Abstract
To develop and validate a radiomics signature for the prediction of gastric cancer (GC) survival and chemotherapeutic benefits. In this multicenter retrospective analysis, we analyzed the radiomics features of portal venous-phase computed tomography in 1591 consecutive patients. A radiomics signature was generated by using the Lasso-Cox regression model in 228 patients and validated in internal and external validation cohorts. Radiomics nomograms integrating the radiomics signature were constructed, demonstrating the incremental value of the radiomics signature to the traditional staging system for individualized survival estimation. The performance of the nomograms was assessed with respect to calibration, discrimination, and clinical usefulness. The radiomics signature consisted of 19 selected features and was significantly associated with DFS (disease-free survival) and OS (overall survival). Multivariate analysis demonstrated that the radiomics signature was an independent prognostic factor. Incorporating the radiomics signature into the radiomics-based nomograms resulted in better performance for the estimation of DFS and OS than the clinicopathological nomograms and TNM staging system, with improved accuracy of the classification of survival outcomes. Further analysis showed that stage II and III patients with higher radiomics scores exhibited a favorable response to chemotherapy. In conclusion, the newly developed radiomics signature is a powerful predictor of DFS and OS, and it may predict which patients with stage II and III GC benefit from chemotherapy.
Keywords: Gastric cancer, Radiomics signature, Prognosis, Chemotherapy
Research in context.
Evidence before this study
We systematically searched PubMed, without date restriction or limitation to English language publications, for research articles with the following terms: “(prognosis OR survival) AND (predictor OR predictive) AND (signature OR model) AND (chemotherapy benefits OR chemotherapeutic benefits) AND (radiomics OR texture feature) AND (gastric cancer OR GC OR stomach cancer OR gastric adenocarcinoma)”. This search did not identify any previous radiomics signatures that predicted prognosis and/or chemotherapeutic benefits in gastric cancer. The current staging system is not adequate for to define prognosis and can't predict whether the gastric cancer patients are likely to benefit from chemotherapy. By extracting high throughput of quantitative descriptors from routinely acquired computed tomography (CT) studies, radiomics enables the noninvasive profiling of tumor heterogeneity. Recent advances in radiomics have provided insights in personalized medicine in oncologic practice associated with tumor detection, prognosis, subtype classification, lymph node metastasis, distant metastasis, and therapeutic response evaluation. Although prognostic signatures have become widely established in cancer research, the development of signatures that can predict response to individual therapies has been a difficult proposition.
Added value of this study
We report, to our knowledge, the first radiomics signature developed in gastric cancer to predict prognosis and benefit from chemotherapy. The results revealed that a 19 feature-based radiomics signature can accurately predict disease-free survival and overall survival after surgery and added incremental value to the TNM staging system and clinical-pathologic risk factors for individual survival estimation. Furthermore, the radiomics signature was able to identify which patients with stage II and III gastric cancer benefit from chemotherapy. Besides, the radiomics nomogram incorporating the radiomics signature and five clinical-pathologic risk factors might facilitate patient counselling and individualise management of patients with gastric cancer.
Implications of all the available evidence
Our findings, together with existing evidence, highlight the potential of radiomics signature in predicting prognosis and survival benefit of chemotherapy, which might facilitate patient counselling and guide individual cure of patients with gastric cancer. Future work should focus on independent validation in additional cohorts, ideally in randomized controlled trials.
Alt-text: Unlabelled Box
1. Introduction
Gastric cancer (GC) is the fifth most common human malignant disease and the third leading cause of cancer-related death worldwide. [44] Staging according to the TNM (tumor, node, and metastasis) system and histological subtype has been the most commonly used benchmark for the prognostic definition and establishment of treatment strategy in GC. According to the new US National Comprehensive Cancer Network guidelines, patients with advanced GC are recommended to receive chemotherapy.([5]; Group et al., [15]; [36]) However, large variations in clinical outcomes have been shown in patients with the same stage and similar treatment regimens. [5,22,36,41] These findings suggest that the present GC staging system provides inadequate prognostic information and does not reflect the biological heterogeneity of GC. [33,36,48] With the rapid development in understanding the molecular biology of GC, various biologic or genetic biomarkers that are related to prognosis or efficacy of chemotherapy have been investigated, [[23], [24], [25],33,41] but the inability to obtain comprehensive information about heterogeneous tumors remains a limitation of these invasive methods. [33,41] Hence, more studies are needed to confirm the clinical roles of these methods, and new prognostic biomarkers are required for personalized medicine.
Radiomics is an emerging field that converts imaging data into a high-dimensional mineable feature space using a large number of automatically applied data-characterization algorithms. [1,13] By extracting large data sets of quantitative descriptors from routinely acquired computed tomography (CT) studies in a high-throughput manner, radiomics enables the noninvasive profiling of tumor heterogeneity. [1,4,8,13,37] Recent advances in radiomics have provided insights for personalized medicine in oncological practice associated with tumor detection, prognosis, subtype classification, lymph node metastasis, distant metastasis, and therapeutic response evaluation. [[1], [2], [3],8,13,19,20] However, research of radiomics with respect to GC survival and chemotherapeutic benefits is still lacking.
The combined analysis of a panel of biomarkers as a signature, rather than individual analyses, is the approach that shows the most promise to change clinical management. [19,20,50,52] The least absolute shrinkage and selection operator (LASSO) method is a popular method for regression of high-dimensional predictors. [42,43,52] Using the LASSO Cox regression model, we previously constructed an immune signature that could effectively predict recurrence, disease-free survival (DFS) and overall survival (OS) in GC. [25,49] Although CT texture assessments have been reported to be associated with prognosis in patients with GC, [12,51] to the best of our knowledge, an optimal approach that combines multiple imaging biomarkers as a predictive signature for survival and chemotherapeutic benefits has not yet been developed.
In this study, we developed and validated a multiple-feature-based radiomics signature to predict DFS and OS and assessed its incremental value to the traditional staging system and clinicopathological risk factors for individual DFS and OS estimation. Furthermore, the radiomics signature might be able to predict which patients with stage II and III GC benefit from adjuvant chemotherapy.
2. Materials and methods
2.1. Study population
The study enrolled 3 independent cohorts of 1591 patients with gastric adenocarcinoma. The training cohort and internal validation cohort that comprised 228 consecutive patients and 186 consecutive patients with total or partial radical gastrectomy were obtained from Nanfang Hospital of Southern Medical University (Guangzhou, China) from January 2007 to April 2011 and from May 2011 to May 2013, respectively. The external validation cohort that comprised 1177 consecutive patients was obtained from Sun Yat-sen University Cancer Center between January 2008 and December 2012 with the same enrollment criteria. Clinicopathological data were retrospectively collected for each patient. The clinical sources of the 1591 patients with GC are listed in Table 1. All these patients satisfied the following inclusion criteria: histologically confirmed gastric adenocarcinoma; standard unenhanced and contrast-enhanced abdominal CT performed <30 days before surgical resection; lymphadenectomy performed and >15 lymph nodes harvested; complete clinicopathological characteristics and follow-up data; no combined malignant neoplasm; and no preoperative chemotherapy. We excluded patients if the tumor lesions could not be identified by CT or if patients received previous treatment with any anticancer therapy. Ethical approval was obtained for this retrospective analysis at every participating center, and the informed consent requirement was waved.
Table 1.
Clinical characteristics of patients according to the radiomics score in the training and validation cohorts.
| Variables | Training cohort (n = 228) |
Internal validation cohort (n = 186) |
External validation cohort (n = 1177) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| low-RS (%) | medium-RS (%) | high-RS(%) | p-value | low-RS(%) | medium-RS(%) | high-RS(%) | p-value | low-RS(%) | medium-RS(%) | high-RS(%) | p-value | |
| Gender | 0.817 | 0.561 | 0.001 | |||||||||
| Male | 77(40.0%) | 30(34.4%) | 34(60.0%) | 74(58.3%) | 24(18.9%) | 29(22.8%) | 152(41.8%) | 134(36.8%) | 78(21.4%) | |||
| Female | 51(34.4%) | 16(34.4%) | 20(65.6%) | 39(66.1%) | 10(16.9%) | 10(16.9%) | 275(33.8%) | 282(34.7%) | 256(31.5%) | |||
| Age(years) | 0.210 | 0.019 | 0.001 | |||||||||
| <60 | 88(60.3%) | 28(19.2%) | 30(20.5%) | 63(55.3%) | 28(24.6%) | 23(20.2%) | 269(40.1%) | 234(34.9%) | 167(24.9%) | |||
| ≧60 | 40(48.8%) | 18(22%) | 24(29.3%) | 50(69.4%) | 6(8.3%) | 16(22.2%) | 158(31.2%) | 182(35.9%) | 167(32.9%) | |||
| Charlson comorbidity index | 0.333 | 0.968 | 0.129 | |||||||||
| 0 | 86(56.6%) | 28(18.4%) | 38(25.0%) | 77(60.2%) | 24(18.8%) | 27(21.1%) | 304(38.5%) | 265(33.5%) | 221(28.0%) | |||
| 1 | 34(55.7%) | 15(24.6%) | 12(19.7%) | 29(63.0%) | 8(17.4%) | 9(19.6%) | 96(30.8%) | 129(41.3%) | 87(27.9%) | |||
| 2 | 7(58.3%) | 1(8.3%) | 4(33.3%) | 6(60.0%) | 2(20.0%) | 2(20.0%) | 21(34.4%) | 18(29.5%) | 22(36.1%) | |||
| 3 | 1(33.3%) | 2(66.7%) | 0(0.0%) | 1(50.9%) | 0(0.0%) | 1(50.0%) | 6(42.9%) | 4(28.6%) | 4(28.6%) | |||
| Tumor size(cm) | 0.073 | 0.410 | 0.001 | |||||||||
| <4 | 60(65.2%) | 14(15.2%) | 18(19.6%) | 56(65.9%) | 14(16.5%) | 15(17.6%) | 192(42.7%) | 153(34.0%) | 105(23.3%) | |||
| ≧4 | 68(50%) | 32(23.5%) | 36(26.5%) | 57(56.4%) | 20(19.8%) | 24(23.8%) | 235(32.3%) | 263(36.2%) | 229(31.5%) | |||
| Tumor location | 0.266 | 0.015 | 0.020 | |||||||||
| Cardia | 29(69%) | 6(14.3%) | 7(16.7%) | 30(88.2%) | 3(8.8%) | 1(2.9%) | 130(32.7%) | 135(34.0%) | 132(33.2%) | |||
| Body | 28(66.7%) | 6(14.3%) | 8(19%) | 17(65.4%) | 3(11.5%) | 6(23.1%) | 97(40.4%) | 92(38.3%) | 51(21.3%) | |||
| Antrum | 55(49.1%) | 26(23.2%) | 31(27.7%) | 53(52%) | 23(22.5%) | 26(25.5%) | 184(37.8%) | 173(35.5%) | 130(26.7%) | |||
| Whole | 16(50%) | 8(25%) | 8(25%) | 13(54.2%) | 5(20.8%) | 6(25%) | 16(30.2%) | 16(30.2%) | 21(39.6%) | |||
| Differentiation | <0.0001 | 0.095 | 0.006 | |||||||||
| Well | 23(82.1%) | 3(10.7%) | 2(7.1%) | 12(37.5%) | 4(37.5%) | 0(0%) | 39(47.0%) | 31(37.3%) | 13(15.7%) | |||
| Moderate | 55(68.8%) | 13(16.3%) | 12(15%) | 36(37.5%) | 6(37.5%) | 10(19.2%) | 82(37.5%) | 90(37.5%) | 51(19.2%) | |||
| Poor and undifferentiated | 50(41.7%) | 30(25%) | 40(33.3%) | 20(37%) | 27(37.5%) | 34(63%) | 306(35.1%) | 295(33.9%) | 270(31.0%) | |||
| Lauren type | <0.0001 | 0.103 | 0.142 | |||||||||
| Intestinal type | 80(71.4%) | 12(10.7%) | 20(17.9%) | 70(67.3%) | 17(16.3%) | 17(16.3%) | 133(33.3%) | 156(39.0%) | 111(27.8%) | |||
| Diffuse or mixed | 48(41.4%) | 34(29.3%) | 34(29.3%) | 43(52.4%) | 17(20.7%) | 22(26.8%) | 294(37.8%) | 260(33.5%) | 223(28.7%) | |||
| CEA | 0.010 | 0.002 | 0.0005 | |||||||||
| Elevated | 11(33.3%) | 12(36.4%) | 10(30.3%) | 11(39.3%) | 4(14.3%) | 13(46.3%) | 69(27.6%) | 88(35.2%) | 93(37.2%) | |||
| Normal | 117(60%) | 34(17.4%) | 44(22.6%) | 102(64.6%) | 30(19%) | 26(16.5%) | 102(38.6%) | 30(35.4%) | 26(26.0%) | |||
| CA199 | <0.0001 | <0.0001 | <0.0001 | |||||||||
| Elevated | 20(35.7%) | 12(21.4%) | 24(42.9%) | 23(44.2%) | 8(15.4%) | 21(40.4%) | 40(22.2%) | 63(35.0%) | 77(42.8%) | |||
| Normal | 108(62.8%) | 34(19.8%) | 30(17.4%) | 90(67.2%) | 26(19.4%) | 18(13.4%) | 387(38.8%) | 353(35.4%) | 257(25.8%) | |||
| Depth of invasion | <0.0001 | <0.0001 | <0.0001 | |||||||||
| T1 | 16(100%) | 0(0%) | 0(0%) | 7(63.6%) | 4(36.4%) | 0(0%) | 74(49.3%) | 55(36.7%) | 21(14%) | |||
| T2 | 12(75%) | 4(25%) | 0(0%) | 11(91.7%) | 1(8.3%) | 0(0%) | 64(48.1%) | 38(28.6%) | 31(23.3%) | |||
| T3 | 3(42.9%) | 0(0%) | 4(57.1%) | 13(76.5%) | 4(23.5%) | 0(0%) | 78(29.9%) | 110(42.1%) | 73(28.0%) | |||
| T4a | 75(65.2%) | 22(19.1%) | 18(15.7%) | 57(68.7%) | 14(16.9%) | 12(14.5%) | 198(36.1%) | 174(31.8%) | 176(32.1%) | |||
| T4b | 22(29.7%) | 20(27%) | 32(43.2%) | 25(39.7%) | 11(17.5%) | 27(42.9%) | 13(15.3%) | 39(45.9%) | 33(38.8%) | |||
| Lymph node metastasis | <0.0001 | <0.0001 | <0.0001 | |||||||||
| N0 | 32(71.1%) | 11(24.4%) | 2(4.4%) | 40(74.1%) | 10(18.5%) | 4(7.4%) | 188(48.0%) | 128(32.7%) | 76(19.4%) | |||
| N1 | 27(75%) | 5(13.9%) | 4(11.1%) | 20(83.3%) | 3(12.5%) | 3(4.2%) | 61(33.9%) | 75(41.7%) | 44(24.4%) | |||
| N2 | 37(55.2%) | 10(14.9%) | 20(29.9%) | 26(63.4%) | 3(7.3%) | 12(29.9%) | 61(29.3%) | 77(37.0%) | 70(33.7%) | |||
| N3 | 32(40%) | 20(25%) | 28(35%) | 27(40.3%) | 18(26.9%) | 22(32.8%) | 117(29.5%) | 136(34.3%) | 144(36.3%) | |||
| Distant metastasis | <0.0001 | <0.0001 | 0.0004 | |||||||||
| M0 | 120(66.3%) | 30(16.6%) | 31(17.1%) | 108(72%) | 24(16%) | 18(12%) | 404(38.1%) | 366(34.5%) | 290(27.4%) | |||
| M1 | 8(17%) | 16(34%) | 23(49%) | 5(13.9%) | 10(27.8%) | 21(58.3%) | 23(19.7%) | 50(42.7%) | 44(37.6%) | |||
| Stage | <0.0001 | <0.0001 | <0.0001 | |||||||||
| I | 18(81.8%) | 4(18.2%) | 0(0%) | 12(75%) | 4(25%) | 0(0%) | 108(52.4%) | 64(31.1%) | 34(16.5%) | |||
| II | 20(87%) | 1(4.3%) | 2(8.7%) | 31(86.1%) | 5(13.9%) | 0(0%) | 125(41.8%) | 105(35.1%) | 69(23.1%) | |||
| III | 82(60.3%) | 25(18.4%) | 29(21.3%) | 65(66.3%) | 15(15.3%) | 18(18.4%) | 171(30.8%) | 197(35.5%) | 187(33.7%) | |||
| IV | 8(17%) | 16(34%) | 23(49%) | 5(13.9%) | 10(27.8%) | 21(58.3%) | 23(19.7%) | 50(42.7%) | 44(37.6%) | |||
RS: radiomic score.
Baseline information for each patient with GC, including age, gender, tumor location, tumor size, differentiation, Lauren type, carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA19-9), TNM stage, postsurgical chemotherapy and follow-up data (follow-up duration and survival), was documented. The TNM staging was reclassified according to the seventh edition of the AJCC Cancer Staging Manual of the American Joint Committee on Cancer (AJCC)/International Union Against Cancer. Patients were postoperatively followed up with abdomen CTs every 6–12 months for the first 2 years and then annually, according to the follow-up protocol of our institution. Follow-up data were collected from hospital records for patients who were lost during follow-up. The follow-up duration was measured from the time of surgery to the last follow-up date, and information regarding the survival status at the last follow-up was collected. The DFS was defined as the time to recurrence at any site, or all-cause death, whichever came first. The OS was defined from the date of surgery to the date of all-cause death or the latest follow-up used for censoring.
2.2. Image acquisition and imaging texture analysis
The details regarding the acquisition parameters, CT image retrieval procedure and imaging texture analysis are presented in Supplementary Materials. The inter- and intra-observer reproducibility for region-of-interest-based texture feature extraction was analyzed by two experienced radiologists (readers 1 and 2 with 11 and 10 years of clinical experience in abdominal CT study interpretation, respectively) (Fig. S1). The radiologists were blind to the clinical and histopathological data but were aware that the patients had gastric cancer.
2.3. Feature selection and radiomics signature building
The LASSO Cox regression model was used to select the most useful prognostic features of all the texture features, and a multiple-feature-based radiomics signature, the radiomics score, was then constructed for predicting survival in the training cohort. [43,52] The “glmnet” package was used to perform the LASSO Cox regression model analysis. The complete details are provided in Supplementary Materials.
2.4. Validation of radiomics signature
The potential association of the radiomics signature with DFS and OS was first assessed in the training cohort and then validated in the validation cohorts by using Kaplan-Meier survival analysis. The patients were classified into high-, medium-, or low radiomics score groups, the thresholds of which were identified using X-tile. [6] Then, the same threshold values were applied to the validation cohorts. Stratified analyses were performed to explore the potential association of the radiomics signature with the DFS and OS using subgroups within the TNM stage and clinicopathological risk factors from all patients. The evaluation of the radiomics signature as an independent biomarker was performed by integrating clinicopathological risk factors into the multivariable Cox proportional hazards model using a backward stepwise approach. [16,26]
2.5. Assessment of incremental value of radiomics signature in individual DFS and OS estimation
To demonstrate the incremental value of the radiomics signature to the TNM staging system and other clinicopathological risk factors for individualized assessment of DFS and OS, both a radiomics nomogram and a clinicopathological nomogram were developed in the training cohort (Fig. S2). The radiomics nomogram incorporated the radiomics signature and the independent clinicopathological risk factors based on the multivariate Cox analysis. The clinicopathological nomogram incorporated only the independent clinicopathological risk factors.
The incremental value of the radiomics signature to the TNM staging system and other clinicopathological risk factors was assessed with respect to calibration, discrimination, reclassification, and clinical usefulness. The performance of the radiomics nomogram was compared to that of both the TNM staging system and the clinicopathological nomogram.
To compare the predicted survival with the actual survival, calibration curves were generated. The agreement between the predicted survival and the actual survival was also analyzed with Bland–Altman plot. To quantify the discrimination performance, Harrell's concordance index (C-index) was measured. [14] To quantify the improvement of usefulness added by the radiomics signature, a net reclassification improvement (NRI) calculation was also applied. [38] Finally, a decision curve analysis determined the clinical usefulness of the radiomics nomogram by quantifying the net benefits at different threshold probabilities. [46]
2.6. Statistical analysis
Differences in distributions between the variables examined were assessed with the unpaired, 2-tailed χ2 test or the Fisher exact test as appropriate. The Kaplan-Meier method and log-rank test were used to estimate DFS and OS. Multivariate analyses were performed using the Cox proportional hazards model. Statistical analysis was conducted with R software (version 3.1.0) and SPSS software (version 19.0). Bonferroni correction was applied to obtain the corrected P value for multiple comparisons. The packages in R that were used in this study are reported in Supplementary Materials. A two-sided P value < 0.05 was considered significant.
3. Results
The clinicopathological characteristics for the training cohort, internal validation cohort and external validation cohort are listed in Table 1. Of the 1591 patients included in the study, 1081 (67.9%) were men, and the median (interquartile range [IQR]) age of all patients was 57 (49–65) years. In the training cohort, the median (IQR) survival time for DFS and OS were 29 (6–70) and 45 (11–74) months, respectively. In the internal validation cohort, the median (IQR) survival times for DFS and OS were 31 (9–48) and 39 (17.0–51.25) months, respectively, and in the external validation cohort, the median (IQR) survival times for DFS and OS were 36.87 (20.32–55.79) and 38.03 (22.9–56.07) months, respectively.
The inter and intraobserver reproducibility of the texture feature extraction was high (Supplementary Materials). Therefore, all outcomes were based on the measurements of the first radiologist.
3.1. Construction of the radiomics score-based radiomics signature
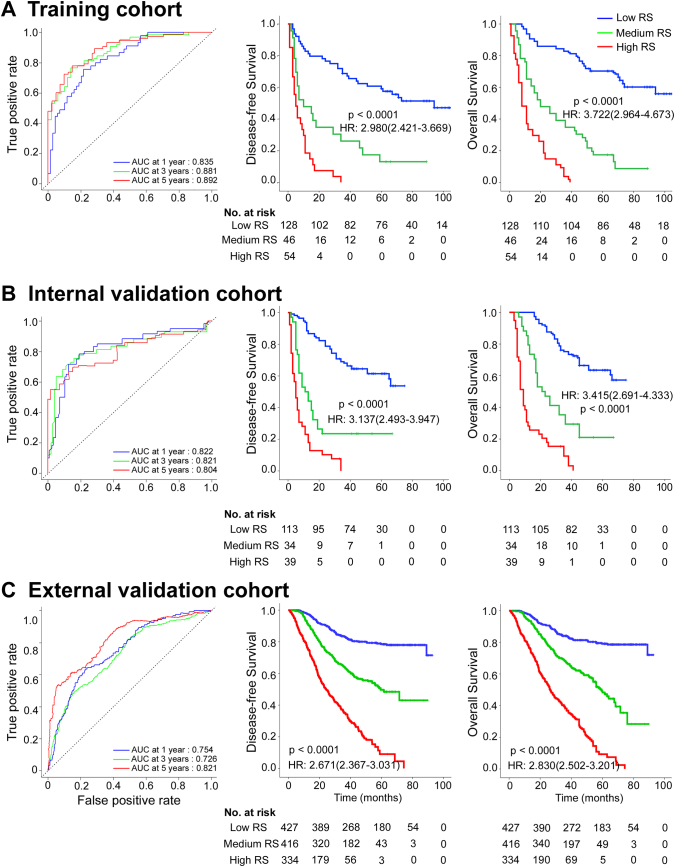
A LASSO Cox regression model was used to build a prognostic classifier, which selected 19 potential predictors from the 269 features identified in the training cohort (Fig. S3). The radiomics signature was constructed, including a radiomics score calculation formula (Supplementary Materials). The optimum cutoffs generated by the X-tile plot were −1.1 and −0.8 (Fig. S4). Accordingly, patients were classified into a low-radiomics score group (radiomics score < −1.1), a medium-radiomics score group (−1.1 ≤ radiomics score < −0.8), and a high-radiomics score group (radiomics score ≥ −0.8). We assessed the prognostic accuracy of the radiomics score in the training cohort using time-dependent receiver operator characteristics (ROC) analysis at different follow-up times (Fig. 1A). The 5-year DFS and OS were 59.4% and 67.2%, respectively, for the low-radiomics score group; 13.0% and 17.4%, respectively, for the medium radiomics score group; and both 0 for the high radiomics score group (hazard ratios[HRs] 2.980 (2.421–3.669) and 3.722(2.964–4.673), respectively; all P < 0.001 and corrected P < 0.001; Fig. 1A). We then performed the same analyses (time-dependent ROC analysis and Kaplan-Meier survival analysis) in the internal validation cohort and similar results were observed (HR 3.137 (2.493–3.947) and 3.415(2.691–4.333), respectively; all P < 0.001 and corrected P < 0.001; Fig. 1B). To confirm that the radiomics signature had an excellent prognostic value in different populations, we further applied it to the external validation cohort, and found similar results (Fig. 1C). When the patients were stratified by clinicopathological risk factors, the radiomics signature remained a clinically and statistically significant prognostic predictor (Fig. S5–8).
Fig. 1.
Radiomics score measured by time-dependent ROC curves and Kaplan-Meier survival in the training, internal and external validation cohorts.
(A) Training cohort. (B) Internal validation cohort. (C) External validation cohort. We used AUCs at 1, 3, and 5 years to assess prognostic accuracy in the training and validation cohorts. We calculated P-values using the log-rank test. Data are the AUC or P-value. ROC = receiver operator characteristic. AUC = area under the curve. HR = hazard ratio.
We also assessed the distribution of radiomics scores, recurrence and survival statuses as well as the expression of 19 radiomics features in the internal and external cohorts (Fig. S9–10). Patients with higher radiomics scores were more likely to have recurrences and deaths. In univariable analysis, low radiomics score patients were associated with significantly poorer OS and DFS (Table S1). Variables demonstrating a significant effect on OS and DFS were included in the multivariable analysis. Multivariate Cox regression analysis after adjustment for clinicopathological variables revealed that the radiomics score remained a powerful and independent prognostic factor for DFS and OS in the training, internal and external validation cohorts (Tables 2 and S2–3).
Table 2.
Multivariable association of the radiomics score and clinicopathological characteristics with disease-free survival and overall survival in the training cohort.
| Variables | Disease-free survival |
Overall survival |
||
|---|---|---|---|---|
| HR (95%CI) | p | HR (95%CI) | p | |
| Radiomics score | 1.744 (1.346–2.261) | <0.0001 | 3.308 (1.752–3.040) | <0.0001 |
| Differentiation status | 0.008 | 0.019 | ||
| Well | Reference | Reference | ||
| Moderate | 2.242 (1.037–4.850) | 0.040 | 2.150 (0.890–5.195) | 0.089 |
| Poor or undifferentiation | 2.835 (1.330–6.044) | 0.007 | 2.904 (1.213–6.954) | 0.017 |
| CA199 (elevated vs. nomal) | 1.829 (1.243–2.692) | 0.002 | 1.963 (1.313–2.934) | 0.001 |
| Depth of invasion | 0.0005 | 0.011 | ||
| T1 | Reference | Reference | ||
| T2 | 1.625 (0.296–8.923) | 0.576 | 1.546 (0.281–8.497) | 0.617 |
| T3 | 5.052 (0.942–27.102) | 0.059 | 4.971 (0.930–26.578) | 0.061 |
| T4a | 3.010 (0.703–12.884) | 0.137 | 2.472 (0.571–10.704) | 0.226 |
| T4b | 6.269 (1.425–27.579) | 0.015 | 4.279 (0.968–18.927) | 0.055 |
| Lymph node metastasis | <0.0001 | 0.0002 | ||
| N0 | Reference | Reference | ||
| N1 | 0.951 (0.478–1.890) | 0.886 | 0.919 (0.441–1.918) | 0.822 |
| N2 | 0.952 (0.517–1.751) | 0.873 | 0.947 (0.494–1.817) | 0.871 |
| N3 | 3.806 (2.110–6.866) | <0.0001 | 2.124 (1.126–4.007) | 0.020 |
| Distant metastasis (yes vs. no) | 6.240 (3.976–9.793) | <0.0001 | 3.518 (2.322–5.330) | <0.0001 |
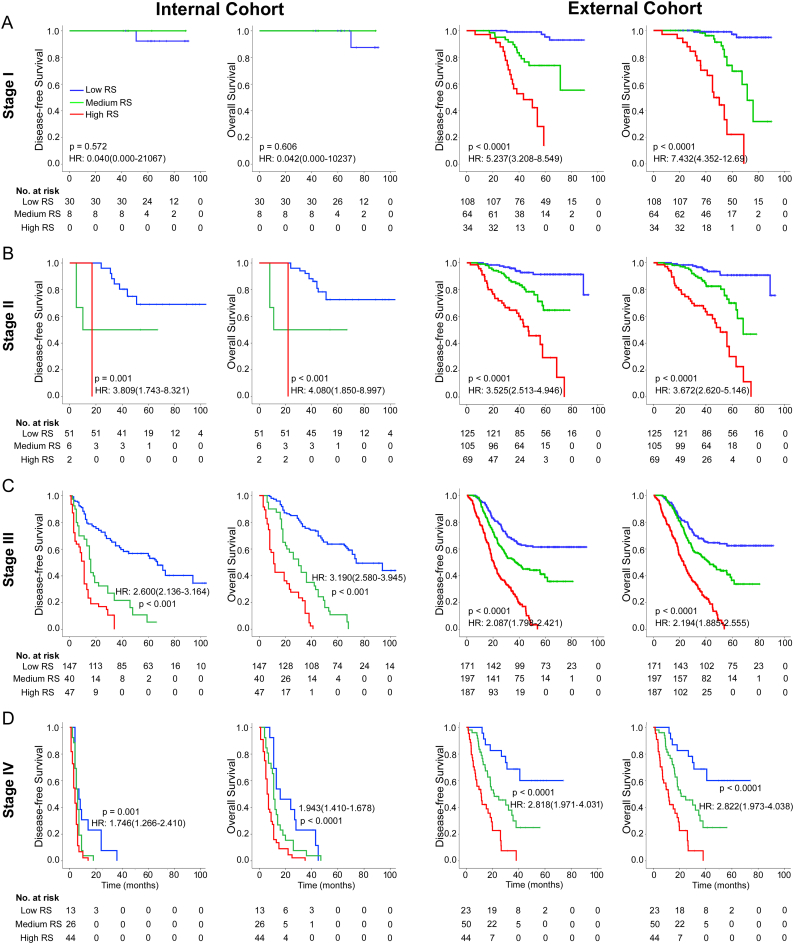
We performed stratified analyses of GC patients with stage I, II, III, and IV disease in the combined internal and external cohorts. Radiomics score could distinguish patients with different prognoses in stage I, II, III, and IV (Fig. 2).
Fig. 2.
Kaplan-Meier survival analysis of disease-free survival and overall survival according to the radiomics score classifier in subgroups of GC patients in the total internal and external cohorts. Total internal cohort (left pane): (A) Stage I (n = 38). (B) Stage II (n = 59). (C) Stage III (n = 234). (D) Stage IV (n = 83). External cohort (right pane): (A) Stage I (n = 206). (B) Stage II (n = 299). (C) Stage III (n = 555). (D) Stage IV (n = 117).
3.2. Assessment of incremental value of radiomics signature in individual DFS and OS performance
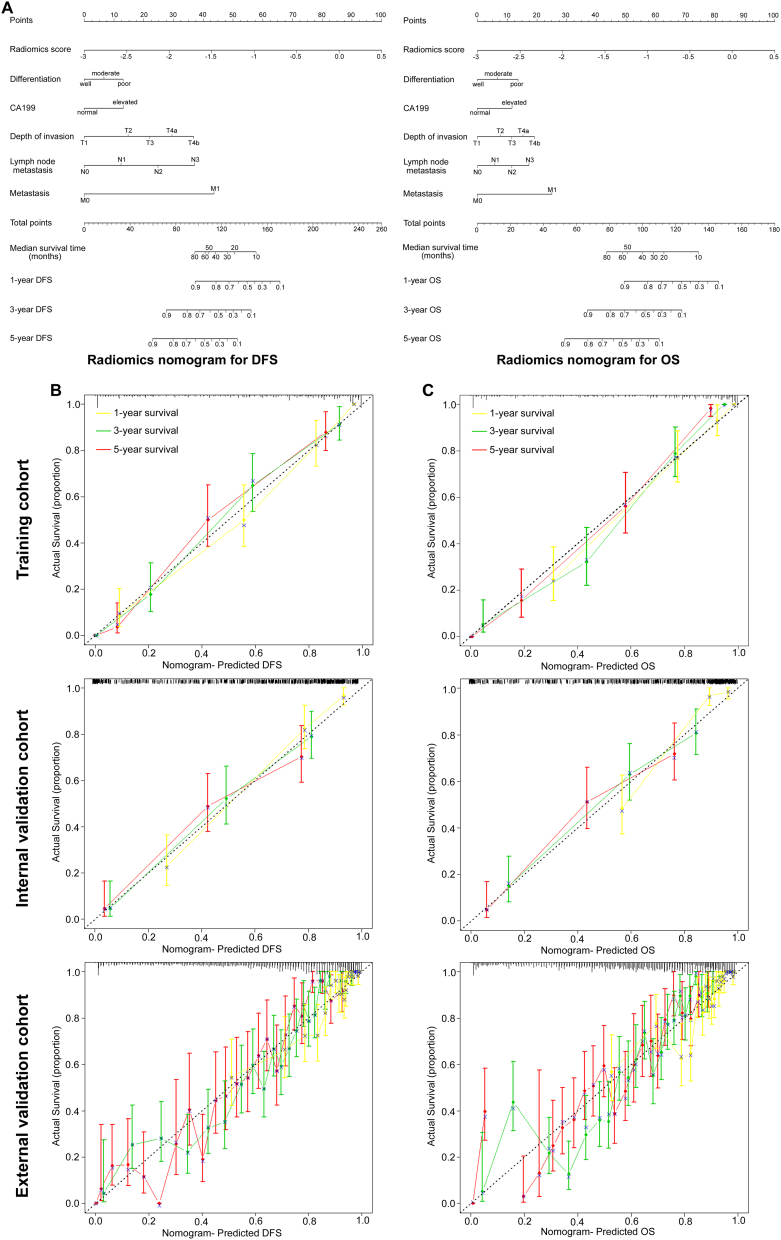
The radiomics nomograms for DFS and OS are presented in Fig. 3A. The calibration curves of the nomograms at 1, 3, or 5 years are shown in Fig. 3B-C, and there was good agreement among the estimations with the radiomics nomogram and actual observations in the training, internal and external cohorts. The Bland-Altman type plots also showed that the radiomics nomograms had a good predictive effect in the training, internal and external validation cohorts, especially for actual survival >20 months (Fig. S11). We also constructed two clinicopathological nomograms for DFS and OS only using these clinicopathological risk factors (Fig. S12). The discrimination performance of the radiomics signature improved when it was integrated into the radiomics nomogram along with the clinicopathological risk factors (C-index for the radiomics nomogram: DFS, 0.850 (95% CI: 0.825, 0.875); OS 0.860 (0.835, 0.885) in the training cohort; Table 3). Compared to either the TNM staging system or the clinicopathological nomogram, the radiomics nomogram showed a better discrimination capability with higher C-indexes in the training, internal and external cohorts (Table 3). Furthermore, the inclusion of the radiomics signature in the clinicopathological nomogram yielded a total NRI of 0.283 (95% CI: 0.086, 0.421; P = 0.01) and 0.317 (0.134, 0.500; P < 0.001; in the training cohort) for DFS and OS, respectively, showing improved classification accuracy for survival outcomes (Table S5 and Fig. S13). Similar results were observed both in the internal and external validation cohorts.
Fig. 3.
Use of the constructed radiomics nomogram to estimate DFS and OS for gastric cancer, along with the assessment of the model calibration. (A) Radiomics nomogram for DFS (left) and OS (right). The patient's radiomics score is located on the radiomics score axis. To determine the number of points toward the probability of DFS and OS the patient receives for his or her radiomics score, a line was drawn straight upward to the point axis, and this process was repeated for each variable. The points achieved for each of the risk factors was then summed. The final sum is located on the total point axis. To find the patient's probability of DFS and OS, a line was drawn straight down. Calibration curves of the radiomics nomogram for DFS (left, (B)) and OS (right, (C)) in the training, internal and external validation cohorts show the calibration of each model in terms of the agreement between the estimated and the observed 1-, 3-, and 5-year outcomes. Nomogram-estimated DFS is plotted on the x-axis, and the observed tumor relapse rate is plotted on the y-axis. Diagonal dotted line represents a perfect estimation by an ideal model, in which the estimated outcome perfectly corresponds to the actual outcome. Solid line represents performance of the nomogram, a closer alignment of which with the diagonal dotted line represents a better estimation. (B) (C): Training cohort (upper panels); Internal validation cohort (middle panels); External validation cohort (lower panels).
Table 3.
Treatment interaction with the radiomics score for disease-free survival and overall survival in patients with stage II and III disease.
| Radiomics score | Chemo | No chemo | Disease-free survival |
Overall survival |
||||
|---|---|---|---|---|---|---|---|---|
| Yes vs No chemo, HR (95% CI) | P | P value for interaction | Yes vs No chemo, HR (95% CI) | P | P value for interaction | |||
| Internal cohort (n = 293) | ||||||||
| High score | 113 | 85 | 0.176(0.083–0.374) | <0.001 | 0.148(0.066–0.333) | <0.001 | ||
| Medium score | 21 | 25 | 0.565(0.300–1.065) | 0.077 | <0.0001 | 0.717(0.381–1.352) | 0.304 | <0.0001 |
| Low score | 27 | 22 | 0.871(0.576–1.318) | 0.514 | 0.806(0.514–1.266) | 0.350 | ||
| External cohort (n = 854) | ||||||||
| High score | 145 | 111 | 0.412(0.306–0.554) | <0.001 | <0.0001 | 0.394(0.293–0.529) | <0.001 | <0.0001 |
| Medium score | 210 | 92 | 0.584(0.412–0.829) | 0.003 | 0.601(0.423–0.854) | 0.005 | ||
| Low score | 207 | 89 | 0.788(0.489–1.269) | 0.327 | 0.807(0.497–1.309) | 0.385 | ||
Chemo: chemotherapy.
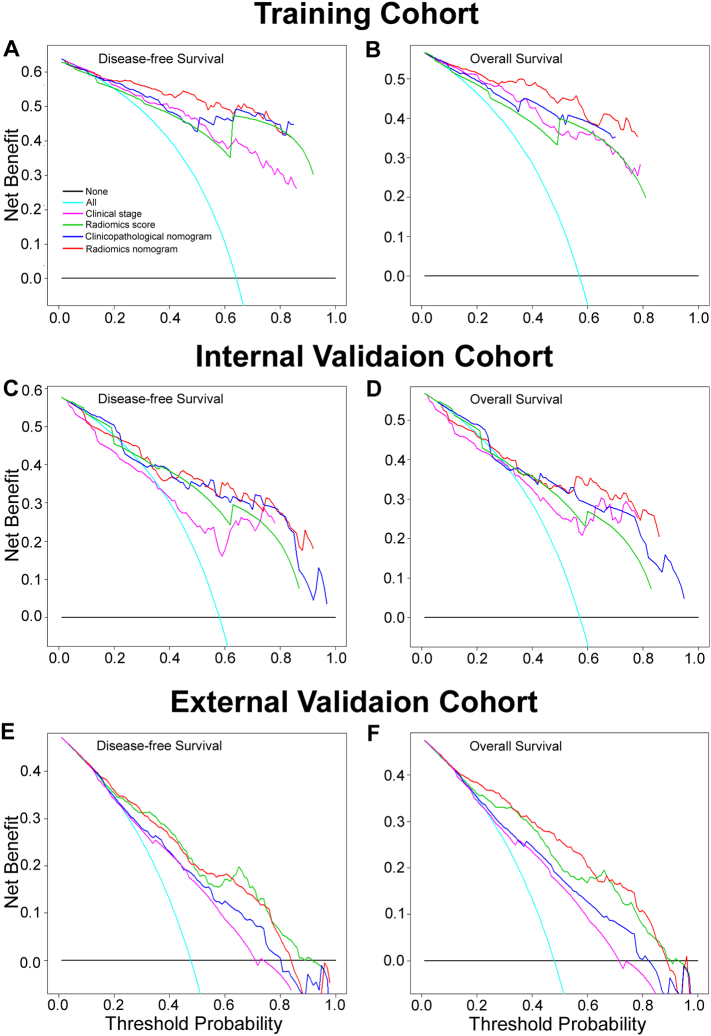
The decision curve analysis showed that the radiomics nomogram had a higher overall net benefit than the clinicopathological nomogram and the TNM staging system across the majority of the range of reasonable threshold probabilities (Fig. 4).
Fig. 4.
Decision curve analysis for each model in the training and validation cohorts. The y-axis measures the net benefit. The net benefit was calculated by summing the benefits (true positive results) and subtracting the harms (false positive results), weighting the latter by a factor related to the relative harm of an undetected cancer compared with the harm of unnecessary treatment. The radiomics model had the highest net benefit compared to both the other models and simple strategies such as follow-up of all patients (green line) or no patients (horizontal black line) across the full range of threshold probabilities at which a patient would choose to undergo imaging follow-up.
3.3. Radiomics signature and adjuvant chemotherapy
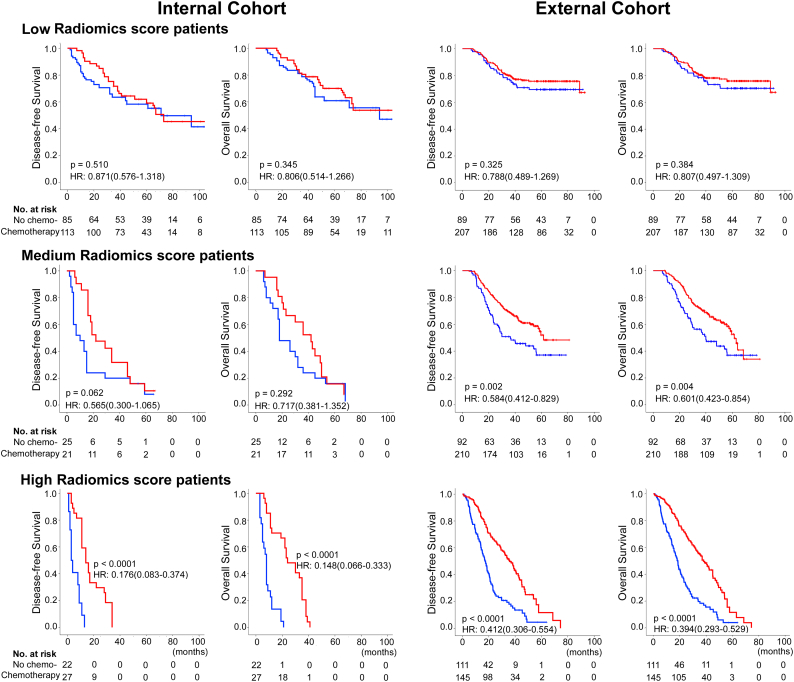
Furthermore, we investigated whether low, medium, or high radiomics score patients with stage II and III GC could benefit from postoperative adjuvant chemotherapy. A test for an interaction between radiomics score and adjuvant chemotherapy indicated that the benefit from adjuvant chemotherapy was superior among patients with high radiomics scores (internal cohort: OS, HR 0.148(0.066–0.333), P < 0.001 and corrected P < 0.001; DFS, 0.176(0.083–0.374), P < 0.001 and corrected P < 0.001; and external cohort: OS, HR 0.394(0.293–0.529), P < 0.001 and corrected P < 0.001; DFS, 0.412(0.306–0.554), P < 0.001 and corrected P < 0.001; all P < 0.0001 for interaction; Table 3) than among those with low and medium scores. The corresponding Kaplan-Meier survival curves, which comprehensively compared low, medium, with high radiomics score by treatment, are shown in Fig. 5. The results from the subset analysis using radiomics score classifier revealed that chemotherapy significantly increased DFS and OS in the high-radiomics score group (internal cohort: P < 0.001 and corrected P < 0.001, P < 0.001 and corrected P < 0.001; and external cohort: P < 0.001 and corrected P < 0.001, P < 0.001 and corrected P < 0.001; respectively), but had no significant effect in the low-radiomics score group (internal cohort: P = 0.510 and corrected P = 1.000, P = 0.345 and corrected P = 0.999; and external cohort: P = 0.325 and corrected P = 0.975, P = 0.384 and corrected P = 0.999; respectively; Fig. 5). Patients with medium radiomics scores also obtained a survival benefit from chemotherapy, although not significantly in the internal cohort (internal cohort: P = 0.062 and corrected P = 0.186, P = 0.292 and corrected P = 0.876; and external cohort: P = 0.002 and corrected P = 0.006, P = 0.004 and corrected P = 0.012; respectively). Furthermore, when the patients were stratified by clinicopathological risk factors, similar results were also observed (Fig. S14–15). Consequently, these results suggested that the radiomics score could successfully identify patients with stage II and III GC who are suitable candidates for chemotherapy.
Fig. 5.
Adjuvant chemotherapy benefit compared using disease-free survival (DFS) and overall survival (OS) for stage II and III gastric cancer patients in the total internal cohort and external cohort. Kaplan-Meier survival curves for patients with stage II and III gastric cancer in different subgroups, which were stratified by the receipt of adjuvant chemotherapy. Total internal cohort (N = 293): left pane; External cohort (N = 854): right pane.
4. Discussion
This study extends the analysis of individual imaging features to an “-omics”-based approach for survival estimation. A multiple-feature-based radiomics signature was identified to be an independent prognosis factor in patients with GC. The radiomics signature successfully stratified those patients into high-, medium-, and low-radiomics score groups with significant differences in DFS and OS. The radiomics nomograms performed better than the traditional staging system and clinicopathological nomograms, demonstrating well the incremental value of the radiomics signature for individualized DFS and OS estimation.
The present radiomics signature, consisting of 19 texture features, provides a noninvasive, fast, low-cost and reproducible method for obtaining phenotypic information, potentially accelerating the development of personalized medicine. The findings from previous studies have supported the hypothesis that phenotypic and proteogenomic information of the tumor can be deduced from radiologic images. [4,28,40] However, interpreting the complex associations between the biological processes and radiomics features still remains an intractable challenge. [45] On the one hand, the biological processes involve multiple interacting components. On the other hand, it is difficult to correlate a single radiomics-based factor with a pathophysiological basis in an intuitive method. Therefore, the construction of multifactor panels is a more common approach for outcome estimation in the “-omics” setting. [4,28,40] Radiomics refers to the comprehensive quantification of tumor phenotypes by applying a large number of quantitative image features. [13] According to the radiomics hypothesis, the genomic heterogeneity may translate to expression in an intratumoral heterogeneity that can be assessed through imaging and that would ultimately exhibit worse prognosis. [9,10,21,29] The radiogenomics analysis by Aerts et al. revealed that a prognostic radiomic signature, capturing intratumour heterogeneity, is associated with underlying gene expression patterns. [1] As demonstrated in the present study, the multifeature radiomics signature effectively predicts survival outcomes, which also supports the hypothesis that the radiomics signature has the potential to capture intratumoral heterogeneity in a noninvasive method. [1,9,27,29,37] In oncology, radiogenomics enable a deeper characterization and understanding of tumor biology in its entirety, including capturing the intrinsic tumor heterogeneity that can drive tumor development. [32,39] Further effort in radiogenomics is needed to elucidate the relationship between tumor genomics characteristics and their imaging appearance as well as develop imaging biomarkers incorporating phenotypic and genotypic metrics that can predict risk and outcomes, thereby better stratifying patients for more precise therapeutic care. [11,32,39]
The complex nature and biological processes of malignancy involve multiple interacting components, which may be better reflected when one takes into account the interactions between different features. [[22], [23], [24], [25],30,37] As the first attempt, to the best of our knowledge, to address the issue of survival estimation using a multicomponent radiomics signature in GC, our study supported the suggestion that multiple variables could provide a more statistically robust approach. [34,35] Although the selected 19 features were observed to be associated with DFS and OS (Figs. S16–17), the radiomics signature developed by the Lasso-Cox regression model showed better prognostic value than any single feature (Fig. S18).
Current guidelines recommend chemotherapy as a standard component for advanced GC therapies, whereas existing studies have shown that a subgroup of patients does not benefit from present chemotherapy strategies. Thus, the accurate identification of subgroups of patients will improve the prognostic system and lead to more personalized therapy. A few studies have assessed the potential of texture analysis for treatment response assessment. Ahn et al. showed that CT texture analysis is useful for the prediction of therapeutic response after cytotoxic chemotherapy in patients with liver metastasis from colorectal cancer. [2] Yoon et al. reported that heterogeneous texture features on CT images are associated with better survival in patients with HER2-positive advanced gastric cancer who received trastuzumab-based treatment. [51] In the present study, we showed that chemotherapy provides a better survival benefit to stage II and III GC patients classified as high radiomics score and that further use of the radiomics score enables a more accurate identification of patients who might benefit from chemotherapy. For patients with low radiomics scores, more effective systemic approaches to improve treatment outcomes need to be identified. Thus, the radiomics score is both a prognostic and predictive tool in stage II and III disease. Thus, patients with higher radiomics scores have a greater likelihood of recurrence and a clear benefit from chemotherapy. The mechanism of the relationship between radiomic features and chemotherapy has not been shown thoroughly, but it may be associated with the strong correlation between intratumor heterogeneity radiomic features and cell cycling pathways [1], and further radiogenomics studies may provide additional information and strategies for treatment. [31]
TNM staging is the most commonly used system to predict outcome for GC patients. However, patients within the same stage show different genetic, cellular, and clinicopathological characteristics, and their survival is not uniform.(Cancer Genome Atlas Research, [7]; [33,41]) As shown in the present study, the radiomics signature successfully identified high-risk patients with poor survival outcomes within stages I, II, III, and IV, for whom more intensified treatment was needed. These results suggested that the radiomics signature reinforced the prognostic ability of TNM stage, thereby adding prognostic value to TNM staging. To provide a more individualized staging system, nomograms have been developed to evaluate a large number of significant clinicopathological predictors to better predict the prognosis of individual patients. The improved prediction of individual outcomes would be useful for counselling patients, personalizing treatment, and scheduling patients' follow-ups. Although there are several GC nomograms available, no particular nomogram has been used widely in clinical settings. [17,18,50] In the present study, we developed and validated two nomograms, which included the radiomics signature, differentiation status, T stage, N stage, M stage, and CA199 level, to improve the accuracy of prognosis for GC patients. These nomograms can be used to better predict an individual patient's probability of 1-, 3-, and 5-year DFS and OS. The nomograms performed well with a higher C-index and positive NRI (P < 0.05). The decision curve analysis demonstrated that the radiomics nomogram was superior to both the clinicopathological nomogram and TNM staging system across the majority of the range of reasonable threshold probabilities, which indicated that the radiomics signature added incremental value for individualized estimation. Compared to previous studies, the radiomics nomograms greatly improved accuracy by integrating the radiomics signature. However, the Bland-Altman type plots showed that the radiomics nomograms didn't have a excellent predictive effect for actual survival <20 months (Fig. S11). It is unclear what is the reason for these large discrepancies for short survival times. One possible explanation is the difference in patient populations. The staging of short survival cases was generally later. These patients' survival time was easily affected by the surrounding environment in China, such as treatment condition, the economy condition, and rural/urban. [22] For example, patients with stage III GC were more likely to obtain greater survival benefit from chemotherapy than stage I or II patients with relatively longer survival, and stage III patients were more likely to receive chemotherapy than stage II patients. [22,25] The discrepancies also suggested that a summary performance metric like concordance does not tell the entire story, more reasonable methods for evaluation of prediction model need to be developed in furture.
The limitations of the present study included the relatively small sample size, and the retrospective nature of the data collection. Although the preferred design would be a prospective longitudinal cohort study, [34] the protracted length of a prospective longitudinal cohort study in GC (on account of the long wait required for survival outcomes) may make the study daunting. [36] Although a large-scale independent prospective multicenter validation cohort is warranted to evaluate the generalizability of the results, the decision curve analysis used in this study, which enables the evaluation of clinical relevance without the necessity for additional validation data in a traditional decision analytic approach, [46,47] demonstrated that the radiomics signature and radiomics nomogram hold great potential for clinical application in postoperative outcome estimation. Furthermore, the use of adjuvant chemotherapy was not within a randomized comparison, and the decision to treat or not to treat patients after surgery was made by the patients and/or clinicians. Therefore, we will develop a multicenter, prospective study to validate these results in a larger population in the future. In addition, other predictive biomarkers may be included to improve the accuracy of the nomograms.
In summary, the radiomics signature can effectively predict survival and add prognostic value to the TNM staging system. Moreover, the radiomics signature may be a useful predictive tool to predict patient benefit from chemotherapy. In addition, the radiomics nomogram may serve as a potential tool to guide individual care.
Funding
This work was supported by grants from: National Natural Science Foundation of China81672446. Natural Science Foundation of Guangdong Province, 2014A030313131. Public welfare in Health Industry, National Health and Family Planning Commission of China (201402015, 201502039). Key Clinical Specialty Discipline Construction Program. Director's Foundation of Nanfang Hospital, 2016B010.
Disclosures of potential conflicts of interest
The author(s) indicated no potential conflicts of interest.
Authors' contributions
Guarantor of the article
Guoxin Li, Yikai Xu, Zhiwei Zhou, Jiang Yu.
Specific author contributions
Conception and design: Guoxin Li, Yikai Xu, Zhiwei Zhou, Yuming Jiang, Tuanjie Li, Xuefan Zha.
Collection and assembly of data: Yuming Jiang, Chuanli Chen, Jingjing Xie, Wenbing Lv, Wei Wang, Hao Chen, Yanfeng Hu, Jiang Yu.
Data analysis and interpretation: Yuming Jiang, Chuanli Chen, Xuefan Zha, Jiang Yu.
Manuscript writing: All authors
Final approval of manuscript: All authors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.007.
Contributor Information
Jiang Yu, Email: balbc@163.com.
Zhiwei Zhou, Email: zhouzhw@sysucc.org.cn.
Yikai Xu, Email: Yikaivip@163.com.
Guoxin Li, Email: gzliguoxin@163.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Aerts H.J., Velazquez E.R., Leijenaar R.T. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn S.J., Kim J.H., Park S.J., Han J.K. Prediction of the therapeutic response after FOLFOX and FOLFIRI treatment for patients with liver metastasis from colorectal cancer using computerized CT texture analysis. Eur J Radiol. 2016;85:1867–1874. doi: 10.1016/j.ejrad.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Altazi B., Fernandez D., Zhang G., Biagioli M., Moros E. Prediction of Cervical Cancer Treatment Response using Radiomics Features based on F18-FDG Uptake in PET Images. Med Phys. 2015;42:3326. [Google Scholar]
- 4.Banerjee S., Wang D.S., Kim H.J. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang Y.J., Kim Y.W., Yang H.K. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379:315–321. doi: 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 6.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coroller T.P., Grossmann P., Hou Y. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davnall F., Yip C.S., Ljungqvist G. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Into Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehn M., Nardini C., Wang D.S. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellingson B.M. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep. 2015;15:506. doi: 10.1007/s11910-014-0506-0. [DOI] [PubMed] [Google Scholar]
- 12.Giganti F., Antunes S., Salerno A. Gastric cancer: texture analysis from multidetector computed tomography as a potential preoperative prognostic biomarker. Eur Radiol. 2017;27:1831–1839. doi: 10.1007/s00330-016-4540-y. [DOI] [PubMed] [Google Scholar]
- 13.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: Images are more than Pictures, they are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonen M., Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965–970. [Google Scholar]
- 15.Group G, Paoletti X., Oba K. Benefit of adjuvant chemotherapy for resectable gastric cancer: a meta-analysis. JAMA. 2010;303:1729–1737. doi: 10.1001/jama.2010.534. [DOI] [PubMed] [Google Scholar]
- 16.Guo X., Liu Y., Huang X. 2017. Serum relaxin as a diagnostic and prognostic marker in patients with epithelial ovarian cancer. Cancer biomarkers: Section a of Disease markers 21; pp. 81–87. [DOI] [PubMed] [Google Scholar]
- 17.Han D.S., Suh Y.S., Kong S.H. Nomogram predicting long-term survival after d2 gastrectomy for gastric cancer. J Clin Oncol. 2012;30:3834–3840. doi: 10.1200/JCO.2012.41.8343. [DOI] [PubMed] [Google Scholar]
- 18.Hirabayashi S., Kosugi S., Isobe Y. Development and external validation of a nomogram for overall survival after curative resection in serosa-negative, locally advanced gastric cancer. Ann Oncol. 2014;25:1179–1184. doi: 10.1093/annonc/mdu125. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y., Liu Z., He L. Radiomics Signature: a potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology. 2016;281:947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y.Q., Liang C.H., He L. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol. 2016;34:2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 21.Jackson A., O'Connor J.P., Parker G.J., Jayson G.C. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clin Cancer Res. 2007;13:3449–3459. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y., Li T., Liang X. Association of Adjuvant Chemotherapy with Survival in patients with Stage II or III Gastric Cancer. JAMA Surg. 2017;152 doi: 10.1001/jamasurg.2017.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y., Liu W., Li T. Prognostic and Predictive Value of p21-activated Kinase 6 Associated support Vector Machine Classifier in Gastric Cancer Treated by 5-fluorouracil/Oxaliplatin Chemotherapy. EBioMedicine. 2017;22:78–88. doi: 10.1016/j.ebiom.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y., Xie J., Han Z. Immunomarker support vector machine classifier for prediction of gastric cancer survival and adjuvant chemotherapeutic benefit. Clin Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-18-0848. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y., Zhang Q., Hu Y. ImmunoScore Signature: a Prognostic and Predictive Tool in Gastric Cancer. Ann Surg. 2018;267:504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 26.Kashani-Sabet M., Nosrati M., Miller J.R., 3rd Prospective Validation of Molecular Prognostic Markers in Cutaneous Melanoma: a Correlative Analysis of E1690. Clin Cancer Res. 2017;23:6888–6892. doi: 10.1158/1078-0432.CCR-17-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar V., Gu Y.H., Basu S. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo M.D., Gollub J., Sirlin C.B., Ooi C., Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vascular Interventional Radiol. 2007;18:821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 29.Lambin P., Rios-Velazquez E., Leijenaar R. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T.J., Jiang Y.M., Hu Y.F. Interleukin-17-Producing Neutrophils link Inflammatory Stimuli to Disease Progression by Promoting Angiogenesis in Gastric Cancer. Clin Cancer Res. 2017;23:1575–1585. doi: 10.1158/1078-0432.CCR-16-0617. [DOI] [PubMed] [Google Scholar]
- 31.Mankoff D.A., Farwell M.D., Clark A.S., Pryma D.A. How imaging can impact clinical trial design molecular imaging as a biomarker for targeted cancer therapy. Cancer J. 2015;21:218–224. doi: 10.1097/PPO.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 32.Mazurowski M.A. Radiogenomics: what it is and why it is important. J Am College Radiol. 2015;12:862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 33.McLean M.H., El-Omar E.M. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664–674. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- 34.Moons K.G., Altman D.G., Reitsma J.B. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 35.Moons K.G., Kengne A.P., Woodward M. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98:683–690. doi: 10.1136/heartjnl-2011-301246. [DOI] [PubMed] [Google Scholar]
- 36.Noh S.H., Park S.R., Yang H.K. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-year follow-up of an open-label randomised phase 3 trial. Lancet Oncol. 2014;15:1389–1396. doi: 10.1016/S1470-2045(14)70473-5. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor J.P., Aboagye E.O., Adams J.E. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169–186. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pencina M.J., D'Agostino R.B., Sr., Steyerberg E.W. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinker K., Chin J., Melsaether A.N., Morris E.A., Moy L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology. 2018;287:732–747. doi: 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- 40.Segal E., Sirlin C.B., Ooi C. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 41.Tan P., Yeoh K.G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149(1153–1162) doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 42.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc B. 2011;73:273–282. [Google Scholar]
- 44.Torre L.A., Bray F., Siegel R.L. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 45.Tran B., Dancey J.E., Kamel-Reid S. Cancer genomics: technology, discovery, and translation. J Clin Oncol. 2012;30:647–660. doi: 10.1200/JCO.2011.39.2316. [DOI] [PubMed] [Google Scholar]
- 46.Vickers A.J., Cronin A.M., Elkin E.B., Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. Medical decision making: an international journal of the Society for. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wadhwa R., Song S., Lee J.S. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M. ImmunoScore predicts gastric cancer postsurgical outcome. Lancet Oncol. 2017;18 doi: 10.1016/S1470-2045(17)30008-6. [DOI] [PubMed] [Google Scholar]
- 50.Woo Y., Son T., Song K. A Novel Prediction Model of Prognosis after Gastrectomy for Gastric Carcinoma: Development and Validation using Asian Databases. Ann Surg. 2016;264:114–120. doi: 10.1097/SLA.0000000000001523. [DOI] [PubMed] [Google Scholar]
- 51.Yoon S.H., Kim Y.H., Lee Y.J. Tumor Heterogeneity in Human Epidermal Growth factor Receptor 2 (HER2)-positive Advanced Gastric Cancer Assessed by CT Texture Analysis: Association with Survival after Trastuzumab Treatment. Plos One. 2016;11 doi: 10.1371/journal.pone.0161278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J.X., Song W., Chen Z.H. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material