Abstract
Imaging mass spectrometry provides a powerful tool for monitoring and discovery of molecular processes in the spatial domain in tissues for research and practical applications in both biology and medicine. This technology directly measures molecular compounds in tissues without the use of target-specific reagents such as antibodies, is applicable to a wide variety of analytes, and can provide spatial resolutions below the single cell level. Importantly, it has paradigm shifting capabilities in clinical applications, especially for anatomic pathology.
Keywords: Image fusion, Molecular analysis, Technology, Tissue imaging mass spectrometry
The last few decades have seen the gathering of an enormous bounty of molecular information concerning the intricacies of the living cell and the organization and specialization of these cells in plants and animals. The acquisition of transformative, paradigm-altering information typically follows the introduction of enabling technologies that allow us to look deeply into the molecular makeup of living systems. Imaging technologies have served science well over many decades because they are exquisitely suited to facilitate human interpretations and understandings of complex molecular systems. One can only marvel at the plethora of imaging technologies that pervade both our private lives and our professional lives, in the latter case, particularly in the basic and applied sciences. One relative newcomer to this field is imaging MS. Although the origins of the use of MS to generate atomic and molecular images goes back several decades, it is only in the past 10–20 years that MS has been shown to be a powerful tool to significantly help unravel the molecular complexities of living systems.
There are many different types of MS technologies that have been reported to find use in producing elemental and molecular images of a sample [1]. Perhaps the oldest of these is secondary ion MS (SIMS) first used to produce elemental images. Today, new ion and atomic and molecular cluster bombardment guns in use in SIMS can routinely produce analyte ions from a variety of biological samples of up to about 1 kDa. Likewise, there are many other types of ionization methods that have been reported to be useful in imaging applications, such as laser desorption ionization (LDI), matrix assisted laser desorption/ionization (MALDI), desorption electrospray ionization (DESI), laser ablation ESI (LAESI), and others [2]. All of these technologies have value and can be shown to have special advantage in some specific applications that best utilizes their capabilities. However, the one technology that stands out for its general effectiveness for imaging of samples from the biological and medical world is MALDI imaging MS [3].
The object of this article is to summarize the incredible advances in the instrumentation and exploding applications of imaging in a world that is deeply focused on understanding health and disease at the molecular level. Obviously, all of the published applications worldwide and the different problems addressed by investigators using various imaging MS approaches cannot be adequately addressed in this brief introductory article. Instead, this article will highlight several areas of work that represent major points of focus for the technology and areas of high impact in the near future. Examples are chosen primarily from the author’s laboratory to illustrate the issues addressed, that is, where the technology is today in terms of enabling the discovery of unknown biology and what its potential is in the near future. This article is not a review of the field, and the work of many laboratories engaged in image analysis by MS is acknowledged. Similarly, the present article is not intended to review various ionization modes and instrumentation platforms. The reader is directed to several recent reviews for information of this kind [1,2].
MALDI imaging MS, here simply abbreviated as IMS, has been shown to have wide application to biological samples for the analysis of metabolites, lipids, peptides, and proteins in tissue samples where molecular mapping is a key task [4]. Early work in this field was somewhat laborious with manual operation of the instrumentation and the use of relatively slow laser technology having a repetition laser at 20 Hz so that the production of an image could take many hours even when imaging even a small piece of tissue. Today, much of sample preparation steps and new instrumentation are highly automated and employ lasers with repetition rates of 10 KHz, leading to very fast image generation. The basic technology is relatively simple: for tissue analysis, a cryostat section is placed on the target (a glass slide, an ITO (indium tin oxide)-coated glass slide or stainless-steel target), robotically coated with a matrix and placed into the source of the mass spectrometer. In a modern TOF mass spectrometer, data acquisition for the analysis of a sagittal mouse brain section would take only a few minutes at a spatial resolution of 10–15 μm. This single data file can then produce several thousand images at each m/z value recorded.
IMS is well suited for the effective mapping of biological compounds in tissue sections for several reasons [1]. It has a wide scope of application among most types of biological molecules, including metabolites, lipids, low-MW carbohydrates, peptides, and proteins, as well as drugs and other xenobiotics [2]. High spatial resolution of (1–<30 μm) can be achieved and is compatible with instruments with high resolving power such as FTICR MS while still maintaining reasonable sensitivity [3]. Sample preparation is straightforward in terms of its compatibility with robotics to coat the tissue sections with matrix compounds [4]. Modern instruments are exceedingly fast, some being equipped with 10 kHz repetition rate lasers and incorporating advanced digital signal acquisition technology. For example, tissue microarrays can hold 100 or more biopsy cores that are typically 1–2 mm in diameter and modern TOF instruments can acquire sufficient information from each core in only a few seconds in order to image that core [5]. The technology is superb for discovery since no target specific reagents are needed, such as antibodies. Finally, [6] reagent costs for the analysis are exceedingly low, amounting to less than a dollar per sample in most cases.
1. Mapping molecules in biology
A great deal has been learned of molecular interactions and metabolic pathways in biology over the years and this work has led to the construction of interaction maps often termed interactomes. These data represent just a fraction of the interactions taking place in the living cell and clearly show the extraordinary complexity of the task of fully understanding these interactive processes. Further, interactomes maps commonly do not account for temporal and spatial cellular processes, that is, that many molecular interactions do not occur at the same time in the life cycle of a cell or in a tissue spatially in the same cell type. This complexity underscores the need for analytical technology that directly measures cellular compounds on a spatial basis. IMS brings such a molecularly specific tool to research that can provide an unbiased view of many cellular compounds through direct analysis, taking advantage of the unique molecular specificity of MS.
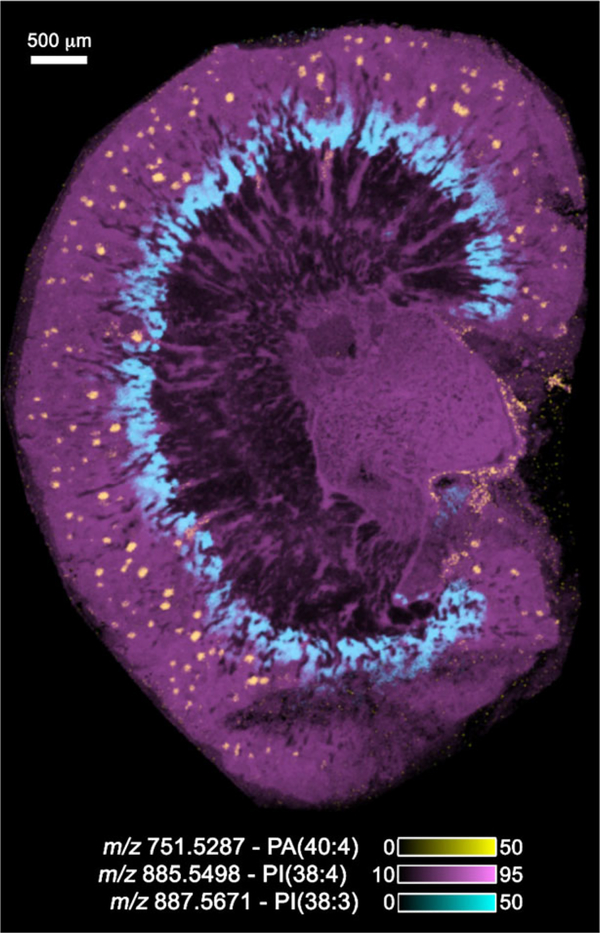
For example, in basic research studies in a mouse model of diabetes, the discovery and identification of advanced glycated end products (AGEs) in diabetic nephropathy were the focus of imaging experiments of the kidney [5, 6]. Damage to the kidney can result from oxidative alterations in essential lipids and proteins induced by hyperglycemia. Reactive carbonyl species such as glyoxal, methylglyoxal, and ROS and even glucose itself can react with certain lipids and amino acid residues in proteins to alter their structure and function. This study reported over 50 AGE modifications of phosphatidylethanolamine and lysyl side chain amino groups. Figure 1 shows the overlay of three lipid images taken from a raster of a mouse kidney section at 15 μm spatial resolution using a 15T solariX FTICR MS. The images were obtained in the negative ion mode at a mass resolving power of 100 000 at m/z 700 and contained a total of 126 509 pixels. Lipid IDs were tentatively assigned through mass accuracy measurements and data comparison with that in the “LipidMaps” database [7]. The figure clearly displays the inner and outer cortices and the medulla of the kidney section with the glomeruli shown in yellow in the figure using the ion at m/z 751.5287 (phosphatidic acid (40:4)) measured with a mass accuracy of 0.69 ppm. The image file acquired from a single raster of the kidney section produced several thousand images at unique m/z values and had a total size of about1TB.
Figure 1.
The IMS image of a mouse kidney taken on a Bruker 15T solariX FTICR MS at a spatial resolution of 15 μm in negative ion mode and at a mass resolving power of 100 000 at m/z 700. The image contains a total number of pixels of 126 509. Tentative lipid IDs were assigned based on exact mass on comparison with data in LipidMaps.
2. Multimodal image analysis and image fusion
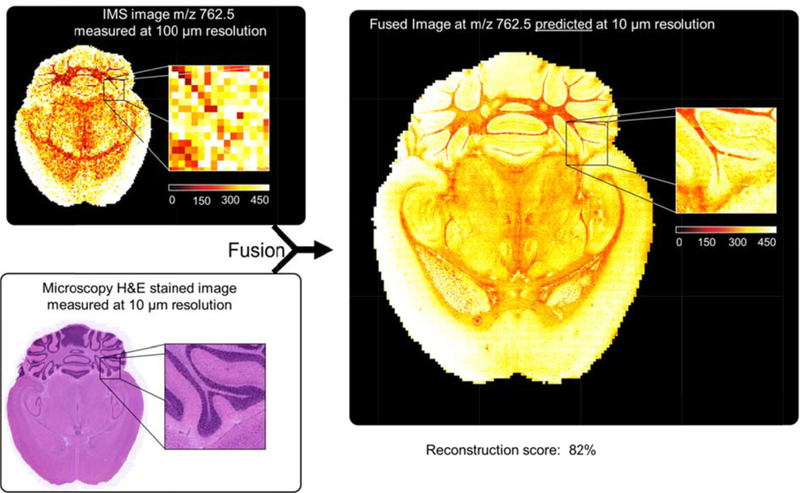
One of the exciting aspects of imaging in general is the potential for image fusion, the process whereby two images are mathematically coordinated pixel by pixel to give a predicted image that embodies the best aspects of each modality. This concept is not new and perhaps is best illustrated from its use in satellite and high-altitude ground image analysis. In this application, two images are fused, one taken in black and white (high spatial resolution but low dimensionality with shades of gray) and the other in color (lower spatial resolution but high dimensionality with millions of colors). The output is a predicted color image having high spatial resolution. In a recent paper [8], this process was applied for the first time to the fusion of IMS and microscopy. The images from IMS have a high level of molecular information but do not have the high-quality spatial resolution found in microscopy. The opposite is true for microscopy, that is, it has low molecular specificity but high spatial resolution. The resulting fused images thus leverage the advantages of both technologies, enabling prediction of a molecular distribution both at high spatial resolution and with high chemical specificity. Rather than a simple overlay of images, the image fusion process uses multivariate regression to model variables in one technology, using variables from the other technology. For example, one of the applications of the approach that benefits the IMS image is image sharpening. Basically, the IMS image can leverage the higher resolution of microscopy and become much sharper and have finer in detail than what is possible from normal image construction. The upside for microscopy is that molecular distributions not inherent in the technology can be integrated into the microscopy image. A specific example of this is shown in Fig. 2, where the higher spatial resolution microscopy image of a mouse brain section (lower left panel) and a lower spatial resolution IMS image (upper left panel) are fused to predict a brain section having both high spatial resolution and hundreds of molecular distributions (e.g., one predicted m/z image is shown in panel on the right).
Figure 2.
A predicted image (right hand panel) generated through the fusion of a microscopy image at 10 um spatial resolution (lower left panel) and an IMS image (upper left panel) taken at 100 μm spatial resolution. (Figure modified from Reference 8, figure 3)
The fusion process also brings several other advantages to image processing besides sharpening such as the prediction of ion distributions in tissue areas that were not measured by IMS and the enrichment of biological signals with the identification of instrumental artifacts that are not easily observed from either microscopy or IMS. The mathematics involved in the fusion process and detailed discussion of these as well as other examples were described in the published work [8].
3. Ultrahigh spatial resolution
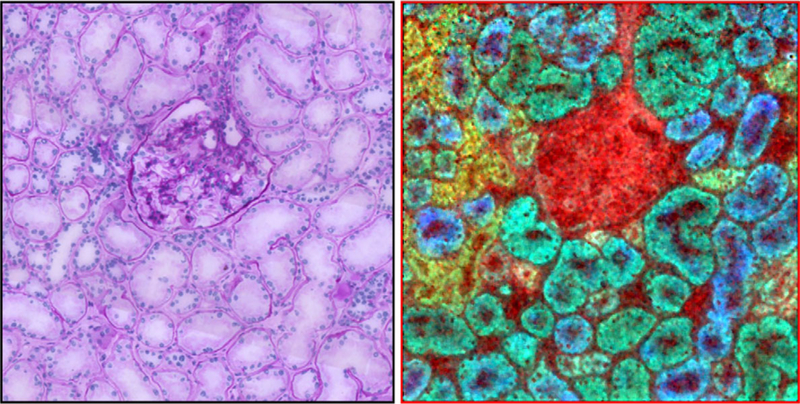
IMS technology today in terms of routine spatial resolution achieved by commercial instruments is in the 10–30 μm range. These instruments use a laser beam or other irradiation beam that impacts the front side of the sample and molecules/ions are ejected in what is geometrically analogous to a reflection-type phenomenon. The fundamental problem is that in order to get a laser beam focused to 1 μm or less, a high-power objective lens is needed that has less than 1 cm in focal length. Thus, the laser optics shares the same physical space in the ion source of the mass spectrometer as does the ion optics. Although some reports have offered some potential solutions, recent work has shown a novel solution in utilizing a transmission geometry source whereby the laser beam enters the source from the backside, penetrates through a glass slide, and ejects ions from the sample on the surface of the slide, a process of linear ablation. This type ion source geometry has been shown to achieve 1 μm [9] and 700 nm spatial resolution (unpublished data). Figure 3 shows an overlay of three ion images obtained from a section of mouse kidney showing taken at 12 μm spatial resolution using a transmission geometry ion source showing the glomerulus with surrounding proximal and distal tubules. The nuclei of the cells in the tubules are seen as dark dots in this image. In terms of laser spot size on target, further gains in spatial resolution can be expected at least down to the wavelength of the laser used. Although this is an exciting technical development, it is important to note that the limiting factor for routine use of very high spatial resolution images will be sensitivity and one’s ability to ionize and record exceedingly small numbers of molecules at <<1 um ablation dimensions in intact cells. This limitation is common to all MS-based imaging approaches.
Figure 3.
IMS ion overlay image of three distinct molecular entities in a section of human kidney (right panel) showing the glomerulus in the center surrounded by proximal and distal tubules. The image was taken at 1 μm spatial resolution using a transmission geometry ion source. An H&E-stained microscopy image is shown for comparison (left panel). The color codes for the IMS image are as follows: red (m/z 750; PE(P-38:4)), yellow (m/z 863; PI (36:1)), green (m/z 885; PI(38:4)), and blue (m/z 1052; SM3 (d18:1/24:0)). (Figure reprinted with permission from Reference 6, Grove thesis)
4. Enhancing diagnostic accuracy in the clinic
Perhaps, the most impactful application of IMS now and in the future is in the medical and clinical arena [10]. Medicine is well into a molecular age for diagnosis, prognosis, and assessment of treatment but there remain areas within this arena that would greatly benefit from the application of advanced IMS. Anatomic pathology is an example of a discipline whereby the direct imaging of a tissue biopsy could be performed yielding spatial distributions of proteins, peptides, lipids, and xenobiotics without the use of antibodies or other target specific reagents. This would allow multiplex signatures of disease to be employed that might entail 10–20 or more biomarkers and significantly improve both sensitivity and specificity. Several examples of the use of IMS for diagnosis of disease have been recently published [11,12].
One example of the application of IMS addressed the diagnosis of Spitzoid neoplasms. These are melanocytic lesions that can be benign or can range in severity to malignant melanomas. The current practice for differentiating benign and metastatic lesions is histopathologic examination. However, for many lesions the diagnoses are not clear and these are typically labeled as “atypical.” Further complicating the diagnosis is the human element in that different pathologists with different experiences can disagree on the diagnosis even when using well-established protocols. In the work cited [12], IMS was used for the histology-directed analysis of formalinfixed paraffin-embedded tissue biopsies in order to assess molecular differences between benign and metastatic lesions. In this approach, pathologists mark spots to be queried on the tissue section using IMS technology. A validated disease specific molecular signature is used for the query based on the analysis of a cohort of patient’s biopsies from a previously analyzed training set of biopsies. From the training set, statistical analysis of the mass spectra identified five peptide markers that distinguished benign from metastatic lesions. The published study encompassed 25 biopsies in the training set and 30 different biopsies in the test set. Mass spectra were taken of areas selected by a pathologist and examined for the presence of the validated biomarkers and then this spot was mapped back to a scanned microscopic image of the tissue. The areas ablated are approximately 200 μm in diameter and the pathologist can assay any number of these spots throughout the biopsy. The results of the test set showed a 97% classification accuracy for benign lesions and 90% for metastatic lesions, a result far better than that obtained from histopathology alone. Further, patient follow-up data showed that none of the lesions classified as benign by IMS recurred or presented as metastatic many years following biopsy.
5. Conclusion and outlook
The concept of a molecular microscope to enable direct molecular analysis of biologicals has been in the minds of both basic scientists and clinical investigators for many decades. However, only in the past decade has the instrumentation been sufficiently advanced so that this concept could become a routine reality. The issue of the technology being routine is ever critical since use by biologists, clinicians, or others not expert in the technical aspects of IMS technology is a key factor in enabling its common use. Enormous improvements in the availability of high repetition rate lasers, electronics capable of acquiring 10 000 mass spectra per second, instruments capable of ion storage and accumulation, and sample preparation protocols to provide high spatial resolution images, and many other innovations have provided the key elements for the production of a highly capable MS-based molecular microscope. The benefits such an instrument brings in today’s molecular age in science are exceptional. First, IMS provides a map of molecules within tissues through direct analysis and is capable of subcellular resolution. Second, it is ideal for discovery since it does not require a target specific reagent such as antibodies. Thus, the specificity is direct and is not complicated by nonspecific interactions. Third, it is extremely rapid—modern MALDI TOF instruments are equipped with high repetition rate lasers and the required high-speed electronics so that a mouse brain image file, containing thousands of individual m/z images, can be acquired in 5 min or less. This also has great importance in projects with extremely high numbers of samples where the necessary data from a 1 mm core biopsy, for example, can be acquired in only a few seconds. Finally, IMS produces images—a medium well accepted in all areas of science and one that lends itself to be correlated and, where advantageous, to be fused with other imaging modalities such as microscopy of many types, MRI, PET imaging, and many others.
The future for IMS is exceedingly bright with applications reaching nearly every discipline where the disposition of molecules within the sample is key to understanding the underlying biology or disease diagnosis. The technology is still evolving in areas including higher spatial resolution, increased sensitivity, robotics for easier sample preparation, more facile molecular identifications, and other areas of technical development. IMS brings the unique advantage of molecular specificity that is second to none, ensuring it a prominent role in both basic science and clinical research and in clinical diagnostic laboratories as we go forward in our quest to one day eliminate the cause and effect of disease in our population.
Acknowledgments
The author thanks the NIH for grant P 41 GM 103391 support and acknowledges Jeffrey Spraggins for the FTICR MS data, Andre Zavalin and Kerri Grove for the transmission geometry data, Raf Van de Plas for assistance with data processing and image fusion data, and Jeremy Norris for help in preparing this article.
Footnotes
The author have declared no conflict of interest.
6 References
- [1].Liam A, McDonnell LA, Heeren RM, Imaging mass spectrometry. Mass Spectrom. Rev 2007, 26, 606–643. [DOI] [PubMed] [Google Scholar]
- [2].Gode D, Volmer D, Lipid imaging by mass spectrometry – a review. Analyst 2013, 138, 1289. [DOI] [PubMed] [Google Scholar]
- [3].Caprioli RM, Farmer TB, Gile J, Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem 1997, 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- [4].Caprioli RM, Imaging mass spectrometry: enabling a new age of discovery in biology and medicine through molecular microscopy. J. Am. Soc. Mass Spectrom 2015, 26, 850–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grove KJ, Voziyan PA, Spraggins JM, Wang S et al. , Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J. Lipid Res 2014, 55, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grove K, Ph.D. thesis, Imaging Mass Spectrometry for the Elucidation of Lipid and Protein Changes in Diabetic Neohropathy and Assessment of Drug Efficacy, Department of Chemistry, Vanderbilt University, 2014. [Google Scholar]
- [7].Fahy E, Subramaniam S, Murphy R, Nishijima M et al. , LIPID MAPS comprehensive classification system for lipids. J. Lipid Res 2009, 50, S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van de Plas R, Yang J, Spraggins J, Caprioli RM, Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nat. Methods 2015, 4, 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zavalin A, Yang J, Hayden K, Vestal M et al. , Tissue protein imaging at 1 m laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal. Bioanal. Chem 2015, 8, 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aichler M, Walch A, MALDI imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Lab. Invest 2015, 95, 422–431 [DOI] [PubMed] [Google Scholar]
- [11].Rauser S, Marquardt C, Balluff B, Deininger S et al. , Classificatiobn of the HER 2 receptor status in breast cancer tissues by MALDI IMS. J. Proteomics Res 2010, 9, 1854–1863. [DOI] [PubMed] [Google Scholar]
- [12].Lazova R, Seeley EH, Keenan M, Gueorguieva R et al. , Imaging mass spectrometry – a new and promising method to differentiate Spitz nevi from Spitzoid malignant melanomas. Am. J. Dermatopathol 2012, 34, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]