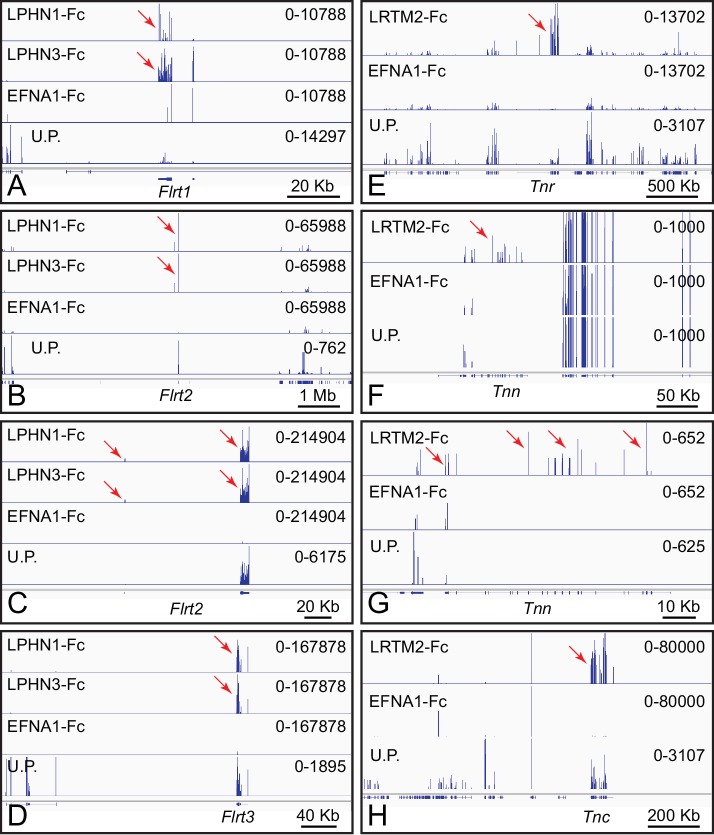
Figure 3. ACAPseq with LPHN1-Fc, LPHN3-Fc, and LRTM2-Fc baits.
(A–D) Flrt1, Flrt2, and Flrt3 mRNAs captured with LPHN1 and LPHN3 baits (red arrows). (B) and (C) are centered on the Flrt2 gene but have different horizontal and vertical scales. (E–H) Tnr, Tnn, and Tnc mRNAs captured with the LRTM2 bait (red arrows). (F) and (G) show the Tnn gene with different horizontal and vertical scales. In (H), the pile up of many reads at a single location in the LRTM2, EFNA1, and total polyribosome tracks is presumed to be a sequencing and/or alignment artifact. Genome browser images from ACAPseq of neonatal mouse brain polyribosomes, as shown in Figure 1B.