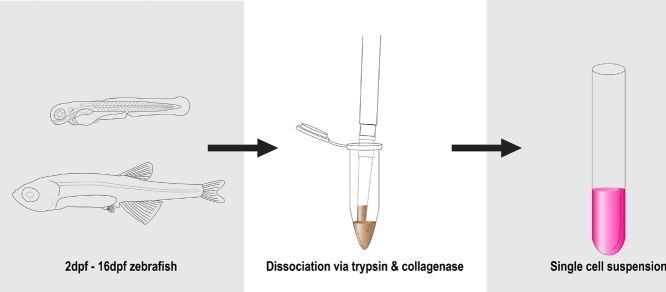
Graphical abstract
Method name: Zebrafish embryo, Larvae dissociation to single cell suspension
Keywords: Zebrafish, Dissociation, Single cell suspension
Abstract
Zebrafish (Danio rerio) has emerged as a powerful animal model to study developmental processes and human diseases. The introduction of CRISPR/Cas9 as a genome editing tool allowed the generation of genetic mutants with high-throughput (Varshney et al., 2015) and has opened the possibility to understand gene function not only during embryonic stages but also in larval stages. Therefore, there is an increasing need to optimize methods for embryo and larvae dissociation that allow the generation of single cell suspension for fluorescence-activated cell sorting (FACS), RNA extraction and single cell RNA-sequencing.
-
•
Here we present a quick and efficient protocol for the dissociation of zebrafish embryos and larvae (2–16 days post fertilization (dpf), Fig. 1). This protocol was built empirically and optimized based on a method protocol from Gallardo and Behra (2013) [1].
-
•
The protocol is divided in three steps:
-
•
Experimental set up (∼10 min)
-
•
Dissociation with trypsin and collagenase (∼10–15 min)
-
•
Preparation of single cell suspension (∼15 min)
-
•
This protocol has been optimized in order to be used for FACS, RNA extraction (for RNA sequencing) and single-cell RNA sequencing.
Fig. 1.
Visual representation of the 3-Step protocol for zebrafish embryos and larvae dissociation.
Specifications Table
| Subject Area | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Developmental biology, zebrafish |
| Method name: | Zebrafish embryo, larvae dissociation to single cell suspension |
| Name and reference of original method |
Our dissociation protocol was built empirically and optimized based on a method protocol from Gallardo et al. Gallardo VE, Behra M. Fluorescent activated cell sorting (FACS) combined with gene expression microarrays for transcription enrichment profiling of zebrafish lateral line cells. Methods. 2013;62(3):226-231. |
| Resource availability |
Reagents:
|
Method details
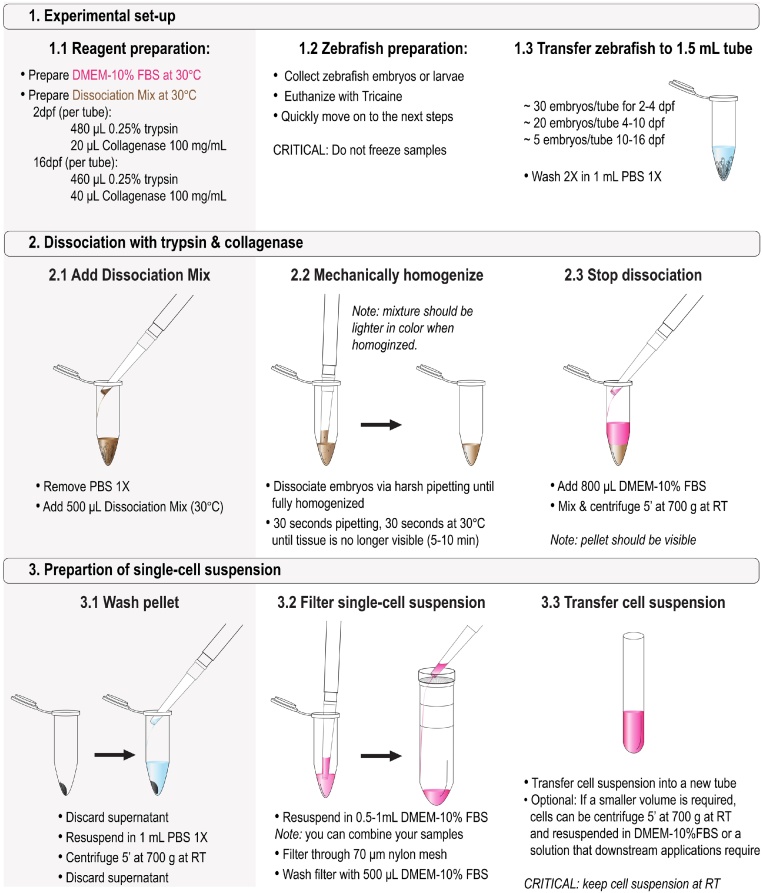
Experimental set-up
Reagent preparation
-
•
Resuspend the collagenase in PBS 1X to prepare a 100 mg/ml stock. Aliquot (40 μl each), store at −20 °C. Use a fresh aliquot for each experiment.
-
•
Prepare DMEM-10%FBS (make fresh every time) and equilibrate at ∼30 °C.
-
•
Prepare the Dissociation Mix* and keep at 30 °C. Each tube requires 500 μl of Dissociation Mix.
*Dissociation Mix
480 μl 0.25% trypsin-EDTA + 20 μl Collagenase 100 mg/ml (for embryos 2–4 dpf)
OR
460 μl 0.25% trypsin-EDTA + 40 μl Collagenase 100 mg/ml (for 5–16 dpf embryos)
Zebrafish preparation
-
•
Collect zebrafish embryos or larvae at the desired stage. Euthanize embryos or larvae with Tricaine and quickly proceed to the next step. CRITICAL: Do not freeze samples.
Transfer zebrafish embryos or larvae into 1.5 ml microcentrifuge tube
∼ 30 embryos/tube for 2–4 dpf
∼ 20 embryos/tube 4–10 dpf
∼ 5 embryos/tube 10–16 dpf
-
•
Wash 2X in 1 ml PBS 1X
Dissociation with trypsin and collagenase
Add dissociation mix
-
•
Remove PBS 1X
-
•
Add 500 μl of Dissociation Mix* (pre-heated at 30 °C) to each 1.5 ml microcentrifuge tube
Mechanically homogenize
-
•
Mechanically dissociate the embryos via harsh pipetting (use a P1000 and then P200). Interval ∼30 s pipetting and ∼30 s in heat-block at 30 °C until tissue is no longer visible (5–10 min).
NOTE: once fully homogenized, the mixture will appear lighter in color.
Stop dissociation
-
•
Add 800 μl of DMEM-10%FBS to the Dissociation Mix
-
•
Mix and centrifuge 5 min at 700 g at room temperature (RT)
NOTE: after centrifugation the pellet should be clearly visible.
Preparation of single cell suspension
Wash the pellet
-
•
Discard the supernatant
-
•
Resuspend the pellet in 1 ml PBS 1X
-
•
Centrifuge 5 min at 700 g at RT
Filter single cell suspension
-
•
Discard the supernatant and resuspend in 0.5–1 ml of DMEM-10%FBS
-
•
NOTE: If multiple samples need to be pooled for downstream applications it can be done at this resuspension step. Resuspend pellet 1 with DMEM-10%FBS and use the resuspension to resuspend subsequent samples (consider 1 ml for up to 3 samples) and then proceed to the next step.
-
•
Filter through 70 μm nylon mesh into 50 ml conical tube
-
•
Wash the filter with 500 μl of DMEM-10%FBS
Transfer the cell suspension
-
•
Transfer the single cell suspension into a new tube and proceed with downstream applications.
-
•
Optional Step: If a smaller volume is required cells can be centrifuge 5 min at 700 g at RT and resuspended in DMEM-10%FBS or a solution that downstream applications require.
CRITICAL: Keep the cell suspension at room temperature and do not store on ice.
Acknowledgements
We would like to thank Dr. Raman Sood and Blake Carrington for helpful discussion and critical reading of the manuscript. The method presented in this manuscript was supported by the Intramural Research Programs at the National Human Genome Research Institute, NIH.
References
- 1.Gallardo V.E., Behra M. Fluorescent activated cell sorting (FACS) combined with gene expression microarrays for transcription enrichment profiling of zebrafish lateral line cells. Methods. 2013;62(3):226–231. doi: 10.1016/j.ymeth.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]