Abstract
Background
Alternative splicing (AS), as a potent and pervasive mechanism of transcriptional regulatory, expands the genome's coding capacity and involves in the initiation and progression of cancer. Systematic analysis of alternative splicing in colorectal cancer (CRC) is lacking and greatly needed.
Methods
RNA-Seq data and corresponding clinical information of CRC cohort were downloaded from the TCGA data portal. Then, a java application, known as SpliceSeq, was used to evaluate the RNA splicing patterns and calculate the Percent Spliced In (PSI) value. Differently expressed AS events (DEAS) were identified based on PSI value between paired CRC and adjacent tissues. DEAS and its splicing networks were further analyzed by bioinformatics methods. Kaplan-Meier, Cox proportional regression and unsupervised clustering analysis were used to evaluate the association between DEAS and patients' clinical features.
Results
After strict filtering, a total of 34,334 AS events were identified, among which 421 AS events were found expressed differently. Parent genes of these DEAS play a important role in regulating CRC-related processes such as protein kinase activity (FDR<0.0001), PI3K-Akt signaling pathway (FDR = 0.0024) and p53 signaling pathway (FDR = 0.0143). 37 DEAS events were found to be associated with OS, and 68 DEAS events were found to be associated with DFS. Stratifying patients according to the PSI value of AT in CXCL12 and RI in CSTF3 formed significant Kaplan-Meier curves in both OS and DFS survival analysis. Unsupervised clustering analysis using DEAS revealed four clusters with distinct survival patterns, and associated with consensus molecular subtypes.
Conclusions
Large differences of AS events in CRC appear to exist, and these differences are likely to be important determinants of both prognosis and biological regulation. Our identified CRC-related AS events and uncovered splicing networks are valuable in deciphering the underlying mechanisms of AS in CRC, and provide clues of therapeutic targets to further validations.
Keywords: CRC, Alternative splicing, RNA-Seq, Prognosis
Research in Context.
Evidence Before This Study
Growing evidence demonstrated that alternative splicing is frequently modified in cancer and is associated with cancer progression. Systematic analysis of alternative splicing signature in colorectal cancer is lacking and greatly needed.
Added Value of This Study
Through our analysis and screening, the genome-wide AS events were profiled, and the CRC-related AS events were identified. Further enrichment analysis confirmed that the parent genes of these AS events (DEAS) have great potential to play vital role in CRC. Interesting splicing correlation network provides novel insights into how these CRC-related AS events were potentially regulated by key splicing factors. In addition, some AS events, which were identified associate with survival, may be most valuable in deciphering the underlying mechanisms of AS in the oncogenesis of CRC, and provide clues of therapeutic targets to further validations.
Implications of All the Available Evidence
Large differences of AS events in CRC appear to exist, and these differences are likely to be important determinants of both prognosis and biological regulation. Here, we provided the most systematic analysis so far of alternative splicing in CRC, which could promote further mechanism and experimental study.
Alt-text: Unlabelled Box
1. Introduction
Despite advances in screening, diagnosis, and curative resection, colorectal cancer (CRC) is still one of the leading causes of cancer-related death worldwide [1]. In addition, it's clinical outcome for individual cases remains unsatisfactory because optimal management and individual therapy strategies still present great challenges [2]. At present, surgical resection is the only potentially curative therapy for CRC. Unfortunately, about 20–25% of patients with newly diagnosed CRC present with distant metastases, and only a small population of these patients can undergo curative operation [3]. More seriously, approximately 50% of CRC patients with resectable tumors will develop recurrence, most within 2 years [4]. Evidence accumulated during the past decades have revealed that cancerogenesis, recurrence and metastasis of colorectal is a complex and tightly regulated process, involving accumulation of numerous genetic alternation over years [5]. These sequential changes not only activate oncogenes or inhibit the action of tumor suppressor genes, but also empower CRC with the ability to invade tissues and metastasize. Thus, further comprehensive understanding of the relationship between the biological mechanism that anticipates in CRC regulation and the corresponding clinicopathological characteristic is the vital step to develop target therapy and improve the prognosis of CRC patients.
The rapid development of the high-throughput technology has opened a new era for cancer genomics study. By applying RNA sequencing(RNA Seq) and microarrays in recent year, gene expression and genomic profiling of CRC have been sufficiently evaluated [[6], [7], [8]], and the arm these studies are mainly focusing on differential gene expression for CRC phenotype classification, but also on gene expression variability according to clinical staging [[9], [10], [11]]. For example, a genome-wide CRC-related genes identification was conducted in our recently published study, which combining sequence data of paired tissue samples(e.g., neoplastic vs normal tissue) and corresponding clinical information to insight into the potential biological and clinical value of transcriptome variation [2]. Additionally, studies involved ncRNA expression, copy number variation, DNA methylation and nucleotide polymorphisms have been widely performed, especially in CRC [12,13]. Results obtained from above-referenced studies not only identified CRC-related alterations in several pathways such as TGF-b, WNT, MAPK, PI3K and p53 signaling, but also dynamically revealed the intricate interwoven relationships that govern CRC biological behavior. More importantly, these results reinforcing the concept that multiple genetic events are required to unleash the malignant progression of CRC, and only a genome-wide approach can interpret complex scenarios in the biology of tumors. These studies, although with promising results, mainly focus on alteration at the level of transcript expression. However, systematic analysis of variation on transcript architecture (alternative splicing) is largely ignored, especially in CRC.
While human cells contain nearly 20,000 protein-coding genes, their transcriptome is tremendously more complex, with 82,141 distinct mRNA sequences indexed on the current version of GENCODE [14,15]. The remarkable discrepancy between the number of protein-coding genes in an organism and its overall cellular complexity indicate that additional mechanisms are acting to expand the coding capacity of the genome and form an intermediate layer of regulation between transcriptional and post-translational networks [16]. Alternative splicing (AS), the process by which a single RNA precursor can be spliced in different arrangements to produce structurally and functionally distinct mRNA and protein variants, may be one of the most extensively applied mechanisms that account for the proteome diversity and cellular complexity [17]. Indeed, AS have a profound effect on the biologic characteristics of the final protein by adding or deleting functional domains, changing its stability, controlling its location, or modifying its protein-protein interactions [18]. More importantly, data from genome-wide studies suggest that 92–95% of multi-exon genes in human undergo AS [19].
In recent years the contribution of AS in human disease, particularly in cancer, has been widely recognized. From the point of view of mechanism, abnormal AS event can directly affect major biogenesis and progression of tumors. For example, the systematic and coordinated alteration of AS in several functionally linked precursor mRNAs could entirely alter the specific process of carcinogenesis. Moreover, increasing evidence suggested that unbalanced expression of splicing variants or the failure to properly express the correct isoforms is another hallmark of cancer [20]. Thus, cancer-specific splice variants may potentially be used as diagnostic, prognostic, and predictive biomarkers as well as therapeutic targets.
Due to the technical limitation, the effect or functions of AS events in CRC have been individually studied in only a few cases. But recently several approaches using high-throughout technique have been applied successfully. For example, Mojica and Hawthorn directly compared exon array-based data from paired tumor and normal tissues from 10 CRC patients, revealing the complexity of AS and pointing out the limitations of current transcript annotation [21]. In another study, genome-wide disruption of pre-mRNA splicing was investigated on 160 CRCs and indicated that AS analogous to genomic instability is a characteristic of CRC [5]. More recently, by using whole transcriptome analysis, Bisognin et al. demonstrate that cancer-specific AS is common in early phases of CRC natural history [22]. Comparing with microarray used in these resent examples [5,21,22], RNA-Seq enabling the quantitative measure of known and novel AS isoforms, and provides better resolution, deeper coverage and higher accuracy, is the most suitable method to apply in AS study. However, the scale of RNA-Seq based studies is small because that the cost of sequencing at deep coverage is still very high. Additionally, as rapidly developing technology, the bioinformatics protocols for processing and quantitatively analyzing RNA-Seq data for AS are still need to be optimized. Furthermore, the lack of corresponding clinical information, few studies annotated the CRC-specific AS events with clinical meaning in a systematic way.
With the rapid accumulation of RNA-seq data in CRC samples, the Cancer Genome Atlas (TCGA) project provides a rich source for the investigation of AS patterns. At present, there are already a total of 677 RNA-seq data obtained from 627 CRC patients that are publicly available online [23]. The detailed clinical information of these data could also be obtained from TCGA. Therefore, it is possible to study the clinical effect of CRC-related AS events in a relative large-scale of populations. Despite the great potential of using RNA-Seq to study AS in cancers, the advantages could not be realized without efficient and reliable bioinformatics processing. However, recently developed analytical tool, known as SpliceSeq, could unambiguously aligns RNA-Seq reads to gene splice graphs, facilitating accurate analysis of complex or low-frequency AS events [24]. That may be particularly advantageous for application to tumors.
To the best of our knowledge, integrating clinical information and large-scale of RNA-seq data to systematic analyses of AS at individual exon resolution have been lacking, and is great needed, especially in CRC. Thus, in this present study, we systematically profiled the genome-wide alternative splicing in CRC cohort from TCGA, and further identified CRC-related AS events and investigated its association with clinical outcome. Our results reveal that such CRC-related AS events as CSTF3-RI, CXCL12-AT et al. are particularly important in CRC, and they can directly affect the major biogenesis and progression of CRC, even serve as reliable prognostic biomarkers.
2. Materials and methods
2.1. Data curation process
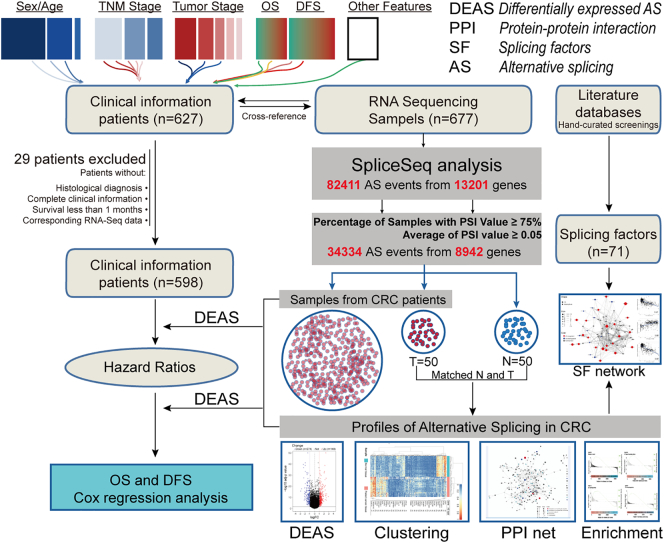
RNA-Seq data and corresponding clinical information of CRC cohort were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/). TCGA data are classified by data type and data level, to allow structured access to this resource with appropriate patient privacy protection. This study meets the publication guidelines provided by TCGA [25]. Then, downloaded samples and corresponding clinical data were cross-referenced by TCGA barcodes. Some patients do not meet the following criteria were eliminated: [1] a histological diagnosis of CRC [2] patients with complete clinical features including sex, age, tumor location, local invasion, lymph node metastasis, distal metastasis, differentiation grade, pathologic stage, survival information; [3] patients were still alive at least 1 month after initial pathologic diagnosis; [4] patients with corresponding RNA-Seq data. To generate the AS profiles for each CRC patient, SpliceSeq, a java application that unambiguously quantify the inclusion level of each exon and splice junction, was used to evaluate the RNA splicing patterns as previous described [26,27]. The Percent Spliced In (PSI) value, rating from zero to one which was commonly used in quantifying AS events, was calculated for each AS events. To generate as reliable a set of AS events as possible we implemented a series of stringent filters (Percentage of Samples with PSI Value ≥75, Average of PSI value ≥0.05). Upset plot generated by UpSetR (version 1.3.3) was used for the quantitative analysis of interactive sets between seven types of AS [28]. Circos Plots generated by Circlize (version 0.3.9) was created to display the detail of AS events and its parent genes in chromosome [29]. Details of this study design are illustrated in Fig. 1 as a flowchart.
Fig. 1.
Flowchart for profiling the alternative splicing of CRC in a large-scale RNA-Seq data. RNA-Seq data and corresponding clinical information of CRC cohort were downloaded from the TCGA data portal. After excluding patients with incomplete clinical data and duplications, we combined these datasets into a large-scale CRC cohort, which was further analyzed by SpliceSeq. Through this process, PSI value was calculated for each AS events. Then, a series of stringent filters (Percentage of Samples with PSI Value ≥75, Average of PSI value ≥0.05) was implemented. Based on the PSI value of each AS events, we identified DEAS between CRC and normal tissues and further investigated the parent genes of these AS events by enrichment analysis. Next, the interaction network of these parent genes and regulation network of DEAS and splicing factors was visualized and analyzed. Finally, to assess the prognostic value of each DEAS event in CRC, their effect on OS and DFS was determined by Cox regression and Kaplan-Meier analysis.
2.2. Identification of differentially expressed AS events (DEAS) and Enrichment Analysis
To identify DEAS between CRC and normal tissues, the PSI value of each AS events was detected from the TCGA CRC cohort (627 CRC tissue and 50 paired normal tissue). The batch effect was removed by using a generalized linear model. The expression differences were characterized by log FC (log 2 fold change) and associated adj.p values. The |log2FC| > 1 and adj.p < .05, represented corresponding AS events were upregulated and downregulated, respectively. The parent genes of these AS events were then subjected to biological function enrichment analysis. GO terms and KEGG pathways with significant enrichment false discovery rate (FDR) values <0.05 were selected for further analysis. The analyses were performed by clusterProfler package (version 3.4.4) [30]. In addition, the parent genes of these AS event were mapped and imported into the Retrieval of Interacting Genes/Proteins (STRING) 9.1 database. The correlation network was then visualized by Cytoscape (version 3.4.0) [31]. Cluster analyses were performed using correlation distance metrics and the average linkage agglomeration algorithm.
2.3. Construction of splicing correlation network
A list of 71 human splicing factors was created by hand-curated screenings of literature and databases [32]. All of these splicing factors were experimentally validated in previous studies, including 13 SR proteins, 27 heterogeneous nuclear ribonucleoprotein (hnRNP) proteins and other 31 proteins belonging to the ELAV, KHDRBS, CELF, Nova and Fox families. The expression of splicing factor were obtained from level 3 mRNA-seq data in TCGA. Correlation between the expression of splicing factor and the PSI value of DEAS were analyzed by WGCNA (version 1.51) [33]. p values were adjusted by Benjamini & Hochberg (BH) correlation and adjusted P value of <0.05 was considered significant. The correlation plots were generated by Cytoscape (version 3.4.0).
2.4. Survival analysis
For DEAS event, CRC patients were divided into two groups based on the PSI value (median cut), and the two-category was modelled as continuous variables to derive more easily interpretable hazard ratios (HRs). Univariate Cox regression followed by multivariate Cox regression was performed based on overall survival (OS) and disease-free survival (DFS) to determine independent prognostic factors respectively. Kaplan-Meier analysis with log-rank test was used to compare patients' survival between subgroups. The effect of each variable on survival was determined by the Cox multivariate regression analysis. Unless otherwise mentioned, all statistical analyses in this study were performed using R software (version 3.2.2), and P value <.05 were considered to be statistically significant.
2.5. Evaluation of the correlation with clinical features
Unsupervised classification of the TCGA CRC cohort was performed using hierarchical clustering (Ward linkage and 1-Pearson correlation coefficient distance used) on the identified DEAS (n = 421). To obtain a robust classification, we used an unsupervised consensus approach implemented in the R package Consensus Cluster Plus [34]. The optimal number of clusters was selected according to the Elbow method and the Gap statistic. Consensus molecular subtyping of CRC was achieved based on gene expression data using the CMScaller package [35], as previously described. The associations between clusters and clinical outcome were assessed using the Chi-squared test and logistic regression as described in Marisa et al. studies [8].
3. Results
3.1. Overview of AS events profiling in CRC
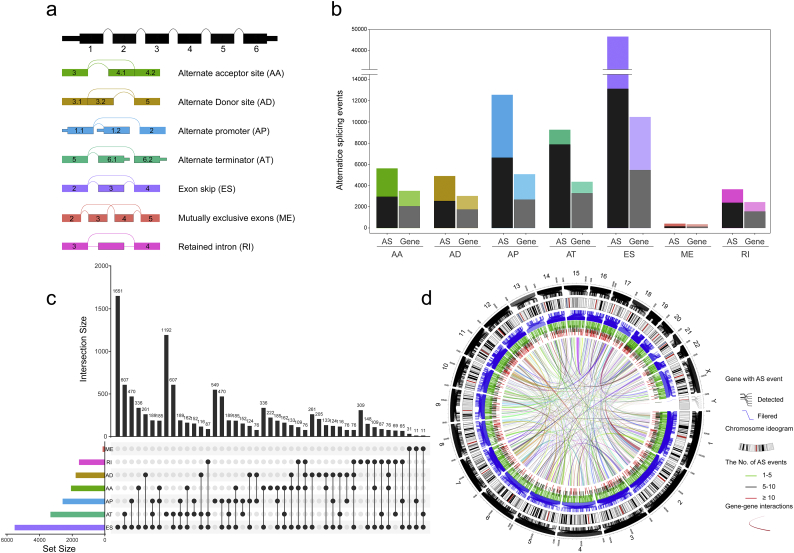
Integrated AS events profiling was established using RNA-Seq data obtained from 627 CRC patients. The included population comprised 334 (53.2%) male and 293 (46.7%) female patients, among which 199 patients (18.9%) developed recurrence and 130 (20.7%) had died, respectively. The median follow-up period after radical resection was 27.5 months (range, 1–147 months). The detailed baseline characteristics and histopathological features of the patients are summarized in Supplementary Table S1. By using SpliceSeq based on the standard protocol as previously reported, we identified a total of 82,411 AS events from 13,201 genes. According to their splicing pattern, these AS events can be roughly divided into seven types, including Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA), which were illustrated in Fig. 2a. Of note is that the majority part of AS events could only be detected in a few samples. In addition, the expression level of certain splicing isoforms was extremely low (PSI value <0.05). In order to generate as reliable a set of AS events as possible we implemented a series of stringent filters (Percentage of Samples with PSI Value ≥75, Average of PSI value ≥0.05). After screening, we obtained 34,334 AS events from 8942 genes, which indicated that one gene might have almost four AS events on average (Fig. 2b). All the detected AS events were well organized in Supplementary Table S2. Considering this, UpSet plot was generated to visualize the intersecting sets of each AS type. As shown in Fig. 2c, most of the AS events were from one gene while one gene might have up to four types of AS events. It is noteworthy that >81.6% of genes contain two or more AS events, different combinations of these AS events provides perhaps the largest potential process for enriching the transcriptome diversity. Finally, in order to intuitively visualized the integrated AS events profiling of CRC, Circos Plots were created to display the detail of AS events and its parent genes in the chromosome (Fig. 2d).
Fig. 2.
Overview of AS events profiling in CRC. (a) Illustrations for seven types of AS events, including Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA). (b) A number of AS events and involved genes from the CRC patients were depicted according to the AS types. Color bar represents the preliminarily detected AS events and involved genes. Black bar represents the AS events and involved genes filtered by stringent criteria. (c) UpSet plot of interactions between the seven types of detected AS events (n = 34,334) in CRC. One gene may have up to four types of alternative splicing. (d) The detail of AS events and its parent genes in chromosome was shown in Circos Plots. The ribbons, in the circos plots, represent the potential interaction between the parent gene of DEAS, and the thickness of the ribbons indicating the extent of the interaction strength. The outer circle is composed of the polyline, each polyline represents an AS event and links to the location of the parent gene in chromosomes. Different color of the short line represent the number of AS events that the corresponding gene have: blue, 1–5 AS events; black, 5–10 AS events; red, greater than or equal to 10 AS events.
3.2. Identification of CRC-related aberrant AS events
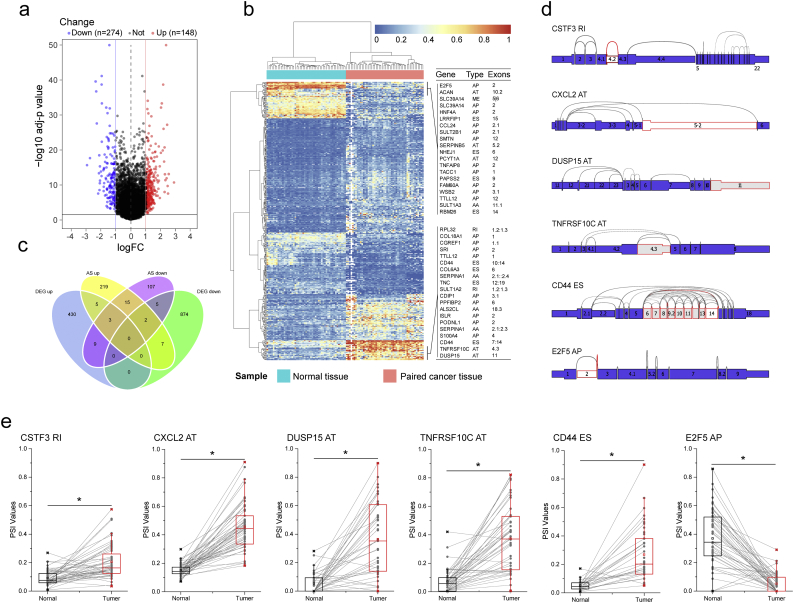
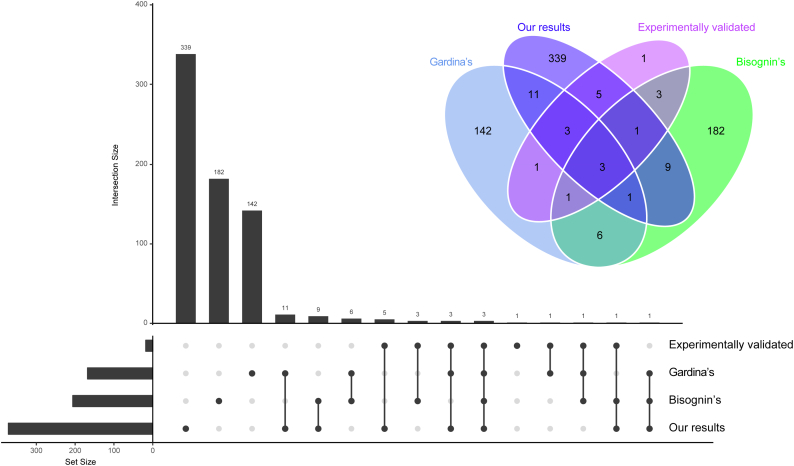
Directly comparing the expression of gene at different pathological state is an effective approach to screen hub genes that involved in the corresponding biological process. This approach has been widely used to identify CRC-related genes in many previous studies. Similarly, significantly different AS events between primary CRC and corresponding adjacent normal tissues could also play a vital role in the whole course of CRC. Thus, AS profiling obtained from 50 paired tumor and normal tissues were integrated to identify differentially expressed alternative splicing (DEAS). Eventually, a total of 421 DEAS were preliminarily screened from 372 genes with the threshold of |log2FC| >1 and adj.P.Value <0.05 (t-test and BH correction), among which there were 174 APs, 81 ESs, 76 ATs, 57 RIs, 18 ADs, 14 AAs and 1 ME (Fig. 3a; Supplementary Table S3). The detailed information of the top 40 most different AS events was listed in Table 1. Compare with recently published results that obtained from microarray [22,36,37], although we applied a series of stringent filters, more AS events were identified in the present study (Supplementary Fig. S1). More importantly, among the AS events (n = 18) that validated by experiment, we detected 12 (66.7%), and that this well above the result from Gardina et al. [37] (n = 8,44.4%) and Bisognin et al. [22] (n = 8,44.4%) (Supplementary Fig. S1). These findings suggested that RNA-seq may be a more suitable method to apply in AS study, and our RNA-seq based results are more robust and reliable.
Fig. 3.
Identification of CRC-related aberrant AS. (a) The difference of AS events between paired colorectal cancer and paracancerous tissue. Volcano Plot visualizing the DEAS identified in CRC. The red and blue points in the plot represent the differentially expressed alternative splicing with statistical significance (adj P value <.05, |logFC| ≥ 1). (b) Heat map of the DEAS. The horizontal axis shows the clustering information of samples which were divided into two major clusters, and the clusters were adjacent normal tissue(N = 50) and paired tumor tissue(N = 50), respectively; the left longitudinal axis showed the clustering information of DEAS. The gradual change of color from green to red represents the expression of DEAS altered from low to high. (c) Venn diagram demonstrated the intersection set of DEAS and DEG. (d) The splice graph of some representative DEAS. The thin exon sections represent untranslated regions (UTR) and the thick exon sections represent coding regions. Exons are drawn to scale and the connecting arcs represent splice paths. (e) The difference of PSI value of AS events between primary CRC and corresponding adjacent normal tissues. Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA). For Fig. 3a, t-test and BH correction was used for data analysis. For Fig. 3e, Student's t-test was used for data analysis. *: P < .0001 compared with adjacent normal tissue.
Table 1.
The detailed information of the top 40 most different AS events.
| Symbol | AS type | exons | From exon | To exon | MeanT | MeanN | logFC | adj.P.Val |
|---|---|---|---|---|---|---|---|---|
| Upregulated | ||||||||
| DUSP15 | AT | 11 | 0.365 | 0.053 | 2.774 | 1.48E-09 | ||
| TNFRSF10C | AT | 4.3 | 0.369 | 0.068 | 2.436 | 1.04E-11 | ||
| CD44 | ES | 7:12.1:13:14 | 5 | 15 | 0.270 | 0.051 | 2.393 | 9.70E-07 |
| S100A4 | AP | 4 | 0.258 | 0.050 | 2.364 | 1.18E-08 | ||
| SERPINA1 | AA | 2.1:2.2:2.3 | 1.1 | 2.4 | 0.644 | 0.128 | 2.334 | 1.30E-24 |
| PODNL1 | AP | 2 | 0.378 | 0.077 | 2.300 | 1.27E-10 | ||
| ISLR | AP | 2 | 0.669 | 0.139 | 2.266 | 2.75E-46 | ||
| ALS2CL | AA | 18.3 | 18.1 | 18.4 | 0.264 | 0.057 | 2.221 | 3.79E-10 |
| PPFIBP2 | AP | 6 | 0.239 | 0.052 | 2.190 | 1.18E-11 | ||
| CDIP1 | AP | 3.1 | 0.254 | 0.056 | 2.170 | 3.97E-07 | ||
| SULT1A2 | RI | 1.2:1.3 | 1.1 | 1.4 | 0.811 | 0.181 | 2.167 | 1.08E-18 |
| TNC | ES | 12:13:14:15:16:19 | 11 | 20 | 0.433 | 0.098 | 2.138 | 6.97E-15 |
| SERPINA1 | AA | 2.1:2.2:2.3:2.4 | 1.1 | 2.5 | 0.709 | 0.165 | 2.101 | 1.61E-23 |
| COL6A3 | ES | 6 | 5 | 7 | 0.614 | 0.145 | 2.079 | 1.45E-35 |
| CD44 | ES | 10:11:12.1:13:14 | 5 | 15 | 0.510 | 0.123 | 2.052 | 6.93E-18 |
| TTLL12 | AP | 1 | 0.779 | 0.196 | 1.991 | 1.95E-30 | ||
| SRI | AP | 2 | 0.729 | 0.190 | 1.939 | 2.64E-29 | ||
| CGREF1 | AP | 1.1 | 0.586 | 0.154 | 1.929 | 3.06E-17 | ||
| COL18A1 | AP | 1 | 0.427 | 0.117 | 1.864 | 1.22E-05 | ||
| RPL32 | RI | 1.2:1.3 | 1.1 | 1.4 | 0.429 | 0.118 | 1.860 | 1.12E-13 |
| Downregulated | ||||||||
| RBM26 | ES | 14 | 13.2 | 15 | 0.066 | 0.221 | −1.744 | 2.23E-07 |
| SULT1A3 | AA | 11.1 | 10 | 11.2 | 0.058 | 0.198 | −1.765 | 1.06E-12 |
| TTLL12 | AP | 12 | 0.221 | 0.804 | −1.861 | 1.95E-30 | ||
| WSB2 | AP | 3.1 | 0.136 | 0.497 | −1.867 | 3.20E-20 | ||
| FAM60A | AP | 2 | 0.130 | 0.478 | −1.873 | 9.08E-38 | ||
| PAPSS2 | ES | 9 | 8 | 10 | 0.170 | 0.626 | −1.878 | 1.45E-35 |
| TACC1 | AP | 1 | 0.058 | 0.215 | −1.884 | 3.37E-11 | ||
| TNFAIP8 | AP | 2 | 0.072 | 0.273 | −1.922 | 2.45E-12 | ||
| PCYT1A | AT | 12 | 0.051 | 0.202 | −1.980 | 4.98E-10 | ||
| NHEJ1 | ES | 6 | 5 | 7 | 0.061 | 0.243 | −1.985 | 1.13E-13 |
| SERPINB5 | AT | 5.2 | 0.062 | 0.248 | −1.994 | 2.29E-09 | ||
| SMTN | AP | 12 | 0.053 | 0.216 | −2.015 | 1.24E-07 | ||
| SULT2B1 | AP | 2.1 | 0.133 | 0.542 | −2.028 | 1.47E-16 | ||
| CCL24 | AP | 2.1 | 0.130 | 0.539 | −2.049 | 6.71E-12 | ||
| LRRFIP1 | ES | 15 | 14 | 16 | 0.055 | 0.234 | −2.100 | 9.70E-07 |
| HNF4A | AP | 2 | 0.108 | 0.491 | −2.187 | 5.65E-25 | ||
| SLC39A14 | AP | 2 | 0.111 | 0.518 | −2.225 | 9.89E-34 | ||
| SLC39A14 | ME | 5|6 | 4 | 7 | 0.122 | 0.587 | −2.263 | 2.60E-36 |
| ACAN | AT | 10.2 | 0.085 | 0.523 | −2.624 | 8.12E-16 | ||
| E2F5 | AP | 2 | 0.052 | 0.368 | −2.815 | 5.46E-14 | ||
MeanT: the mean PSI value in colorectal cancer tissue; MeanN: the mean PSI value in paired normal tissue; logFC: log2 fold change. The Adj.P value was calculated by t-test and adjusted through BH correction.
Supplementary Fig. S1.
UpSet and Venn diagram displaying the overlap of AS events. The overlap of the AS events that respectively obtained from the studies of Gardina, Bisognin et al., hand-curated screenings of literature (experimentally validated) and our own results were displayed by the UpSet and Venn diagram.
Intriguingly, the proportion of AS types between DEAS and the entire AS was inconsistent. As the largest number of AS types, ES events (36.8%) only contribute 17.9% DEAS. Moreover, using unsupervised hierarchical clustering based on these DEAS, the samples of tumor and normal were clearly separated into two discrete groups which indicated that the DEAS screened above were credible (Fig. 3b). Aberrant AS events may directly affect the expression of its parent RNA, especially when AT or AP events occurred. In order to investigate the relationship between DEAS and differently expressed genes (DEG), we analyzed the DEAS that occurred in DEG. Fig. 3c summarized the results and detailed information were provided in Supplementary Table S4. As we expected, 94.4% DEAS in its corresponding DEG is AT (44.4%) and AP (50.0%). Fig. 3d depicts part of identified DEAS in splice graph, which summarises the transcript variations into a directed acyclic graph, and represents exons as rectangular nodes and splice junctions as edges. Furthermore, for intuitively showing the difference of these AS events between primary CRC and corresponding adjacent normal tissues, we generate graphs in which scatter plot is overlaid with the boxplot(Fig. 3e). Considering all of these evidence, it suggested that, like CRC-related genes, CRC-related AS events play a vital role in CRC biological and need further research.
3.3. Enrichment and interaction analysis of DEAS
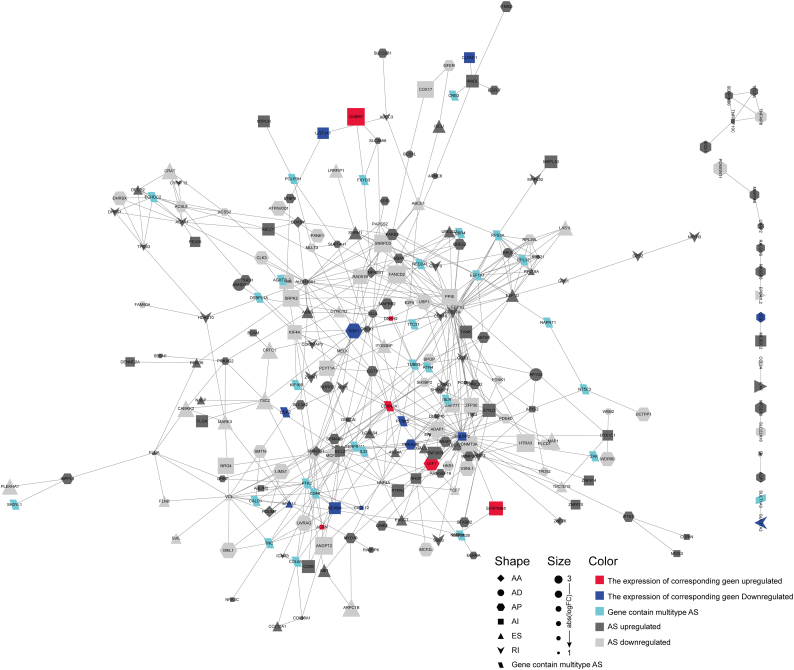
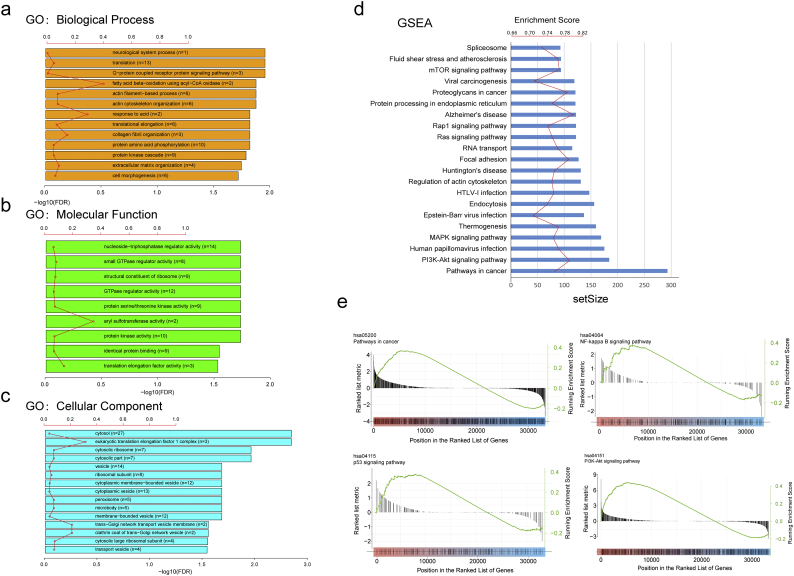
It was evident that AS could directly affect the protein function through several mechanisms. Thus, we can shed light on the potential influence of DEAS by analyzing its corresponding protein. Supplementary Fig. S1 shows that specific GO categories closely associated with CRC, such as identical protein binding (Fisher's Exact Test,FDR = 0.0046), cell morphogenesis (Fisher's Exact Test,FDR = 0.0058), GTPase regulator activity (Fisher's Exact Test,FDR = 0.0040), and protein kinase activity (Fisher's Exact Test,FDR<0.0001), were significantly enriched in these genes that occurred DEAS (Supplementary Fig. S2a-c). Additionally, some KEGG pathways well known to be involved in CRC metastasis and recurrence were enriched, including Pathways in cancer (Fisher's Exact Test,FDR = 0.0036), PI3K-Akt signaling pathway (Fisher's Exact Test,FDR = 0.0024), p53 signaling pathway (Fisher's Exact Test,FDR = 0.0143) and NF-kappa B signaling pathway (FDR = 0.0021) (Supplementary Fig. S2d, e). Taken together, above results indicated that the parent genes of DEAS play an important role in regulating the CRC-related biological process. While variation on the structure of these genes' transcript (alternative splicing) may inevitably affect its protein translation, and further modify protein feature. Thus, it is necessary to investigate these AS events from the protein network point of view. PPI network analysis based on the DEAS associated genes not only demonstrated the interactive relationship in normal condition, but also revealed the potential influence of AS events to the entire network (Fig. 4).
Fig. 4.
Protein-protein interaction analysis of identified DEAS. Interactome of the 372 genes showing 249 nodes and 514 edges in the PPI network in CRC. Genes were denoted as nodes in the graph and the interactions between them were presented as edges. The shape, size and color of node respectively represent AS type, the value of logFC and change pattern. Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA).
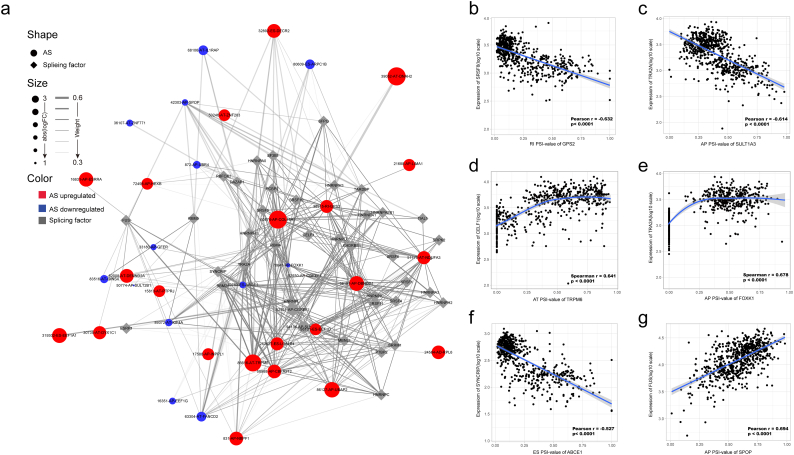
3.4. DEAS evens correlation network of splicing factors
AS events are mainly regulated by splicing factors, which bind to pre-mRNAs and influence exon selection and splicing site choice and more importantly, dysregulated AS within tumor microenvironment may be orchestrated by a limited number of splicing factors [38]. Therefore, an important issue is that whether a significant fraction of these DEAS events is potentially regulated by a few key splicing factors in CRC. By hand-curated screenings of literature and databases, we firstly identified 71 splicing factors (Supplementary Table S5) which were all experimentally validated in the previous study. The expression of these splicing factors was obtained from RNA sequencing data of TCGA CRC cohort. Next, correlation analyses between expression levels of these 71 splicing factors and the PSI values of each DEAS event were performed in the CRC cohort with splicing regulatory network built among the significant correlations (|R| > 0.5; t-test, P < .05). In the splicing correlation network shown in Fig. 5A, 33 DEAS events, including 22 upregulated AS events (red dots) and 11 downregulated AS events (blue dots), were significantly correlated with the 37 splicing factors (gray dots). Interestingly, the majority of splicing factors (gray dots) were correlated with more than one AS events, among which some were playing opposite roles in the regulation of difference AS events(Fig. 5a). In addition, from the network, we can see that different splicing factors are in competition for the same binding sites (AS event), which at least partly, account for the reason why transcripts can yield several different splicing isoforms. Representative correlations between splicing factors and specific AS events were presented in the dot plots (Fig. 5b–g). For example, expression of TRA2A was negatively correlated with AT of SULT1A3 (Fig. 5C), whereas positively correlated with AP of FOXK1(Fig. 5E).
Fig. 5.
Splicing correlation network in CRC. (a) DEAS evens correlation network of splicing factors. The correlation analyses between expression levels of these 71 splicing factors and the PSI values of each DEAS event were performed in the CRC cohort with splicing regulatory network built among the significant correlations. The shape, size and color of node respectively represent a type (AS event or splicing factor), the value of logFC and change pattern (upregulated or downregulated). The breadth of the line represents the extent of interaction strength. (b-g) Representative dot plots of correlations between expression of splicing factors and PSI values of AS events. Spearman's rank correlation analysis was used for non-normal distribution data (Fig. 3d and e). Person correlation was used for continuous variables that meet normal distribution (Fig. 3b, c, f and g). All the absolute value of r coefficient > 0.5 and all P < .0001.
3.5. The prognostic value of DEAS in CRC
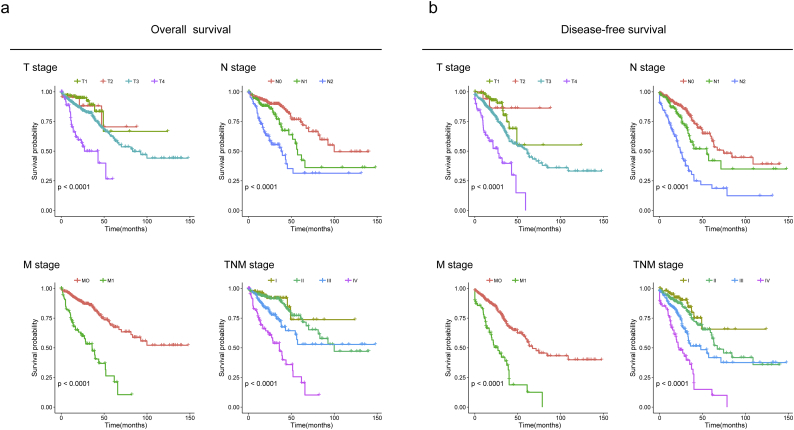
Before studying the prognostic value of DEAS events, univariate survival tests were conducted to assess the relationship between clinicopathological features and outcome in the TCGA CRC cohort. As revealed in Supplementary Table S6, T stage (HR = 2.603, 95%CI: 1.96–3.44; Likelihood-ratio test, P < .001), N stage (HR = 1.903, 95%CI: 1.59–2.77; Likelihood-ratio test, P < .001) and TNM stage (HR = 3.041, 95%CI: 2.44–3.78; Likelihood-ratio test, P < .001) were significantly associated with OS. Meanwhile, T stage (HR = 1.992, 95% CI: 1.45–2.73; Likelihood-ratio test, P < .001) and TNM stage (HR = 2.311, 95% CI: 1.06–5.03; Likelihood-ratio test, P = .035) was significantly associated with DFS. More important, survival rates predicted by TNM stage depicted significant distinction between the Kaplan-Meier curves in both OS and DFS (Supplementary Fig. S3). The results of this preliminary assessment revealed that the survival data for the TCGA CRC cohort, although containing censored data, were informative and appropriate for use in further survival analysis.
Supplementary Fig. S3.
Kaplan-Meier estimates of OS and DFS stratified by the Tumor, Node, Metastasis system and TNM stage in the TCGA CRC cohort. (a) Overall survival. (b) Disease-free survival. Local invasion stage(T1, T2, T3, T4), lymph node metastasis (N0, N1, N2), Distant metastasis (M0, M1); The differences between the two curves were determined by the two-sided log-rank test. Overall log-rank test, p-value <.0001.
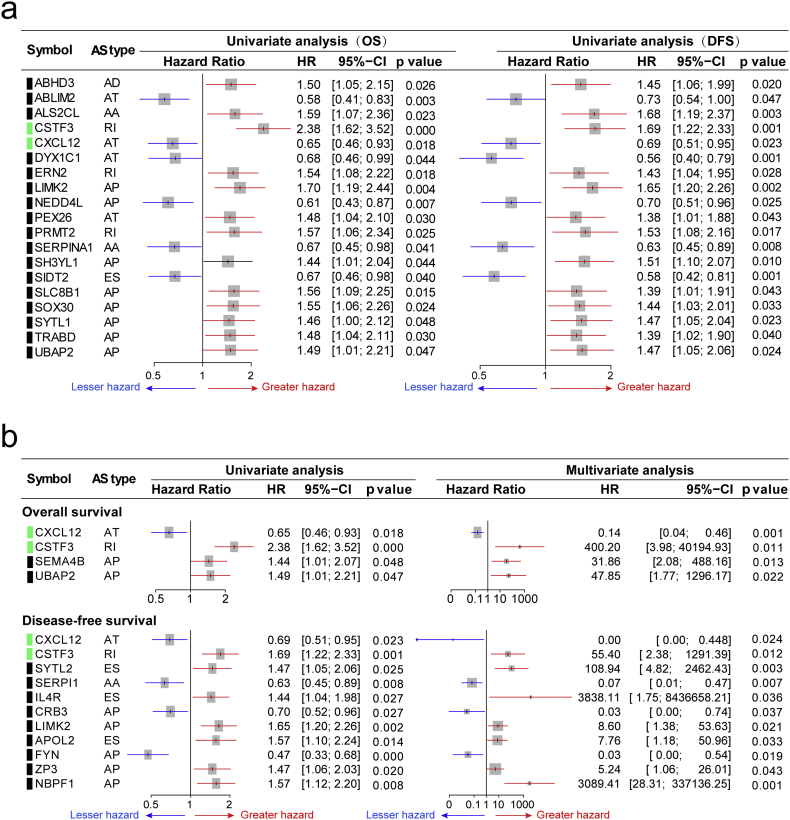
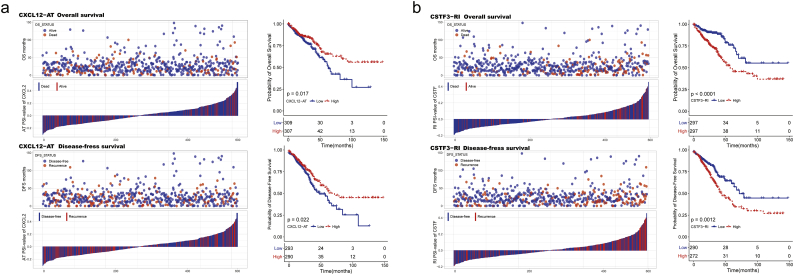
The relationship between our identified DEAS and CRC prognosis was investigated in the TCGA cohort. For each DEAS events, CRC patients were divided into two groups based on the PSI value (median cut). Then univariate survival analyses for OS and DFS were conducted respectively (Supplementary Table S7). As results, a total of 37 DEAS events were found to be associated with OS, and 68 DEAS events were found to be associated with DFS. Among these prognosis related DEAS events that simultaneously associated with OS and DFS were shown in Fig. 6a. The DEAS events that significantly correlated with survival in the univariate analysis were further assessed by multivariate analysis to identify independent prognostic indicator in CRC (Supplementary Table S8). As shown in Fig. 6b, there were 4 and 11 DEAS events could be recognized as an independent prognostic indicator for OS and DFS, respectively. From the results of the above analysis, we unexpectedly found that AT in CXCL12 and RI in CSTF3 was an independent prognostic indicator for both OS and DFS in CRC cohort. Actually, as shown in Fig. 7a and b, stratifying patients according to the PSI value (median of AT in CXCL12 and RI in CSTF3) formed significant Kaplan-Meier curves in both OS and DFS survival analysis. Above results directly suggested that DEAS events are of not only important biological meaning but also potential clinical value.
Fig. 6.
The prognostic value of DEAS in CRC. (a)The DEAS events that simultaneously associated with OS and DFS. Univariate analysis of DEAS on overall and disease-free survival, respectively. Unadjusted HRs (boxes) and 95% confidence intervals (horizontal lines) limited to AS events with P-value <.05. Box size is inversely proportional to the width of the confidence interval. (b) The DEAS events that present with independent prognostic value on survival. Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA). Cox regression (univariate and multivariate) was used for data analysis. Likelihood-ratio test was used to determine the P value. All P < .05.
Fig. 7.
Kaplan-Meier curves for OS and DFS according to PSI value of AT in CXCL12 (a) and RI in CSTF3 (b). Exon Skip (ES), Mutually Exclusive Exons (ME), Retained Intron (RI), Alternate Promoter (AP), Alternate Terminator (AT), Alternate Donor site (AD), and Alternate Acceptor site (AA). Kaplan-Meier analysis and log-rank test were applied to compare the survival outcomes. All P < .05.
3.6. AS clusters associated with prognosis and molecular subtypes
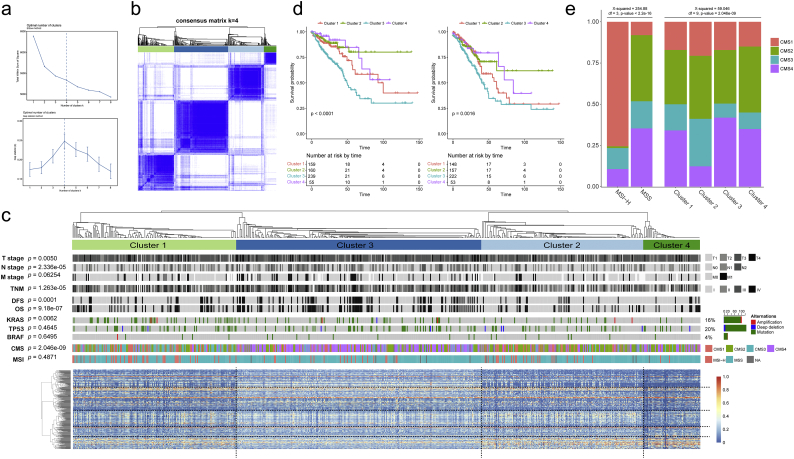
Our findings demonstrated that the expression of each DEAS varied considerably at the individual level and partly reflected the prognosis in patient with CRC. We wonder whether distinct patterns of AS could be discerned based on the 421 DEAS by performing consensus unsupervised analysis of all samples. The optimal number of clusters was determined by combining Elbow method and Gap statistic method, and the more balanced partition, as suggested, appeared to be for k = 4 (Fig. 8a). Consequently, four clusters of samples were determined as follow: C1 (n = 164, 26.1%), C2 (n = 161, 25.6%), C3 (n = 242, 38.5%), C4 (n = 57, 9.8%) (Fig. 8b). The consensus matrix heatmap revealed the identified four clusters with significant interconnectivity, among which C2, C3 and C4 appeared as well individualized clusters, whereas there was more classification overlap between C1 and C3 (Fig. 8b). The association of clusters with anatomoclinical and DNA alterations data are shown in Fig. 8c, which revealed that on the whole the distribution of different CMS, TNM stage, KRASm and survival status (OS and DFS) in CRC samples between clusters was not random. For example, tumors classified as C3 was more frequently KRAS mutant and enriched CMS4 and advanced TNM stage. As the previously defined consensus molecular subtypes of CRC, CMS4 has been proved to comprise the more mesenchymal-like cancers, with high stromal infiltration and poor patient prognosis [7,39]. Thus, Kaplan-Meier analysis was conducted to assess the relationship between cluster and prognosis. The results suggested that clusters were associated with distinct patterns of survival (Fig. 8d), among which C3 were both associated with poor outcome in OS and DFS analysis. In addition, no association between clusters and MSI was found, but CMS1 comprises most tumors with MSI (Fig. 8e). Collectively, these findings suggest that there is considerable variability in the nature of the AS across CRC —partly determined by molecular characteristics of a primary tumor—and that this influences clinical outcome.
Fig. 8.
AS clusters associated with prognosis and molecular subtypes. (a) Elbow and Gap statistic analysis for different numbers of clusters (k = 2 to 8). (b) Consensus matrix heatmap defined four clusters of samples for which consensus values range from 0 (in white, samples never clustered together) to 1 (dark blue, samples always clustered together). (c) Heatmap of the 421 DEAS ordered by cluster, with annotations associated with each cluster, data analyzed using chi-squared test. (d) Kaplan-Meier survival analysis of patients within different clusters on both OS and DFS. Depicted P-values are from log-rank tests. (e) Spine plots of the relationship between molecular subtypes and MSI hypotype, AS cluster respectively. P-values are from Kruskal-Wallis tests.
4. Discussion
The majority of genes in the human genome consists of multiple exons interspersed with introns that undergo splicing to form mature mRNA and protein products [18]. Although AS provides cells with a method to diversify the proteome, increasing evidence suggests that AS plays critical roles in the initiation or maintenance of human disease, including CRC [37,40,41]. The cancerogenesis of CRC is a complex multistep process, involving the unbalanced expression of splicing variants or the failure to properly express the correct isoforms. The adenoma-carcinoma process classically illustrates the multistep etiology of CRC. As the frequently involved genes, APC, K-Ras, and TP53 are all express splice isoforms whose functional properties are distinct and even antagonistic. For example, several AS events of APC have been described that result in proteins with molecular weights ranging from 90 to 300 kDa [20]. Using nested 5′ and 3′ primer to examined 24 patients who with a germline mutation in the APC gene, De Rosa and colleagues identified nine novel splice isoforms [42]. One of the transcripts contained an additional exon termed exon 1A; its inclusion leads to a premature stop codon in exon 2. The inclusion of exon 1A was found to be 3.5-fold higher in CRC versus paired normal tissue. The potential significance is that greater inclusion of exon 1A could effectively result in less APC protein being expressed because less productive mRNAs are synthesized overall. Additionally, many previous studies also reported the biological effect of AS events in K-Ras, especially alternation splicing of the fourth coding exon [43]. Splicing variant contains exon 4A results in a proapoptotic protein, while the inclusion of exon 4B lead to produce an antiapoptotic K-Ras. Both the two K-Ras variants are coexpressed in many tissues, but their ratio is changed in sporadic CRC favoring the antiapoptotic 4B isoform [44]. Moreover, the TP53 gene was thought for a long time to encode a single protein, but it is now abundantly clear that it is extensively alternatively spliced [45]. A remarkably complex series of splice isoforms have been described, and several of these can regulate the transcriptional activity of p53 [46,47]. Besides the three hub genes (APC, K-Ras, and TP53), research of aberrant AS in CRC-related genes, such as CD44,KLF6,NFS2, VEGF and Bcl-x et al., have been frequently reported in recent years [48]. More importantly, increasing data from genome-wide studies suggest that >90% of human genes undergo AS. Taken together, these results confirmed that not only the expression of genes, but also the balance of its splice isoforms need to be better investigated. Because this may immensely expand the scope in the screening of potential biomarkers and therapeutic targets.
With the advancement of high throughput technology in the last decades, great success has been made in the research of diversity of AS profiling in CRC. For example, Gardina et al. analyzed 20 paired tumor-normal colon cancer samples using microarray, and reported 189 putative AS events occurred in 168 genes. More recently, by using whole transcriptome analysis, Bisognin et al. revealed that 206 genes with probable AS events in CRC development and progression were identified, and that are involved in processes and pathways relevant to tumor biology, as cell-cell and cell-matrix interactions [22]. Similar to what we observed in the present study, 372 genes with 421 AS events were identified associated with CRC initiation or/and maintenance. Some specific GO categories and KEGG pathways, such as protein binding, cell morphogenesis and PI3K-Akt signaling pathway, were enriched in these genes that occurred DEAS.
Despite a series of stringent filters (Percentage of Samples with PSI Value ≥75, Average of PSI value ≥0.05; |log2FC| >1 and adj.P.Value <0.05) were implemented in the process of screening, more AS events were identified in the present study (Supplementary Fig. S1). Additionally, when compared the results that obtained from microarray (Gardina's [37] and Bisognin's [22]), we found that there was little overlap between the gene that occurred DEAS in CRC (Supplementary Fig. S1). More importantly, through hand-curated screenings of literature, we retrieved 18 AS events that were experimentally validated. Among these AS events, we detected 12 (66.7%), and that this well above the result from Gardina et al. [37] (n = 8,44.4%) and Bisognin et al. [22] (n = 8,44.4%) (Supplementary Fig. S1). These findings suggested that RNA-seq may be a more suitable method to apply in AS study.
Potential explanation for this results may be that RNA-Seq enabling the quantitative measure of known and novel AS isoforms, and provides better resolution, deeper coverage and higher accuracy. Recent comparisons of RNA-seq with qPCR for differential expression analysis have found good overall agreement [49]. However, measurement performance of RNA-seq depends in large part on analysis pipeline [19,49,50]. Primary results from the Sequencing Quality Control project revealed that appropriate choice of analysis pipeline could allow RNA-Seq with superior performance in many applications [49]. In this present study, a recent developed analysis pipeline (integration tool), known as SpliceSeq, which could facilitate accurate analysis of complex or low-frequency AS events, was applied to detect AS events. Collectively, although this present study is a computational work that solely based on RNA-seq data, we consider the results the most robust and reliable currently available for CRC.
Actually, we preliminarily analysis detected a total of 82,411 AS events from 13,201 genes, which accounts for approximately 66% of human genes. More importantly, deep sequencing co-operates with reliable bioinformatics approach could provide more detail on the structural variation of the splice isoform. Based on these information, AS events can be roughly divided into seven types and clearly depicted by the splice graph. According to our results, the vast majority of detected AS events belong to ES (36.8%). However, ES events only account for 17.9% of DEAS. This results is similar to many previous studies [26,27,51] and revealed to some extent that alternative splicing is not caused by transcription errors. Aberrant expression of trans-acting factors is known to trigger differences in AS patterns [52]. Thus, integrating the expression of splicing factors and DEAS provided an approach to address the underlying mechanism of the splicing pathway involved in CRC. The splicing correlation networks in CRC showed distinguished interactions between splicing factors and DEAS events. It is worth noting that the majority of splicing factors (gray dots) were correlated with more than one DEAS events, among which some were playing opposite roles in the regulation of difference AS events (Fig. 5a). This interesting phenomenon brought the hypothesis that the regulation effect of some splicing factors, such as TRA2A in CRC, relies on the joint action of splicing factors itself and its cis-acting regulatory elements.
Due to the potential significance of AS in cancer biology, its clinical relevance in malignancies has aroused increasing attention. The diagnostic implication of AS has been systematically demonstrated in cancers of ovarian and lung [26,27]. In addition, the prognostic value of CD44v6, one of the splice isoform of CD44, in CRC has also been revealed [53]. Moreover, it has been clarified recently that cancer-specific splice variants may potentially be used as diagnostic, prognostic, and predictive biomarkers as well as therapeutic targets. However, previous studies have mainly focused on single genes, survival associated AS in CRC remains largely unstudied.
To the best of our knowledge, the current study is the first to perform a systematic identification and analysis of survival associated AS events in primary CRC tissues. As results, a total of 37 DEAS events were found to be associated with OS, and 68 DEAS events were found to be associated with DFS (P < .05, Supplementary Table S6). The genes with survival associated AS events, including CD44, CXCL12, EGFL7, IL4R and TCF7, which itself play critical roles in cancer biology [[54], [55], [56], [57]]. More importantly, this genome-wide approach provides numerous prognostic AS events which might have an underlying mechanism in CRC initiation or maintenance. For example, a survival (OS and DFS) associated AS event involving exon 5 of CXCL12 (CXCL12-AT, Fig. 3d) could directly change the nucleic acid sequences and the activity of its protein. This protein functions as the ligand for the G-protein coupled receptor and plays a critical role in determining the metastatic destination of CRC [58]. Despite six different isoforms of CXCL12 have been reported, most studies focus only on the CXCL12-ES (exon: 2.2:3.1:5.1; Supplementary Table S3) or do not distinguish among isoforms [59]. Interestingly, according to our results, only CXCL12-AT, instead of the most common AS events (CXCL12-ES), was prognostic for OS and showed the same trend for DFS. This finding suggested that the expression changes of low abundance AS events may also have important biological meaning in CRC, and need more attention. Another AS events that were identified as an independent prognostic for both OS and DFS is CSTF3-RI. The protein encoded by CSTF3 is one of three (including CSTF1 and CSTF2) cleavage stimulation factors that combine to form the cleavage stimulation factor complex (CSTF) [60]. The retained intron (CSTF3-RI) could directly change the nucleic acid sequences of 3′-untranslated region. This could influence the function of CSFT protein while result in profound effect on the downstream pathway through the mechanism of competitive endogenous [61,62]. As far as we know, the exact role of RI in CSTF3 and its potential biological value has not been reported so far. Our results suggested that deciphering the underlying mechanisms of CSTF3-RI may provide valuable clues for seeking the possible therapeutic targets of CRC.
Another interesting finding of the present study was that the distribution of different CMS, TNM stage, KRASm and survival status (OS and DFS) between AS clusters was not random. CRC sample classified as C3 was more frequently KRAS mutant and enriched CMS4 and advanced TNM stage. Moreover, C3 were both associated with poor outcome in OS and DFS analysis. In the consensus molecular subtypes, CMS1-immune comprises most tumors with microsatellite instability (MSI) and is characterized by infiltration of activated immune cells [35]. CMS4 comprises the more mesenchymal-like cancers, with high stromal infiltration and poor patient prognosis [35]. These findings suggested that there is considerable variability in the nature of the DEAS across CRC —partly determined by molecular characteristics of the primary tumor—and that this influences clinical outcome. In addition, as shown in Fig. 7c and e, CMS1 comprises most tumors with MSI, which is in accordance with previously published conclusion [7] and partly validate the reliability of our results.
Finally, despite the rigorous quality controls result in the exclusion of the vast majority of AS events, this allowed us to be confident that the screened splice events are ubiquitous in CRC patient. Based on that, 421 AS events were identified with significant difference between CRC and paired normal tissue. Further enrichment analysis confirmed that the parent genes of these AS events have great potential to play a vital role in CRC. In addition, interesting splicing correlation network provides novel insights into how these CRC-related AS events were potentially regulated by key splicing factors. More importantly, the survival associated AS events, which may be most valuable in deciphering the underlying mechanisms of AS in the oncogenesis of CRC, provide clues of therapeutic targets to further validations.
The following are the supplementary data related to this article.
Supplementary Fig. S2.
Enrichment analysis of the parent gene of DEAS. (A-D) GO and KEGG pathway analyses of parent genes of identified DEAS. The vertical axis represents Go or KEGG pathway annotations. The horizontal axis represents the number of genes assigned to the corresponding annotation. (a) Biological process; (b) cellular components; (c) molecular function; and (d) KEGG pathway. (e) Enrichment plots for significant pathways.
Clinical features for the CRC patients in the TCGA cohort.
The detailed information of all the detected AS events.
The detailed information of the detected differently expressed AS events.
The relationship between DEAS and DEG.
The detail of the 71 experimentally validated splicing factors.
Univariate analysis of clinicopathological features for OS and DFS.
Univariate survival analysis of DEAS for OS and DFS.
Multivariate analysis of CRC prognosis-related DEAS events.
Acknowledgments
Acknowledgements
We thank the two anonymous reviewers for constructive comments. We are grateful to our colleagues Wenxian You, Xingye Wu and Jinglai Wei, as well as to members of our laboratory including Lixue Chen and Jun He.
Declaration of interest
The authors declare no potential conflicts of interest.
Funding sources
This work was financially supported by grants from the National Natural Science foundation of China (Grant No. 81572319; Project recipient: ZhongXue Fu).
Authors' contributions
Conception and design: Y. Xiong, L. Si, Z. Fu. Development of methodology: Ying Deng. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Y. Xiong, Y. Deng, K. Wang, H. Zhou. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y. Xiong, X. Zheng, Ying Deng. Writing, review, and/or revision of the manuscript: Y. Xiong, Y. Deng. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Z. Fu and Y. Xiong.
Contributor Information
Liangyi Si, Email: Si_Liangyi@outlook.com.
Zhongxue Fu, Email: fzx19990521@126.com.
References
- 1.Siegel R.L., Miller K.D., Fedewa S.A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Xiong Y., You W., Wang R., Peng L., Fu Z. Prediction and Validation of Hub Genes Associated with Colorectal Cancer by Integrating PPI Network and Gene Expression Data. Biomed Res Int. 2017;2017 doi: 10.1155/2017/2421459. 2421459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibutani M., Maeda K., Nagahara H. Tumor-infiltrating Lymphocytes Predict the Chemotherapeutic Outcomes in patients with Stage IV Colorectal Cancer. In Vivo. 2018;32(1):151–158. doi: 10.21873/invivo.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kindler H.L., Shulman K.L. Metastatic colorectal cancer. Curr Treat Options Oncol. 2001;2(6):459–471. doi: 10.1007/s11864-001-0068-7. [DOI] [PubMed] [Google Scholar]
- 5.Sveen A., Agesen T.H., Nesbakken A., Rognum TO, Lothe R.A., Skotheim R.I. Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med. 2011;3(5):32. doi: 10.1186/gm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Y., Wang R., Peng L. An integrated lncRNA, microRNA and mRNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2017;8(49):85463–85478. doi: 10.18632/oncotarget.20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guinney J., Dienstmann R., Wang X. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marisa L., de Reynies A., Duval A. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5) doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J.-X., Song W., Chen Z.-H. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 10.Salazar R., Roepman P., Capella G. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. Journal of Clinical Oncology. 2011;29(1):17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T., Kobunai T., Sakamoto E. Gene expression signature for recurrence in stage III colorectal cancers. Cancer. 2009;115(2):283–292. doi: 10.1002/cncr.24023. [DOI] [PubMed] [Google Scholar]
- 12.Smolle M., Uranitsch S., Gerger A., Pichler M., Haybaeck J. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. Int J Mol Sci. 2014;15(8):13993–14013. doi: 10.3390/ijms150813993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roock W.D., Vriendt V.D., Normanno N., Ciardiello F., Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12(6):594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 14.Frankish A., Uszczynska B., Ritchie G.R. Comparison of GENCODE and RefSeq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genomics. 2015;16(Suppl. 8):S2. doi: 10.1186/1471-2164-16-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrow J., Frankish A., Gonzalez J.M. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biamonti G., Catillo M., Pignataro D., Montecucco A., Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol. 2014;32:30–36. doi: 10.1016/j.semcdb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Climente-Gonzalez H., Porta-Pardo E., Godzik A., Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017;20(9):2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Lee S.C., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22(9):976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng H., Qin Z., Zhang X. Opportunities and methods for studying alternative splicing in cancer with RNA-Seq. Cancer Lett. 2013;340(2):179–191. doi: 10.1016/j.canlet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol. 2013;2013 doi: 10.1155/2013/463786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mojica W., Hawthorn L. Normal colon epithelium: a dataset for the analysis of gene expression and alternative splicing events in colon disease. BMC Genomics. 2010;11:5. doi: 10.1186/1471-2164-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bisognin A., Pizzini S., Perilli L. An integrative framework identifies alternative splicing events in colorectal cancer development. Mol Oncol. 2014;8(1):129–141. doi: 10.1016/j.molonc.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary oncology (Poznan Poland) 2015;19(1a):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan M.C., Cleland J., Kim R., Wong W.C., Weinstein J.N. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28(18):2385–2387. doi: 10.1093/bioinformatics/bts452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Jensen M.A., Zenklusen J.C. A Practical Guide to the Cancer Genome Atlas (TCGA) Methods Mol Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 26.Zhu J., Chen Z., Yong L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol Oncol. 2018;148(2):368–374. doi: 10.1016/j.ygyno.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Sun N., Lu Z. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017;393:40–51. doi: 10.1016/j.canlet.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 30.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics : a journal of integrative biology. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su G., Morris J.H., Demchak B., Bader G.D. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47:8.13.1–24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giulietti M., Piva F., D'Antonio M. SpliceAid-F: a database of human splicing factors and their RNA-binding sites. Nucleic Acids Res. 2013;41(Database issue):D125–D131. doi: 10.1093/nar/gks997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkerson M., Hayes D. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eide P.W., Bruun J., Lothe R.A., Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7(1):16618. doi: 10.1038/s41598-017-16747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorsen K., Sorensen K.D., Brems-Eskildsen A.S. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Molecular & Cellular Proteomics. 2008;7(7):1214–1224. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Gardina P.J., Clark T.A., Shimada B. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X., Coulombe-Huntington J., Kang S. Widespread expansion of Protein Interaction Capabilities by Alternative Splicing. Cell. 2016;164(4):805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dienstmann R., Vermeulen L., Guinney J., Kopetz S., Tejpar S., Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17(2):79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 40.Moore M.J., Wang Q., Kennedy C.J., Silver P.A. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142(4):625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126(1):37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 42.De Rosa M., Morelli G., Cesaro E. Alternative splicing and nonsense-mediated mRNA decay in the regulation of a new adenomatous polyposis coli transcript. Gene. 2007;395(1–2):8–14. doi: 10.1016/j.gene.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 43.Patek C.E., Arends M.J., Rose L. The pro-apoptotic K-Ras 4A proto-oncoprotein does not affect tumorigenesis in the ApcMin/+ mouse small intestine. BMC Gastroenterol. 2008;8:24. doi: 10.1186/1471-230X-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo F., Ye H., Hamoudi R. K-ras exon 4A has a tumour suppressor effect on carcinogen-induced murine colonic adenoma formation. J Pathol. 2010;220(5):542–550. doi: 10.1002/path.2672. [DOI] [PubMed] [Google Scholar]
- 45.Marcel V., Hainaut P. p53 isoforms - a conspiracy to kidnap p53 tumor suppressor activity? Cellular and molecular life sciences. CMLS. 2009;66(3):391–406. doi: 10.1007/s00018-008-8336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcel V., Khoury M.P., Fernandes K., Diot A., Lane D.P., Bourdon J.C. Detecting p53 isoforms at protein level. Methods Mol Biol. 2013;962:15–29. doi: 10.1007/978-1-62703-236-0_2. [DOI] [PubMed] [Google Scholar]
- 47.Bourdon J.C. p53 and its isoforms in cancer. Br J Cancer. 2007;97(3):277–282. doi: 10.1038/sj.bjc.6603886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura K., Fujibuchi W., Unno M. Splice isoforms as therapeutic targets for colorectal cancer. Carcinogenesis. 2012;33(12):2311–2319. doi: 10.1093/carcin/bgs347. [DOI] [PubMed] [Google Scholar]
- 49.Consortium SM-I A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32(9):903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C., Wu P.Y., Tong L., Phan J.H., Wang M.D. The impact of RNA-seq aligners on gene expression estimation. Acm Conference. 2015;2015:462–471. doi: 10.1145/2808719.2808767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J., Li H., Shen S., Sun L., Yuan Y., Xing C. Alternative splicing events implicated in carcinogenesis and prognosis of colorectal cancer. J Cancer. 2018;9(10):1754–1764. doi: 10.7150/jca.24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J., Weiss W.A. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34(1):1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 53.Todaro M., Gaggianesi M., Catalano V. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14(3):342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 54.von Pawel J., Spigel D., Ervin T. Randomized phase II Trial of Parsatuzumab (Anti-EGFL7) or Placebo in Combination with Carboplatin, Paclitaxel, and Bevacizumab for First-Line Nonsquamous Non-Small Cell Lung Cancer. Oncologist. 2018;23(6) doi: 10.1634/theoncologist.2017-0690. 654-e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zboralski D., Hoehlig K., Eulberg D., Frömming A., Vater A. Increasing Tumor-Infiltrating T Cells through Inhibition of CXCL12 with NOX-A12 Synergizes with PD-1 Blockade. Cancer Immunol Res. 2017;5(11):950–956. doi: 10.1158/2326-6066.CIR-16-0303. [DOI] [PubMed] [Google Scholar]
- 56.Ingram N., Northwood E.L., Perry S.L. Reduced type II interleukin-4 receptor signalling drives initiation, but not progression, of colorectal carcinogenesis: evidence from transgenic mouse models and human case-control epidemiological observations. Carcinogenesis. 2013;34(10):2341–2349. doi: 10.1093/carcin/bgt222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakuma K., Sasaki E., Kimura K. HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition. Gut. 2018;67(6):1103–1111. doi: 10.1136/gutjnl-2016-312927. [DOI] [PubMed] [Google Scholar]
- 58.Akishima-Fukasawa Y., Nakanishi Y., Ino Y., Moriya Y., Kanai Y., Hirohashi S. Prognostic significance of CXCL12 expression in patients with colorectal carcinoma. Am J Clin Pathol. 2009;132(2):202–210. doi: 10.1309/AJCPK35VZJEWCUTL. [quiz 307] [DOI] [PubMed] [Google Scholar]
- 59.Zhao S., Chang S.L., Linderman J.J., Feng F.Y., Luker G.D. A Comprehensive Analysis of CXCL12 Isoforms in Breast Cancer1,2. Translational oncology. 2014;7(3):429–438. doi: 10.1016/j.tranon.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takagaki Y., Manley J.L., MacDonald C.C., Wilusz J., Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4(12a):2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 61.Karreth F., Pandolfi P. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomson D., Dinger M. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical features for the CRC patients in the TCGA cohort.
The detailed information of all the detected AS events.
The detailed information of the detected differently expressed AS events.
The relationship between DEAS and DEG.
The detail of the 71 experimentally validated splicing factors.
Univariate analysis of clinicopathological features for OS and DFS.
Univariate survival analysis of DEAS for OS and DFS.
Multivariate analysis of CRC prognosis-related DEAS events.