Abstract
Background
Disordered folliculogenesis is a key feature of polycystic ovary syndrome (PCOS), but the underlying molecular mechanism remains unclear.
Methods
Long non-coding RNA (lncRNA) expression in luteinized granulosa cells (hLGCs) derived from women with and without PCOS were analyzed using microarray and qRT-PCR. Immortalized human granulosa cell lines were cultured for proliferation assays after transfection with the LINC-01572:28 over-expression vector in the presence or absence of p27 siRNA. Protein expression analysis, rescue assays, and RNA immunoprecipitation (RIP) were used to confirm the LINC-01572:28 substrate.
Findings
LINC-01572:28 and p27 protein were elevated whereas proliferating cell nuclear antigen protein was decreased in the hLGCs of women with PCOS. LINC-01572:28 expression was positively correlated with basal testosterone levels. Over-expression of LINC-01572:28 inhibited cell proliferation and impeded G1/S transition, which were partially reversed by siRNA-mediated p27 knockdown.
Interpretation
Our findings, therefore, suggest that LINC-01572:28 suppresses cell proliferation and cell cycle progression by reducing the degradation of p27 protein via SKP2 binding.
Keywords: Polycystic ovary syndrome, lncRNA, Granulosa cells, Proliferation
Research in context.
Evidence before this study
Polycystic ovary syndrome (PCOS), is a leading cause of female infertility. Although women with PCOS exhibit more follicles than women without PCOS, none of these follicles develop into a dominant follicle, leading to abnormal ovulation. The granulosa cell layers surrounding these follicles show signs of atresia, degradation, and hypertrophy, indicating abnormal proliferation and/or apoptosis. The granulosa cells are essential for providing the oocyte with nutrients and growth regulators during oocyte development. Their dysfunction, therefore, may contribute to the aberrant folliculogenesis observed in PCOS.
Previous studies have demonstrated that lncRNAs may be involved in follicular development. For example, Yerushalmi et al. found that lncRNA Neat1 knockout (KO) mice were unable to become pregnant due to corpus luteum dysfunction and low progesterone levels. Furthermore, Huang et al. demonstrated different microarray expression patterns of lncRNAs and mRNAs in cumulus cells isolated from patients with and without PCOS. Liu et al. also found differential expression of lncRNA-HCG26 in women with PCOS, which may influence the proliferation and steroidogenesis of the granulosa cells. Despite these findings, however, the molecular mechanism underlying the involvement of lncRNAs in disordered folliculogenesis in PCOS remains unclear.
In this study, we conducted microarray analysis to identify differentially expressed protein-coding genes and lncRNAs expression profiles in luteinized granulosa cells obtained from women with and without PCOS. Diagnosis of PCOS was based on the following revised Rotterdam diagnostic criteria for PCOS [35], which requires the presence of at least 2 of the following criteria for a PCOS diagnosis: (i)oligo-ovulation and/or anovulation; (ii) clinical and/or biochemical signs of hyperandrogenism; and (iii) polycystic ovaries. Diagnoses of PCOS were made after exclusion of other etiologies for hyperandrogenemia and ovulatory dysfunction (eg. 21-hydroxylase deficiency, congenital adrenal hyperplasia, Cushing syndrome, androgen-secreting tumors, thyroid disease, and hyperprolactinemia). All subjects in the control group had regular menstrual cycles (26–35 days) and normal ovarian morphology; total testosterone was evaluated to exclude hyperandrogenism. Peripheral blood samples were collected from all subjects during days 2 to 4 of spontaneous cycles or after cessation of bleeding after a 12-h overnight fast.
Added value of this study
In the current study, we demonstrated that the abundance of LINC-01572:28 was elevated in the hLGCs of women with PCOS compared to those from control women using microarray and qRT-PCR. The elevation of LINC-01572:28 impeded the interaction between SKP2 and p27 via competitive binding to SKP2, which resulted in the accumulation of p27. The accumulation of p27 in turn induced cell cycle arrest and inhibited granulosa cell proliferation. Furthermore, the increase in PCNA protein and decrease in p27 protein were also observed in hLGCs of women with PCOS but not in those from control women.
Previous studies reported hyperandrogenemia could impede the proliferation of granulosa cells in PCOS animal models, which may induce the aberrant folliculogenesis. The result of our study was in consistent with that. As an intergenic lncRNA, LINC-01572:28 is highly expressed in testis and positive related with the concentration of testosterone in serum of women with PCOS. Thus, we think LINC01572:28 might be a mediator or treatment target in this pathway.
Implications of all the available evidence
In the current study, our findings showed that LINC-01572:28 was higher in granulosa cells of women with PCOS, and that its up-regulation was associated with hyperandrogenemia. LINC-01572:28 may have suppressed the proliferation of granulosa cells partly by decreasing the degradation of the p27 protein. Our study not only demonstrated the role of LINC01572:28 involved in the etiology of PCOS, but also provided a novel interaction mode for lncRNAs, meanwhile, a potential target for the treatment of PCOS.
Alt-text: Unlabelled Box
1. Introduction
Polycystic ovary syndrome (PCOS), a common endocrine disease among reproductive-aged women [21], is characterized by hyperandrogenemia, chronic oligo/anovulation, and polycystic ovarian morphology [7,35]. PCOS is a leading cause of female infertility. Although women with PCOS exhibit more follicles than women without PCOS [11], none of these follicles develop into a dominant follicle, leading to abnormal ovulation. The granulosa cell layers surrounding these follicles show signs of atresia, degradation, and hypertrophy, indicating abnormal proliferation and/or apoptosis [1]. The granulosa cells are essential for providing the oocyte with nutrients and growth regulators during oocyte development [22,25]. Their dysfunction, therefore, may contribute to the aberrant folliculogenesis observed in PCOS.
Microarray analysis of tissue from women with and without PCOS identifies a significant proportion of the differentially expressed transcripts in PCOS as non-coding RNAs. Non-coding RNA, especially long non-coding RNA (lncRNA), have important potential regulatory effects on gene expression. lncRNAs, which are defined as transcripts longer than 200 nucleotides without coding potential [33], play a crucial role in cell development, differentiation, proliferation, and apoptosis via interactions with RNA-binding proteins, chromatin modification, and ceRNA networks [6,32]. Previous studies have demonstrated that lncRNAs may be involved in follicular development. For example, Yerushalmi et al. found that lncRNA Neat1 knockout (KO) mice were unable to become pregnant due to corpus luteum dysfunction and low progesterone levels [27]. Furthermore, Huang et al. demonstrated different microarray expression patterns of lncRNAs and mRNAs in cumulus cells isolated from patients with and without PCOS [10]. Liu et al. also found differential expression of lncRNA-HCG26 in women with PCOS, which may influence the proliferation and steroidogenesis of the granulosa cells [20]. Despite these findings, however, the molecular mechanism underlying the involvement of lncRNAs in disordered folliculogenesis in PCOS remains unclear.
In this study, we conducted microarray analysis to identify differentially expressed protein-coding genes and lncRNAs expression profiles in luteinized granulosa cells obtained from women with and without PCOS. We identified the novel lncRNA LINC-01572:28, a 473-nt transcript located at chromosome 16q22 that is elevated in patients with PCOS. We investigated the effect of an increase in LINC-01572:28 in the development of PCOS. The mechanisms underlying LINC-01572:28 were further characterized in human luteinized granulosa SVOG and KGN cell lines.
2. Materials and methods
2.1. Subjects
Ovarian granulosa cells were collected from patients who underwent in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) at the Center for Reproductive Medicine of Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine. The study was approved by the ART Ethnics committee of Ren Ji hospital, school of medicine, shanghai Jiao Tong university (number: 2017041411), and informed consent was obtained from all participants. Anthropometric variables, such as age, body mass index (BMI), and select endocrine and biochemical parameters were recorded and are presented in Table 1. A diagnosis of PCOS was based on the revised Rotterdam diagnostic criteria for PCOS [35][35]. All patients in the non-PCOS control group had regular menstrual cycles (26–35 days) and normal ovarian morphology by ultrasound examination. Follicle-stimulating hormone (FSH), luteinizing hormone (LH), testosterone (T), estradiol (E2), and anti-Müllerian hormone (AMH) levels were determined using either chemiluminescent assay (Beckman Access Health Company, Chaska, MN) or ELISA (Kangrun, Guangzhou, China) kits.
2.2. Collection of follicular fluid and culture of primary ovarian granulosa, KGN, and SVOG cells
Ovarian stimulation and oocyte retrieval were performed under a gonadotropin-releasing hormone (GnRH) antagonist or agonist protocol. After adequate follicle development, as detected by both ovarian ultrasound and serum estradiol assay, human chorionic gonadotropin (Lvzhu, Zhuhai, China) was administered to trigger ovulation. Ultrasound-guided oocyte retrieval was performed 36 h later.
Human luteinized granulosa cells (hLGCs) were retrieved from the follicular fluid as previously described [43]. The follicular fluid was pooled and centrifuged at 2500 rpm for 15 min, then the pellets were re-suspended in phosphate-buffered saline (PBS) and dispersed in 0.1% hyaluronidase (Sigma Chemical Co., St. Louis, MO, USA) at 37 °C for 15 min. Granulosa cells then were purified by Ficoll-Paque (GE Healthcare Bio-Science, Uppsala, Sweden). The isolated granulosa cells were stored at −80 °C or used after 3 days in culture. SVOG and KGN cells were obtained from Shandong University. All the hLGCs and SVOG and KGN cells were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) (Gibco, Grand Island, NY) containing 10% charcoal-stripped fetal bovine serum (Biological Industries, US origin) and 1% penicillin-streptomycin-neomycin (PSN, Gibco) at 37 °C in a humidified atmosphere with 5% CO2. KGN and SVOG cells were passaged every 3 days.
2.3. Microarray analysis
The hLGCs from three women with PCOS and three BMI, age, and treatment-matched reproductively normal, control women were selected for microarray analysis (Affymetrix HTA 2.0 Array). The microarray contained >6,000,000 probes covering 44,699 coding genes and 22,829 non-coding genes. Both the design and annotation of the probes were derived from authoritative databases, including RefSeq, Ensembl, UCSC (known genes and lincRNA transcripts), Mammalian Gene Collection (MC v10), NONCODE, lncRNA db, the Broad Institute Human Body Map lincRNAs, and the TUCP catalog (Broad Institute). The microarray analyses were performed by Biotechnology Corporation (Shanghai, China) as described in the supplementary materials. Significant differentially expressed transcripts were filtered by both p-value and fold change (P < .05 and fold change >1.5, respectively) between PCOS and control samples.
2.4. Transfection of cells
KGN cells, SVOG cells, and hLGCs (2 × 105) were seeded into six-well plates and cultured for 1 day and then transfected with siRNAs and/or plasmid DNA using Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. After transfection, the cells were incubated for 48 or 72 h before further treatment.
2.5. RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from cells by TRIzol (Invitrogen) and then reverse transcribed into cDNA (PrimeScript™ RT reagent Kit, Takara, Dalian, China). Target gene expression was detected using quantitative real-time polymerase chain reaction (rRT-PCR). The relative expression of RNA was calculated using the formula 2-△△Ct. Beta-actin (ACTB) was employed as an internal control for the quantification of target genes, the first ∆Ct normalization was to β-actin, and the 2nd ∆Ct normalization was to the control group or un-transfected cells. The primer sequences of the tested genes are shown in Supplemental Table S1.
2.6. Western blot assay
Cells were harvested and lysed in ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer (Shenggong, Shanghai, China) containing a protease inhibitor cocktail (Shenggong) and phosphatase inhibitor (Shenggong). The protein was extracted from cells and quantified as previously described [43] using 10–15% SDS-polyacrylamide gel. Relative band density was detected by Gel-Pro Analyzer (Media Cybernetics, L.P.) and normalized to β-actin Ab (Proteintech, Wuhan Hubei, P.R.C). Especially, the relative phosphorylation of target protein was calculated by the ratios of phosphorylated protein to target protein. The antibodies used in the Western blot analysis are summarized in Supplementary Table S2.
2.7. Cell proliferation assay
After transfection for 24 h, cells were harvested by trypsinization. The CCK-8 assay was used according to the manufacturer's protocol to determine cell viability. The absorbance of the solution was measured at 450 nm. Ethynyl-2-deoxyuridine (EdU) assays were performed using an EdU Cell Proliferation Assay Kit (RiboBio, Guangzhou, China) according to the manual book. Cell proliferation was analyzed under a fluorescent microscope (Zeiss, Jena, Germany).
2.8. Flow cytometry analysis
For cell cycle analysis, KGN cells were harvested by trypsinization 48 h after transfection with siRNAs and/or plasmid DNA, then centrifuged and suspended in 1 mL ice-cold PBS. The cells were washed twice by PBS, and then the cells were re-suspended using 70% ice-cold ethanol and kept at 4 °C overnight. The fixed cells were centrifuged and the supernatant fluid was discarded. The cell sediment then was re-suspended in 500 μL propidium iodide with RNase A. After incubation at room temperature for 15 min, the cells were filtered once through 400 mesh sieves and then subjected to flow cytometry analysis. The results were analyzed by ModFIT LT 2.0.
2.9. RNA immunoprecipitation
RNA immunoprecipitation (RIP) experiments were performed using a Magna Nuclear RIP™ (Cross-Linked) Nuclear RNA-Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA, USA) according to the manufacturer's instructions. EZH2 and SKP2 antibodies (Proteintech, Wuhan Hubei, P.R.C) were used for the immunoprecipitation.
2.10. Co-immunoprecipitation
Co-immunoprecipitation (co-IP) was performed in lysates prepared from transfected cells (100 μg total protein) using either the SKP2 antibody or normal rabbit IgG at 4 °C overnight. On the next morning, the protein-antibody complex was incubated with 15 μL magnetic protein A + G beads for 1 h at 4 °C with gentle rotation. The antibody-protein-bead complexes then were washed three times with co-IP buffer. The protein in the complex then was eluted with 20 μL 1× loading buffer and boiled before running on a 15% SDS-polyacrylamide gel. The proteins were transferred to nitrocellulose membranes, and SKP2-associated p27 protein was immunoblotted using antibodies against p27.
2.11. Statistical analysis
All calculations were performed using the SPSS statistical software package (version 19.0) (IBM Corp., Armonk, NY, USA). The data are presented as the mean ± standard deviation (SD). The Kolmogorov-Smirnov test was used to assess whether the data were of normal distribution. Statistical comparisons between women with and without PCOS were performed using two-tailed Student's t-test. The Spearman's correlation and linear regression analyses were used to examine the association between gene expression levels and the distributions of relevant variables. Statistical comparisons between two groups of cells (the experimental group and the control group) were performed using the Student's t-test. P < .05 was considered statistically significant. Statistical significance was evaluated using data from at least three independent experiments.
3. Results
3.1. Evaluation of clinical characteristics
A total of 132 subjects (89 women with PCOS and 43 normal women) were included in this study (3 PCOS patients and 3 controls for microarray analysis and the others for qRT-PCR). The major anthropometric variables and endocrine parameters of the women are presented in Table 1. Women with PCOS had significantly higher serum levels of LH and testosterone compared with the control group. These findings are consistent with the typical features of PCOS.
Table 1.
Clinical and biochemical profiles of women with and without PCOS
| Cohort 1 | Cohort 2 | Cohort 3 | Cohort 4 | ||||
|---|---|---|---|---|---|---|---|
| Variable | PCOS | Control | PCOS | Control | PCOS | Control | PCOS |
| No. | 3 | 3 | 10 | 10 | 30 | 30 | 46 |
| Age(years) | 28±1 | 26±2 | 28±2 | 27±3 | 28±3 | 29±3 | 27±3 |
| BMI (Kg/m2) | 21.0±1.4 | 19.3±0.7 | 23.2±1.0 | 22.3±1.0 | 23.3±2.9 c | 21.8±2.4 | 23.0±3.6 |
| Basal LH(IU/L) | 8.1±2.1 c | 4.1±1.3 | 8.5±1.6c | 4.8±0.5 | 7.4±4.2 b | 5.0±1.7 | 8.3±4.2 |
| Basal FSH(IU/L) | 5.3±0.4 | 5.7±0.4 | 5.9±0.4 | 6.4±0.3 | 6.17±1.33 | 6.3±1.6 | 6.0±1.3 |
| Basal T (ng/dL) | 69.8±13.5 c | 29.7±2.7 | 51.2±9.0 b | 19.8±1.2 | 43.0±23.4 a | 19.3±7.8 | 46.8±37.5 |
| AMH (ng/mL) | 11.9±3.9 | 4.7±1.4 | 9.2±1.1b | 5.5±0.9 | 10.1±5.4 a | 5.6±3.3 | 12.1±4.8 |
| Hormones on hCG day | |||||||
| E2(pg/mL) | 6523±2045 | 2043±415 | 4382±482 c | 2737±251 | 4007±1669 b | 2666±832 | 3827±2094 |
Cohort 1 represents the GCs subjects tested by microarray. Cohort 2 represents the GCs subjects tested by quantitative real-time PCR for 6 selected lncRNAs. Cohort 3 represents the GCs tested by quantitative real-time PCR for LINC01572:28. Cohort 4 represents the GCs which are used for clinical characters analysis. Data are presented as mean±SD, BMI: Body mass index, LH: luteinizing hormone, FSH: follicle stimulating hormone, T: testosterone. a p<0.001, b p<0.01, c p<0.05
3.2. Expression profiles of differentially expressed transcripts
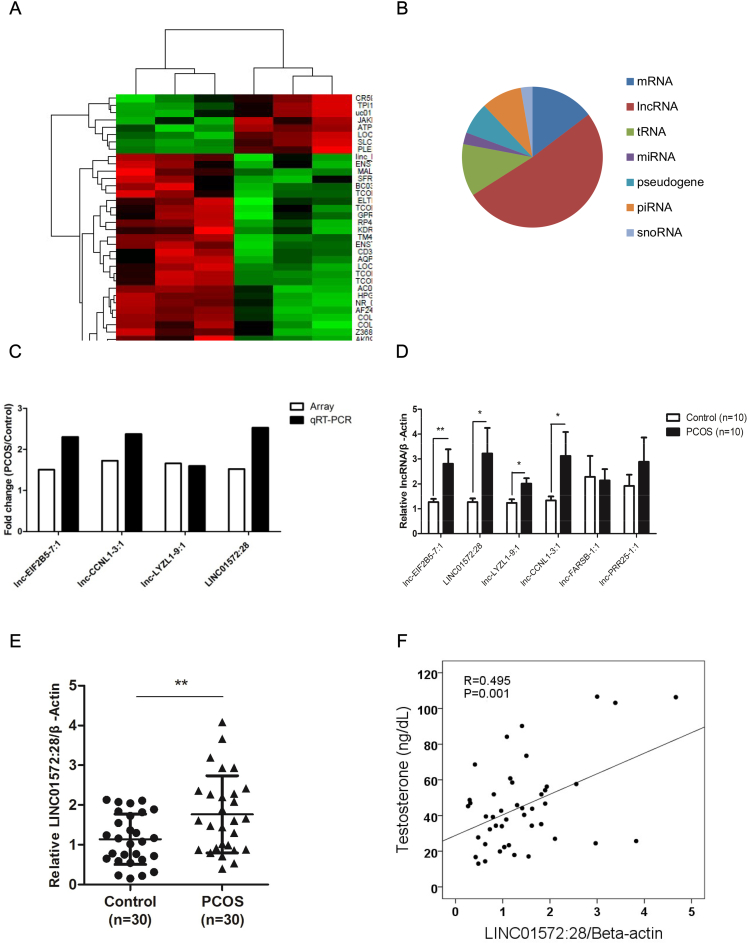
To identify transcripts that are potentially involved in the etiology of PCOS, we evaluated whole-transcript expression profiles in hLGCs by microarray analysis. As shown in Fig. 1A, most of the non-coding transcripts in the expression profile were not annotated. A total of 128 non-coding transcripts and 22 protein-coding transcripts exhibited differential expression between patients with and without PCOS. Among the 128 non-coding transcripts, 77 were lncRNAs, 14 were piRNAs, 4 were snoRNAs, 4 were miRNAs, and 18 were tRNAs (18) (Fig. 1B).
Fig. 1.
LINC01572:28 is upregulated in granulosa cells of the PCOS patients.
(A) Heat-map showing the hierarchical clustering of differentially expressed genes in patients with and without PCOS. red: upregulated genes; green: downregulated genes.
(B) Graph showing the biotypes of non-coding RNAs profiled.
(C) The differential expression of lncRNAs from microarrays and qRT-PCR respectively.
(D) Graph showing the levels of the selected lncRNAs in 10 PCOS patients and 10 healthy controls. The expression levels were detected via qRT-PCR and normalized againstβ-actin.
(E) Graph showing the level of the LINC01572:28 in 30 PCOS patients and 30 healthy controls. The expression level was detected via qRT-PCR and normalized againstβ-actin.
(F) Graph showing the correlation between the expression level of LINC01572:28 in hLGCs and the serum concentration of testosterone in patients with PCOS. The expression level was detected via qRT-PCR. The first ∆Ct normalization was to β-actin, and the 2nd ∆Ct normalization was to the control group in cohort 3.
Error bars represent SDs of at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
To validate the microarray results, six differentially expressed lncRNAs were selected and analyzed in a cohort of 10 women with PCOS and 10 treatment-matched reproductively normal control women using qRT-PCR (Fig. 1C, D). The expression levels of LINC-01572:28, lnc-EIF2B5–7-1, lnc-SIK2–1, lnc-CCNL1–3-1, and lnc-LYZL-9-1 were increased in patients with PCOS (P < .05 for all lncRNAs), which was consistent with the microarray results. The top 20 differentially expressed lncRNAs and mRNAs are reported in Supplementary Table S2 and the microarray data have been uploaded to Gene Expression Omnibus database (accession number: GSE114419).
3.3. Elevation of novel lncRNA LINC-01572:28 in hLGCs from women with PCOS
LINC-01572:28 was one of the up-regulated lncRNAs identified from the hLGCs of patients with PCOS. To further validate the increase in LINC-01572:28, we investigated the expression level of LINC-01572:28 in another cohort of 30 women with PCOS and 30 controls (Fig. 1E). We also analyzed the correlation between clinical characteristics of PCOS and the expression level of LINC-01572:28 in a separate group of patients with PCOS (46 samples) (Fig. 1F). As shown in Fig. 1F, LINC-01572:28 was significantly positively correlated with testosterone concentration in women with PCOS (Spearman's rho = 0.459, P = .001). No significant correlations between LINC-01572:28 and other clinical characteristics were observed.
3.4. Overexpression of LINC-01572:28 suppressed granulosa cell proliferation and cell-cycle progression in vitro.
To investigate the role of LINC-01572:28 in the development of PCOS, we first identified whether LINC-01572:28 is a potential coding gene. The PhyloCSF score for LINC-01572:28 was −154.2917, indicating that LINC-01572:28 is 1015.42917 times more likely to be a non-coding sequence than a coding sequence [19]. Consistently, evaluation with the Coding-Potential Assessment Tool identified a 2.53% probability that LINC-01572:28 is a coding sequence. Thus, these findings indicate that LINC-01572:28 is a non-coding RNA sequence [40].
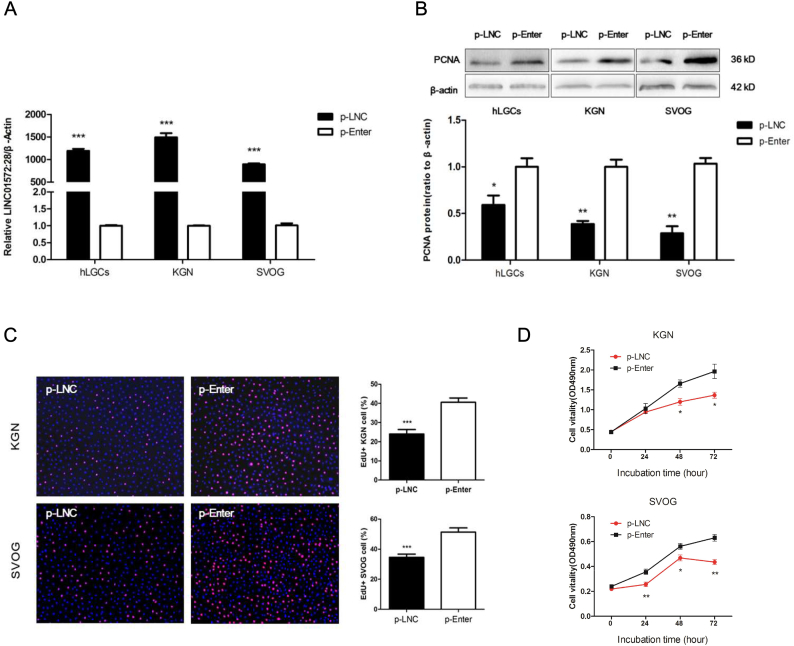
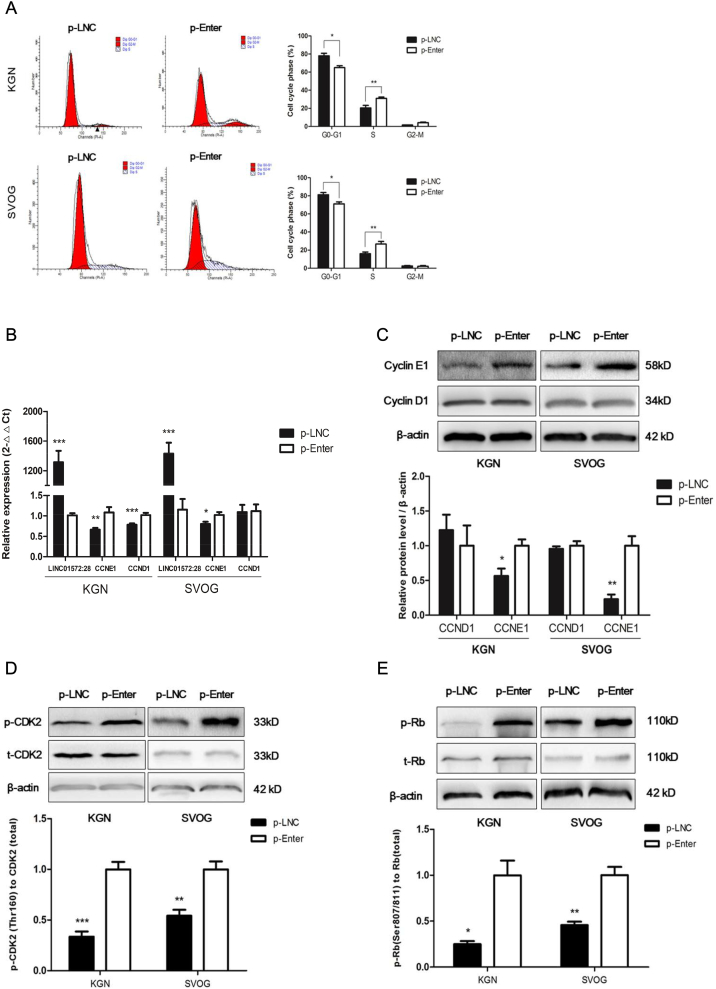
To evaluate the effects of LINC-01572:28 on cell biological function, we measured cell proliferation after transfection of the LINC-01572:28 overexpression vector into KGN cells, SVOG cells, and hLGCs. A blank loaded vehicle vector (p-Enter) was used as the control. The efficiency of the vector transfection was confirmed by qRT-PCR (Fig. 2A). We found that the abundance of PCNA protein was reduced in the overexpressed LINC-01572:28 granulosa cells (Fig. 2B). EdU and CCK-8 assays consistently showed that over-expressed LINC-01572:28 decreased the proportion of proliferating cells in total cells (Fig. 2C) and inhibited cell proliferation ability (Fig. 2D). We also examined whether the cell cycle was affected by LINC-01572:28 with propidium iodide staining via flow cytometry analysis and found that overexpression of LINC-01572:28 significantly induced G0-G1 phase arrest and decreased the percentage of cells in the S phase (Fig. 3A). Western blot analyses consistently showed that overexpression of LINC-01572:28 had no effect on the expression of cyclin D1, a factor involved in G0/G1 transition [24,41], but decreased the abundance of cyclin E1, a factor involved in G1/S transition [39], in KGN and SVOG cells (Fig. 3B and C). Furthermore, the phosphorylation of CDK2 and Rb, two factors involved in cell cycle progression, also were reduced upon overexpression of LINC-01572:28 in KGN and SVOG cells (Fig. 3D and E).These data suggest that LINC-01572:28 may inhibit cell proliferation via G0-G1 phase arrest in granulosa cells.
Fig. 2.
Enforced expression of LINC01572:28 inhibited granulosa cell proliferation in vitro.
(A) Graph showing the level of LINC01572:28 in granulosa cells (human primary granulosa cells, KGN, and SVOG cells) treated with LINC01572:28 overexpression vector (p-LNC) or negative control vector (p-Enter). The expression level was detected via qRT-PCR and normalized againstβ-actin.
(B) Cells were transfected with either LINC01572:28 overexpression vector (p-LNC) or negative control vector (p-Enter); protein samples were collected 48 h after transfection, and then subjected to immunoblot analysis. Data are representative of at least 3 independent experiments.
(C) Graphs showing the proportion of proliferation cells via Edu assay in granulosa cells treated as in (A). The amount of cells was detected by staining with Hochest (blue), as the proliferated cells were detected by staining with EdU (red). The result was analyzed by fluorescence microscope. Percentages of EdU-positive cells were graphed. (D) Cell proliferation assays determined by cell counting Kit-8 at the indicated time points after transfection were performed from granulosa cells treated as in (A)
Error bars represent SDs from at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
Fig. 3.
LINC01572–28 induced cell cycle arrest at G0-G1 phase in granulosa cells.
(A) The granulosa cells were transfected with p-LNC or p-Enter for 48 h; the effect on cell cycle was assessed by PI staining followed by Modfit analysis.
(B) qRT-PCR analysis for CCNE1 and CCND1 mRNA level in p-LNC and p-Enter KGN and SVOG cells. The expression level was normalized againstβ-actin.
(C) Western blot analysis using Cyclin E1 and Cyclin D1 antibodies on protein extracts form granulosa cells after transfected with vectors for 48 h.
(D and E) The levels of phosphorylated CDK2 and Rb in p-LNC and p-Enter granulosa cells were assessed by immunoblotting.
Error bars represent SDs from at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
3.5. Involvement of p27 in LINC-01572:28-induced proliferation arrest of granulosa cells
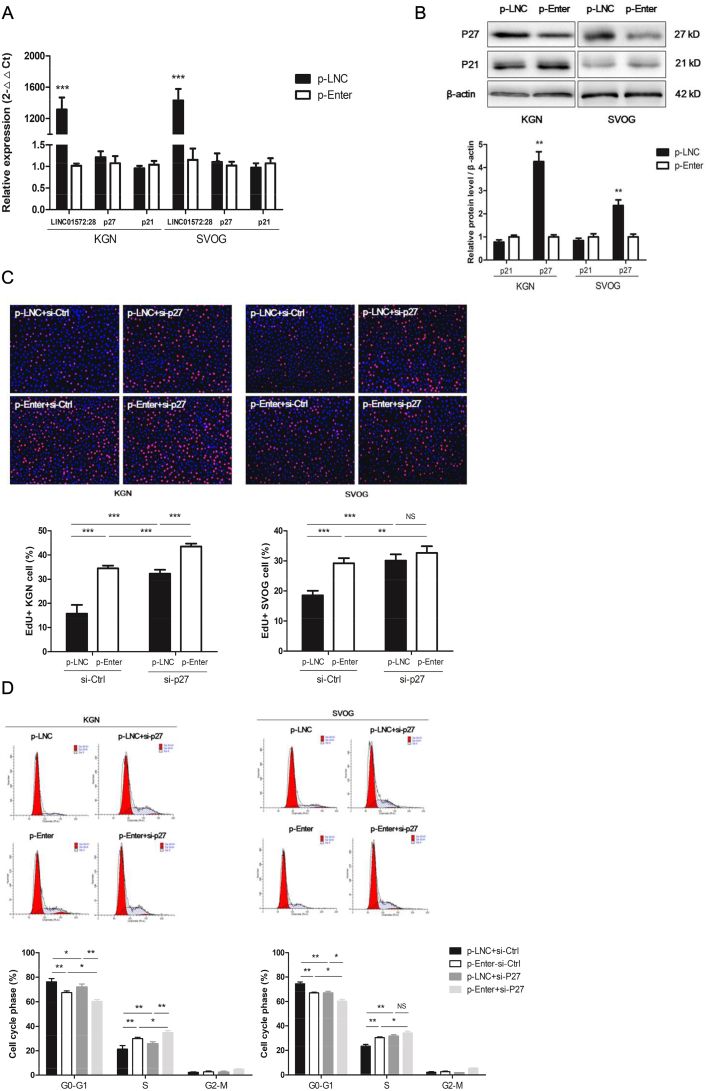
Cyclin-dependent kinase inhibitor 1A (p21) and 1B (p27) are two factors related to the G1/S transition that may inhibit the activity of the cyclin E/CDK2 complex. We examined whether the overexpression of LINC-01572:28 affects the expression of these two factors. LINC-01572:28 had no effect on p21 (CDKN1A) mRNA and protein or p27 (CDKN1B) mRNA expression (Fig. 4A), but significantly induced an abundance of p27 protein in KGN and SVOG cells (Fig. 4B).
Fig. 4.
Involvement of p27 in LINC01572:28-induced proliferation arrest of granulosa cells.
(A and B) The mRNA and protein level of p27 and p21 in granulosa cells after transfection with vectors for 48 h.
(C) p-LNC and p-Enter cells were treated with si-Ctrl or si-p27 for 48 h; the proportion of proliferation cells was assessed by EdU assay in granulosa cells.
(D) p-LNC and p-Enter cells were treated with si-Ctrl or si-p27 for 48 h; the effect on cell cycle was assessed by PI staining followed by Modfit analysis.
Error bars represent SDs from at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
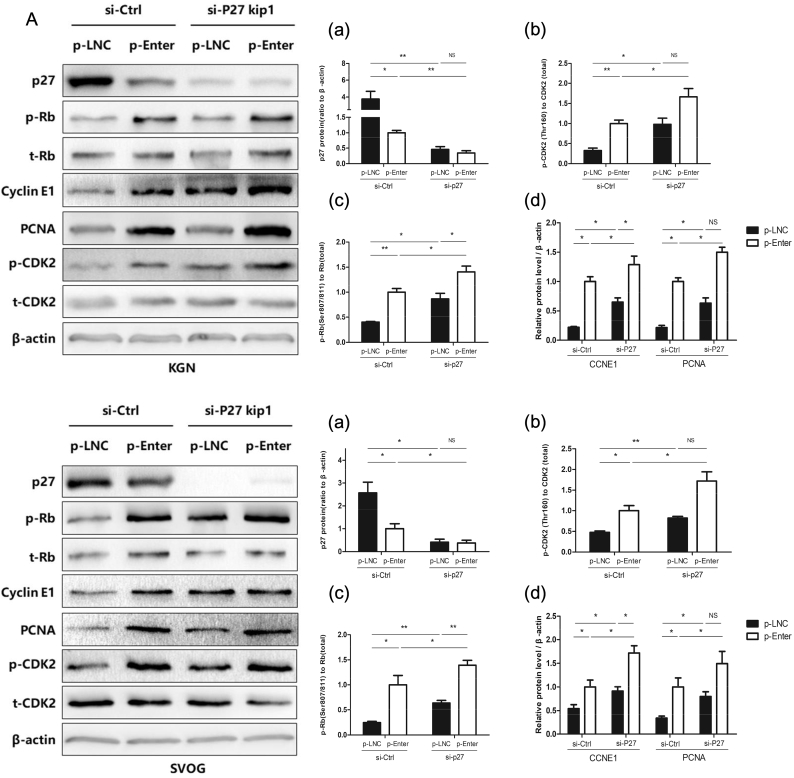
Next, we conducted rescue assays to determine whether p27 is involved in the LINC-01572:28-induced cell proliferation arrest. siRNA-mediated knockdown of p27 expression significantly increased the proliferation rate (Fig. 4C), reduced the G0-G1 phase arrest (Fig. 4D), increased the abundance of PCNA and cyclin E1, and increased the phosphorylation of CDK2 and Rb (Fig. 5) in KGN and SVOG cells. The efficiency of the p27 knockdown is shown in Supplementary Fig. S1. Furthermore, the effects of the overexpression of LINC-01572:28 on the cell proliferation rate (Fig. 4C), G0-G1 phase arrest (Fig. 4D), PCNA and cyclin E1 abundance, and CDK2 and Rb phosphorylation were rescued by the siRNA-mediated knockdown of p27 in KGN and SVOG cells (Fig. 5). These results suggest that LINC-01572:28 inhibits granulosa cell proliferation at least in part due to an increase in p27 protein abundance.
Fig. 5.
The Over-expressed LINC01572:28 induced cell proliferation arrest is partially rescued by knocking down p27 in granulosa cells.
(A) p-LNC and p-Enter cells were treated with si-Ctrl or si-p27 for 48 h; and immunoblotting for p27 (a), PCNA (d), cyclin E1 (d), and the level of phosphorylated CDK2 and Rb (b and c) protein was performed.
Error bars represent SDs from at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
3.6. LINC-01572:28 stabilized p27 by interacting with SKP2 in granulosa cells
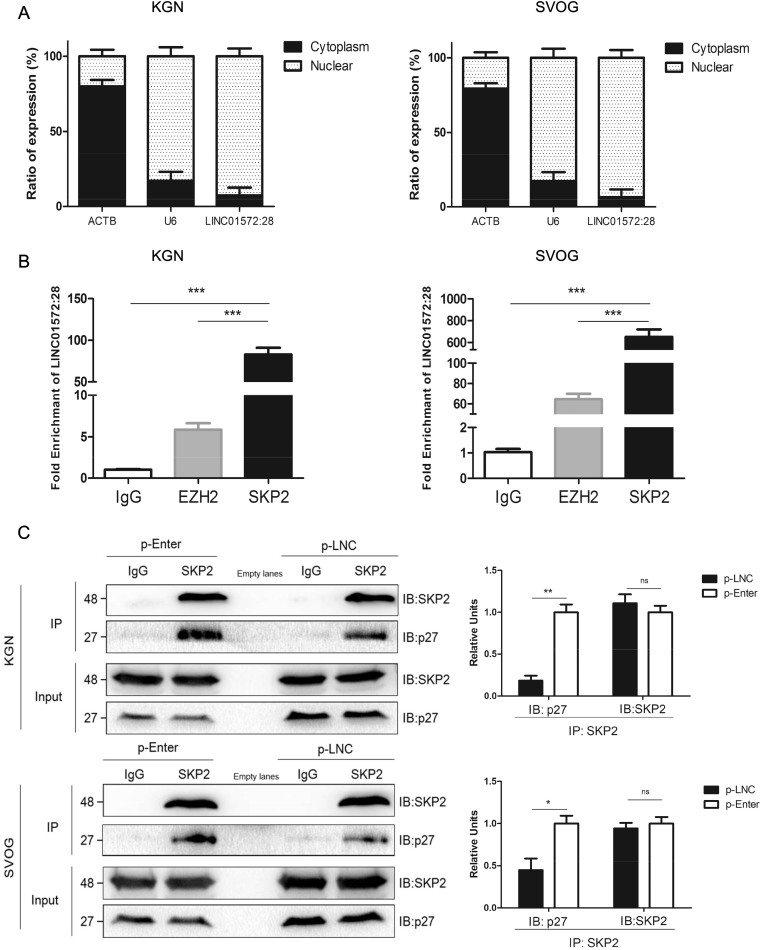
To determine the molecular mechanism underlying the regulation of p27 protein by LINC-01572:28, we first detected the subcellular location of LINC-01572:28 in granulosa cells. The cytoplasmic and nuclear RNA were separated and reverse transcribed, which revealed that LINC-01572:28 was located predominantly in the nucleus in both KGN and SVOG cells (Fig. 6A). ACTB and U6 were used as cytoplasm and nucleus controls, respectively.
Fig. 6.
LINC01572:28 stabilize p27 by interacting with SKP2 in granulosa cells.
(A) qRT-PCR for LINC01572:28 from nuclear and cytoplasmic fractions of untreated granulosa cells. The cytoplasmic β-Actin and nuclear U6 were used as controls.
(B) The enrichment of LINC01572:28 was measured by qRT-PCR from SKP2 RNA IPs (RIP) performed from formalde-crosslinked granulosa cells. Immunoglobulin G (IgG) IP was used as negative controls and EZH2 IP was used as positive controls.
(C) SKP2-p27 interaction was examined in KGN and SVOG cells by immunoprecipitation SKP2 from whole-cell lysates followed by immunoblotting for p27 and SKP2. About 5% of cell lysis used for coIP were loaded as the inputs. IgG IP was used as control.
Error bars represent SDs from at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
Because overexpression of LINC-01572:28 did not affect p27 mRNA expression, we hypothesized that LINC-01572:28 may inhibit the degradation of the p27 protein. Previous studies have demonstrated that SKP2, a key component of the SCFSKP2 ubiquitin-ligase complex, may mediate the cyclin E/CDK2 dependent ubiquitination and degradation of p27 by binding to p27 [23,30,37]. LINC-01572:28 was shown to possess the ability to bind to SKP2 using the online algorithm RPISeq. Thus, we hypothesized that LINC-01572:28 may stabilize p27 by interacting with SKP2 in granulosa cells. Khail et al. reported that approximately 20% of lncRNAs bind to PRC2, which includes EZH2. We therefore used the EZH2-HOTAIR interaction pair as a positive control (interaction probability = .75) [26]. Using RPISeq, we found that the LINC-01572:28-SKP2 interaction pair had a higher score (interaction probability = .9) than LINC-01572:28-EZH2 (interaction probability = .75). Furthermore, RIP results showed a more significant enrichment of LINC-01572:28 using the SKP2 antibody compared with both the EZH2 antibody and the non-specific IgG control antibody (Fig. 6B) in both KGN and SVOG cells. In addition, the co-IP results demonstrated that the overexpression of LINC-01572:28 reduced the interaction of SKP2 with p27 in both KGN and SVOG cells (Fig. 6C). These data suggest that LINC-01572:28 may increase p27 protein abundance by reducing the degradation of p27 protein via an interaction with SKP2 in granulosa cells.
3.7. Decrease in PCNA expression and increase in p27 expression in granulosa cells in patients with PCOS
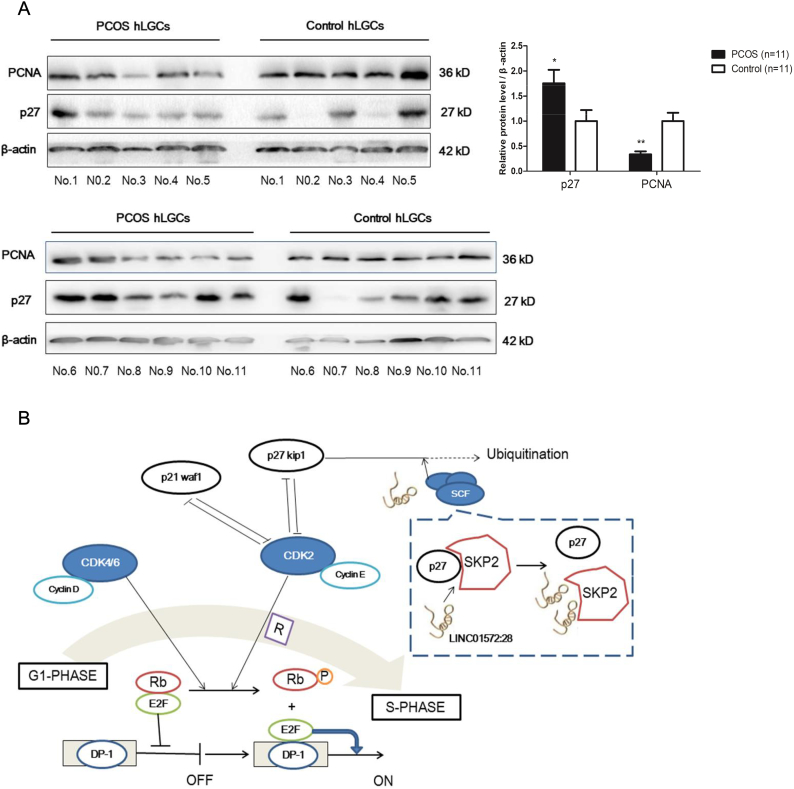
We further tested the PCNA and p27 expression in the luteinized granulosa cells of patients with and without PCOS. As shown in Fig. 7A, the PCNA protein level was decreased whereas the p27 protein was increased in the granulosa cells obtained from women with PCOS.
Fig. 7.
Decrease in PCNA expression while increase in p27 expression in granulosa cells of the PCOS patients.
(A) The protein level of PCNA and p27 in the luteinized granulosa cells of PCOS women (n = 11) and healthy controls (n = 11).
(B) Scheme graph of LINC01572:28 induced proliferation arrest in granulosa cells. LINC-01572:28 suppresses cell proliferation and cell cycle progression by reducing the degradation of p27 protein via competitive SKP2 binding.
Error bars represent SDs from at least 3 independent experiments. ***: p < .001, **: p < .01, *: p < .05, and ns (not significant; P > .05) correspond to two–tailed Student's tests.
4. Discussion
In the current study, we demonstrated that the abundance of LINC-01572:28 was elevated in the hLGCs of women with PCOS compared to those from control women using microarray and qRT-PCR. The elevation of LINC-01572:28 impeded the interaction between SKP2 and p27 via competitive binding to SKP2, which resulted in the accumulation of p27. The accumulation of p27 in turn induced cell cycle arrest and inhibited granulosa cell proliferation. Furthermore, the decrease in PCNA protein and increase in p27 protein were also observed in hLGCs of women with PCOS but not in those from control women.
The arrest of follicle growth is considered the main cause of chronic oligo/anovulation in women with PCOS who exhibit disordered folliculogenesis [4,18,29]. Granulosa cells, which play an important role in folliculogenesis by providing nutrients and growth factors to developing oocytes, are regulated by FSH during the antral follicle phase [5,17,28]. Previous studies have suggested that FSH stimulates the proliferation of granulosa cells via the up-regulation of PCNA protein [42]. In addition, PCNA is down-regulated in the granulosa cell layers of rat and bovine PCOS [2,3,13,36], potentially impairing granulosa cell proliferation. Consistent with these animal models of PCOS, in the present study, we detected a decrease in PCNA protein levels in the luteinized granulosa cells of women with PCOS.
p27 is a cyclin-dependent kinase inhibitor, which negatively regulates G1 phase progression. Previous studies have reported that p27 may act as a tumor suppressor by reducing the phosphorylation of Rb (a negative regulator of the cell cycle which can be inactivated after phosphorylated) via binding to cyclin E-CDK2 or cyclin D-CDK4 complexes [16,38]. Previous studies also have shown that FSH may induce granulosa cell proliferation by regulating p27 expression via the PI3K/Akt and MAPK/ERK pathways [12,14]. p27 also was shown to negatively regulate the activation of murine primordial oocytes in ovarian granulosa cells [8]. Moreover, Robker et al. reported that LH could terminate follicular growth via the up-regulation of p27 and p21 in rhesus monkeys [34]. In the present study, we demonstrated that the siRNA-mediated knockdown of p27 consistently induced granulosa cell proliferation, suggesting that p27 plays a critical role in granulosa cell proliferation. In addition, p27 was more abundant in women with PCOS, suggesting that p27 appears to be involved in the arrest of follicle progression characteristic of PCOS.
In this study, we found that LINC-01572:28 could regulate p27 protein level due to its high affinity to SKP2, a key component of the SCFSKP2 ubiquitin-ligase complex that mediates the cyclin E-CDK2 dependent ubiquitination and degradation of p27 [23,30,37]. Previous studies of proliferation-related lncRNAs, which were performed mainly in tumor cells, showed that lncRNAs could function via an interaction with RNA-binding proteins to influence the transcriptional activity of the target gene, either epigenetically or through the direct stabilization of the mRNA or protein [9,31]. In the present study, we found that lncRNA also could regulate the target gene via competitive binding to a key component of the ubiquitination complex, resulting in a decrease in the degradation of the target protein. This finding thus identifies a new interaction mode for lncRNAs providing additional insight into their function.
LINC01572:28 is an intergenic lncRNA, which is highly expressed in testis[15]. Meanwhile, hyperandrogenemia is not only a key character of PCOS, but also a leading cause of subfertility or infertility. Furthermore, we found that the abundance of LINC01572:28 in the granulosa cells of women with PCOS are positive related with the concentration of testosterone in serum. All of these clues indicated that LINC01572:28 might play a role in the etiology of PCOS, thus, we chose this lncRNA as the studying object. However, we could not elucidate the relationship between the LINC01572:2 and hyperandrogenemia due to the unavailability of sufficient human theca cell, the main source of androgen in ovary. What's more, it seems that overexpressed LINC01572:28 did not influence the abundance of CYP19A1 protein (Supplementary Fig.6), an enzyme responsible for a key step in the biosynthesis of estrogens, is responsible for the aromatization of androgens into estrogens. So, the relationship between the elevation of LINC01572:28 in granulosa cells of women with PCOS and androgen excess was still vague and needed to be elucidated in the future study.
The relationship between hyperandrogenemia and granulosa cell proliferation arrest had been reported several times [2,3], and LINC01572:28 might be a mediator or treatment target in this pathway. As FSH could stimulate granulosa cells to grow, we also detected the abundance of FSHR and LHCGR mRNA in the LINC01572:28 overexpressed granulosa cells, and it showed that there was no difference between these two groups (Supplementary Fig.5). Besides the dysfunction of p27/CDK2 and Cyclin E/Rb axis in the LINC01572:28 overexpressed cell, we also found that mRNA level of Cyclin D1 is downregulated, but there is no change of Cyclin D1 protein level. These indicate LINC01572:28 may influence the degradation of Cyclin D1 protein at the same time, so we could not observe a consistent change in mRNA and protein level of Cyclin D1 after overexpression of LINC01572:28 in KGN cells. Of note, Liu et al. detected the transcripts in a cohort which includes 7 PCOS patients and 7 controls and found that a total of 862 lncRNA were differentially expressed [20], however, we only identified 77 differentially expressed lncRNA between control and PCOS patients in this current study, we think the difference may come from the sample size, the clinical characters and treatment protocol of the PCOS patients, and the different microarray platforms from two companies.
In summary, our findings showed that LINC-01572:28 was higher in granulosa cells of women with PCOS, and that its up-regulation was associated with hyperandrogenemia. LINC-01572:28 may have suppressed the proliferation of granulosa cells partly by decreasing the degradation of the p27 protein. Our study provides a new perspective regarding the molecular mechanisms underlying the disordered folliculogenesis characteristic of PCOS and identifies LINC-01572:28 as a novel target for the treatment of PCOS.
Authors' contributions
Yanzhi Du and Zi-Jiang Chen conceived and supervised the study. Jieying Xu assisted with microarray analysis. Jun Zhao, Wangsheng Wang, Xiaojing Liu and Jiansheng Liu designed and conducted the study, including acquisition, analysis, and interpretation of data. Han Zhao, Yun Sun and Hongbiu Liu participated in the sample collecting and kindly provided cell lines. Jun Zhao and Wangsheng Wang drafted the manuscript. Andrea Dunaif and Yanzhi Du commented on and revised the drafts of the manuscript. All authors critically reviewed, edited and approved the manuscript.
Funding
This research was supported by grants from the National Key Research and Development Program of China (2017YFC1001002), the National Natural Science Foundation (81490743, 81671413 and 81671414), National Institutes of Health (1R01HD085527), Shanghai Municipal Education Commission–Gaofeng Clinical Medicine (20152510) and Shanghai Key Laboratory for Assisted Reproduction and Reproductive Genetics (17DZ2271100).
Acknowledgements
This research was supported by grants from the National Key Research and Development Program of China (2017YFC1001002), the National Natural Science Foundation (81490743, 81671413 and 81671414), National Institutes of Health (1R01HD085527), Shanghai Municipal Education Commission–Gaofeng Clinical Medicine (20152510) and Shanghai Key Laboratory for Assisted Reproduction and Reproductive Genetics (17DZ2271100).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.043.
Contributor Information
Yanzhi Du, Email: duyz@sjtu.edu.cn.
Zi-Jiang Chen, Email: chenzijiang@hotmail.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Broekmans F.J., Fauser B.C. Diagnostic criteria for polycystic ovarian syndrome. Endocrine. 2006;30:3–11. doi: 10.1385/ENDO:30:1:3. [DOI] [PubMed] [Google Scholar]
- 2.Chen H., Guo J.H., Zhang X.H., Chan H.C. Defective CFTR-regulated granulosa cell proliferation in polycystic ovarian syndrome. Reproduction. 2015;149:393–401. doi: 10.1530/REP-14-0368. [DOI] [PubMed] [Google Scholar]
- 3.Chen M.J., Chou C.H., Chen S.U., Yang W.S., Yang Y.S., Ho H.N. The effect of androgens on ovarian follicle maturation: Dihydrotestosterone suppress FSH-stimulated granulosa cell proliferation by upregulating PPARgamma-dependent PTEN expression. Sci Rep. 2015;5 doi: 10.1038/srep18319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewailly D., Catteau-Jonard S., Reyss A.C., Maunoury-Lefebvre C., Poncelet E., Pigny P. The excess in 2-5 mm follicles seen at ovarian ultrasonography is tightly associated to the follicular arrest of the polycystic ovary syndrome. Hum Reprod. 2007;22:1562–1566. doi: 10.1093/humrep/dem060. [DOI] [PubMed] [Google Scholar]
- 5.Edson M.A., Nagaraja A.K., Matzuk M.M. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn R.A., Chang H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodarzi M.O., Dumesic D.A., Chazenbalk G., Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol. 2011;7:219–231. doi: 10.1038/nrendo.2010.217. [DOI] [PubMed] [Google Scholar]
- 8.Hirashima Y., Moniruzzaman M., Miyano T. p27(Kip1) negatively regulates the activation of murine primordial oocytes. J Reprod Dev. 2011;57:217–222. doi: 10.1262/jrd.10-119h. [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X., Hao C., Bao H., Wang M., Dai H. Aberrant expression of long noncoding RNAs in cumulus cells isolated from PCOS patients. J Assist Reprod Genet. 2016;33:111–121. doi: 10.1007/s10815-015-0630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughesdon P.E. Morphology and morphogenesis of the Stein-Leventhal ovary and of so-called hyperthecosis. Obstet Gynecol Surv. 1982;37:59–77. doi: 10.1097/00006254-198202000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Hunzicker-Dunn M.E., Lopez-Biladeau B., Law N.C., Fiedler S.E., Carr D.W., Maizels E.T. PKA and GAB2 play central roles in the FSH signaling pathway to PI3K and AKT in ovarian granulosa cells. Proc Natl Acad Sci U S A. 2012;109:E2979–E2988. doi: 10.1073/pnas.1205661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isobe N., Yoshimura Y. Immunocytochemical study of cell proliferation in the cystic ovarian follicles in cows. Theriogenology. 2000;54:1159–1169. doi: 10.1016/s0093-691x(00)00423-4. [DOI] [PubMed] [Google Scholar]
- 14.Kayampilly P.P., Menon K.M. Follicle-stimulating hormone inhibits adenosine 5′-monophosphate-activated protein kinase activation and promotes cell proliferation of primary granulosa cells in culture through an Akt-dependent pathway. Endocrinology. 2009;150:929–935. doi: 10.1210/en.2008-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolupaeva V., Basilico C. Overexpression of cyclin E/CDK2 complexes overcomes FGF-induced cell cycle arrest in the presence of hypophosphorylated Rb proteins. Cell Cycle. 2012;11:2557–2566. doi: 10.4161/cc.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar T.R., Wang Y., Lu N., Matzuk M.M. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat Genet. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Li R., Yu H., Zhao S., Yu Y., Qiao J. The relationship between serum anti-Mullerian hormone levels and the follicular arrest for women with polycystic ovary syndrome. Syst Biol Reprod Med. 2015;61:103–109. doi: 10.3109/19396368.2014.973123. [DOI] [PubMed] [Google Scholar]
- 19.Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y.D., Li Y., Feng S.X., Ye D.S., Chen X., Zhou X.Y. Long Noncoding RNAs: Potential Regulators Involved in the Pathogenesis of Polycystic Ovary Syndrome. Endocrinology. 2017;158:3890–3899. doi: 10.1210/en.2017-00605. [DOI] [PubMed] [Google Scholar]
- 21.March W.A., Moore V.M., Willson K.J., Phillips D.I., Norman R.J., Davies M.J. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum Reprod. 2010;25:544–551. doi: 10.1093/humrep/dep399. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda F., Inoue N., Manabe N., Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 23.Montagnoli A., Fiore F., Eytan E., Carrano A.C., Draetta G.F., Hershko A. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller H., Lukas J., Schneider A., Warthoe P., Bartek J., Eilers M. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci U S A. 1994;91:2945–2949. doi: 10.1073/pnas.91.8.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munakata Y., Kawahara-Miki R., Shiratsuki S., Tasaki H., Itami N., Shirasuna K. Gene expression patterns in granulosa cells and oocytes at various stages of follicle development as well as in in vitro grown oocyte-and-granulosa cell complexes. J Reprod Dev. 2016;62:359–366. doi: 10.1262/jrd.2016-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muppirala U.K., Honavar V.G., Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics. 2011;12:489. doi: 10.1186/1471-2105-12-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oktem O., Urman B. Understanding follicle growth in vivo. Hum Reprod. 2010;25:2944–2954. doi: 10.1093/humrep/deq275. [DOI] [PubMed] [Google Scholar]
- 29.Orisaka M., Hattori K., Fukuda S., Mizutani T., Miyamoto K., Sato T. Dysregulation of ovarian follicular development in female rat: LH decreases FSH sensitivity during preantral-early antral transition. Endocrinology. 2013;154:2870–2880. doi: 10.1210/en.2012-2173. [DOI] [PubMed] [Google Scholar]
- 30.Pagano M., Tam S.W., Theodoras A.M., Beer-Romero P., Del Sal G., Chau V. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 31.Qiu M., Xu Y., Wang J., Zhang E., Sun M., Zheng Y. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 33.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robker R.L., Richards J.S. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol Endocrinol. 1998;12:924–940. doi: 10.1210/mend.12.7.0138. [DOI] [PubMed] [Google Scholar]
- 35.Rotterdam, E. & ASRM-SPONSORED, P. 2004. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS), Human Reproduction (OxfordEngland), 19, 41. [DOI] [PubMed]
- 36.Salvetti N.R., Panzani C.G., Gimeno E.J., Neme L.G., Alfaro N.S., Ortega H.H. An imbalance between apoptosis and proliferation contributes to follicular persistence in polycystic ovaries in rats. Reprod Biol Endocrinol. 2009;7:68. doi: 10.1186/1477-7827-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheaff R.J., Groudine M., Gordon M., Roberts J.M., Clurman B.E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 38.Slingerland J., Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J Cell Physiol. 2000;183:10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z.A., Kalderon D. Cyclin E-dependent protein kinase activity regulates niche retention of Drosophila ovarian follicle stem cells. Proc Natl Acad Sci U S A. 2009;106:21701–21706. doi: 10.1073/pnas.0909272106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong Y., Menninger J., Beach D., Ward D.C. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics. 1992;13:575–584. doi: 10.1016/0888-7543(92)90127-e. [DOI] [PubMed] [Google Scholar]
- 42.Yu F.Q., Han C.S., Yang W., Jin X., Hu Z.Y., Liu Y.X. Role of ERK1/2 in FSH induced PCNA expression and steroidogenesis in granulosa cells. Front Biosci. 2005;10:896–904. doi: 10.2741/1584. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Q., Zuo R., He Y., Wang Y., Chen Z.J., Sun Y. Local Regeneration of Cortisol by 11beta-HSD1 Contributes to Insulin Resistance of the Granulosa Cells in PCOS. J Clin Endocrinol Metab. 2016;101:2168–2177. doi: 10.1210/jc.2015-3899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material