C/EBP-ATF response element
- Cartpt
Cocaine- And Amphetamine-Regulated Transcript Protein
- Chac1
Cation transport regulator 1
- Creb3l1
Cyclic AMP-Responsive Element-Binding Protein 3-Like Protein
- CRISPR
Clustered regularly-interspaced short palindromic repeats
- Ddit3
DNA Damage Inducible Transcript 3
- Defb1
Defensin Beta 1
- Eif4ebp1
Eukaryotic Translation Initiation Factor 4E Binding Protein 1
- ER
endoplasmic reticulum
- Gpt2
Glutamate Puruvate Transaminase 2
- HA
hemagglutinin
- HFD
high fat diet
- IP
immunoprecipitation
- PA
palmitate
- PDX1
Pancreatic and duodenal homeobox 1
- Rad51l1
RAD51-Like 1
- Slc7a1
Solute Carrier Family 7 Member 1
- Tg
thapsigargin
- Trib3
Tribbles Pseudokinase 3
- TSS
transcriptional start site
- UCSC
University of California Santa Cruz
- UPR
Unfolded Protein Response
1. Introduction
Diabetes results from the failure of insulin producing β cells to compensate for increased metabolic demand and stress (ER, oxidative, inflammatory), leading to the decline of β cell function and survival. Therefore, understanding the complex mechanisms and underlying regulators of the stress response and of cell fate choices is critical to develop effective prevention and therapeutics for this condition.
The human diabetes gene and homeodomain protein PDX1 is of well described importance for the function and survival of β cells [1], [2], [3], [4], [5]. Our previously published work has established a role for PDX1 in β cell survival in response to high fat diet (HFD) [6]. While PDX1 expression is not itself induced by stress, it regulates islet compensation for a HFD-induced insulin resistance, in part through direct transcriptional regulation of Atf4 and Wsf1 [6], [7]. PDX1 preferentially binds to the consensus sequence (5′-CTCTAAT (T/G)AG-3′), however enrichment of PDX1 has also been found over other genomic motifs [8]. In previous work, we identified PDX1 de novo motifs that bore resemblance to the C/EBP-ATF response element (CARE) site sequence (TGATGXAAX) under Pdx1 enrichment peaks associated with the Eif4ebp1 and Atf5 genes [8].
Activating transcription factors are a family of transcription factors known to bind CARE motifs [9]. Activating Transcription Factor 4 (ATF4) is a member of the survival and homeostasis regulatory CREB/ATF family of DNA binding basic leucine zipper domain containing transcription factors [10]. ATF4 has been extensively studied in cellular stress responses and its downstream targets include both negative and positive regulators of translation and protein synthesis [11], [12]. ATF4 is one member of a set of privileged mRNAs, that also includes family member ATF5, that are specifically translated in the context of stress when there is global translation arrest of most other mRNAs [13], [14]. ATF5 plays tissue specific adaptive and maladaptive roles in cell survival in response to a variety of stresses, such as serum deprivation, oxidative stress, and ER stress [10], [15], [16], [17]. In primary β cells ATF5 deficiency reduces cell survival, mediated at least in part by its regulation of translational arrest in response to stress through Eif4ebp1 [18].
Here we identify previously undiscovered stress responsive transcriptional complex (es) containing PDX1, ATF4 and ATF5 that regulates expression of stress and apoptosis genes to influence β cell fate decisions during stress.
2. Materials and methods
2.1. Animals
Wild type (WT) CD1 males were purchased from Charles River Laboratories. Experiments were performed on islets isolated from 8 to 10-week-old mice housed in a 12hr light/dark cycle with ad libitum access to food. The University of Pennsylvania Institutional Animal Care and Use Committee approved all mouse studies.
2.2. Cell culture and lentiviral infection
P20-30 Min6 cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS. For shRNA experiments, 293T cells were nucleofected by lipofectamine 2000 with pLenti CRISPR/Cas9 vectors targeting Pdx1, Atf4, or Atf5, pLenti Ultramir shRNA vectors targeting Pdx1, Atf4, Atf5, or Gpt2, or a nontargeting control, and viral packaging component vectors (psPax2 and PMD2G). Virus was harvested at 48 and 72 h post-transfection, concentrated by centrifugation, and then used to infect Min6 cells or isolated mouse islets. Infected cells or islets were harvested for protein or RNA 72–96 h post-infection.
2.3. Islet isolation, culture, and lentiviral infection
Islets were isolated from 8 to 10 week old mice by collagenase digestion as described [18]. After three rounds of hand-picking under a light microscope, islets were allowed to recover in media for 2 h. Islets were incubated with 250 mg/mL trypsin for 2 min and mixed gently twice. FBS was added to stop the trypsinization and islets were centrifuged to remove FBS/trypsin. Concentrated lentiviral media was added to the islets and they were incubated overnight in the tissue culture incubator. Islets were recovered from the islet media by resuspending the islets and handpicking the intact islets twice into new plates with fresh islet media. This protocol is adapted from [19]. Islets recovered in culture in RPMI 1640 for 48 h and were then picked to size-match and treated with vehicle (DMSO), thapsigargin (Tg) (1uM) (Sigma, St. Louis, MO, USA), or 500uM palmitate for the denoted times. Palmitate was dissolved in 50% ethanol (150 mM) and conjugated to 10% BSA (7 mM). The Image-iT LIVE Red Caspase-3 Detection Kit (Thermofisher) was used as per the provided protocol to stain active caspase-3 in islets. The islets were then dissociated into single cells, spun onto a slide by cytospin, and analyzed by a Keyence BZ-X710 fluorescent microscope and quantitated with the Keyence BZ-X Analyzer software.
2.4. Western blot analysis
Protein lysates were prepared using NP-40 lysis buffer (0.4% NP-40, 150 mM NaCl, 50 mM Tris base, pH 7.6). Protein concentration was determined using BCA (ThermoFisher, Saint Louis, Missouri). Proteins were separated by SDS-PAGE and immunoblotted with the following primary antisera: rabbit anti-cleaved caspase-3 (Cell Signaling, Danvers, MA, USA), mouse anti-tubulin (Sigma Aldrich), rabbit anti-Atf4 (Santa Cruz), guinea pig anti-Pdx1 (Chris Wright), and anti-HA-HRP conjugated (Roche, San Francisco, CA, USA). HRP conjugated antibodies were incubated for 2 h at room temperature. Luminate Crescendo HRP substrate (Millipore) was used to visualize blots with film or a Chemi-doc Touch Imaging system (Bio-Rad, Hercules, California, USA). Use and dilution information for anti-sera is found in Table 1.
2.5. Immunoprecipitation and chromatin immunoprecipitation
For immunoprecipitation, Min6 cells or isolated mouse islets were washed 3 times with cold 1x PBS, then nuclear extract was collected as previously described [20] except buffer C did not contain glycerol. Nuclear lysate was incubated overnight with antibodies for denoted proteins or control IgG and then incubated for 2 h with G Protein Dynabeads (ThermoFisher). For ChIP, Min6 cells (∼1 × 107 cells) or 200 isolated mouse islets were cross-linked with 1% formaldehyde in 1xPBS for 10 min at room temperature and quenched with glycine to a final concentration of 0.125 M. Chromatin was sonicated in a Bioruptor (Diagenode, Denville, NJ) and precleared with normal mouse IgG (Santa Cruz) overnight at 4 °C. After removal of an aliquot for input, precleared chromatin was divided equally for IP with either rabbit anti-ATF4 (Santa Cruz), anti-HA conjugated agarose (Sigma), or normal IgG (Santa Cruz) for 3 h at 4 °C. Use and dilution information for anti-sera is found in Table 1.
2.6. RNA isolation, RNA-sequencing, and qPCR
Min6 cells and isolated islets were harvested and stored in Trizol (Invitrogen) and processed according to the manufacturer's instructions. Min6 RNA was extracted using phenol chloroform extraction and ethanol precipitation [21]. Islet RNA was extracted using the RNeasy Mini Kit applied to the aqueous phase after the phenol extraction step of the TRIZOL protocol (QIAGEN, Gaithersburg, MD, USA). Min6 samples were reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Wilmington, DE, USA) and random primers. Islet samples were reverse transcribed using SuperScript III (Invitrogen) and oligo (dT) primers. Transcript was analyzed by quantitative PCR (BioRad CFX384) and normalized to Hprt transcript. Primer sequences are found in Table 2.
2.7. Statistics
To calculate the binding potential profiles, we ordered the Pdx1 peaks by peak strength and put them into 8 groups of decreasing strength. For each motif of interest we identified matches using a PWM and retained any match that had a log-likelihood ratio score of at least 6. To compute the binding profile of the motif for a set of Pdx1 peaks, we converted the genomic motif match coordinates to its position relative to the nearest Pdx1 peak. We used a sliding window of 200 motif matches and divided the total log-likelihood ratio score of matches in the window by the distance from the first to the last match in the window to give a local average binding potential. The median position of the matches was used as the average location of the matches in the window. For display purposes the profile was smoothed with the R function smoothspline with df = 200 to perform very gentle smoothing. For the plot the position coordinates where transformed using the square root to emphasize the binding potential profile near to the Pdx1 peak. Plots were made using the R Studio R gplot package.
The hypergeometric test was calculated using R Studio.
Data are presented as mean ± SEM. Differences between compared groups were achieved by 2-tailed Student's t-tests and considered significant when p-values were less than 0.05.
3. Results
3.1. PDX1 binds CARE sites of stress and apoptosis genes
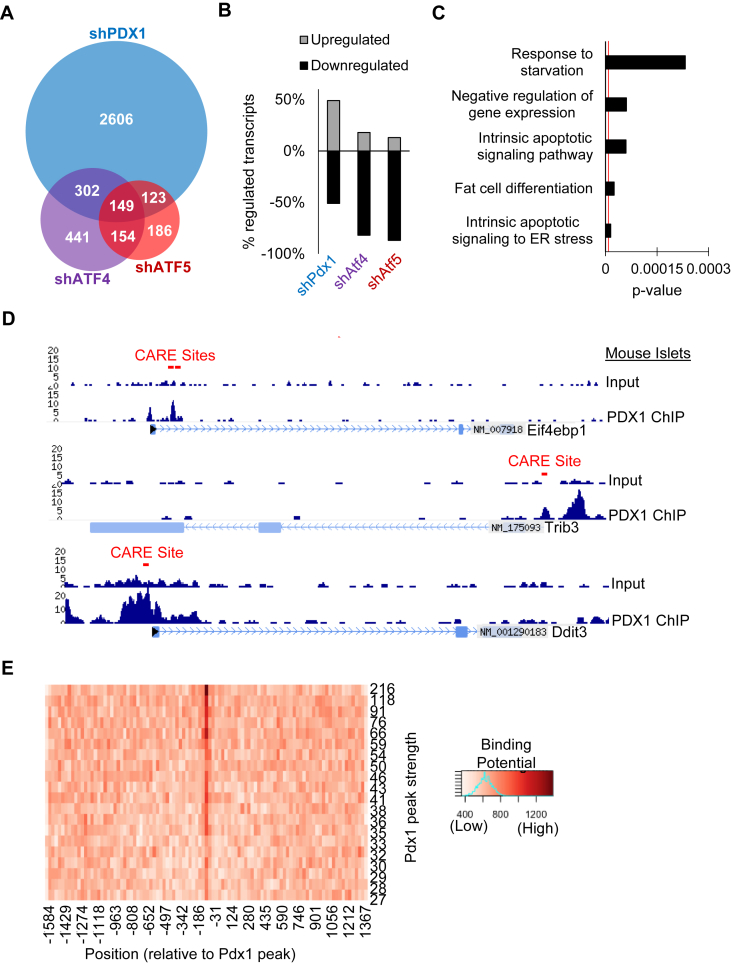
Comparison of RNAseq transcript profiles derived from Min6 cells deficient in Pdx1, Atf4, or Atf5, revealed a set of 149 genes dysregulated by all three deficiency conditions (Figure 1A, Supplemental Figure 1A). This extent of overlap was statistically significant as per calculation by the SuperExactTest R package [22]. An equal number of transcripts were upregulated and downregulated with deficiency of Pdx1; whereas Atf4 and Atf5 deficiency were each associated with predominantly down regulated transcripts (Figure 1B). In contrast, the Pdx1 target genes co-regulated with Atf4 and/or Atf5 were largely downregulated. Given the fraction of Pdx1 target genes that are downregulated, the number of genes downregulated in the Pdx1-Atf subpopulation of genes is significantly over-represented as calculated by a hypergeometric test (Supplemental Figure 1B) (Pdx1-Atf4-Atf5, p = 0.04; Pdx1-Atf4, p = 1.5 × 10−5; Pdx1-Atf5, p = 1.1 × 10−6).
Figure 1.
PDX1 enrichment at CARE sites of stress and apoptosis genes. (A) Venn diagram demonstrating the overlap of RNAseq transcript profiles from Min6 cells deficient in PDX1, ATF4, or ATF5. (B) Percentage dysregulation direction of total regulated transcripts after shRNA mediated deficiency of Pdx1, Atf4, or Atf5 in Min6 cells. (C) p-values of top categories from gene ontology analysis of the subset of 149 genes commonly regulated by deficiency of Pdx1, Atf4, and Atf5. (D) PDX1 enrichment of CARE sites associated with Eif4ebp1, Trib3, and Ddit3 in mouse islet chromatin, demonstrated by ChIPseq tracks. (E) Calculation of PDX1 binding potential to CARE sites genome wide by peak strength and position related to PDX1 enrichment peaks.
Gene ontology (GO) analysis of these 149 genes commonly dysregulated by Pdx1, Atf4, and Atf5 deficiency revealed highly significant categories associated with apoptosis and signaling in response to stress (Figure 1C). Similar GO analysis of the genes dysregulated in both Pdx1 and Atf4 deficient cells (451 transcripts) and Pdx1 and Atf5 deficient cells (272 transcripts) also revealed significant categories in stress response and apoptosis (Supplementary Figure 1C, D). Taken together, the results support the possibility of a coordinated positive regulation of apoptosis and stress related genes by PDX1, ATF4 and ATF5.
Activating transcription factors, such as ATF4 and ATF5, can bind composite ATF consensus sites, known as CARE sites, upon dimerization with certain binding partners. To investigate whether PDX1 enrichment is associated with CARE sites, we examined results from ChIPseq analysis of isolated mouse islets [8]. Intriguingly, we found PDX1 enrichment at CARE sites of a subset of the genes commonly regulated by Pdx1, Atf4, and Atf5. This list included Eif4ebp1 (Figure 1D). We recently identified a role for Eif4ebp1, regulated by ATF5, in β cell survival in response to stress at least in part through the modulation of recovery from global translational arrest [18]. Other loci with canonical roles in stress response, Ddit3 and Trib3, also exhibited PDX1 enrichment of an associated CARE site (Figure 1D).
To characterize PDX1 enrichment of CARE motifs in an unbiased manner, we visualized the locations of matches to the CARE motif by annotating the genome within 2000 bp on both sides of each Pdx1 peak for matches to the CARE motif. We then sorted the Pdx1 binding sites in order of decreasing binding strength and grouped them into 8 groups of peaks with roughly equal strength. Within each group we computed the total motif score in 133 position bins. We found that CARE motifs are typically located within the Pdx1 peak. Stronger Pdx1 peaks are more likely to have a CARE motif. The CARE binding potential is significant and about 2 times the background level (Figure 1E). Weaker Pdx1 peaks have progressively weaker CARE motif binding potential with the weakest peaks being about 10% higher than background.
To further characterize PDX1 enrichment of CARE sites, we analyzed the distribution of this enrichment in relation to the transcriptional start site (TSS) of associated nearby genes. The cumulative distribution demonstrates about 25% upstream, 35% near the TSS, and 40% downstream, presumably in the gene body (Supplementary Figure S1E). As the CARE site has base pair variability at the 3’ end of the motif (XAAX), we examined whether any of the possible base pair combinations were over represented in the subset of commonly regulated genes with PDX1 enrichment. Comparison of the possible combinations revealed no particular pattern and a relatively even distribution with slightly more representation found for AAAC (15%) and AAAT (15%) (Supplementary Figure S1F). Taken together, these results demonstrate that CARE site motifs follow a distinct pattern of enrichment by PDX1 and that this novel binding site for PDX1 is associated with apoptosis and stress response loci.
3.2. Stress inducible interactions among PDX1, ATF4, and ATF5
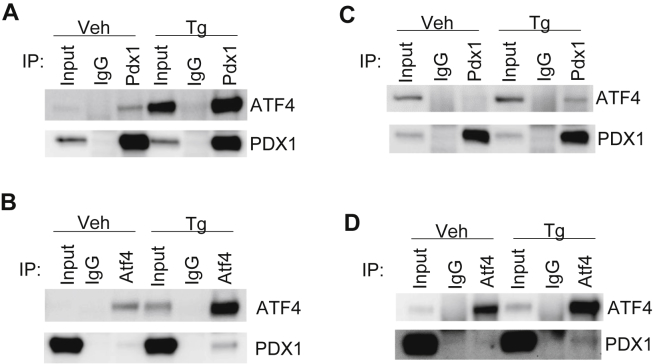
The identification of multiple non-homeobox motifs under PDX1 enrichment peaks supports the notion that PDX1 works cooperatively with other transcription factors in the regulation of gene expression, possibly in a context specific manner. We previously established that PDX1 transcriptionally regulates expression of Atf4 and Atf5 [6], [18], thus overlapping transcript targets were not unexpected; however, the discovery of PDX1 enrichment of CARE sites suggested the possibility of interaction at the protein level. Indeed, we clearly demonstrate interaction between endogenous PDX1 and ATF4 in the Min6 β cell line by co-immunoprecipitation (Figure 2A,B). The interaction is more pronounced in the context of ER stress (Figure 2A,B; right side of panels: thapsigargin (Tg)). To assess whether this interaction is conserved in primary islets, we performed co-immunoprecipitation of nuclear lysates prepared from primary isolated islets. Similar to our results in Min6 cells, immunoprecipitation of PDX1 demonstrated a stress responsive co-IP of ATF4 and vice versa in primary islets (Figure 2C,D).
Figure 2.
Stress-induced interactions among PDX1, ATF4 and ATF5. (A, B) Immunoprecipitation (IP) of nuclear lysates obtained from Min6 cells after treatment with vehicle or Tg (1uM) for 6 hrs with anti-PDX1, anti-ATF4, anti-HA, or IgG control as denoted. (C, D) IP of nuclear lysates obtained from primary isolated mouse islets cells after treatment with vehicle or Tg (1uM) for 6 hrs with anti-PDX1, anti-ATF4, or IgG control as denoted. Results are representative of at least 3 independent experiments.
In the absence of a suitable antiserum for endogenous ATF5, we created a Min6 cell line with stable overexpression of a C-terminally hemagglutinin (HA)-tagged ATF5. HA-ATF5 showed a robust interaction with ATF4 that was increased by stress, whereas a weak interaction of HA-ATF5 with PDX1 was not enhanced during stress (Supplemental Figure 2). It is possible that the interaction between PDX1 and ATF5 is impeded by the epitopes recognized by the antisera used for immunoprecipitation; however, these data could also be indicative of the formation of two separate complexes (i.e. PDX1-ATF4 and ATF4-ATF5). Our current data do not differentiate between the formation of dyad complexes or a single triad complex; indeed, both situations are possible. Thus, our results show previously undiscovered stress inducible interactions among PDX1, ATF4, and ATF5 in the pancreatic β cell.
3.3. ATF4 and ATF5 enrichment at CARE sites of stress inducible gene targets
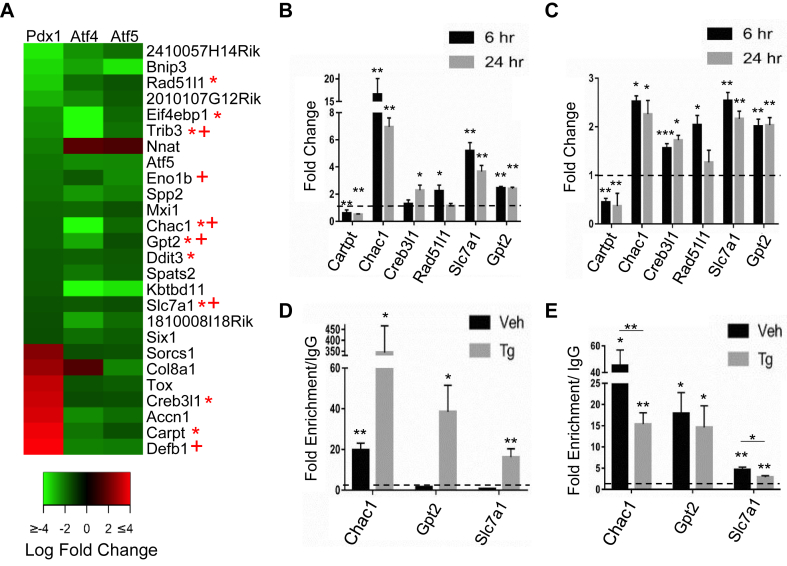
Twenty-six of the 149 genes commonly dysregulated in the setting of Pdx1, Atf4, and Atf5 deficiency also have PDX1 enrichment of an associated CARE site, determined through ChIPseq analysis of mouse islets ([8]; Supplemental Figure 3). A majority of these genes (19 of 26) were downregulated by deficiency of any of the three transcription factors indicating positive regulation by PDX1, ATF4, and ATF5 (Figure 3A). Compellingly, the list of commonly regulated targets contained several genes with either known pro-apoptotic functions (Bnip3, Rad51l1, Trib3, Chac1, Ddit3) or pro-survival functions (Eif4ebp1, Atf5, Creb3l1, Defb1). The combination of both adaptive and maladaptive gene targets suggests a checkpoint role for the PDX1 transcriptional complex (es) in the determination of β cell fate decisions. Further data mining revealed that nine of these genes shared conserved PDX1 occupancy of a CARE site containing region near the human orthologs of these genes (identified using the UCSC human LiftOver tool) (Figure 3A, (*)) [8].
Figure 3.
ATF4 and ATF5 enrichment at CARE sites of stress inducible gene targets. (A) Heatmap demonstrating the direction and magnitude of change in expression of denoted transcripts after deficiency of either Pdx1, Atf4, or Atf5 in Min6 cells. * indicates genes with PDX1 enrichment of CARE sites near the human orthologs, identified using the UCSC Genome Browser LiftOver tool. + denotes that induction of the transcript by palmitate was observed in human islets (Cnop, 2014) (B) Min6 or (C) primary isolated mouse islets treated with Tg (1uM) or vehicle for 6 or 24hrs then harvested for RNA. qPCR of Cartpt, Chac1, Creb3l1, Rad51l1, Slc7a1, and Gpt2. ChIP PCR analysis of (D) ATF4 or (E) ATF5 enrichment at the CARE sites associated with Chac1, Gpt2, and Slc7a1. Dashed line indicates value of normalized untreated control. Bars show the mean and error bars the standard error of mean (S.E.M.) of 3 independent experiments. p-values were calculated with Student's T-Test (*, p < 0.05; **, p < 0.01).
For several gene targets with no previously identified roles in β cell stress or survival, we examined the effect of stress on transcript expression. In each case, induction of ER stress affected gene expression in both Min6 cells and in primary isolated islets (Figure 3B,C). Chac1 was the most robustly induced transcript. While most of the examined transcripts were significantly induced by stress, Cartpt expression was unique in that its expression was reduced under stress conditions. Three of the transcripts (Chac1, Gpt2, Slc7a1) were upregulated at both acute (6hrs) and chronic (24hrs) time points (Figure 3B,C).
Notably, Chac1, Gpt2, Slc7a1 exhibited PDX1 enrichment of a CARE site at an orthologous human region as well as transcript induction by palmitate in human islets (Figure 3A, *+) [8], [23]. These targets were prioritized for their high likelihood of conserved function in β cell stress and survival in human islets. ChIP PCR was used to determine whether ATF4 and ATF5 enrichment is also present at the CARE motifs near these genes. Results indicate that ATF4 enrichment is stress induced at these CARE sites (Figure 3D), whereas significant ATF5 enrichment is present basally and significantly decreased with stress for Chac1 and Slc7a1, but not Gpt2 (Figure 3E). This decrease could reflect competition for binding with ATF4, which is highly recruited to the site upon stress treatment. These observations are in line with the enrichment pattern of ATF5 and ATF4 we previously found at the Atf5 and Eif4ebp1 CARE sites [18]. Our results have identified stress inducible targets with potential roles in β cell survival and marked by significant enrichment of PDX1, ATF4, and ATF5.
3.4. PDX1, ATF4, and ATF5 are required for basal and stress induced gene transcription
From our differential expression analysis, we found that deficiency of Pdx1, Atf4, and Atf5 reduced expression of Chac1, Gpt2, and Slc7a1 under basal/unstressed conditions. To determine whether chromatin enrichment at the CARE motifs of these genes was associated with a functional role in stress induced gene expression, CRISPR/Cas9 genome editing was used to assess the effects of Pdx1, Atf4, and Atf5 deficiency on stress induction of these transcripts in β cells. Reduction efficiency by CRISPR/Cas9 genome editing of PDX1 and ATF4 was assessed by immunoblot (Supplemental Figure 4). ATF5 levels could not be assessed as no ATF5 antisera suitable for Western blot analysis are currently available.
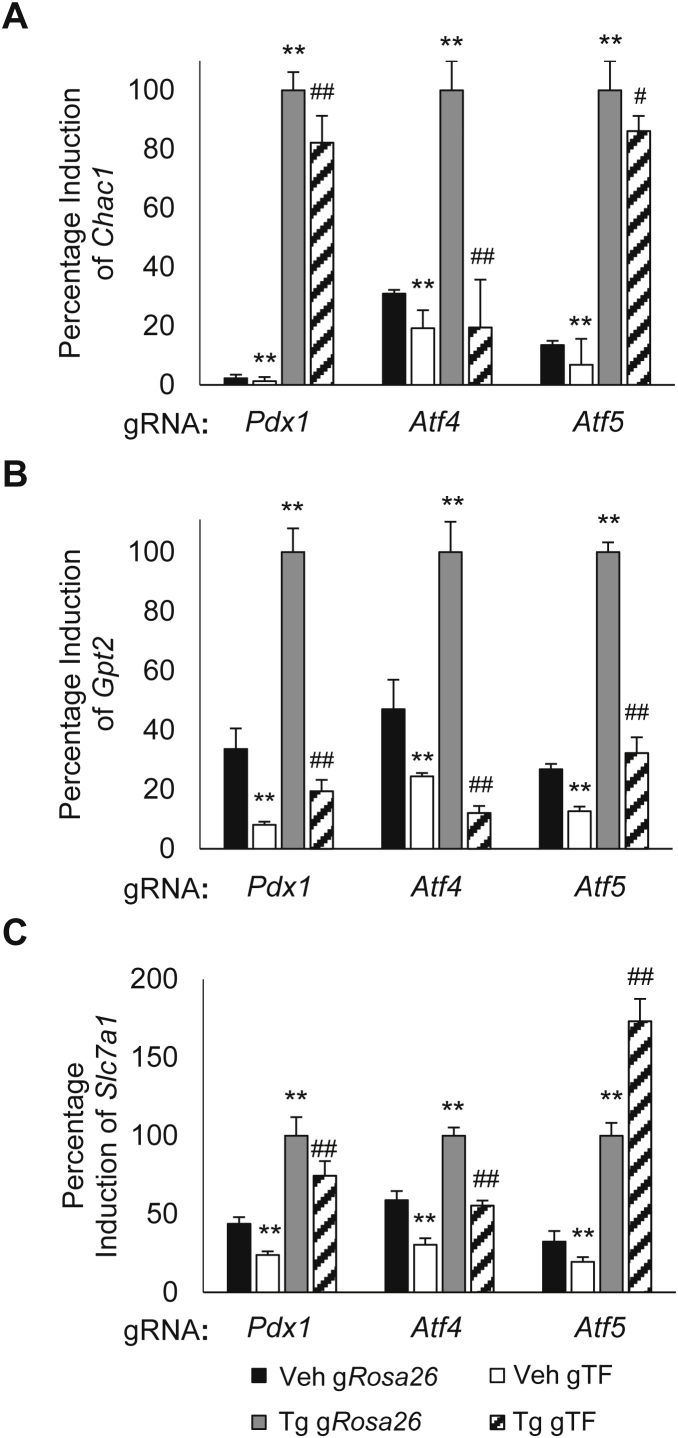
Basal transcript expression of Chac1, Gpt2, and Slc7a1 was reduced by deficiency of Pdx1, Atf4, and Atf5 (Figure 4A,B, C). Stress induction of the most robustly induced transcript, Chac1, was significantly reduced by deficiency of all three transcription factors; however, Atf4 deficiency resulted in the largest decrease (Figure 4A). Induction of Gpt2 transcript was the most uniformly reduced by deficiency of all three transcription factors: Pdx1 (80%), Atf4 (90%), and Atf5 (70%) (Figure 4B). Induction of Slc7a1 transcript was significantly reduced by deficiency of Pdx1 and Atf4, but increased by Atf5 deficiency during stress (Figure 4C). These diverse patterns of regulation for Chac1, Gpt2, and Slc7a1 support the possibility of distinct configurations of the transcriptional complex (es) containing PDX1, ATF4 and ATF5. Our results clearly indicate roles in both basal and stress induced transcriptional gene regulation by a PDX1 transcriptional complex or complexes.
Figure 4.
PDX1, ATF4, and ATF5 are required for basal and stress induced gene transcription. Percentage fold induction of (A) Chac1, (B) Gpt2, or (C) Slc7a1 after treatment with either Tg (1uM) or vehicle control for 6 hrs in Min6 cells with CRISPR/Cas9 mediated reduction of Pdx1, Atf4, Atf5, or Rosa26 control. Cells were harvested for RNA and analyzed by qPCR. Bars show the mean and error bars ±standard error of mean (S.E.M.) of 3 independent experiments. p-values were calculated with Student's T-Test (Comparison to Veh Rosa26: *, p < 0.05; **, p < 0.01; Comparison to Tg Rosa26: #, p < 0.05; ##, p < 0.01).
3.5. Gpt2 deficiency protects β cells from apoptosis due to stress
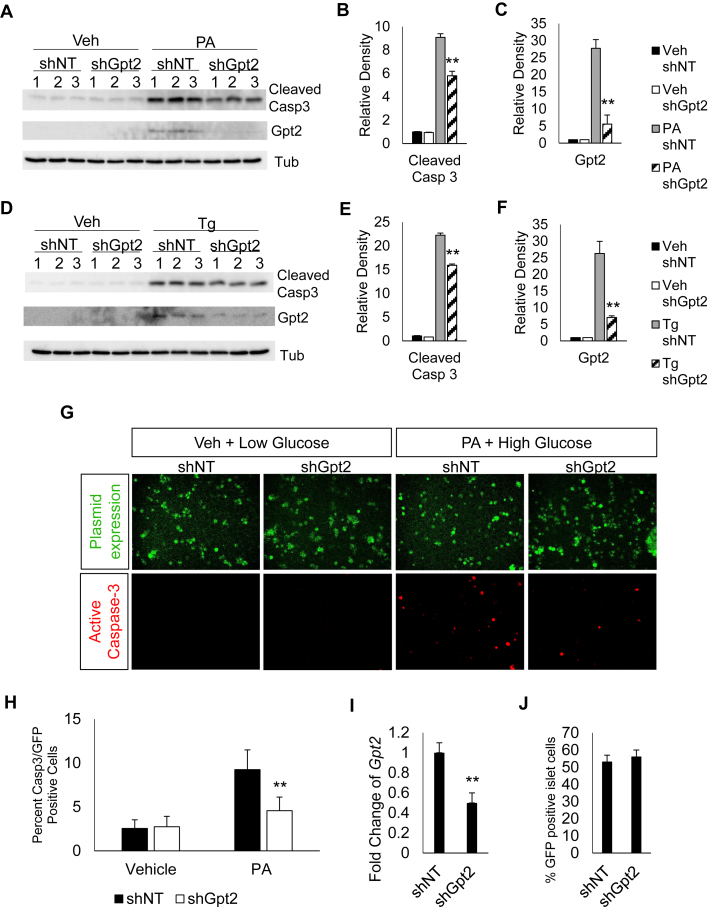
To evaluate whether Gpt2 plays a role in β cell survival during stress, we used shRNA mediated reduction of Gpt2 in Min6 cells treated with either thapsigargin (Tg) or the diabetes associated stress palmitate (PA). An ∼80% reduction in Gpt2 transcript and protein reduced the activation of caspase-3 by palmitate (Figure 5A,B, C). Similarly, treatment of Min6 cells with Tg in the context of Gpt2 deficiency resulted in a significant reduction in the activation of caspase-3 (Figure 5D,E, F).
Figure 5.
Gpt2 deficiency protects β cells from apoptosis due to stress. Min6 cells infected with lentivirus expressing shRNA targeting Gpt2 or non-targeting (NT) control, treated 96hrs post-infection with (A) PA (500uM) or vehicle control for 30 h, then harvested for protein and analyzed by western blot for cleaved caspase-3, GPT2, and tubulin. Quantitation of relative density for (B) cleaved caspase-3 or (C) GPT2. (D) Treatment with Tg (1uM) or vehicle control for 6 h, then harvested for protein and analyzed by western blot for cleaved caspase-3, GPT2, and tubulin. Quantitation of relative density for (E) cleaved caspase-3 or (F) GPT2. (G) Primary isolated mouse islets infected with lentivirus expressing GFP and shRNA targeting Gpt2 or non-targeting (NT) control, treated 48hrs post-infection with PA (500uM) + 25 mM glucose or control (Vehicle + 11 mM glucose). Stained for active caspase-3, dissociated into single cells and cytospun onto a slide. (H) Cell counts of caspase-3 positive/GFP positive cells. Bars show the mean and error bars the standard error of mean (S.E.M.) of 3 independent experiments. p-values were calculated with Student's T-Test (**, p < 0.01; compared to shNT + stress).
To examine the role of Gpt2 in β cell survival in primary β cells, we employed lentiviral delivery of shRNA constructs with a GFP expression cassette to primary isolated mouse islets. Treatment of primary isolated islets with the pathophysiologically relevant glucolipotoxic stress (PA + high glucose) increased the number of cells containing activated caspase-3 ∼2.5 fold compared to controls; this was markedly attenuated by Gpt2 deficiency (Figure 5G,H). We achieved highly efficient deletion of Gpt2 in ∼55% of islet cells (Figure 5I,J). A multitude of studies have characterized the susceptibility of beta cells to glucolipotoxicity in beta cell lines, rodent islets, and human islets [24], [25], [26], [27]. Importantly, it has been established that alpha cells are resistant to palmitate induced apoptosis [28]; thus, the cells positive for active caspase-3 in response to palmitate treatment are likely β cells. Taken together, these results indicate that the PDX1 transcriptional complex gene target, Gpt2, has a distinct role in β cell survival in response to stress.
4. Discussion
Our results show that the transcription factors PDX1, ATF4, and ATF5 form a stress responsive complex or complexes and co-occupy CARE sites to regulate apoptosis and stress response genes. We discovered a multitude of downstream targets regulated by the stress responsive PDX1 transcriptional complex, including several pro-apoptotic genes (Bnip3, Rad51l1, Trib3, Chac1, Ddit3) and several pro-survival genes (Eif4ebp1, Atf5, Creb3l1, Defb1). The pro-apoptotic gene targets function through various mechanisms including BCL2 binding (Bnip3) [29], [30], DNA damage sensing (Rad51l1) [31], and glutathione degradation (Chac1) [32], [33], transcriptional regulation (Ddit3, Trib3) [34], [35], [36], [37], [38], [39], [40], whereas the pro-survival gene targets function through the regulation of translation (Eif4ebp1, Atf5) [11], [18], transcription (Atf5, Creb3l1) [18], [41], and secreted peptide signaling (Defb1) [42].
Several gene targets (Gpt2, Chac1, and Slc7a1) were raised to a higher priority by the conservation of stress induction and PDX1 enrichment of CARE sites in human islets, suggesting a conserved role in human islet biology. SLC7A1 is a cationic amino acid transporter located in the plasma membrane of the cell that may function to regulate arginine transport [43]. CHAC1 is a gamma-glutamylcyclotransferase that degrades glutathione [32]. Interestingly, Chac1 expression is also a marker for a recently identified iron dependent, necrosis-like form of cell death called ferroptosis [44].
Here, we show that upregulation of Gpt2 contributes to stress induced apoptosis in β cells. GPT2 is a rapid equilibrium transaminase that catalyzes a glutaminolysis reaction converting glutamate and pyruvate into α-ketoglutarate and alanine. The production of α-ketoglutarate feeds into the tricarboxylic acid (TCA) cycle, resulting in synthesis of ATP [45], [46]. There are several ATP dependent steps necessary for apoptotic signaling (caspase activation and chromatin condensation); thus limitation of ATP synthesis by Gpt2 deficiency could delay or inhibit apoptotic signaling [47]. Notably, replenishment of TCA cycle intermediates (TCA anaplerosis) is of particular importance for cancer cells due to the glutamine dependence of cancer cells for survival and proliferation [48], [49]. Recent reports implicate Gpt2 upregulation as a key step in the reprogramming of glutamine metabolism of cancer cells [50], [51]. We speculate that Gpt2 also reprograms glutamine metabolism in β cells.
The identification of Pdx1 targets with negative roles in cell survival appears incongruous with its in vivo role to promote β cell survival [6], [52]. We draw comparison to ATF4, which is well recognized for its dichotomy of function in regulating both pro-survival and pro-apoptotic pathways. ATF4 regulates targets necessary for recovery from stress (amino acid transport, protection from oxidative stress, and protein homeostasis) [53], [54], [55], [56] as well targets involved in cell death (Ddit3, Trib3) [35], [36], [37], [40]. Rather than promoting cell survival, the PDX1-ATF4 complex may function as a gatekeeper, integrating positive and negative signals to determine β cell fate. Our identification of this complex and its targets introduces a new avenue for therapeutic intervention to prevent or ameliorate diabetes by improving β cell survival.
Author contributions
CAJ and DAS designed experiments and analyzed data. CAJ, JY, CEC, AG, and MWH conducted experiments, acquired data, and analyzed data. CAJ and DAS wrote and edited the manuscript.
Acknowledgements
This work was supported by an Innovative Basic Science grant from the American Diabetes Association 1-18-1BS-052 (to DAS), National Institutes of Health Grants P01 DK49210 (to DAS) and F32-DK103454 (to CAJ). We thank Jonathan Schug and Shilpa Rao of the Next Generation Sequencing Core (NGSC) at the University of Pennsylvania for support in motif and high throughput statistical analysis. We gratefully acknowledge support from the Penn Diabetes Research Center (DK019525).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2018.07.007.
Conflict of interest
The authors have no conflict of interest to declare.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplemental Figure 1: (A) Min6 cells infected with lentivirus expressing shRNA targeting Pdx1, Atf4, Atf5 or non-targeting (NT) control, collected 96hrs post-infection for RNA and qPCR analysis. (B) Percentage of regulated transcripts (Figure 1A) that are concordantly downregulated/ upregulated or discordantly mixed for genes commonly regulated by PDX1-ATF4-ATF5, PDX1-ATF4, PDX1-ATF5, and ATF4-ATF5. (C) p-values of top categories from GO analysis of the 451 genes commonly regulated by PDX1 and ATF4. (D) p-values of top categories from GO analysis of the 272 genes commonly regulated by PDX1 and ATF5. (E) Distribution of PDX1 enrichment in relation to the transcriptional start site (TSS) of associated genes. (F) Pie chart representing the base pair combinations of the variable portion of the CARE site sequence (XAAX) and the percentage with PDX1 enrichment from the list of commonly regulated genes. Supplemental Figure 2: Immunoprecipitation of nuclear lysates obtained from Min6 cells stably expressing HA-ATF5 after treatment with vehicle or Tg (1uM) for 6 hrs with anti-HA or IgG control. Supplemental Figure 3: (A) ChIPseq tracks from mouse islets showing PDX1 enrichment at CARE sites associated with Cartpt, Creb3l1, Chac1, Slc7a1, Gpt2, Rad51b. Supplemental Figure 4: (A) Min6 cells infected with lentivirus expressing CRISPR/ Cas9 mediated reduction of Pdx1, Atf4, or Rosa26 control. 96 post-infection cells were treated with Tg (1uM) or vehicle control for 6 h and then collected for protein. WB analysis of ATF4, PDX1, and Tubulin. * denotes a non-specific band. Quantitation of relative density for (B) ATF4 or (C) PDX1.
References
- 1.Brissova M., Shiota M., Nicholson W.E., Gannon M., Knobel S.M., Piston D.W. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. Journal of Biological Chemistry. 2002;277(13):11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 2.Kulkarni R.N., Jhala U.S., Winnay J.N., Krajewski S., Montminy M., Kahn C.R. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. The Journal of Clinical Investigation. 2004;114(6):828–836. doi: 10.1172/JCI21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohlsson H., Karlsson K., Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. The EMBO Journal. 1993;12(11):4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen H.V., Serup P., Leonard J., Michelsen B.K., Madsen O.D. Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(22):10465–10469. doi: 10.1073/pnas.91.22.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raum J.C., Gerrish K., Artner I., Henderson E., Guo M., Sussel L. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Molecular and Cellular Biology. 2006;26(15):5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sachdeva M.M., Claiborn K.C., Khoo C., Yang J., Groff D.N., Mirmira R.G. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proceedings of the National Academy of Sciences. 2009;106(45):19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sachdeva M.M., Stoffers D.A. Minireview: meeting the demand for insulin: molecular mechanisms of adaptive postnatal ß-cell mass expansion. Molecular Endocrinology. 2009;23(6):747–758. doi: 10.1210/me.2008-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khoo C., Yang J., Weinrott S.A., Kaestner K.H., Naji A., Schug J. Research resource: the Pdx1 cistrome of pancreatic islets. Molecular Endocrinology. 2012;26(3):521–533. doi: 10.1210/me.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fawcett T.W., Martindale J.L., Guyton K.Z., Hai T., Holbrook N.J. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. The Biochemical Journal. 1999;339(Pt 1):135–141. [PMC free article] [PubMed] [Google Scholar]
- 10.Persengiev S.P., Green M.R. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003:225–228. doi: 10.1023/a:1023633704132. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi S., Ishihara H., Yamada T., Tamura A., Usui M., Tominaga R. ATF4-Mediated induction of 4E-BP1 contributes to pancreatic β cell survival under endoplasmic reticulum stress. Cell Metabolism. 2008;7(3):269–276. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Han J., Back S.H., Hur J., Lin Y.-H., Gildersleeve R., Shan J. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biology. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences. 2004;101(31):11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatano M., Umemura M., Kimura N., Yamazaki T., Takeda H., Nakano H. The 5’-untranslated region regulates ATF5 mRNA stability via nonsense-mediated mRNA decay in response to environmental stress. The FEBS Journal. 2013;280(18):4693–4707. doi: 10.1111/febs.12440. [DOI] [PubMed] [Google Scholar]
- 15.Dluzen D., Li G., Tacelosky D., Moreau M., Liu D.X. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. The Journal of Biological Chemistry. 2011;286(9):7705–7713. doi: 10.1074/jbc.M110.207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumi S., Saito A., Kanemoto S., Kawasaki N., Asada R., Iwamoto H. The endoplasmic reticulum stress transducer BBF2H7 suppresses apoptosis by activating the ATF5-MCL1 pathway in growth plate cartilage. The Journal of Biological Chemistry. 2012;287(43):36190–36200. doi: 10.1074/jbc.M112.373746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou D., Palam L.R., Jiang L., Narasimhan J., Staschke K.A., Wek R.C. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. The Journal of Biological Chemistry. 2008;283(11):7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 18.Juliana C.A., Yang J., Rozo A.V., Good A., Groff D.N., Wang S.-Z. ATF5 regulates β-cell survival during stress. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(6):1341–1346. doi: 10.1073/pnas.1620705114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jimenez-Moreno C., de Gracia Herrera-Gomez I., Lopez-Noriega L., Lorenzo P., Cobo-Vuilleumier N., Fuente-Martin E. A simple high efficiency intra-islet transduction protocol using lentiviral vectors. Current Gene Therapy. 2015;15(4):436–446. doi: 10.2174/1566523215666150630121557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber E., Miller M.M., Chaffner W., Mathias P. In: Tissue specific gene expression. Renkawitz R., editor. 1989. pp. 33–54. [Google Scholar]
- 21.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Wang M., Zhao Y., Zhang B. Efficient test and visualization of multi-set intersections. Scientific Reports. 2015;5(1):16923. doi: 10.1038/srep16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cnop M., Abdulkarim B., Bottu G., Cunha D.A., Igoillo-Esteve M., Masini M. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63(6):1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 24.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 25.El-Assaad W., Buteau J., Peyot M.-L., Nolan C., Roduit R., Hardy S. Saturated fatty acids synergize with elevated glucose to cause pancreatic β-cell death. Endocrinology. 2003;144(9):4154–4163. doi: 10.1210/en.2003-0410. [DOI] [PubMed] [Google Scholar]
- 26.Lupi R., Dotta F., Marselli L., Del Guerra S., Masini M., Santangelo C. Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that beta-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes. 2002;51(5):1437–1442. doi: 10.2337/diabetes.51.5.1437. [DOI] [PubMed] [Google Scholar]
- 27.Maedler K., Spinas G.A., Dyntar D., Moritz W., Kaiser N., Donath M.Y. Distinct effects of saturated and monounsaturated fatty acids on beta-cell turnover and function. Diabetes. 2001;50(1):69–76. doi: 10.2337/diabetes.50.1.69. [DOI] [PubMed] [Google Scholar]
- 28.Marroqui L., Masini M., Merino B., Grieco F.A., Millard I., Dubois C. 2015. Pancreatic α cells are resistant to metabolic stress-induced apoptosis in type 2 diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B., Xiao J.-L., Ling Y.-H., Meng X.-J., Wu B., Yang X.-Y. BNIP3 upregulation by ERK and JNK mediates cadmium-induced necrosis in neuronal cells. Toxicological Sciences. 2014;140(2):393–402. doi: 10.1093/toxsci/kfu091. [DOI] [PubMed] [Google Scholar]
- 30.Diao H., Liu B., Shi Y., Song C., Guo Z., Liu N. MicroRNA-210 alleviates oxidative stress-associated cardiomyocyte apoptosis by regulating BNIP3. Bioscience Biotechnology and Biochemistry. 2017;81(9):1712–1720. doi: 10.1080/09168451.2017.1343118. [DOI] [PubMed] [Google Scholar]
- 31.Thacker J. The role of homologous recombination processes in the repair of severe forms of DNA damage in mammalian cells. Biochimie. 1999;81(1–2):77–85. doi: 10.1016/s0300-9084(99)80041-8. [DOI] [PubMed] [Google Scholar]
- 32.Crawford R.R., Prescott E.T., Sylvester C.F., Higdon A.N., Shan J., Kilberg M.S. Human CHAC1 protein degrades glutathione, and mRNA induction is regulated by the transcription factors ATF4 and ATF3 and a bipartite ATF/CRE regulatory element. The Journal of Biological Chemistry. 2015;290(25):15878–15891. doi: 10.1074/jbc.M114.635144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A., Tikoo S., Maity S., Sengupta S., Sengupta S., Kaur A. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Reports. 2012;13(12):1095–1101. doi: 10.1038/embor.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teske B.F., Fusakio M.E., Zhou D., Shan J., McClintick J.N., Kilberg M.S. CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Molecular Biology of the Cell. 2013;24(15):2477–2490. doi: 10.1091/mbc.E13-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. The Journal of Clinical Investigation. 2002;109(4):525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X.Z., Lawson B., Brewer J.W., Zinszner H., Sanjay A., Mi L.J. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Molecular and Cellular Biology. 1996;16(8):4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes & Development. 1998;12(7):982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang N., Zhang W., Xu S., Lin H., Wang Z., Liu H. TRIB3 alters endoplasmic reticulum stress-induced β-cell apoptosis via the NF-κB pathway. Metabolism. 2014;63(6):822–830. doi: 10.1016/j.metabol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Qian B., Wang H., Men X., Zhang W., Cai H., Xu S. TRIB3 [corrected] is implicated in glucotoxicity- and endoplasmic reticulum-stress-induced [corrected] beta-cell apoptosis. The Journal of Endocrinology. 2008;199(3):407–416. doi: 10.1677/JOE-08-0331. [DOI] [PubMed] [Google Scholar]
- 40.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. The EMBO Journal. 2005;24(6):1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asada R., Kanemoto S., Kondo S., Saito A., Imaizumi K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. Journal of Biochemistry. 2011:507–518. doi: 10.1093/jb/mvr041. [DOI] [PubMed] [Google Scholar]
- 42.Sun C.Q., Arnold R., Fernandez-Golarz C., Parrish A.B., Almekinder T., He J. Human beta-defensin-1, a potential chromosome 8p tumor suppressor: control of transcription and induction of apoptosis in renal cell carcinoma. Cancer Research. 2006;66(17):8542–8549. doi: 10.1158/0008-5472.CAN-06-0294. [DOI] [PubMed] [Google Scholar]
- 43.Erős D., Őrfi L., Csuka I., Kéri G., Hrabák A. Binding specificity of the l-arginine transport systems in mouse macrophages and human cells overexpressing the cationic amino acid transporter hCAT-1. Amino Acids. 2009;36(3):483–492. doi: 10.1007/s00726-008-0106-x. [DOI] [PubMed] [Google Scholar]
- 44.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W., Li H., Ogando D.G., Li S., Feng M., Price F.W. Glutaminolysis is essential for energy production and ion transport in human corneal endothelium. EBioMedicine. 2017;16:292–301. doi: 10.1016/j.ebiom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Molecular Cell. 2015;59(2):298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eguchi Y., Shimizu S., Tsujimoto Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Research. 1997;57(10):1835–1840. [PubMed] [Google Scholar]
- 48.Hosein A.N., Beg M.S. Pancreatic cancer metabolism: molecular mechanisms and clinical applications. Current Oncology Reports. 2018;20(7):56. doi: 10.1007/s11912-018-0699-5. [DOI] [PubMed] [Google Scholar]
- 49.Muir A., Danai L.V., Gui D.Y., Waingarten C.Y., Lewis C.A., Vander Heiden M.G. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. eLife. 2017;6 doi: 10.7554/eLife.27713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao Y., Samuels Y., Li Q., Krokowski D., Guan B.-J., Wang C. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nature Communications. 2016;7:11971. doi: 10.1038/ncomms11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith B., Schafer X.L., Ambeskovic A., Spencer C.M., Land H., Munger J. Addiction to coupling of the Warburg effect with glutamine catabolism in cancer cells. Cell Reports. 2016;17(3):821–836. doi: 10.1016/j.celrep.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson J.D., Ahmed N.T., Luciani D.S., Han Z., Tran H., Fujita J. Increased islet apoptosis in Pdx1+/- mice. The Journal of Clinical Investigation. 2003;111(8):1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wortel I.M.N., van der Meer L.T., Kilberg M.S., van Leeuwen F.N. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends in Endocrinology and Metabolism: TEM. 2017;28(11):794–806. doi: 10.1016/j.tem.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 55.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular Cell. 2003;11(3):619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 56.Chan J.Y., Luzuriaga J., Maxwell E.L., West P.K., Bensellam M., Laybutt D.R. The balance between adaptive and apoptotic unfolded protein responses regulates β-cell death under ER stress conditions through XBP1, CHOP and JNK. Molecular and Cellular Endocrinology. 2015;413:189–201. doi: 10.1016/j.mce.2015.06.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: (A) Min6 cells infected with lentivirus expressing shRNA targeting Pdx1, Atf4, Atf5 or non-targeting (NT) control, collected 96hrs post-infection for RNA and qPCR analysis. (B) Percentage of regulated transcripts (Figure 1A) that are concordantly downregulated/ upregulated or discordantly mixed for genes commonly regulated by PDX1-ATF4-ATF5, PDX1-ATF4, PDX1-ATF5, and ATF4-ATF5. (C) p-values of top categories from GO analysis of the 451 genes commonly regulated by PDX1 and ATF4. (D) p-values of top categories from GO analysis of the 272 genes commonly regulated by PDX1 and ATF5. (E) Distribution of PDX1 enrichment in relation to the transcriptional start site (TSS) of associated genes. (F) Pie chart representing the base pair combinations of the variable portion of the CARE site sequence (XAAX) and the percentage with PDX1 enrichment from the list of commonly regulated genes. Supplemental Figure 2: Immunoprecipitation of nuclear lysates obtained from Min6 cells stably expressing HA-ATF5 after treatment with vehicle or Tg (1uM) for 6 hrs with anti-HA or IgG control. Supplemental Figure 3: (A) ChIPseq tracks from mouse islets showing PDX1 enrichment at CARE sites associated with Cartpt, Creb3l1, Chac1, Slc7a1, Gpt2, Rad51b. Supplemental Figure 4: (A) Min6 cells infected with lentivirus expressing CRISPR/ Cas9 mediated reduction of Pdx1, Atf4, or Rosa26 control. 96 post-infection cells were treated with Tg (1uM) or vehicle control for 6 h and then collected for protein. WB analysis of ATF4, PDX1, and Tubulin. * denotes a non-specific band. Quantitation of relative density for (B) ATF4 or (C) PDX1.