Abstract
BACKGROUND
Surveillance, Epidemiology, and End Results Cancer Registries began collecting human papillomavirus (HPV) status for upper aerodigestive tract cancers in 2010. However, classification of p16-testing was not included in Collaborative Stage coding guidelines, potentially leading to inconsistent coding.
METHODS
HPV values for Iowa oropharyngeal patients (n=824) were recoded based on modified guidelines that included p16 test results, and compared to original guidelines.
RESULTS
Forty percent of patients were recoded to a different value, and the HPV-testing rate increased from 45% to 55%; 56% received p16-testing only. Of those originally coded as HPV-type 16 (n=187), 89% were recoded to HPV-NOS. When comparing high-level positive/negative/not done categories, original coding captured 81% of HPV-positive patients.
CONCLUSIONS
p16 was the most common HPV test, but was inconsistently coded as HPV-testing. p16-positivity was also erroneously equated with HPV-type 16. Adding a separate p16 variable would improve consistency and accuracy of HPV coding.
Keywords: oropharyngeal cancer; head and neck cancer; human papillomavirus (HPV)-related cancer; Surveillance, Epidemiology, and End Results (SEER); American Joint Committee on Cancer (AJCC) staging
INTRODUCTION
Human papillomavirus (HPV) infections are the most common sexually transmitted infections in the U.S. It is estimated more than 90% of sexually active men and 80% of sexually active women will be infected with HPV in their lifetimes.(1) Approximately half of these are infected with a high-risk HPV-type, which is known to be a major risk factor for developing cancer.(1) Incidence of oropharyngeal squamous cell carcinoma (OPSCC) has been rising for the past several years, with 70% of patients testing positive for HPV-DNA.(2) Oropharyngeal cancer accounts for 78% of all HPV-associated cancers among men and 12% among women.(2)
In 2008, the National Comprehensive Cancer Network (NCCN) first suggested that HPV-testing should be performed for patients with an oropharyngeal or occult primary, because HPV-positive OPSCC patients have been shown to have a substantially higher 5-year survival (75%) compared to HPV-negative patients (14%).(3, 4) The only HPV tests recommended at that time were polymerase chain reaction (PCR) and in situ hybridization (ISH). Beginning with patients diagnosed in 2010, Surveillance, Epidemiology, and End Results (SEER) Registries have collected HPV-testing as a site-specific factor (SSF-10) for upper aerodigestive tract cancers (oropharynx, oral cavity, nasopharynx, hypopharynx, and larynx).
In 2013, the College of American Pathologists (CAP) recommended that HPV-testing be added to their standard pathologic evaluation of OPSCC and in 2016 CAP convened a panel to develop a clinical practice guideline for HPV-testing for oropharyngeal cancer patients.(5) HPV-testing is expected to be routinely performed, but no standard method of testing has been specified so test type remains at the discretion of the laboratories.(6) These tests have a variety of targets, including HPV-DNA, HPV-RNA, cellular proteins, and HPV-specific serum antibodies. For many reasons, it has become common to test for p16, a surrogate marker of HPV activity, via immunohistochemistry (IHC). The presence of p16 is highly correlated with HPV-driven squamous cell carcinoma.(7–10) The latest NCCN guidelines (2016) now suggest HPV-testing via PCR, ISH, or p16 IHC.(11)
The evolving use of p16 as a surrogate HPV test has potentially led to some inconsistencies in the interpretation of the Collaborative Stage (CS) guidelines for the HPV variable collected by cancer registrars since 2010. CS guidelines for SSF-10 mention that HPV results can be found in IHC staining in pathology reports and that p16 is the standard method of testing.(12, 13) However, guidance from the American Joint Committee on Cancer (AJCC) stated that patients tested with only p16 IHC marker should be coded as 999 (unknown).(13) Furthermore, p16-testing is not specifically mentioned in any of the code descriptions for SSF-10.
The objective of this study was to review HPV test status for all OPSCC patients to determine the following: 1) accuracy of SSF-10, 2) types of HPV-testing being performed in Iowa, 3) impact of excluding p16 from the definition of HPV-testing, and 4) patient, tumor and facility characteristics associated with HPV-testing.
MATERIALS AND METHODS
Data Source
A secondary data analysis of Iowa Cancer Registry data, extracted from their SEER Data Management System (SEER*DMS), was conducted. The Iowa Cancer Registry is one of the original nine, out of the now eighteen, population-based cancer registries that are a part of the National Cancer Institute’s SEER program. Since 1973, the Iowa Cancer Registry has been capturing cancer diagnoses among Iowans. Together, all eighteen registries provide cancer surveillance information for about 28% of the United States population. The types of information captured by these registries include patient demographics, tumor characteristics, specific cancer markers, stage at diagnosis, first course of treatment, and patient survival.(14) This information is captured from hospitals, pathology laboratories, radiology facilities, physician offices, and other facilities. Records for all eligible patients were manually reviewed to determine if HPV-testing was completed, and if so, which pathology tests were performed.
Pathology reports were sought for each case when the SEER*DMS text was unclear regarding HPV-testing. If a pathology report was not available, additional medical record information was sought from the hospital that provided the greatest amount of care. This study was granted human subject exemption status by the University of Iowa Institutional Review Board.
Study Population
We initially identified 1,064 patients with invasive or in situ upper aerodigestive tract cancer who were Iowa residents and diagnosed between 2010-2014. ICD-O-3 topographies included those listed in the CS Schema for HPV status that SEER Registries were instructed to follow, which includes: tongue base C01.9, C02.4; soft palate C05.1-C05.2; oropharynx C09.0-C09.1, C09.8-C09.9, C10.0, C10.2-C10.4, C10.8-C10.9; pharyngeal tonsil C11.1; nasopharynx C11.0-C11.3, C11.8-C11.9; hypopharynx C12.9, C13.0-C13.2, C13.8-C13.9; pharynx other C14.0-C14.2, C14.8). This dataset was then limited to patients with OPSCC (cancer of the soft palate, tongue base, and oropharynx, NOS; ICD-O-3 histologies: 8032, 8050-052, 8070-8076, 8078, 8082-8084, 8801). Patients were excluded if they were not in SEER*DMS, we were unable to discern HPV test type or results, or did not have oropharyngeal cancer or a histology of squamous cell carcinoma (N=240).
Modified Guidelines Used in Recoding HPV Status
A set of modified guidelines for determining HPV status was developed that accounted for p16-testing and, thereafter, was applied to this cohort. These values were based on the supporting text of SEER*DMS and pathology reports. If HPV-DNA testing was the only testing performed, then coding values were applied based on the existing CS SSF-10 definitions.(15) If p16-testing (IHC) was the only test performed, p16-positive patients were coded as “070: HPV positive – types not stated (NOS),” or if it was stated as high risk, then the case was coded as “060: HPV positive-high risk types, NOS.” p16-negative patients were coded as “000: negative.” If both p16-testing and HPV-DNA testing were performed, results were classified according to the HPV-DNA results.
Study Variables
Outcome variables included documentation of HPV-testing and HPV results. We conducted two separate comparisons of SSF-10 values based on different coding guidelines. In order to examine the accuracy of the HPV variable and determine the rate of HPV-testing if p16 were included as HPV-testing, we compared the original CS SSF-10 values with our modified guideline values described above. Accuracy was defined as the agreement between the original CS SSF-10 values and our modified guideline values. Next, in order to determine the impact of excluding p16-testing from the HPV-testing variable, we compared CS SSF-10 values with those recoded according to the AJCC guidance in which patients who received only p16-testing were given an HPV status value of “999: unknown”.
Patient demographic variables included age at diagnosis, race/ethnicity, sex, insurance status, marital status, and year of diagnosis. Tumor characteristics included HPV test type performed, HPV test results, CS schema, stage, grade, and cancer sequence. Treatment variables included surgery, radiation, and chemotherapy provided or planned as first-course therapy. Facility characteristics included American College of Surgeons Commission on Cancer program designation and bed size. In states with multiple SEER cancer registries, registries were combined: California includes the Greater California, Los Angeles, and Greater Bay Area (San Francisco-Oakland and San Jose-Monterey) registries; Georgia includes Atlanta, Greater Georgia, and Rural Georgia registries.
Statistical Analysis
Differences in patient, tumor, treatment, and facility characteristics were compared between patients who received HPV-testing and those who did not using Chi-square tests. Logistic regression was used to determine characteristics associated with receipt of HPV-testing. All variables listed above were considered for inclusion into the model. Analyses were conducted using SAS version 9.4 (SAS Institute, Cary, NC) and SEER*Stat version 8.3.4.(16)
RESULTS
There were 824 OPSCCs that met inclusion criteria. Based on CS guidelines, 45% (n=372) were coded as having had HPV-testing (codes 000-070 and 997). After applying our modified guidelines, 55% (n=450) had HPV-testing (Table 1). Applying our modified guidelines among all those tested, 73% (n=328) tested positive for HPV and/or p16. The majority received p16-testing only (56%), followed by p16 and HPV-DNA testing (23%) and HPV-DNA testing only (20%). Those who had p16-testing only had a higher rate of HPV-positive results (80%) compared to those who had HPV-DNA testing only (62%) or both (65%) (p<0.0001). Among those who had both p16 and HPV-DNA testing, 65% (n=68) were positive for both p16 and HPV, and 25% (n=26) had discordant results (Table 1). Discordant results refer to patients who were either negative for HPV, but positive for p16 or positive for HPV, but negative for p16.
Table 1.
HPV testing among patients with oropharyngeal squamous cell carcinoma based on recoding with modified guidelines*, Iowa, 2010-2014.
| Modified Guideline Test Results*
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Tested | All | Positive | Negative | Discordant† | ||||
|
| ||||||||
| No. of patients | % | No. of patients | % | No. of patients | % | No. of patients | % | |
| Yes | 450 | 55 | 328 | 73 | 96 | 21 | 26 | 6 |
| p16 only | 254 | 56 | 203 | 80 | 51 | 20 | – | – |
| HPV-DNA only | 92 | 20 | 57 | 62 | 35 | 38 | – | – |
| p16 + HPV-DNA | 104 | 23 | 68 | 65 | 10 | 10 | 26 | 25 |
| No | 374 | 45 | ||||||
| Total | 824 | |||||||
p16 only - if p16 (IHC) testing was the only test performed, p16-positive patients were coded as “070: HPV positive, types not stated” (n=194), or if it was stated as high risk, then the case was coded as “060: HPV positive, high-risk types, type not stated” (n=9); p16-negative patients were coded as “000: Negative” (n=51). P16 + HPV-DNA - if both p16 testing and HPV-DNA testing were performed, results were classified according to the HPV-DNA results.
Includes those patients that were tested for both p16 and HPV-DNA, but had discordant results (p16+, HPV-DNA-; p16-, HPV-DNA+).
Dashes indicate cells that are not applicable to those patients.
Abbreviations: HPV-Human papillomavirus; IHC-Immunohistochemistry
Applying our modified guidelines, 44% (n=359) of patients were recoded to a different SSF-10 value than initially documented using CS guidelines (Table 2A). Of the recoded patients, 52% (n=187) were originally coded as “030: HPV positive-type 16 only” and 70% (n=250) were recoded as “060: HPV positive-high risk types, type NOS” or “070: HPV positive-NOS.” Of the originally coded “030: HPV positive-type 16 only” patients, 89% (n=187) were recoded; 94% (n=175) of these were recoded as 060 or 070. After recoding it was found that 18% (n=83) of patients originally coded as “998: test not done” or “999: unknown” had some form of HPV-testing performed. Of these patients, 83% (n=69) received only p16-testing or a combination of p16 and HPV-DNA testing. When collapsing all of the SSF-10 values into the following three categories: HPV-positive, HPV-negative, and not tested, and then comparing the categories based on CS guidelines vs. our modified guidelines, the initial coding based on the CS guidelines accurately identified 81% of HPV-positive patients, and 68% of HPV-negative patients (Table 2B).
Table 2.
Comparison of HPV SSF-10 values initially recorded for Iowa oropharyngeal squamous cell carcinoma cases (2010-2014) using Collaborative Stage guidelines to recoded values based on our modified guidelines.
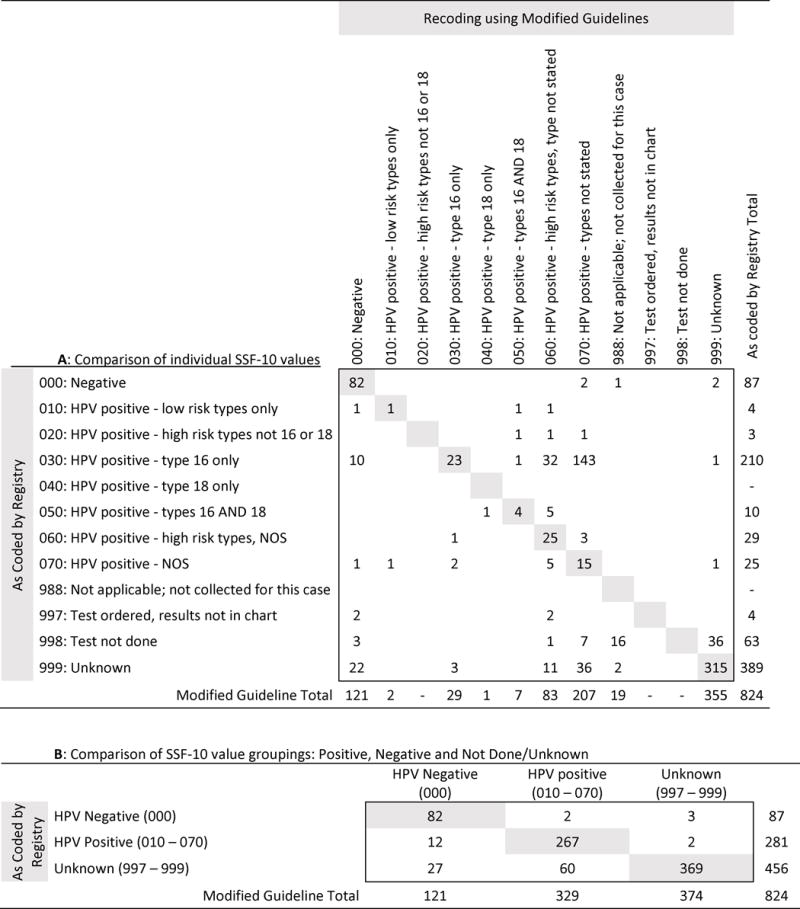
|
Numbers in gray represent HPV status values that did not change according to our modified guidelines.
Numbers in white represent HPV status values that did change according to our modified guidelines.
Blanks indicate there were no patients with that combination of SSF-10 codes.
Abbreviations: HPV-Human papillomavirus; NOS-types not stated
Recoding according to the AJCC guidance that excluded p16 as an HPV test (i.e., coding p16-testing only as “999: unknown”) resulted in 69% (n=144) of patients who were originally coded as “030: HPV positive-type 16 only” and 48% (n=42) of patients who were originally coded as “000: Negative” to be reclassified as “999:unknown” (Table 3A). Overall, based on this recoding according to the AJCC guidance, 55% (n=203) of patients who were originally tested for “HPV” were reclassified as “999: unknown” (Table 3B). This led to 76% (n=628) of the 824 cases being classified as unknown.
Table 3.
Comparison of HPV SSF-10 values initially recorded using current Collaborative Stage guidelines and recoded values based on AJCC guidance to exclude p16 from the definition of HPV testing for patients with oropharyngeal squamous cell carcinoma, Iowa, 2010-2014.
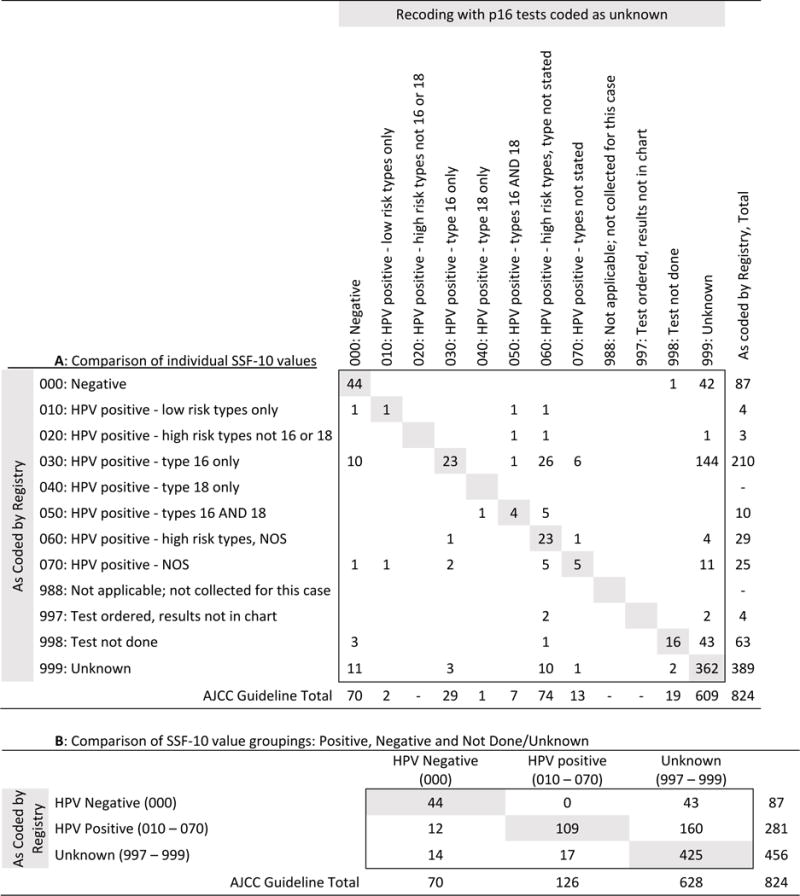
|
Excluding p16 from the definition of HPV testing (p16 patients coded as 999: Unknown)
Numbers in white are those patients whose HPV status value changed according to the AJCC guidance.
Blanks indicate there were no patients with that combination of SSF-10 codes.
Abbreviations: HPV-Human papillomavirus; NOS-types not stated
We examined characteristics associated with HPV-testing using our modified guidelines. Table 4 displays the frequencies, percentages, and p-values corresponding to Chi-square tests for patient, tumor, and treatment characteristics by HPV-testing status. Odds ratios (OR) and 95% confidence intervals (CI) are also displayed from a logistic model in which the dependent variable was HPV-testing and the independent variables included all variables listed in Table 4, except test type and test results (N=799 due to 25 patients missing values for facility characteristics). Univariate analysis demonstrated that all variables, except race/ethnicity and sex, were significantly associated with HPV-testing. HPV-testing rates varied the most by age (65% for those <50 vs. 41% 75+), diagnosis year (29% in 2010 vs. 75% in 2014), and site (60% oropharynx vs. 24% soft palate). The following variables were significantly associated with HPV-testing in the logistic model (p<0.05): later diagnosis year, cancer of the oropharynx, poorly differentiated grade (vs. well differentiated), receiving surgery, and being treated at a hospital with 500+ beds.
Table 4.
Patient, tumor, treatment, and facility characteristics by HPV testing (based on modified guidelines) and odds of receiving testing among patients with oropharyngeal squamous cell carcinoma of the head and neck diagnosed in Iowa, 2010-2014.
| HPV Tested
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| All | Yes | No | Adjusted* (N=799) | ||||||
|
|
|
||||||||
| No. of patients | %† | No. of patients | %† | No. of patients | %† | p-value | OR | CI | |
|
|
|
||||||||
| All | 824 | 100 | 450 | 55 | 374 | 45 | N/A | ||
| Patient Characteristics | |||||||||
|
| |||||||||
| Age at diagnosis | |||||||||
| <50 | 91 | 11 | 59 | 65 | 32 | 35 | 0.001 | 1.43 | (0.65, 3.14) |
| 50-64 | 437 | 53 | 254 | 58 | 183 | 42 | 1.16 | (0.61, 2.18) | |
| 65-74 | 211 | 26 | 102 | 48 | 109 | 52 | 1.03 | (0.55, 1.96) | |
| 75+ | 85 | 10 | 35 | 41 | 50 | 59 | 1.00 | Referent | |
| Race/Ethnicity | |||||||||
| White | 799 | 97 | 433 | 54 | 366 | 46 | 0.172 | 1.00 | Referent |
| Black/Other | 25 | 3 | 17 | 68 | 8 | 32 | 1.78 | (0.61, 5.16) | |
| Sex | |||||||||
| Male | 670 | 81 | 367 | 55 | 303 | 45 | 0.843 | 1.00 | Referent |
| Female | 154 | 19 | 83 | 54 | 71 | 46 | 1.28 | (0.83, 1.96) | |
| Insurance Status | |||||||||
| Private Insurance | 474 | 58 | 282 | 59 | 192 | 41 | 0.002 | 1.00 | Referent |
| Not Insured | 24 | 3 | 17 | 71 | 7 | 29 | 2.1 | (0.73, 6.07) | |
| Medicaid | 89 | 11 | 42 | 47 | 47 | 53 | 0.69 | (0.39, 1.24) | |
| Medicare | 218 | 26 | 99 | 45 | 119 | 55 | 0.76 | (0.48, 1.18) | |
| Unknown | 19 | 2 | 10 | 53 | 9 | 47 | 2.11 | (0.59, 7.60) | |
| Marital Status | |||||||||
| Married | 504 | 61 | 293 | 58 | 211 | 42 | 0.037 | 1.00 | Referent |
| Unmarried | 303 | 37 | 148 | 49 | 155 | 51 | 0.74 | (0.52, 1.07) | |
| Unknown | 17 | 2 | 9 | 53 | 8 | 47 | 0.70 | (0.18, 2.79) | |
| Diagnosis Year | |||||||||
| 2010 | 153 | 19 | 45 | 29 | 108 | 71 | <0.0001 | 1.00 | Referent |
| 2011 | 155 | 19 | 74 | 48 | 81 | 52 | 2.29 | (1.34, 3.91) | |
| 2012 | 150 | 18 | 84 | 56 | 66 | 44 | 3.46 | (2.00, 5.96) | |
| 2013 | 194 | 24 | 118 | 61 | 76 | 39 | 4.68 | (2.75, 7.96) | |
| 2014 | 172 | 21 | 129 | 75 | 43 | 25 | 11.4 | (6.35, 20.31) | |
|
| |||||||||
| Tumor Characteristics | |||||||||
|
| |||||||||
| Test Type† | |||||||||
| p16 | 254 | 31 | 254 | 56 | – | – | <0.0001 | – | – |
| HPV-DNA | 92 | 11 | 92 | 20 | – | – | – | – | |
| p16+HPV-DNA | 104 | 13 | 104 | 23 | – | – | – | – | |
| Not tested | 374 | 45 | – | – | 374 | 100 | – | – | |
| Test results (Missing=374)† | |||||||||
| Positive | 328 | 40 | 328 | 40 | – | – | NA | – | – |
| Negative | 96 | 12 | 96 | 12 | – | – | – | – | |
| Other‡ | 26 | 3 | 26 | 3 | – | – | – | – | |
| CS schema | |||||||||
| Oropharynx | 520 | 63 | 311 | 60 | 209 | 40 | <0.0001 | 1.00 | Referent |
| Soft Palate | 38 | 5 | 9 | 24 | 29 | 76 | 0.49 | (0.20, 1.23) | |
| Tongue Base | 266 | 32 | 130 | 49 | 136 | 51 | 0.61 | (0.42, 0.88) | |
| Stage | |||||||||
| 1 Localized | 67 | 8 | 25 | 37 | 42 | 63 | 0.001 | 1.00 | Referent |
| 2 Localized | 50 | 6 | 22 | 44 | 28 | 56 | 0.88 | (0.36, 2.17) | |
| 3 - Regional | 138 | 17 | 71 | 51 | 67 | 49 | 1.34 | (0.62, 2.89) | |
| 4A - Regional | 379 | 46 | 239 | 63 | 140 | 37 | 1.34 | (0.64, 2.80) | |
| 4B - Regional | 55 | 7 | 28 | 51 | 27 | 49 | 0.97 | (0.37, 2.51) | |
| 4C - Distant Metastasis | 35 | 4 | 16 | 46 | 19 | 54 | 0.82 | (0.28, 2.46) | |
| 4 NOS | 53 | 6 | 29 | 55 | 24 | 45 | 1.41 | (0.55, 3.65) | |
| Unknown | 47 | 6 | 20 | 43 | 27 | 57 | 1.03 | (0.41, 2.59) | |
| Grade | |||||||||
| Well Differentiated | 46 | 6 | 18 | 39 | 28 | 61 | <0.0001 | 1.00 | Referent |
| Moderately Differentiated | 243 | 29 | 115 | 47 | 128 | 53 | 1.25 | (0.57, 2.73) | |
| Poorly Differentiated | 370 | 45 | 241 | 65 | 129 | 35 | 2.41 | (1.12, 5.23) | |
| Undifferentiated | 15 | 2 | 11 | 73 | 4 | 27 | 4.66 | (0.88, 24.77) | |
| Unknown | 150 | 18 | 65 | 43 | 85 | 57 | 0.91 | (0.40, 2.06) | |
| Cancer sequence | |||||||||
| First cancer | 643 | 78 | 370 | 58 | 273 | 42 | 0.001 | 1.00 | Referent |
| Second or higher cancer | 181 | 22 | 80 | 44 | 101 | 56 | 0.65 | (0.42, 1.02) | |
| Surgery | |||||||||
| Yes | 345 | 42 | 215 | 62 | 130 | 38 | 0.0002 | 1.82 | (1.23, 2.68) |
| No | 479 | 58 | 235 | 49 | 244 | 51 | 1.00 | Referent | |
| Radiation | |||||||||
| Yes | 678 | 82 | 389 | 57 | 289 | 43 | 0.001 | 1.41 | (0.82, 2.43) |
| No | 146 | 18 | 61 | 42 | 85 | 58 | 1.00 | Referent | |
| Chemotherapy | |||||||||
| Yes | 560 | 68 | 327 | 58 | 233 | 42 | 0.002 | 1.41 | (0.88, 2.25) |
| No | 264 | 32 | 123 | 47 | 141 | 53 | 1.00 | Referent | |
|
| |||||||||
| Facility Characteristics | |||||||||
|
| |||||||||
| CoC Program Designation (Missing=25) | |||||||||
| Academic Comprehensive | 275 | 33.4 | 161 | 59 | 114 | 41 | 0.034 | 0.89 | (0.42, 1.90) |
| Comprehensive Community/VA | 329 | 39.9 | 184 | 56 | 145 | 44 | 1.58 | (0.92, 2.70) | |
| Community | 33 | 4 | 18 | 55 | 15 | 45 | 1.29 | (0.53, 3.14) | |
| Other or No CoC Designation | 162 | 19.7 | 72 | 44 | 90 | 56 | 1.00 | Referent | |
| Bed size (Missing=25) | |||||||||
| <100 | 78 | 9.47 | 43 | 55 | 35 | 45 | <0.0001 | 0.82 | (0.34, 1.97) |
| 100-299 | 162 | 19.7 | 69 | 43 | 93 | 57 | 0.33 | (0.17, 0.63) | |
| 300-499 | 213 | 25.9 | 101 | 47 | 112 | 53 | 0.31 | (0.18, 0.56) | |
| 500+ | 346 | 42 | 222 | 64 | 124 | 36 | 1.00 | Referent | |
Model adjusted for the following variables: age, insurance status, diagnosis year, CS schema, grade, cancer sequence, surgery, radiation, chemotherapy, CoC program designation, and bed size
Percentages are row percentages except test type and test results are reported as column percentages
Includes those patients that were tested for both p16 and HPV-DNA and had discordant results (p16+, HPV-DNA-; p16−, HPV-DNA+).
Dashes indicate that variable was not included in model.
Abbreviations: HPV-Human papillomavirus; NOS-not otherwise specified; CS-Collaborative Stage; NOS-not otherwise specified; CoC-Commission on Cancer; VA-Veterans’ Administration
When comparing the 26 patients with discordant results to the study population who had concordant p16 and HPV results (n=78), no significant differences in patient, tumor, or facility characteristics were detected (data not shown). However, there were differences in facility characteristics when looking at whether a patient received only p16-testing, only HPV-DNA testing, or received both tests. Patients who received only p16-testing more frequently went to Academic Comprehensive designated hospitals and major teaching hospitals (51% and 55%, respectively), while those who received HPV-DNA testing only or both tests more frequently went to Comprehensive Community and Veterans’ Administration designated hospitals (56% and 63%, respectively) and minor teaching hospitals (38% and 56%, respectively) (data not shown).
While our analysis focused solely on Iowa cancer patients, we wanted to assess how the Iowa registry compared to the other SEER Registries to determine the variation in HPV coding across registries. Figure 1 depicts the HPV-testing rates for OPSCC patients among the SEER-18 Registries (excluding Alaska due to their small number of patients) diagnosed from 2010 to 2014. HPV-testing rates varied from 33% to 57% (Seattle; Connecticut). While Iowa originally had a testing rate of 45%, after recoding its rates exceeded those of other registries (55%), except Connecticut and Utah (57%, 55%). Compared to other code values, “030: HPV positive-type 16 only” was the most common code for those with positive results. After recoding in Iowa, the 030 rate decreased from 26% to 4% among those with positive results, while the NOS values (060, 070) rose from 7% to 35%.
Figure 1.
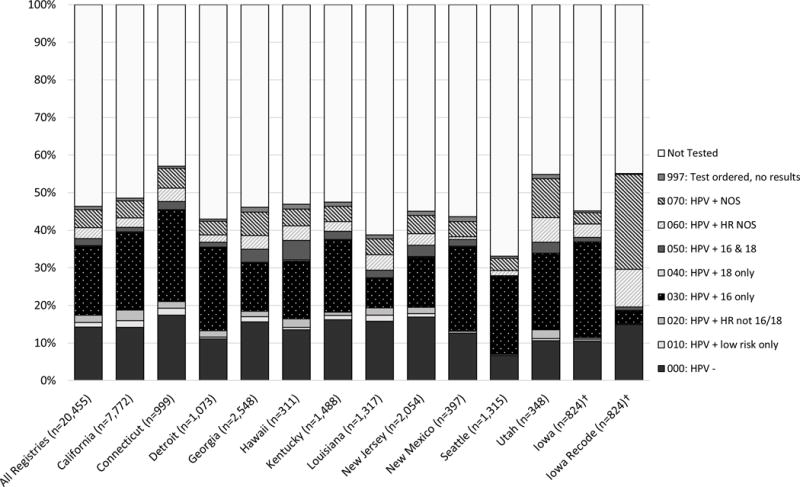
*Alaska was excluded due to small number of patients. †The Iowa data are as presented in Table 2A, while the remaining SEER Registries’ HPV SSF-10 values were derived from SEER*Stat. Abbreviations: HPV-Human papillomavirus; NOS-not otherwise specified; HR-high risk
DISCUSSION
Review of Iowa OPSCC patients revealed issues with consistency and accuracy of the HPV-testing SSF-10 variable. Consistent application of our modified guidelines, which included p16-testing, led to a substantial increase in the reported testing rate from 45% to 55% during 2010-2014. It also led to a large shift in the distribution of values from “030: HPV positive-type 16 only” to HPV NOS (060, 070), likely because registrars were erroneously equating p16-positivity with HPV-type 16. Among those assessed for HPV status, over half of Iowa patients received only p16-testing and an additional 23% received p16-testing first, followed by an HPV-DNA test. Given that p16 is the predominant test to assess HPV status in Iowa, completely excluding it as an HPV test would cause currently reported testing rates in Iowa to decline from 45% to 21%. Based on our findings, it is imperative that coding options and guidelines for SSF-10 be updated to improve the accuracy, consistency, and usefulness of this variable.
This study has identified an inconsistency in the coding of HPV status in Iowa for oropharyngeal tumors, under the CS schema used for national cancer databases including SEER and the National Cancer Database (NCDB). Though we are not aware that a similar inconsistency has previously been reported in data collected in other registries, we presume it most likely that other cancer registries would find similar results. It is difficult to estimate the effect this inconsistency in coding has on the data currently available to the public from these national databases, and the bias this introduces in observational studies. In Iowa, 18% of patients with HPV-associated tumors were not originally identified as such. This omission certainly affects epidemiologic estimates of the incidence of p16-positive oropharyngeal cancers, and decreases the available sample size of patients for inclusion in studies of HPV-associated oropharyngeal cancers. It would also affect studies that make comparisons between the HPV-positive subset and the unknown-status subset of patients. Separately and similarly, the coding of discordant p16-positive HPV-negative patients into “000: negative” also creates a systematic bias in studies that examine outcomes for HPV-negative patients. While our findings do not change how clinicians interpret patient records, clinicians should be aware of this issue when interpreting literature derived from cancer registry data.
Published studies utilizing SSF-10 to stratify outcomes demonstrate an unawareness of the inconsistency in registrar coding of p16 as a surrogate marker for HPV. For instance, a study based on NCDB data from 2009-2011 comparing HPV-negative to HPV-positive oropharynx cancers states that SSF-10 defines HPV-positive patients based on ISH tests for HPV-16 or -18, and/or positive p16 staining by IHC.(17) The definition of SSF-10 in the CS schema does not mention p16 staining by IHC. The data in this present study suggest that the assumption by Amini et al. regarding p16-testing being included is partially inaccurate, though whether this affects the study’s analysis and conclusions is difficult to judge.
For maximum clarity and completeness, the creation of a new and separate SSF for p16 (SSF-11 “p16 status”) would be ideal. This variable could easily capture if an IHC test for p16 was negative (000), positive (010), or not done/unknown (998, 999). This would be a clean and simple way to capture the p16 status of patients and subsequent code values would not have to be added to current SSFs. Given the major changes in the AJCC eighth edition staging manual in which a separate staging algorithm has been developed for HPV-associated oropharyngeal cancer based on p16 status, p16 status will be a critical prognostic for cancer registries to collect.(18)
The provision of more education around p16 in the guidelines, as well as clarification that p16 is not testing for HPV-type 16, would likely be beneficial in assisting registrars to code this variable accurately and consistently. The SSF-10 code distribution among the Registries displayed in Figure 1 suggests that staff in other registries were also erroneously classifying p16-positive as HPV-type 16 positive, as the 030 code was the most commonly recorded among those with positive results, while our recoding in Iowa suggests that 030 should be far less common than the 060 and 070 codes.
While a new SSF for p16 and more comprehensive guidelines and education focused on p16 and HPV status would improve the quality of data for upper aerodigestive tract cases collected by the Registries in the future, there still are potential uses of the HPV data already collected (diagnosis years 2010-2014) despite the aforementioned coding issues. The overwhelming majority (81%) of HPV-positive cases were captured by the original Registry coding based on CS guidelines. Thus, certain research questions could be potentially addressed using the data for those cases already collected based on CS guidelines (e.g., epidemiology of OPSCC within the HPV-positive population irrespective of HPV-type). However, research questions focused on evaluating patients based on presence or absence of HPV-testing or on specific types of HPV (e.g., HPV-type 16) could be problematic among cases diagnosed in 2010-2014.
This study has limitations that should be considered when interpreting results. For analytic purposes, patients who received both p16 and HPV-DNA testing were classified according to their HPV-DNA results and 26 of these patients had discordant results. As previously mentioned, p16 results may actually be a better indicator of prognosis. Further study of these patients could lead to refinements in survival estimates obtained from SEER data.
Another limitation involved the inconsistencies in the structure of, and language used in, pathology reports from many different institutions. Due to the unstructured nature of some of the reports, it is possible that classification of test types were incorrect in some patients. Two members of the study team are pathologists and assisted with interpretation of terminology used in the reports to ensure maximum accuracy in classifying test types. Finally, the models developed in this study were based only on patients residing in Iowa at the time of diagnosis, so results may not generalize to patients in other areas.
This study also has important strengths. HPV-testing rates in Iowa across patient, tumor, treatment, and facility characteristics were assessed using data from a large population-based registry. This is one of the first studies to identify and report the type of HPV-testing being performed, which showed large inconsistencies in coding. It is also one of the first studies providing a population-based estimate of HPV-testing among oropharyngeal cancer patients treated in a wide variety of hospital sizes and locations.
The nature of oropharyngeal cancer across the population has changed over the past two decades due to the effect of HPV. As such, there is an ongoing need for accurate epidemiologic data to inform clinical practice and investigation. Our data show that capturing p16 as a synonymous marker for HPV-testing is required for optimal accuracy. Given the large difference in survival outcomes between HPV-positive and HPV-negative oropharyngeal cancer, accurate registry assessment of HPV status is required to assess changes in survival over time. Such information is also important to understand treatment decisions: for example, the ongoing ECOG3311 and NRG-HN002 trials of treatment of HPV-associated oropharyngeal cancer specify the use of p16 IHC for registration and not HPV-DNA testing. If HPV status is eventually used to steer patients down different treatment paths, it is imperative that it is captured correctly in cancer registries so that practice patterns and survival implications associated with these different treatment paths can be assessed over time in the population. Important distinctions about clinical findings and outcomes would go uncaptured without modification of SSF-10 guidelines.
Acknowledgments
Grant Support: This work was supported in part under NIH/NCI contract number HHSN261201300020I (MEC, ARK, CFL).
Footnotes
Presentations: This study was presented at the North American Association of Central Cancer Registries conference in Albuquerque, New Mexico, June 2017.
References
- 1.National Cancer Institute. HPV and Cancer. 2015 https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hpv-fact-sheet.
- 2.Saraiya M, Unger ER, Thompson TD, et al. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst. 2015;107(6):djv086. doi: 10.1093/jnci/djv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology v.2.2008: Head and Neck Cancers. 2008 doi: 10.6004/jnccn.2004.0021. [DOI] [PubMed] [Google Scholar]
- 4.Weinberger PM, Yu Z, Haffty BG, et al. Molecular classification identifies a subset of human papillomavirus–associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24(5):736–47. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- 5.Seethala RR, Weinreb I, Carlson DL, et al. Protocol for the Examination of Specimens From Patients With Carcinomas of the Pharynx Pharynx 3.3.0.0. 2013 http://www.cap.org/ShowProperty?nodePath=/UCMCon/Contribution%20Folders/WebContent/pdf/pharynx-13protocol-3300.pdf.
- 6.Robinson M, Sloan P, Shaw R. Refining the diagnosis of oropharyngeal squamous cell carcinoma using human papillomavirus testing. Oral Oncol. 2010;46(7):492–6. doi: 10.1016/j.oraloncology.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Jordan RC, Lingen MW, Perez-Ordonez B, et al. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol. 2012;36(7):945–54. doi: 10.1097/PAS.0b013e318253a2d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Ihloff AS, Gorogh T, et al. p16(INK4a) overexpression predicts translational active human papillomavirus infection in tonsillar cancer. Int J Cancer. 2010;127(7):1595–602. doi: 10.1002/ijc.25174. [DOI] [PubMed] [Google Scholar]
- 9.Lewis JS., Jr p16 Immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head Neck Pathol. 2012;6(Suppl 1):S75–82. doi: 10.1007/s12105-012-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116(9):2166–73. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 11.National Comprehensive Cancer Network. NCCN Clinical Prectice Guidelines in Oncology V.2.2016: Head and Neck Cancers. 2016 [Google Scholar]
- 12.Collaborative Stage Work Group of the American Joint Committee on Cancer. Collaborative Stage Data Collection System User Documentation and Coding Instructions, version 02.03.02. Chicago, IL: American Joint Committee on Cancer; 2011. [Google Scholar]
- 13.American Joint Committee on Cancer. CAnswer forum; Collaborative Stage; Pharynx; Oropharynx. http://cancerbulletin.facs/org/forums.
- 14.U.S. Department of Health and Human Services. SEER Surveillance, Epidemiology, and End Results. 2012 https://seer.cancer.gov/about/factsheets/SEER_brochure.pdf.
- 15.Collaborative Stage Work Group of the American Joint Committee on Cancer. Oropharynx: CS Site-Specific Factor 10 Human Papilloma Virus (HPV) Status. 2013 http://web2.facs.org/cstage0205/oropharynx/Oropharynx_spd.html.
- 16.Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 18 Regs Custom Data Oral Cavity and Pharynx (with SSF 10 - in-house), Nov 2016 Sub (2010-2014) - Linked To County Attributes - Total U.S., 1969-2015 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released May 2017, based on the November 2016 submission. In.
- 17.Amini A, Jasem J, Jones BL, et al. Predictors of overall survival in human papillomavirus-associated oropharyngeal cancer using the National Cancer Data Base. Oral Oncol. 2016;56:1–7. doi: 10.1016/j.oraloncology.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Lydiatt WM, Patel SG, O’Sullivan B, et al. Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(2):122–137. doi: 10.3322/caac.21389. [DOI] [PubMed] [Google Scholar]


