Highlights
-
•
GATA1 and FLI1 switch on the endogenous platelet factor 4 (Pf4) gene in fibroblasts.
-
•
Pf4 expression is maintained and increases over time.
-
•
PF4 protein is secreted and readily collected.
-
•
A new technology for the g0065neration of important biomolecules in heterologous cells.
Keywords: Platelet factor 4, Reprogramming, Megakaryocyte, Fibroblast
Abstract
The ability of transcriptional regulators to drive lineage conversion of somatic cells offers great potential for the treatment of human disease. To explore the concept of switching on specific target genes in heterologous cells, we developed a model system to screen candidate factors for their ability to activate the archetypal megakaryocyte-specific chemokine platelet factor 4 (PF4) in fibroblasts. We found that co-expression of the transcriptional regulators GATA1 and FLI1 resulted in a significant increase in levels of PF4, which became magnified over time. This finding demonstrates that such combinations can be used to produce potentially beneficial chemokines in readily available heterologous cell types.
Since Yamanaka’s pioneering discovery of cellular reprogramming of fibroblasts into induced pluripotent stem cells [1,2], attention in the field has been aimed towards uncovering combinations of factors that can directly convert differentiated cells from one lineage to another. However, the downstream applications of transdifferentiation remain restricted due to low conversion efficiency, safety concerns and phenotypic differences between target and reprogrammed tissues [3]. Recognizing the challenges and limitations of cellular reprogramming, we decided to investigate whether defined factors can be used to initiate a specific heterologous gene expression program with the aim of producing secretable, beneficial biomolecules such as growth factors, cytokines or hormones.
To explore this strategy, we have developed a model cell-based assay system to assess candidate reprogramming factors for their ability to induce heterologous gene expression in fibroblasts. For the assay, we chose platelet factor 4 (PF4/CXCL4), an archetypal megakaryocyte chemokine with multiple roles in megakaryopoiesis (reviewed in [4]). Having selected a pool of candidate factors with key roles in megakaryocyte differentiation, we defined specific combinations capable of initiating a switch from a fibroblast to a more megakaryocyte-like gene expression program – namely, transcriptional regulators GATA1 and FLI1. We found that these factors induced significant levels of platelet factor 4, and furthermore showed that platelet factor 4 was abundantly secreted into the medium from the reprogrammed cells.
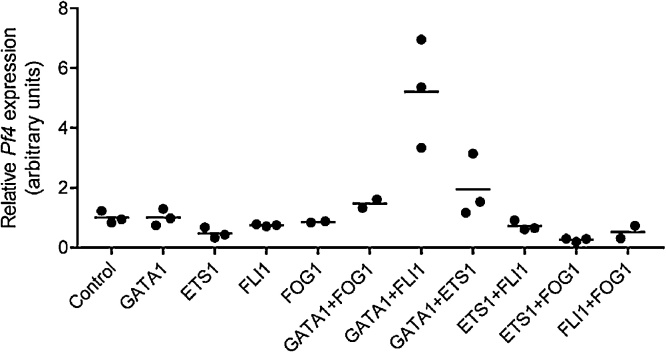
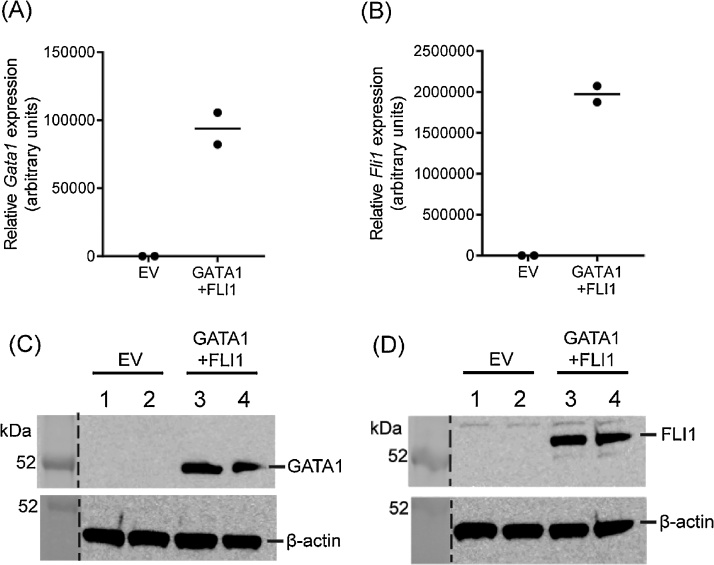
To identify factors capable of inducing Pf4 gene expression, we used a candidate approach by selecting a pool of hematopoietic transcriptional regulators with known roles in megakaryopoiesis [[5], [6], [7], [8], [9]]. Retroviral vectors encoding GATA1 (pMSCV-Hygro; Clontech), FLI1 and ETS1 (pMSCVPuro; Clontech) were constructed by inserting the coding sequence (GATA1) or open reading frame (FLI1 and ETS1) into the multiple cloning site. A lentiviral vector containing the coding sequence for FOG1 (pLV411; provided by Prof Thomas Gonda) was created using the Gateway PCR Cloning System and an LR Clonase II Enzyme mix kit (Life Technologies), according to manufacturers’ instructions. Retroviral and lentiviral constructs were transduced into murine embryonic fibroblasts (MEFs) with single and two factor combinations. Cells were maintained in DMEM containing 10% serum, 1000 U/L penicillin, 1 μg/mL streptomycin, 2 mM L-glutamine and 50 ng/mL recombinant human thrombopoietin (TPO) in addition to 2.5 μg/mL puromycin or 500 μg/mL hygromycin where appropriate. To evaluate the potency of these factors, we extracted total RNA as described previously [10] and used real time RT-PCR to assay for Pf4 gene expression [11] (Fig. 1). Whilst no single factor enhanced Pf4 expression, through forced combinatorial expression we discovered that co-transduction of GATA1 and FLI1 resulted in a significant five-fold boost in Pf4 gene activity (primer sequences can be found in Table 1). To confirm this observation, we used antibiotic selection to generate stable cell lines expressing these factors and demonstrated high mRNA and protein expression of GATA1 and FLI1 (Fig. 2A–D).
Fig. 1.
Co-transduction of GATA1 and FLI1 promote Pf4 gene expression in fibroblasts. Real time RT-PCR was used to assess mRNA levels of Pf4 in MEFs transduced with candidate transcription factors or empty vector (control). Expression levels were normalised to 18S rRNA and are shown relative to cells transduced with empty vector, with the empty vector mean value being set to 1.0. The mean for two to three independent experiments is shown as a horizontal line.
Table 1.
Real time RT-PCR primer sequences.
| Gene name | 5’–3’ sequence | 3’-5’ sequence |
|---|---|---|
| 18S rRNA | CACGGCCGGTACAGTGAAAC | AGAGGAGCGAGCGACCAA |
| Gata1 | AGCATCAGCACTGGCCTACT | AGGCCCAGCTAGCATAAGGT |
| Fli1 | CAACCAGCCAGTGAGAGTCA | GCCCACCAGCTTGTTACATT |
| Pf4 | GCGGTTCCCCAGCTCATAG | CCGGTCCAGGCAAATTTTC |
Fig. 2.
Stable expression of Gata1 and Fli1 in fibroblasts. MEFs were stably transduced with pMSCV-Hygro-Gata1 or pMSCV-Puro-Fli1 and expression of Gata1 (A) and Fli1 (B) were confirmed by real time RT-PCR, normalised to levels of 18S rRNA and with empty vector (EV) set to 1. Horizontal lines represent the mean of two independent experiments. Nuclear extracts were also prepared from stably transduced cells to assess GATA1 (D) and FLI1 (E) protein levels by Western blot with normalisation to β-actin. Anti-GATA1 N6 (Santa Cruz) and anti-FLI1 C-19 (Santa Cruz) antibodies were used to probe for each protein respectively. Two independent cell lines of EV (lanes 1 and 2) and GATA1/FLI1-transduced MEFs (lanes 3 and 4) were used for Western blots.
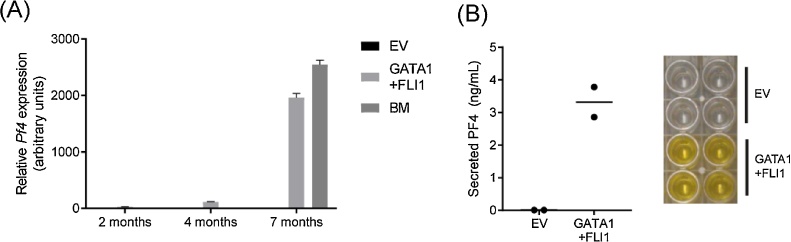
Having established stably-expressing cell lines we examined Pf4 expression over time and observed a significant progressive elevation of Pf4 mRNA, comparable to expression seen in bone marrow (Fig. 3A). To assess whether sustained Pf4 activation translated into expression and secretion of protein, we assayed platelet factor 4 levels in cell culture media of control and GATA1/FLI1 MEFs using ELISA according to the manufacturer’s instructions (Quantikine, R&D Systems). Fig. 3B shows that while no PF4 is detected in the media of control cells, the addition of GATA1 and FLI1 factors resulted in the secretion of high levels of the protein (>3 ng/mL).
Fig. 3.
Increasing long-term expression of platelet factor 4 in GATA1/FLI1 stable cell lines. (A) Pf4 gene expression was determined by real time RT-PCR at the indicated time points, following stable transduction of MEFs with pMSCV-Hygro-Gata1 and pMSCV-Puro-Fli1. Bone marrow (BM) was used as a positive control. Expression levels were normalised to 18S rRNA and are shown relative to cells transduced with EV (set to 1 at each time point). Error bars represent standard error of the mean. (B) Secreted platelet factor 4 levels in GATA1/FLI1-transduced MEFs were compared to EV controls by quantitative ELISA (R&D Systems). Means are representative of two independent samples, shown by a horizontal line.
Additionally, microarrays were performed on GATA1/FLI1 cells and control cells four and seven months post-transduction (GEO accession no. GSE108983), to observe the extent of lineage reprogramming away from fibroblasts towards a megakaryocytic signature. These time points were chosen given that expression of our biomolecule of interest PF4 was most significantly up-regulated following long-term overexpression of GATA1/FLI1. RNA at 50 ng/uL (total of 500 ng for each sample) was provided to the Ramaciotti Centre for Genomics (UNSW Sydney) for hybridization to Affymetrix GeneChip® Mouse Gene 1.0 ST arrays and scanning, and analysis was carried out using Partek® Genomics SuiteTM software. Following co-transduction of GATA1/FLI1, we observed an increase in various megakaryocytic genes including Pf4 and Gp9, encoding the secreted chemokine platelet factor 4 and the megakaryocyte-specific surface marker CD42a, respectively. On the other hand, genes that were mostly down-regulated in the GATA1/FLI1 cells included many fibroblast-specific genes such as Fbln5, Fbn1, Col5a, and Fgf2, suggesting that the GATA1/FLI1-expressing cells departed from a fibroblast-specific gene program. We compared up-regulated genes in four- and seven-month GATA1/FLI1-transduced cells to a set of megakaryocytic-enriched genes derived from the Haemopedia Mouse RNA-Seq. This dataset is accessible at the Haemosphere online database (haemosphere.org) and contains the 100 most highly enriched genes in bone marrow-derived megakaryocytes cultured in TPO [22]. We found that several of the most highly enriched megakaryocyte genes were up-regulated in our GATA1/FLI1-transduced cells, indicating partial conversion and confirming an increase in expression of platelet factor 4 (Table 2). While we did not see comprehensive reprogramming towards the megakaryocytic lineage as others have observed [[12], [13], [14]], we did identify a shift in gene expression away from the fibroblastic profile including, importantly, distinct up-regulation of PF4.
Table 2.
Upregulated genes in GATA1/FLI1-transduced cells corresponding to the top 100 most highly-enriched genes from bone marrow-derived megakaryocytes (Haemopedia Mouse RNA-Seq).
| Gene symbol (four months) | FCa | Gene symbol (seven months) | FCa |
|---|---|---|---|
| Podxl | 11.5 | Gm10419 | 12.3 |
| Casp4 | 4.5 | Dusp4 | 10.3 |
| Dusp4 | 4.3 | Podxl | 10.2 |
| Gm10419 | 3.9 | Casp4 | 6.8 |
| Tmem40 | 2.8 | Clu | 4.8 |
| Pf4 | 4.3 | ||
| Tmem40 | 4.2 | ||
| Gp9 | 3.6 |
FC: fold change (GATA1/FLI1-transduced compared to empty vector).
Having determined that forced expression of GATA1 and FLI1 can redirect fibroblasts to express platelet factor 4, we decided to investigate the potential of replacing GATA1 with a naturally occurring mutant form of the protein that lacks the N-terminal activation domain but retains the ability to bind DNA and interact with the GATA1 cofactor FOG [15]. While GATA1 promotes megakaryocyte differentiation, this short form of GATA, termed GATA1s, has been shown to maintain megakaryocyte progenitor proliferation [16,17]. In defining factors capable of switching on the expression of cytokines in heterologous cells, we reasoned that the inclusion of a gene regulatory protein that promotes progenitor proliferation might ultimately prove beneficial in enhancing the population size of transdifferentiating cells.
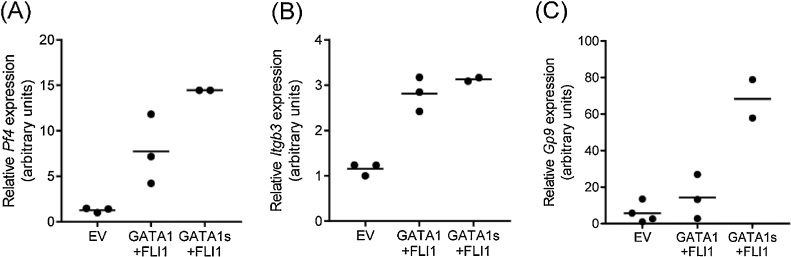
To confirm that GATA1s can substitute for GATA1 in our experiments, we first transduced MEFs with GATA1s and FLI1 to generate stable cell lines and compared expression of critical megakaryocyte genes such as Pf4, Itgb3 and Gp9 in these cells with lines expressing only GATA1 and FLI1. We found that substitution of GATA1 by GATA1s resulted in a significant increase in Pf4 and Gp9 mRNA, while leading to similar levels of Itgb3, encoding the megakaryocyte surface marker CD61 (Fig. 4A–C). These data demonstrate that GATA1s has the potential to substitute for GATA1 to work in combination with FLI1 to initiate a shift in the cellular gene expression programme in fibroblasts leading to upregulation of platelet factor 4.
Fig. 4.
GATA1s can substitute for GATA1 in driving megakaryocyte gene expression. (A) Expression levels of Pf4 (A), Itgb3 (B) and Gp9 (C) after three weeks in MEFs transduced with either pMSCV-Hygro-Gata1 and pMSCV-Puro-Fli1 or pMSCV-Hygro-Gata1s and pMSCV-Puro-Fli1 were normalised to levels of 18S rRNA. EV cell lines were transduced with pMSCV-Hygro or pMSCV-Puro and set to 1, with means represented by horizontal lines.
Here, we have screened candidate factors for their potential to reprogram murine embryonic fibroblasts to produce the megakaryocyte-specific chemokine platelet factor 4. In refining our choice of reprogramming factors, we found that GATA1s, a short mutant form of the protein [18], can substitute for GATA1, again working synergistically with FLI1 to drive expression of megakaryocyte-specific genes in fibroblasts. Given that GATA1s promotes lineage differentiation while still maintaining cellular proliferation [17], such alternative forms of reprogramming factors could be preferable in strategies aimed at establishing sizeable pools of biomolecule-producing cells.
In addition to up-regulating Pf4 gene activity, we found that forced expression of GATA1 and FLI1 in our fibroblast lines was accompanied by increased expression of other megakaryocyte genes, such as Itgb3 and Gp9, in line with partial reprogramming toward a megakaryocyte-like phenotype. This shift towards the megakaryocyte lineage corroborates recent research proposing a role for GATA1 and FLI1 in megakaryocyte forward programming [12]. However, it is apparent in our case that reprogramming is incomplete. We did not observe definitive morphological changes consistent with a megakaryocyte-like phenotype (data not shown), indicating that genetic barriers to further reprogramming remain in place. This suggests that additional or alternative combinations of factors are required to complete transdifferentiation of fibroblasts into megakaryocyte-like cells. However, we have shown that targeted addition of transcription factors can shift fibroblasts towards a more functional megakaryocytic state with the capacity to produce platelet factor 4 en masse.
The candidate factors used in this screen were chosen largely due to known roles either in directing haematopoietic progenitors toward the megakaryocyte lineage, in megakaryocyte progenitor proliferation or in megakaryocyte differentiation. While GATA1 and FLI1 have been reported as inducers of megakaryocyte gene expression [19], our study is the first to identify that these factors drive the expression of platelet factor 4, a megakaryocyte-restricted biomolecule.
In this study, we have determined that combinatorial forced expression of the transcriptional regulators GATA1 and FLI1 is capable of megakaryocyte-specific reprogramming, which can have beneficial implications. The ability to switch on the Pf4 gene to manipulate levels of this chemokine may have future therapeutic applications, given its multiple roles in maintaining hemostasis and managing thrombosis [4]. Furthermore, PF4 has recently been shown to be a component of a negative paracrine regulatory circuit that controls haematopoietic stem cell numbers within specific niches in the bone marrow [20]. The identification of factors that drive increased Pf4 expression may therefore prove useful in the treatment of disease states where low levels of the chemokine are associated with deregulated hematopoietic stem cell proliferation and blood cancer [21]. It is encouraging for future applications that we were able to confirm that gene regulatory proteins with known roles in megakaryopoiesis are capable of activating megakaryocyte-specific gene expression in fibroblasts. These findings provide a proof of concept for the use of reprogramming factors to readily generate accessible beneficial biomolecules, and the success of this strategy suggests that continued research in this area may define a wide range of combinatorial factors capable of driving expression of a variety of beneficial biomolecules in heterologous cell types.
Conflict of interest
All authors declare that there is no conflict of interest.
Acknowledgements
This work was supported by grants from the Australian National Health and Medical Research Council (APP1025873). CMA and AJK were supported by Australian Postgraduate Awards. All authors declare that there is no conflict of interest.
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41(Suppl. 1):51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma T., Xie M., Laurent T., Ding S. Progress in the reprogramming of somatic cells. Circ. Res. 2013;112:562–574. doi: 10.1161/CIRCRESAHA.111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowalska M.A., Rauova L., Poncz M. Role of the platelet chemokine platelet factor 4 (PF4) in hemostasis and thrombosis. Thromb. Res. 2010;125:292–296. doi: 10.1016/j.thromres.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira R., Ohneda K., Yamamoto M., Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol. Cell. Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J., Pazin M.J., Ravid K. Properties of ets-1 binding to chromatin and its effect on platelet factor 4 gene expression. Mol. Cell. Biol. 2004;24:428–441. doi: 10.1128/MCB.24.1.428-441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lulli V., Romania P., Morsilli O., Gabbianelli M., Pagliuca A., Mazzeo S., Testa U., Peschle C., Marziali G. Overexpression of Ets-1 in human hematopoietic progenitor cells blocks erythroid and promotes megakaryocytic differentiation. Cell Death Differ. 2006;13:1064–1074. doi: 10.1038/sj.cdd.4401811. [DOI] [PubMed] [Google Scholar]
- 8.Shivdasani R.A. Molecular and transcriptional regulation of megakaryocyte differentiation. Stem Cells. 2001;19:397–407. doi: 10.1634/stemcells.19-5-397. [DOI] [PubMed] [Google Scholar]
- 9.Tijssen M.R., Ghevaert C. Transcription factors in late megakaryopoiesis and related platelet disorders. J. Thromb. Haemost. 2013;11:593–604. doi: 10.1111/jth.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock D., Funnell A., Jack B., Johnston J. Introducing undergraduate students to real-time PCR. Biochem. Mol. Biol. Educ. 2010;38:309–316. doi: 10.1002/bmb.20414. [DOI] [PubMed] [Google Scholar]
- 11.Ravid K., Beeler D.L., Rabin M.S., Ruley H.E., Rosenberg R.D. Selective targeting of gene products with the megakaryocyte platelet factor 4 promoter. Proc. Natl. Acad. Sci. U. S. A. 1991;88:1521–1525. doi: 10.1073/pnas.88.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau T., Evans A.L., Vasquez L., Tijssen M.R., Yan Y., Trotter M.W., Howard D., Colzani M., Arumugam M., Wu W.H., Dalby A., Lampela R., Bouet G., Hobbs C.M., Pask D.C., Payne H., Ponomaryov T., Brill A., Soranzo N., Ouwehand W.H., Pedersen R.A., Ghevaert C. Large-scale production of megakaryocytes from human pluripotent stem cells by chemically defined forward programming. Nat. Commun. 2016;7:11208. doi: 10.1038/ncomms11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ono Y., Wang Y., Suzuki H., Okamoto S., Ikeda Y., Murata M., Poncz M., Matsubara Y. Induction of functional platelets from mouse and human fibroblasts by p45NF-E2/Maf. Blood. 2012;120:3812–3821. doi: 10.1182/blood-2012-02-413617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pulecio J., Alejo-Valle O., Capellera-Garcia S., Vitaloni M., Rio P., Mejía-Ramírez E., Caserta I., Bueren Juan A., Flygare J., Raya A. Direct conversion of fibroblasts to megakaryocyte progenitors. Cell Rep. 2016;17:671–683. doi: 10.1016/j.celrep.2016.09.036. [DOI] [PubMed] [Google Scholar]
- 15.Wechsler J., Greene M., McDevitt M.A., Anastasi J., Karp J.E., Le Beau M.M., Crispino J.D. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 16.Kuhl C., Atzberger A., Iborra F., Nieswandt B., Porcher C., Vyas P. GATA1-mediated megakaryocyte differentiation and growth control can be uncoupled and mapped to different domains in GATA1. Mol. Cell. Biol. 2005;25:8592–8606. doi: 10.1128/MCB.25.19.8592-8606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muntean A.G., Crispino J.D. Differential requirements for the activation domain and FOG-interaction surface of GATA-1 in megakaryocyte gene expression and development. Blood. 2005;106:1223–1231. doi: 10.1182/blood-2005-02-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng R., Blobel G.A. GATA transcription factors and cancer. Genes Cancer. 2010;1:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Crispino J.D., Letting D.L., Nakazawa M., Poncz M., Blobel G.A. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 2002;21:5225–5234. doi: 10.1093/emboj/cdf527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruns I., Lucas D., Pinho S., Ahmed J., Lambert M.P., Kunisaki Y., Scheiermann C., Schiff L., Poncz M., Bergman A., Frenette P.S. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aivado M., Spentzos D., Germing U., Alterovitz G., Meng X.Y., Grall F., Giagounidis A.A., Klement G., Steidl U., Otu H.H., Czibere A., Prall W.C., Iking-Konert C., Shayne M., Ramoni M.F., Gattermann N., Haas R., Mitsiades C.S., Fung E.T., Libermann T.A. Serum proteome profiling detects myelodysplastic syndromes and identifies CXC chemokine ligands 4 and 7 as markers for advanced disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1307–1312. doi: 10.1073/pnas.0610330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Graaf Carolyn A., Choi J., Baldwin Tracey M., Bolden Jessica E., Fairfax Kirsten A., Robinson Aaron J., Biben C., Morgan C., Ramsay K., Ng Ashley P., Kauppi M., Kruse Elizabeth A., Sargeant Tobias J., Seidenman N., D’Amico A., D’Ombrain Marthe C., Lucas Erin C., Koernig S., Morelli Baz, A Wilson, Michael J., Dower Steven K., Williams B., Heazlewood Shen Y., Hu Y., Nilsson Susan K., Wu L., Smyth Gordon K., Alexander Warren S., Hilton Douglas J. Haemopedia: an expression atlas of murine hematopoietic cells. Stem Cell Rep. 2016;7:571–582. doi: 10.1016/j.stemcr.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]