Abstract
Background
Oculocutaneous albinism (OCA) is a group of autosomal recessive disorders characterized by reduced melanin that are caused by mutations in the gene encoding tyrosinase (TYR), which is the rate-limiting enzyme in the production of the pigment melanin. Many studies or meta-analyses have suggested an association between the TYR T373K SNP and OCA1, but there is limited biochemical and genetic evidence to support this association.
Methods
We overexpressed TYR-WT and TYR-T373K mutants on HK293T cells and tested the changes of melanin production and tyrosinase activity. Then we generated TYR-K373T knock-in (KI) rabbits by microinjection of ssDNA and synthesized RNAs targeting C1118A using CRISPR/Cas9-HDR to observe the formation of melanin.
Findings
We demonstrated that the T373K mutation in TYR can reduce tyrosinase activity, leading to an absence of melanin synthesis at the cell-level. The gene-edited TYR-K373T rabbits exhibited rescued melanin production in hair follicles and irises, as inferred from the evident decrease in pigmentation in TYR-T373K rabbits, thus providing functional validation of the albinism-associated T373K SNP at the animal level.
Interpretation
Our study provides the first animal-level functional validation of the albinism-associated TYR K373T SNP in rabbits, and these results will facilitate gene therapy of OCA1 in pre-clinical settings in the future.
Fund
The National Key Research and Development Program of China Stem Cell and Translational Research, the Strategic Priority Research Program of the Chinese Academy of Sciences, the Guangdong Province Science and Technology Plan Project, and the Program for JLU Science and Technology Innovative Research Team.
Keywords: Tyrosinase, OCA, T373K, Point mutation, Rabbit, CRISPR/Cas9
Research in context.
Evidence before this study
Oculocutaneous albinism (OCA) is a group of autosomal recessive disorders characterized by hypopigmented hair, skin and eyes. According to the human gene mutation database (HGMD), 249 missense/nonsense mutations affecting 177 amino acids in TYR are associated with OCA1. Many studies or meta-analysis suggested the association between the TYR T373K SNP and oculocutaneous albinism. However, information regarding the molecular mechanisms underlying the frequent T373K mutation is limited. In this study, we providing functional validation of the albinism-associated T373K SNP at the cell and animal level and explored the possibility that T373K SNP (C1118A) is response for the reduced pigment in OCA1 patients.
Added value of this study
The prevalence of OCA is often estimated as being 0.05–1% of the population, and OCA is the most common cause of depigmentation worldwide. Notably, there is no suitable and effective clinical treatment for OCA currently. We are the first group to explore functional validation of the albinism-associated T373K SNP at animal level. More importantly, new trends in disease research show that gene therapy has emerging clinical potential in cancer therapy. But the biochemical function and genetic evidence to support this association of T373K SNP and oculocutaneous albinism is lacking. Hence, findings in the presents study could provide novel insights into clinical treatment of OCA.
Implications of all the available evidence
Here, we identified functional validation of the albinism-associated TYR K373T SNP in rabbits, which could be the targets for gene therapy of OCA1 in pre-clinical settings in the future. Furthermore, decreased tyrosinase activity has been targeted for the prevention of skin hyperpigmentation, such as melasma and age spots. Therefore, the T373K locus could be a potential therapeutic target for further research, and restoration of tyrosinase production and inhibition of tyrosinase could be used to treat skin disorders.
Alt-text: Unlabelled Box
1. Introduction
Oculocutaneous albinism (OCA) is a group of autosomal recessive disorders characterized by hypopigmented hair, skin and eyes [1,2]. Distinctive ocular changes occur in all types of OCA, including nystagmus and reduced pigmentation of the iris and retina [3]. The prevalence of OCA is often estimated as being 0.05–1% of the population, and OCA is the most common cause of depigmentation worldwide [4]. Moreover, all individuals with OCA are at greater risk of developing squamous cell carcinoma of sun-exposed skin than the general population [5]. Notably, there is no suitable and effective clinical treatment for OCA currently [2].OCA type 1 (OCA1) is the most severe and common form of OCA, characterized by complete absence of pigment in the skin, hair, and eyes, which is caused by mutations in the tyrosinase gene (TYR) [6].
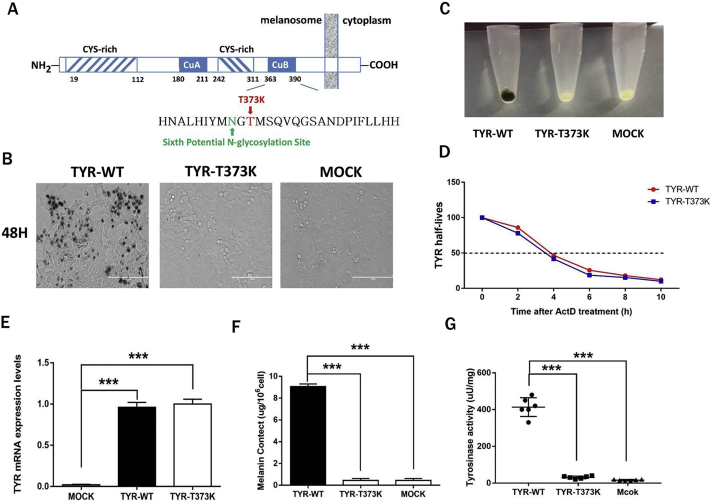
Tyrosinase is a copper-containing enzyme that catalyses the first two steps of the melanin biosynthesis pathway, converting tyrosine to L-dihydroxy-phenylalanine (DOPA) and subsequently to DOPA-quinone [1,6]. According to the human gene mutation database (HGMD) (http://www.hgmd.org/) [7], 249 missense/nonsense mutations affecting 177 amino acids in TYR are associated with OCA1 [8], including the frequent human albino substitution (c. C1118A; p. Thr373Lys) in codon 373, which leads to a putative amino acid change from threonine (ACA) to lysine (AAA) in exon 3 of the TYR gene. Previous research has shown that the T373K mutation influenced melanogenesis in vitro experiment [9]. Tyrosinase has two conserved sequence motifs to bind two copper ions, termed CuA and CuB, and also has six N-glycosylation sequences sites. Interestingly, the T373K mutation was located on CuB and interfered the sixth potential N-glycosylation site (Fig. 1A). Previous study has confirmed that the CuA and CuB domains of human tyrosinase are important for copper-binding and catalytic activity [[10], [11], [12], [13]], and mutations at glycosylation sites critical to the operation of the quality control mechanism lead to a reduction in the activity of the expressed protein [[14], [15], [16], [17]]. However, information regarding the molecular mechanisms underlying the frequent T373 K mutation is limited. Thus, biochemical functional assays and elucidation of the genetic mechanism at the animal level are required to confirm the association between the T373K SNP and OCA1.
Fig. 1.
Reduced melanin synthesis in TYR-T373K mutant cells.
(A) Schematic representation of the sequence of human tyrosinase CYS-rich, regions rich in conserved cysteine residues; CuA and CuB, proposed binding sites for the two copper atoms based on sequence comparisons with other copper-binding proteins. (B) Overexpression of the WT-TYR and TYR-T373K mutant genes in HEK293T cells. (C) The colours of the transfected cell pellets were compared between the WT-TYR- and TYR-T373K-transfected cells. (D) mRNA half-lives were compared between WT-TYR and TYR-T373K. (E) Gene mRNA expression was compared between WT-TYR and TYR-T373K. (F) Melanin content was compared between the WT-TYR- and TYR-T373K-transfected cells. (G) Tyrosinase activity was compared between WT-TYR and TYR-T373K. Mock, negative control. ***, p < .001.
To date, most of the preliminary functional validation of albinism-associated tyrosinase mutations has been carried out in albino mice, which are widely used animal models for pathogenesis and pre-clinical studies [[18], [19], [20], [21]]. However, mice with the TYR (C85S) mutation have been frequently used for albinism studies, and no genetic evidence for the T373K mutation has been reported to date. Interestingly, the identical mutation (T373K) was identified in both albino rabbits and humans OCA1 [22]. Furthermore, rabbits are phylogenetically closer to primates than mice and are physically large enough to permit non-lethal monitoring of physiological changes.
The CRISPR/Cas9 system has been shown to be an efficient gene-targeting technology in mammalian cells [23,24] and animals [21,[25], [26], [27]]. In addition, the combination of CRISPR/Cas9 with single-stranded DNA (ssDNA) donors has been developed as an efficient genome engineering tool to introduce site-directed point mutations [28]. To date, several gene knockout (KO) rabbit models have been generated using the CRISPR/Cas9 system to study human hereditary diseases [27,29,30]; however, the rate of site-directed point mutation using CRISPR/Cas9-mediated homology-directed repair (HDR) remains to be further improved [31].
In this study, we analysed the molecular mechanisms underlying the effects of T373K, a frequent SNP associated with OCA1, demonstrating that reduction of tyrosinase activity by the T373K mutation is responsible for reduced pigmentation and inhibits melanogenesis in OCA1. Moreover, we achieved base replacement (A-C) by microinjecting synthesized RNAs with ssODN donors, leading to rescued melanin production in the skin and irises of TYR-K373T rabbits.
2. Materials and methods
2.1. Ethics statement
The New Zealand white rabbits and Lianshan black rabbits used in this study were obtained from the Laboratory Animal Center of Jilin University. All animal studies were conducted according to experimental practices and standards approved by the Animal Welfare and Research Ethics Committee at Jilin University.
2.2. Vectors construction and cell culture
The coding sequences (CDSs) of TYR-WT and TYR-T373K were amplified by RT-PCR from the Lianshan black rabbit and New Zealand white rabbit cDNAs, respectively. The purified PCR products were cut with EcoR I and Nhe I and then cloned into the backbone of the PIRES-CMV vector to obtain the PIRES-TYR-WT and PIRES-TYR-T373K vectors, which were verified by Sanger sequencing. The primers used are listed in Table S1.
Human kidney epithelial cells (HEK293T) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum (HyClone), 2 mM GlutaMAX (Life Technologies), 100 U/ml penicillin, and 100 mg/ml streptomycin and incubated at 37 °C in an atmosphere containing 5% CO2. The cells were seeded into 6-well plates at a density of 120,000 per well and transfected using Turbofect TM in vitro transfection reagent (Fermentas) according to the manufacturer's instructions. The PIRES-TYR-T373K and the PIRES-TYR-WT vectors were transiently transfected into the HEK293T cells, and melanin formation was determined by fluorescence microscopy (NikonTS100, Tokyo, Japan).
2.3. Melanin content determination
Transfected cells were seeded at a density of 2 × 105 in 60-mm dishes. After overnight incubation, the cells were cultured in the presence or absence of arenarol for 72 h. The cells were washed twice with phosphate-buffered saline (PBS) and harvested by treatment with 0.05% trypsin/0.02% EDTA. Then, the cell pellets were dissolved in 1 N NaOH, followed by incubation at 60 °C for 2 h. The amount of melanin in the solution was determined by measuring the absorbance at 405 nm. The melanin content was expressed as delta absorbance (405 nm)/lg protein.
2.4. mRNA stability assay
For the mRNA stability assay, HEK293T cells were transfected with the PIRES-TYR-WT and PIRES-TYR-T373K vectors in 6-cm dishes. Twenty-four hours after transfection, actinomycin D (ActD; 10 mg/ml) was added to the culture medium to block mRNA transcription, and then, the cells were harvested at selected time points. Total RNA was extracted using TRIzol.
Total RNA concentrations for the WT and mutant were quantified by spectrophotometry. The cDNAs were obtained by reverse transcription with the PrimeScriptTM RT Reagent Kit (TaKaRa, Japan) from 500 ng of total RNA. Total mRNA levels were normalized to GAPDH expression. All samples were analysed in triplicate, and each mRNA quantification represents an average of at least three measurements. All the data are expressed as the mean ± SEM.
2.5. Intracellular tyrosinase activity
Intracellular tyrosinase activity was determined as previously described [32]. Briefly, the cells were washed twice with PBS and homogenized with 50 mM PBS (pH 7.5) containing 1.0% Triton X-100 and 0.1 mM phenylmethyl-sulfonyl fluoride (PMSF). Cellular extracts (100 μl) were mixed with freshly prepared L-DOPA solution (5.0 mM in 50 mM PBS, pH 6.8) and incubated at 37 °C for 30 min. The absorbance at 490 nm was measured with a Gen5™ microplate reader (BioTek Instruments, Winooski, VT, USA) to measure the production of dopachrome.
2.6. DNA constructs and microinjection
The method for the design of targeted sgRNAs has been described previously (http://crispr.mit.edu/) [25]. The sgRNAs were transcribed using the T7 RNA Synthesis Kit (Ambion) and purified using the miRNeasy Mini Kit (Qiagen) according to the manufacturers' instructions. The concentration and quality of the synthesized mRNAs were determined by a NanoDrop 2000 and agarose gel electrophoresis, respectively. The Cas9 expression construct, namely, 3xFLAG-NLS-SpCas9-NLS, was synthesized and cloned into the pCS2-vector. The construct was linearized with NotI and transcribed in vitro using the mMESSAGE mMACHINE SP6 Kit (Ambion, USA) and the RNeasy Mini Kit (Qiagen).
The protocol for microinjection of pronuclear-stage rabbit zygotes has been described in detail in our previous publication [33]. Briefly, mixtures of in vitro-transcribed mRNA derived from the gRNAs (25 ng/μl), Cas9 (100 ng/μl) and ssODN (5 ng/μl) were injected into the cytoplasm of pronuclear stage embryos. Then, the injected embryos were transferred to embryo culture medium for 30–60 min, followed by transfer into the oviduct of the recipient mother (approximately 30–50 embryos).
2.7. Mutation detection in blastocysts and pups by PCR
Each injected zygote was collected at the blastocyst stage and incubated in embryo lysis buffer at 50 °C for 20 min and 90 °C for 5 min in a Bio-Rad PCR machine. Genomic DNA from the mutant and WT rabbits was isolated using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) according to the manufacturer's instructions. The PCR primers used for mutation detection are listed in Table S1. The PCR conditions were as follows: 98 °C for 3 min; 35 cycles of 98 °C for 10 s, 60 °C for 20 s, and 72 °C for 10 s; and extension at 72 °C for 5 min, after which the reaction was held at 12 °C. The PCR products were gel purified and cloned into the pGM-T vector (Tiangen, Beijing, China). Ten positive plasmid clones were sequenced, and DNAman was used for sequence analysis.
2.8. Histology and western blotting
Haematoxylin and eosin (HE) staining was performed as previously described [33]. Briefly, the skin and irises from WT and T373K knock-in (KI) rabbits were fixed with 4% paraformaldehyde for 48 h, embedded in paraffin wax, sectioned, mounted on slides, stained with HE and analysed by microscopy (Nikon ts100).
Silver staining was performed as previously described [34]. The skin was fixed in 10% buffered formaldehyde and routinely processed. Paraffin sections (4 μm) were stained with potassium permanganate and silver solutions and analysed by microscopy (Nikon ts100).
For western blotting, the skin from WT and T373K mutant rabbits were homogenized in 150 ml of lysis buffer, and protein concentrations were measured using the BCA Protein Assay Kit (Beyotime). Anti-TYR polyclonal antibody (1:2000; Abcam) was used as the primary antibody, and anti-GAPDH monoclonal antibody (1:2000; Beyotime) was used as an internal control. The image was quantified using ImageJ software (NIH), and all the data are expressed as the mean ± SEM.
2.9. Off-target site assay
Potential off-target sites (POTSs) of the sgRNAs were predicted using the CRISPR online design tool (http://tools.genomeengineering.org). The top 10 POTSs that were most likely to produce off-target mutations were selected and subjected to PCR and sequence analysis. Vector NTI and DNAman were used for sequence analysis. The primers used are shown in Table S2.
2.10. T7 endonuclease I (T7EI) assay
A T7 endonuclease I (T7EI) assay was performed as described previously [28]. Briefly, the genomic DNA of each Cas9/gRNA-injected blastocyst and new-born pups was extracted as described above. The PCR products were denatured and annealed under the following conditions: 95 °C for 5 min, 95 °C for 5 min, 95 to 85 °C at −2 °C/s, 85 to 25 °C at −0.1 °C/s, hold at 4 °C. The annealed samples were digested with T7EI (NEB M0302 L), separated and measured on an ethidium bromide-stained 10% polyacrylamide TAE gel.
3. Statistical analyses
Data are expressed as the mean ± SD, with at least three individual measurements in all experiments. The data were analysed with one-way ANOVA using GraphPad Prism 6.0 software. A probability of p < .05 was considered statistically significant.
4. Results
4.1. Reduced melanin synthesis in TYR-T373K mutated cells
A previous study showed that the T373K SNP is associated with OCA1 and is one of the most frequently observed mutations in the clinical [8]. To elucidate the effect of the TYR-T373K SNP on melanin formation, the TYR-WT and TYR-T373K vectors were transiently transfected into HEK293T cells. After 48 h, a marked increase in melanin production was observed in the TYR-WT-transfected cells but not in the MOCK- and TYR-T373K-transfected cells (Fig. 1B). Functional integrity was confirmed by comparison of the colours of the collected cell pellets (Fig. 1C). To determine whether the reduced melanin levels due to the T373K SNP was caused by variations in mRNA stability, the transfected cells were treated with ActD and subjected to an mRNA stability assay. Unexpectedly, no significant difference in mRNA stability was observed between the TYR-WT- and TYR-T373K-transfected cells (Fig. 1D), which was also confirmed by qRT-PCR (Fig. 1E).
Absence of tyrosinase activity is associated with OCA in clinical studies [2]. We next examined whether the genetic mutations affected tyrosinase activity. The melanin content (Fig. 1F) and tyrosinase activity (Fig. 1G) were determined. In contrast to the high activity of the TYR-WT-transfected cells, the TYR-T373K-transfected cells exhibited significantly low melanin content and tyrosinase activity, similar to the MOCK-transfected cells. Thus, these results showed that the T373 K mutation could reduce tyrosinase activity, leading to OCA1 in humans, which is characterized by inactive tyrosinase and the total absence of pigmentation.
4.2. Generation of TYR-K373T rabbits by the CRISPR/CAS9 system
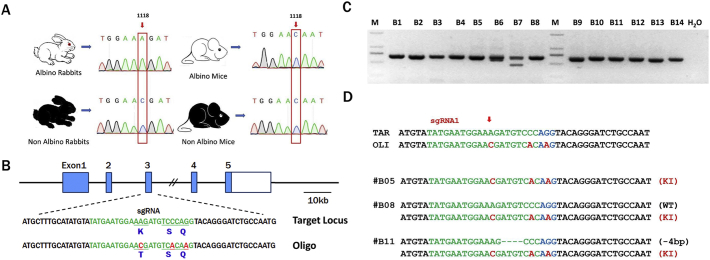
In previous studies and our cell-based experiments, the TYR mutation virtually abolished tyrosinase activity and melanin production [5], so we asked whether repair of the mutation site could restore melanin production. Next, we tested our hypothesis regarding the association of the T373K mutation with OCA1 at the animal level. While no T373K mutation was found in the natural black and white mice, as confirmed by Sanger Sequence analysis (Fig. 2A), we verified the genotype-to -phenotype model using New Zealand white rabbits, which have an inherent T373K SNP and albinism phenotype (white hair and red eyes). Hence, C1118A (K373T) rabbits were generated by ssDNA- and CRISPR/Cas9-mediated homologous recombination (HDR).
Fig. 2.
Editing of the TYR gene via the Cas9/gRNA system in zygotes.
(A) Schematic diagram depicting the TYR-K373T SNP in albino rabbits and mice.
The K373T SNP is shown in the red box and is indicated by the arrows to be present in rabbits but not mice. (B) Diagram of the sgRNA and donor ssODNs used in this study. The sgRNA is marked in green, and the point mutation is shown in red. The base C was replaced with T, producing a single precision point mutation, resulting in K373T (AAG-ACG). Two synonymous mutations near the protospacer adjacent motif (PAM) were introduced in the PAM sequence (red). The long sequence shown at the bottom is that of an ssODN with the desired mutations, synonymous mutations and homologous arms (45 bp to the left and right). (C) Genome editing of the TYR gene in blastocysts by PCR. B1–B14 represent the different blastocysts used in this study, M, DNA marker D2000. (D) Editing of the TYR gene in KI blastocysts was verified by T-cloning and Sanger sequencing. The WT sequence is shown above the target sequence. The sgRNA sequence is marked in green, and the PAM sequences are marked in red and underlined. WT, wild type; deletion, “−”; insertion, “+”.
To explore the feasibility of generating point mutants of TYR C1118A (K373 T), a 90-nt ssDNA donor with nucleotide C instead of A at position 1118 (K373 T) was designed for HDR-mediated genome mutation (Fig. 2B). To reduce the possibility of cutting the sequences successfully knocked-in by the redundant sgRNA, two synonymous nucleotide changes near the protospacer adjacent motif (PAM) were added to the ssDNA donor. A pair of primers was designed and used for the detection of mutations via PCR (Table S1). A mixture of 200 ng/μl Cas9 RNA, 20 ng/μl sgRNA and 50 mM ssODN was co-microinjected into rabbit pronuclear-stage zygotes. Out of 18 injected embryos, 14 developed to the blastocyst stage and were then sequenced to determine mutation efficiency. As shown in Fig. 2C–D and S1, TYR mutations were found in all the blastocysts, and 3 of the 14 blastocysts carried the desired TYR-K373T mutation (B5, B8 and B11), demonstrating that the TYR-K373T mutation can be achieved via this method in zygotes.
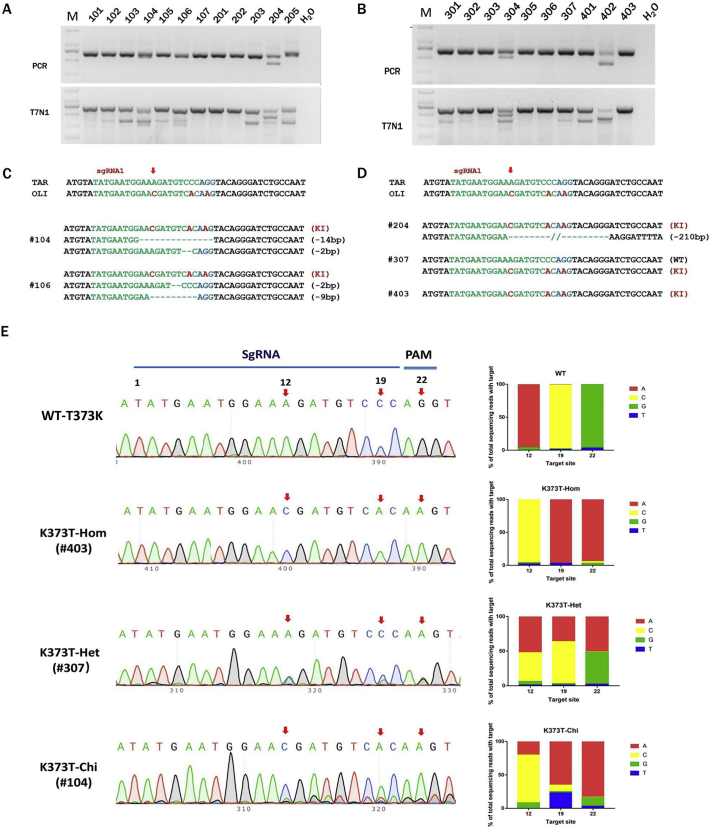
To generate TYR-K373T mutant rabbits, 158 injected rabbit zygotes were transferred into four pseudo-pregnant recipient females, who gave birth to 22 live pups (Table 1). As expected, the gene mutation was detected in 20 rabbits (91%) (Fig. S2), and 5 of the rabbits had the base substitution (K373T), as determined by the T7E1 assay and Sanger sequencing, with efficiencies of 20% (Fig. 3A–D). Notably, one of the K373T rabbits was a homozygous mutant (#403, K373T-Hom), and the other were monoallelic mutants (#204 and #307, K373 T-Het). Interestingly, chimaeric mutations were found in two K373T rabbits with multiple genotypes (#104 and #106, K373T-Chi), indicating that K373T mutations of the TYR gene can be achieved via the CRISPR/Cas9-mediated HDR system with high efficiency in rabbits. GUIDE-seq results confirmed the complexity of base change or mutation on the ssODN-targeted site in TYR-K373T mutant rabbits (Figs. 3E and S3).
Table 1.
Generation of genetically targeted rabbits using CRISPR/Cas9.
| Recipient | gRNA/Cas9 (ng/μl)/ssDND (mM) | Embryos injected | Embryos transferred (% microinjected) | Pregnant | Pups obtained (% transferred) | Pups with mutations (% pups) | Pups with K373T (% pups) | Pups with phenotype |
|---|---|---|---|---|---|---|---|---|
| 1 | 25/100/50 | 50 | 42 (84%) | Yes | 7 (19.5%) | 5 (71.4%) | 2 (28.6%) | 2 |
| 2 | 25/100/50 | 45 | 40 (89%) | Yes | 5 (12.5%) | 4 (80.0%) | 1 (20.0%) | 1 |
| 3 | 25/100/50 | 48 | 40 (83.3%) | Yes | 7 (17.5%) | 5 (71.4%) | 1 (14.3%) | 1 |
| 4 | 25/100/50 | 45 | 36 (80%) | Yes | 3 (8.33%) | 3 (66.6%) | 1 (33.3%) | 1 |
Fig. 3.
Generation of TYR-K373T mutant rabbits by CRISPR/Cas9.
(A, B) Mutation detection in founder rabbits by PCR and the T7E1 assay. M, DNA marker D2000. 101–403 represent the different pups used in this study. (C, D) Mutation detection in founder rabbits by T-cloning and Sanger sequencing. The PAM sites are underlined and highlighted in red; the target sequences are shown in green; deletions (−) and insertions (+) are indicated. WT, wild type. (E) GUIDE-seq results for sgRNA-targeted genomic sites in TYR-K373T mutant rabbits. Chromatogram sequence analysis of sgRNA-targeted genomic sites. The red arrows represent the mutation sites, and the numbers represent the sgRNA base position from the 5′ end. The percentage of sequencing reads harbouring the mutation relative to the total reads is shown. The numbers represent the sgRNA base position from the 5′ end. Values and error bars reflect the means and SDs of n = 3 biologically independent samples.
Furthermore, Sanger sequencing and the T7E1 assay showed that no off-target mutations were present at these potential sites in the TYR-K373T rabbits, indicating that the sgRNAs used in this study were locus specific (Fig. S4).
4.3. Increased melanin production in TYR-K373T rabbits
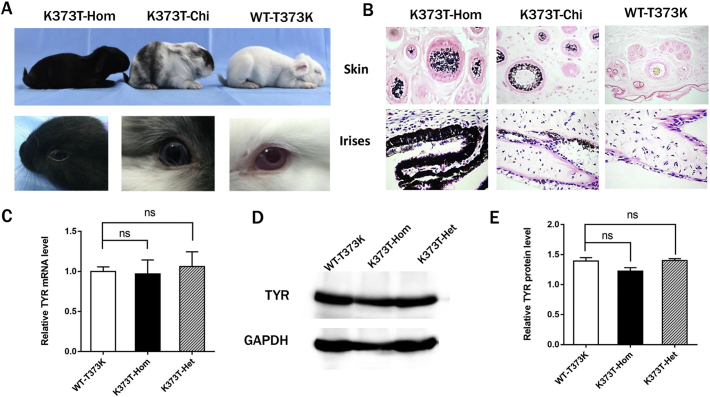
To verify the effect of the K373T mutation on melanogenesis, melanin deposition in the skin and iris was monitored. As shown in Fig. 4A, the K373T rabbits, including the K373T-Hom and K373T-Het rabbits, exhibited completely black hair and eyes, in contrast to the typical symptoms of albinism observed in WT-T373K rabbits. Interestingly, mosaic coat pigmentation (black and white) was observed in two K373T-Chi rabbits with dark eyes. Furthermore, histological HE and silver staining confirmed a significant increase in melanin levels in the hair follicles and irises of K373T-Hom and K373T-Chi rabbits (Fig. 4B), indicating the occurrence of melanogenesis in K373T rabbits.
Fig. 4.
Increased melanin production in TYR-K373T rabbits.
(A) Black hair and eyes in TYR-K373T rabbits. (B) Pathological analysis of the skin and irises from WT-T373K and TYR-K373T rabbits. (C) Gene expression levels of TYR were determined by qRT-PCR. (D) Tyrosinase protein levels were determined by western blotting. (E) Grey-scale analysis of the tyrosinase protein by ImageJ software. All data are expressed as the mean ± SEM. K373T-Hom, homozygous mutation of K373T; K373T-Chi, chimaeric mutation of K373T; WT-T373K, New Zealand white rabbit with a natural T373K mutation.
We further examined whether the gene mutations caused an increase in gene expression. Consistent with the results of the cell-based study, no significant difference in gene expression in the back skin of the WT-T373K and K373T rabbits was observed by RT-PCR (Fig. 4C) and western blotting (Fig. 4D–E). Thus, the results showed the first functional validation of the albinism-associated tyrosinase T373K SNP in rabbits, suggesting that the K373T mutation can rescue melanin production in albinism.
4.4. Heritability of the K373T precision mutation
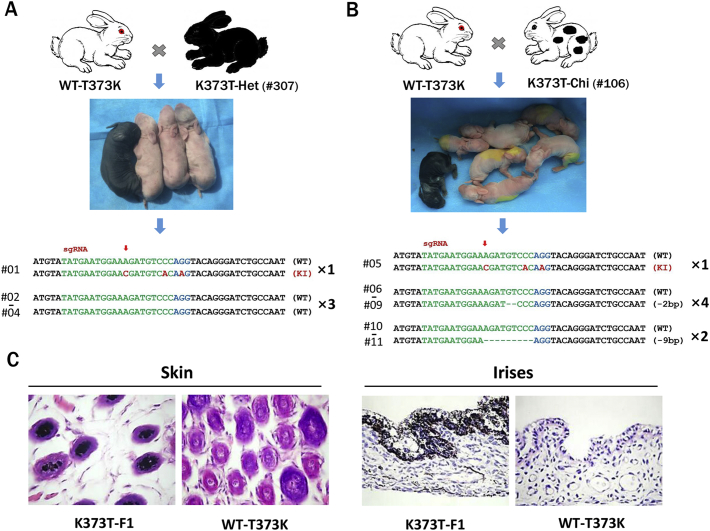
To determine whether the genotype and phenotype of the K373T precision mutation could be stably transmitted to offspring, male K373T-Het rabbits (#307) were mated with female New Zealand white rabbits, which carried a natural T373K mutation (WT-T373K). One F1 rabbit exhibited typical dark pigmentation of the skin and eyes (Fig. 5A). T-cloning and PCR sequencing results confirmed that the K373T mutation of TYR was present in pup #01. Furthermore, similar results were also observed in the offspring of K373T-Chi (#106) rabbits mated with WT-T373K rabbits, and the K373T mutation was detected in pup #05.
Fig. 5.
Heritability of the TYR-K373T mutation in rabbits.
(A) A male K373T-Het rabbit (#307) was mated with female wild-type white rabbits (WT-T373K), and F1 pups with black skin colour were obtained. (B) A male K373T-Chi rabbit (#106) was mated with female WT-T373K white rabbits, and F1 pups with black skin colour were obtained. T-cloning sequence analysis of the F1 rabbits. The PAM sites are highlighted in blue; the target sequences are shown in green; the point mutation sequences are shown in red. Deletions (−) and insertions (+) are indicated. (C) HE staining of the skin and irises from WT-T373K and K373T- F1 rabbits. K373T-Het, heterozygous mutation of K373T; K373T-Chi, chimaeric mutation of K373T; WT-T373K, New Zealand white rabbit with a natural T373 K mutation.
In addition, histological HE staining showed an increase in melanin levels in the hair follicles and irises of K373T-F1 rabbits but not the WT-T373K littermates (Fig. 5C), demonstrating that the K373T mutation and melanogenesis phenotype are heritable and that genome modifications induced by CRISPR/Cas9 are transmitted to the germ line.
5. Discussion
Pigmentation is critically dependent on the functional integrity of tyrosinase, the rate-limiting enzyme in melanin synthesis. The genotype-to-phenotype association between OCA1 and TYR mutations, including mutation of the CDS, 3′UTR and 5′ region, was reported in a recent study [[35], [36], [37]]; however, studies on point mutations, which are the most frequently reported mutations in OCA1 patients, have been limited [8]. Here, we propose a strategy to functionally validate the association between T373K and OCA1. The data show that reduced melanin synthesis in TYR-T373K -mutated cells rescued melanin production in TYR-K373T rabbits. To the best of our knowledge, this is the first study to investigate the biochemical function of the albinism-associated TYR SNP (T373K) at both the cell and animal levels.
Recently, CRISPR-based editing systems that directly perform nucleotide conversion without the introduction of double-stranded breaks (DSBs) have been developed; these systems, named cytosine base editors (CBEs) and adenine base editors (ABEs), enable C·G to T·A or A·T to G·C base pair conversion in organisms with high efficiency [38,39]. However, neither CBEs nor ABEs are suitable for editing bases outside the narrow range, and these systems are limited to inducing base transversion (purine-pyrimidine and pyrimidine-purine) mutations. Therefore, although targeted mutation by BEs is highly efficient, these systems were not suitable for our study (A1118C). Therefore, ssDNA and CRISPR/Cas9-mediated HDR was used in this study. In mice, high efficiency of HDR has been reported with ssODN donors and the CRISPR/Cas9 system [31,40,41]. It has been suggested that the larger a DNA fragment is, the more difficult it is to integrate the fragment into target sites via HDR, although the exact repair mechanisms remain unknown [42].
In this study, 90-bp ssODN donors with substitution mutations were identified, suggesting that CRISPR/Cas system-mediated HDR could be used for SNP exchange and site-specific insertion with high efficiency in rabbits. Interestingly, two chimaeric TYR-KI rabbits with mosaic coat patterns (black and white) were generated in this study, confirming that the CRISPR/Cas9-mediated zygote gene mutation could result in genetically mosaic individuals [21,26], as observed in zebrafish [43], mice [44] and Drosophila [45]. Thus, microinjection of the Cas9 protein instead of RNA may reduce mosaicism and the frequency of undesired mutagenic repair in future studies [21,44].
Tyrosinase is a copper-containing enzyme that catalyses several reactions in the biosynthesis of melanin pigments and is absent in patients with OCA1 [4,5]. tyrosinase is thought to bind two copper ions, one at each of two conserved sequence motifs, termed CuA and CuB (Fig. 1A), each containing three histidine residues that coordinate the binding of Cu(II) ions and are structurally juxtaposed to form the binuclear Cu catalytic site [4,46,47]. Moreover, the tyrosinase enzyme also has six N-glycosylation sites, which are conserved in human and mouse tyrosinases, suggesting that glycosylation may be functionally relevant [48]. A previous study showed that mutations at glycosylation sites that are critical for the quality control mechanism lead to reduction in the activity of the expressed protein [[14], [15], [16], [17]]. Notably, the T373K mutation, which occurs at a conserved glutamate residue near CuB (Fig. 1A), also leads to elimination of the sixth potential N-glycosylation site [48]. In our study, tyrosinase activity and melanin production were found to have been virtually abolished in TYR-T373K-transfected cells. Therefore, we hypothesize that the destruction of the sixth potential N-glycosylation site by the T373K mutation virtually abolishes the ability of tyrosinase to bind copper and similarly destroys the catalytic activity of tyrosinase. Furthermore, decreased tyrosinase activity has been targeted for the prevention of skin hyperpigmentation, such as melasma and age spots [4,6,49]. Therefore, the T373K locus could be a potential therapeutic target for further research, and restoration of tyrosinase production and inhibition of tyrosinase could be used to treat skin disorders.
In summary, this study provides the first genotype-to-phenotype model of the biochemical function of the albinism-associated tyrosinase T373K SNP. Moreover, functional analysis of T373K, in both cell and animal models, provided insights into the mechanisms of albinism pathogenesis as well as strategies for diagnosis or pre-clinical treatment.
Funding
This work was financially supported by the National Key Research and Development Program of China Stem Cell and Translational Research (2017YFA0105101), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030501, XDA16030503), the Guangdong Province Science and Technology Plan Project (2014B020225003), and the Program for JLU Science and Technology Innovative Research Team (2017TD-28).
Competing interests
The authors declare no competing financial interests.
Acknowledgments
Acknowledgements
The authors thank Peiran Hu for providing excellent technical assistance at the Embryo Engineering Center.
Author contributions
Conceptualization: Yuning Song and Yuxin Zhang; Methodology: Jichao Deng, Mao Chen., and Zhanjun Li; Investigation: Yuning Song, Yuxin Zhang, Jichao Deng, Mao Chen, and Zhanjun Li; Writing Original Draft: Yuning Song and Yuxin Zhang; Writing Review & Editing: Yuning Song and Yuxin Zhang; Funding Acquisition: Liangxue Lai; Resources: Tingting Sui and Zhanjun Li; Supervision: Tingting Sui and Zhanjun Li.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.041.
Contributor Information
Liangxue Lai, Email: lai_liangxue@gibh.ac.cn.
Zhanjun Li, Email: lizj_1998@jlu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Cooksey C.J., Garratt P.J., Land E.J., Pavel S., Ramsden C.A., Riley P.A. Evidence of the indirect formation of the catecholic intermediate substrate responsible for the autoactivation kinetics of tyrosinase. J Biol Chem. 1997;272:26226–26235. doi: 10.1074/jbc.272.42.26226. [DOI] [PubMed] [Google Scholar]
- 2.Gronskov K., Ek J., Brondum-Nielsen K. Oculocutaneous albinism. Orphanet J Rare Dis. 2007;2:43. doi: 10.1186/1750-1172-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saluja G., Azad S.V., Pujari A., Temkar S. Oculocutaneous albinism with iridofundal coloboma. BMJ Case Rep. 2018;2018 doi: 10.1136/bcr-2017-223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai X., Wichers H.J., Soler-Lopez M., Dijkstra B.W. Structure and function of human tyrosinase and tyrosinase-related proteins. Chemistry. 2018;24:47–55. doi: 10.1002/chem.201704410. [DOI] [PubMed] [Google Scholar]
- 5.Lekalakala P.T., Khammissa R.A., Kramer B., Ayo-Yusuf O.A., Lemmer J., Feller L. Oculocutaneous albinism and squamous cell carcinoma of the skin of the head and neck in Sub-Saharan Africa. J Skin Cancer. 2015;2015:167847. doi: 10.1155/2015/167847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K B., Purohit R. Mutational analysis of TYR gene and its structural consequences in OCA1A. Gene. 2013;513:184–195. doi: 10.1016/j.gene.2012.09.128. [DOI] [PubMed] [Google Scholar]
- 7.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shaw K., Cooper D.N. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi0113s39. Chapter 1. Unit1 13. [DOI] [PubMed] [Google Scholar]
- 8.Kamaraj B., Purohit R. Mutational analysis of oculocutaneous albinism: a compact review. Biomed Res Int. 2014;2014 doi: 10.1155/2014/905472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halaban R., Svedine S., Cheng E., Smicun Y., Aron R., Hebert D.N. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc Natl Acad Sci U S A. 2000;97:5889–5894. doi: 10.1073/pnas.97.11.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oetting W.S., King R.A. Analysis of mutations in the copper B binding region associated with type I (tyrosinase-related) oculocutaneous albinism. Pigment Cell Res. 1992;5:274–278. doi: 10.1111/j.1600-0749.1992.tb00549.x. [DOI] [PubMed] [Google Scholar]
- 11.Oetting W.S., King R.A. Analysis of tyrosinase mutations associated with tyrosinase-related oculocutaneous albinism (OCA1) Pigment Cell Res. 1994;7:285–290. doi: 10.1111/j.1600-0749.1994.tb00629.x. [DOI] [PubMed] [Google Scholar]
- 12.Oetting W.S., Fryer J.P., King R.A. Mutations of the human tyrosinase gene associated with tyrosinase related oculocutaneous albinism (OCA1). Mutations in brief no. 204. Online. Hum Mutat. 1998;12:433–434. doi: 10.1002/(SICI)1098-1004(1998)12:6<433::AID-HUMU14>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 13.Linzen B., Soeter N.M., Riggs A.F., Schneider H.J., Schartau W., Moore M.D. The structure of arthropod hemocyanins. Science. 1985;229:519–524. doi: 10.1126/science.4023698. [DOI] [PubMed] [Google Scholar]
- 14.Muller G., Ruppert S., Schmid E., Schutz G. Functional analysis of alternatively spliced tyrosinase gene transcripts. EMBO J. 1988;7:2723–2730. doi: 10.1002/j.1460-2075.1988.tb03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witkop C.J., Jr. Albinism. Clin Dermatol. 1989;7:80–91. doi: 10.1016/0738-081x(89)90059-x. [DOI] [PubMed] [Google Scholar]
- 16.Giebel L.B., Strunk K.M., King R.A., Hanifin J.M., Spritz R.A. A frequent tyrosinase gene mutation in classic, tyrosinase-negative (type IA) oculocutaneous albinism. Proc Natl Acad Sci U S A. 1990;87:3255–3258. doi: 10.1073/pnas.87.9.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park K.C., Chintamaneni C.D., Halaban R., Witkop C.J., Jr., Kwon B.S. Molecular analyses of a tyrosinase-negative albino family. Am J Hum Genet. 1993;52:406–413. [PMC free article] [PubMed] [Google Scholar]
- 18.Inui M., Miyado M., Igarashi M., Tamano M., Kubo A., Yamashita S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep. 2014;4:5396. doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakso M., Sauer B., Mosinger B., Jr., Lee E.J., Manning R.W., Yu H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossant J., Spence A. Chimeras and mosaics in mouse mutant analysis. Trends Genet. 1998;14:358–363. doi: 10.1016/s0168-9525(98)01552-2. [DOI] [PubMed] [Google Scholar]
- 21.Singh P., Schimenti J.C., Bolcun-Filas E. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aigner B., Besenfelder U., Muller M., Brem G. Tyrosinase gene variants in different rabbit strains. Mammal Genome. 2000;11:700–702. doi: 10.1007/s003350010120. [DOI] [PubMed] [Google Scholar]
- 23.Huang X., Wang Y., Yan W., Smith C., Ye Z., Wang J. Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33:1470–1479. doi: 10.1002/stem.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang P., Xu Y., Zhang X., Ding C., Huang R., Zhang Z. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Xu F., Zhu C., Ji J., Zhou X., Feng X. Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci Rep. 2014;4:7581. doi: 10.1038/srep07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y., Yuan L., Wang Y., Chen M., Deng J., Lv Q. Efficient dual sgRNA-directed large gene deletion in rabbit with CRISPR/Cas9 system. Cell Mol Life Sci. 2016;73:2959–2968. doi: 10.1007/s00018-016-2143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 29.Song Y., Liu T., Wang Y., Deng J., Chen M., Yuan L. Mutation of the Sp1 binding site in the 5′ flanking region of SRY causes sex reversal in rabbits. Oncotarget. 2017;8:38176–38183. doi: 10.18632/oncotarget.16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan L., Yao H., Xu Y., Chen M., Deng J., Song Y. CRISPR/Cas9-mediated mutation of alphaA-crystallin gene induces congenital cataracts in rabbits. Invest Ophthalmol Vis Sci. 2017;58:BIO34–BIO41. doi: 10.1167/iovs.16-21287. [DOI] [PubMed] [Google Scholar]
- 31.Yang D., Song J., Zhang J., Xu J., Zhu T., Wang Z. Identification and characterization of rabbit ROSA26 for gene knock-in and stable reporter gene expression. Sci Rep. 2016;6 doi: 10.1038/srep25161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J.Y., Koo J.H., Song Y.G., Kwon K.B., Lee J.H., Sohn H.S. Stimulation of melanogenesis by scoparone in B16 melanoma cells. Acta Pharmacol Sin. 2006;27:1467–1473. doi: 10.1111/j.1745-7254.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Fan N., Song J., Zhong J., Guo X., Tian W. Generation of knockout rabbits using transcription activator-like effector nucleases. Cell Regener. 2014;3:3. doi: 10.1186/2045-9769-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwint O.A., Labraga M., Cervino C.O., Haffar M., Sequeiros P.H., Marcos H.J. A modification of the staining technique of reticular fibres for image analysis of the cardiac collagen network. Cardiovasc Pathol. 2004;13:213–220. doi: 10.1016/S1054-8807(03)00153-4. [DOI] [PubMed] [Google Scholar]
- 35.Yuan L., Sui T., Chen M., Deng J., Huang Y., Zeng J. CRISPR/Cas9-mediated GJA8 knockout in rabbits recapitulates human congenital cataracts. Sci Rep. 2016;6 doi: 10.1038/srep22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y., Xu Y., Deng J., Chen M., Lu Y., Wang Y. CRISPR/Cas9-mediated mutation of tyrosinase (Tyr) 3' UTR induce graying in rabbit. Sci Rep. 2017;7:1569. doi: 10.1038/s41598-017-01727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seruggia D., Fernandez A., Cantero M., Pelczar P., Montoliu L. Functional validation of mouse tyrosinase non-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis. Nucleic Acids Res. 2015;43:4855–4867. doi: 10.1093/nar/gkv375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 39.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X.M., Eden E.R., Tosi I., Neuwirth C.K., Wile D., Naoumova R.P. Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet. 2005;14:1161–1169. doi: 10.1093/hmg/ddi128. [DOI] [PubMed] [Google Scholar]
- 41.Brouwer D.A., van Doormaal J.J., Muskiet F.A. Clinical chemistry of common apolipoprotein E isoforms. J Chromatogr B Biomed Appl. 1996;678:23–41. doi: 10.1016/0378-4347(95)00256-1. [DOI] [PubMed] [Google Scholar]
- 42.Staessen C., Janssenswillen C., De Clerck E., Van Steirteghem A. Controlled comparison of commercial media for human in-vitro fertilization: Menezo B2 medium versus Medi-Cult universal and BM1 medium. Hum Reprod. 1998;13:2548–2554. doi: 10.1093/humrep/13.9.2548. [DOI] [PubMed] [Google Scholar]
- 43.Sung Y.H., Kim J.M., Kim H.T., Lee J., Jeon J., Jin Y. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014;24:125–131. doi: 10.1101/gr.163394.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrimon N. Creating mosaics in Drosophila. Int J Dev Biol. 1998;42:243–247. [PubMed] [Google Scholar]
- 46.Lai X., Wichers H.J., Soler-Lopez M., Dijkstra B.W. Structure of human tyrosinase related protein 1 reveals a binuclear zinc active site important for melanogenesis. Angew Chem. 2017;56:9812–9815. doi: 10.1002/anie.201704616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Setaluri V. Sorting and targeting of melanosomal membrane proteins: signals, pathways, and mechanisms. Pigment Cell Res. 2000;13:128–134. doi: 10.1034/j.1600-0749.2000.130302.x. [DOI] [PubMed] [Google Scholar]
- 48.Branza-Nichita N., Negroiu G., Petrescu A.J., Garman E.F., Platt F.M., Wormald M.R. Mutations at critical N-glycosylation sites reduce tyrosinase activity by altering folding and quality control. J Biol Chem. 2000;275:8169–8175. doi: 10.1074/jbc.275.11.8169. [DOI] [PubMed] [Google Scholar]
- 49.Spritz R.A., Ho L., Furumura M., Hearing V.J., Jr. Mutational analysis of copper binding by human tyrosinase. J Invest Dermatol. 1997;109:207–212. doi: 10.1111/1523-1747.ep12319351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material