Abstract
Surgical site infections (SSIs) are the most common and costly of hospital acquired infections. An important step in reducing SSIs is accurate SSI detection, which enables measurement and quality improvement, but currently remains expensive through manual chart review. Building off of previous work for automated and semi-automated SSI detection using expert-derived “strong features” from clinical notes, we hypothesized that additional SSI phrases may be contained in clinical notes. We systematically characterized phrases and expressions associated with SSIs. While 83% of expert-derived original terms overlapped with new terms and modifiers, an additional 362 modifiers associated with both positive and negative SSI signals were identified and 62 new base observations and actions were identified. Clinical note queries with the most common base terms revealed another 49 modifiers. Clinical notes contain a wide variety of expressions describing infections occurring among surgical specialties which may provide value in improving the performance of SSI detection algorithms.
Keywords: Surgical Wound Infection, Quality and Safety, Text-mining
Introduction
Heathcare associated infections (HAIs) are a significant problem among hospitals worldwide. Surgical site infections (SSIs) are unfortunately the most common and costly of HAIs. SSIs increase post-operative morbidity and mortality. The overall prevalence of SSIs is estimated to be 2-5% for inpatient surgical cases in the United States [1–3]. Prevalence is significantly higher in certain specialities such as colon and rectal surgery (approximately 13-15%) [4].
An SSI can be classified into three categories (i.e., superficial, deep, and organ space) according to specific definitions which include time frame following surgery. Several classification systems designate an infection event as an SSI only if the occurence is within 30 days of the index operation. A superficial SSI involves only the skin or subcutaneous tissue and requires documentation of one or more of the following: purulent drainage from the wound, aseptically obtained wound culture with isolated organisms, opening of the wound by a physician with clinical symptoms of infection, or diagnosis by a physician [5]. A deep SSI involves the muscle or fascia and requires: purulent drainage from the deep incision, opening of the wound, positive wound culture, or spontaneous dehiscing of the wound and clincial signs or symptoms of a wound infection, or an abscess or other evidence of infection diagnosed by pathology or imaging. An organ space SSI involves the space deep to the muscle or fascia, identification of involvement in a specific organ system, and either purulent fluid from a organ space drain, an organism identified by culture, or an abscess or other evidence of infection diagnosed by pathology or imaging.
Another factor adding to the complexity of documenting SSIs is the question of a pre-existing infection. Careful documentation is required if there is an infection present at the time of surgery (PATOS). In the setting of PATOS, a post-operative infection would be excluded from being counted as an SSI if the site of infection is the same in both instances [5]. For example, if an abdominal wall abscess recurs after surgery performed for drainage of an abdominal wall abscess, this infection is not considered an SSI. However, if a patient undergoes surgery for intraabdominal abscesses from diverticulitis then develops a superficial infection of the wound, this is an SSI because the infection sites are different.
Many hospitals in the United States use the American College of Surgeons National Surgery Quality Improvement Program (NSQIP) as a quality improvement database to track post-operative complications such as SSIs. NSQIP is recognized as a national leader of post-operative complication measurement and quality improvement measures [6]. Currently, a surgical clinical reviewer who is usually a registered nurse employed by the hospital manually reviews the charts of post-operative patients and makes positive or negative SSI determintations by clinical judgement based on the SSI definition. This process results in high quality outcomes data for quality improvement and benchmarking efforts, but is prohibitively expensive for some centers [7]. Methods to automate or semi-automate SSI detection are of high interest since they may significantly reduce the burden of manual chart review and decrease the costs of quality improvement initiatives like NSQIP.
We previously developed supervised machine learning algorithms for SSI detection utilizing structured and unstructured clinical data [8]. SSI determination with our algorithms is based on a list of “strong features” identified for each type of SSI. The algorithm creates a score for each record correlating with the probability of an acquired SSI [9].
While the performance of our algorithms is good, improvements in the algorithm can be made particularly for intermediate scoring records and potentially in improving the feature set for unstructured text, which have to-date been based upon keywords and concepts derived from expert consensus (surgeons and hospital surgical clinical reviewers at our center). We hypothesized that there may be additional signals in the form of expressions directly describing or otherwise associated with SSIs in clinical notes. The study’s objective was therefore to characterize expressions associated with SSI determinations from clinical notes in a systematic manner.
Methods
Records from surgical patients included in the University of Minnesota Medical Center’s NSQIP database were extracted from the University of Minnesota’s clinical data repository. For our initial analysis, we included patients from 2014-2015 identified as having an SSI occurrence by the NSQIP surgical clinical reviewer along with patients having a high probability score (>40) for having an SSI from our SSI detection algorithm which used the following set of “strong” text features (Table 1). In all cases, the index operation was identified and all clinical notes within the 30 day time window after the operation were reviewed including all inpatient and ambulatory notes.
Table 1.
Original Surgical Site Infection Features
| Surgical Site Infection Features | |||
|---|---|---|---|
| abdominal abscess |
empyema | antibiotics | phlegmon |
| abscess | erythema | joint abscess | presacral abscess |
| anastomotic dehiscence |
evisceration | leak | purulent |
| cellulitis/ cellulitic |
extraluminal | interventional radiology |
rectal stump blowout |
| cloudy | extravasation | malodorous | rim enhancing |
| dehiscence | fistula | murky | wet to dry |
| demarcated/ demarcation |
foul-smelling | open wound | wound dehiscence |
| drain care | Hartmann’s blowout |
packing/ packing change |
wound infection |
| drainage | induration/ indurated |
pelvic abscess |
wound packing |
| drain placement |
infected/ infection |
pelvic collection |
vac dressing |
| dressing/ dressing change |
intra- abdominal abscess |
pelvic sepsis | |
In the review of each chart, one of two surgical residents (SS and EA) reviewed each post-operative note in chronological order from the index operation. All terms including those identified by experts previously within clinical notes contributory to an SSI determination were recorded along with information about misspellings, discrepancies and inaccuracies, and the associated note type containing the information. Documentation of repeated factors for an individual record was not recorded. All terms were carefully categorized into observations of the patient/patient data, actions performed by the clinical team, antibiotics, organisms, or clinical plans. These terms were also analyzed for modifiers and overall compared to the original set of terms.
Following categorization and initial analysis, we performed a validation of three “base terms” (i.e., “fluid collection”, “drainage”, and “infection”) which were the most frequent and contained the greatest number of modifiers. From this analysis, we sought to validate the associated modifiers identified from our initial analysis. For this, we examined a separate cohort of patients with and without SSI (25 records each) from the year 2015. These were also patients within the the institution’s NSQIP database, but had not been assessed for SSI-related phrases. We utilized the Natural Language Processing-Patient Information Extraction for Research (NLP-PIER) clinical research clinical note search engine for each base term [10]. Each of the encountered modifiers within the search engine were recorded and added to our representation model in our evaluation where applicable.
Institutional review board approval was obtained and informed consent waived for this minimal risk study. Interrater reliability was assessed on 160 (ten percent) of SSI phrases by both physician-raters (SS and EA) to assess agreement on the whether the phrase was associated with SSI and whether the term was positive or negative (i.e., not indicating an SSI). Agreement was 100% for association with SSI and 0.94 with a kappa of 0.82 for positive or negative designation agreement.
Results
A total of 54 positive SSI patient cases from the NSQIP database (n=41) or with a high probabilty SSI score by algorithm (n=13) were reviewed. After reaching 45 patients, saturation of our corpus was assessed by tracking new terms. After assessing 9 additional patients, only 8 new terms were identified and our cohort was completed with 54 total patients. Demographics and surgical specialty of these patient cases are displayed in Table 2. The total number of notes reviewed was 3,232. Multiple surgical services were represented.
Table 2.
Summary of 54 SSI patient cases
| Demographics | |
|---|---|
| Median Age (Range) | 55(25-92) |
| Gender (%) | Male (48%) |
| Median Length of Stay in days (Range) | 10(1-43) |
| Median Number of Notes per chart (Range) |
54(10-218) |
| Surgical Service | N (%) |
| General | 11(20%) |
| Colorectal | 11(20%) |
| Vascular | 6(11%) |
| Transplant | 6(11%) |
| Orthopedic | 6(11%) |
| Plastic | 4(7%) |
| Otolaryngology | 4(7%) |
| Neurologic | 4(7%) |
| Urology | 2(4%) |
Overall, 1,536 distinct phrases were identified that were important for the designation of a SSI. There was a median of 25.5 (range 9-64) unique phrases identified in each chart. The majority of phrases 1,304 (85%) were identified from inpatient encounters. Outpatient encounters accounted for the remaining 15%, with 232 phrases. The majority of the SSI-related phrases were found in the progress notes of clinical teams following the patients in the hospital and is summarized in Table 3.
Table 3.
Location of SSI-Related Terms
| Note Type | Terms | % of Total |
|---|---|---|
| History & Physical | 78 | 5% |
| Operative Note | 98 | 6% |
| Consultation | 158 | 10% |
| Progress Note | 869 | 57% |
| Primary Team | 794 | 52% |
| Consult Team | 75 | 5% |
| Discharge Summary | 93 | 6% |
| Office Visit | 119 | 8% |
| Telephone Encounter | 45 | 3% |
| Emergency Visit | 76 | 5% |
While most of SSI-related phrases had a positive correlation with an SSI occurrence, 161 (10%) phrases offered evidence against a surgical site infection (e.g., “no obvious purulence”, “improving of erythema”, “wound c/d/i”). These phrases “protective” of an SSI generally occurred in the early post-operative period, or late in the course of the infection, signifying potential recovery.
SSI-Related Base Term Classification
Observations were categorized by isolating the “base term” that was being observed. There were 63 unique base terms that were recorded from review of 1,536 SSI-related phrases (Table 4).
Table 4.
SSI Base Terms
| Observation Base Terms | ||
|---|---|---|
| incision | redness | fasciitis |
| debris | fluid collection | pain |
| skin | air | sepsis |
| succus | swelling | ecchymosis |
| drain | abscess | necrosis |
| induration | film | warmth |
| discharge | wound | tissue |
| output | purulence/pus | mesh |
| infection | seroma | fever |
| drainage | cellulitis | rigors |
| fluid | aspirate | edema |
| erythema | fistula | inflammation |
| dehiscence | thickening | blistering |
| leak | peritonitis | petechiae |
| hematoma | blood | purpura |
| odor | site | fluctuance |
| firmness | culture | separation |
| material | gas | osteomyelitis |
| dressings | stool | evisceration |
| tunneling | eschar | amputation |
| gangrene | exudate | colon |
There were a few “actions” found to be pertinent to SSIs in clinical notes. Sixteen unique verbs were found relating to SSIs. Most verbs were found in multiple tenses. These verbs (Table 5) were documented when used to explain procedures relevant to surgical site infections.
Table 5.
SSI Related Actions
| SSI Actions | ||
|---|---|---|
| open | place | examine |
| incise | evacuate | close |
| remove | drain | culture |
| probe | washout | debride |
| aspirate | change | I&D |
| irrigate | ||
Directives of the clinical plan also included some phrases related to SSIs. Each group of phrases had some variability, but there were six main themes: computed tomography (CT) requests, wound culture/gram stain orders, specific wound care plan, consulting infectious disease (ID) and interventional radiology (IR), tentative plans for operative intervention, and antibiotic changes.
Other SSI-Related Phrases
Antibiotics were common SSI-related phrases included in the clinical note. Antibiotics comprised 241 (16%) of the recorded SSI-related phrases. The use of antibiotics was not consistent in the treatment of SSIs. Antibiotics were included in the analysis if started empirically (concern for SSI but no definitive evidence) or if being used to treat an SSI. There was a wide range of antibiotics used for treating SSIs due to the multiple organ systems represented by each different surgical service. Documentation in the clinical notes included both the trade and generic names of the antibiotics (Table 6). General terms, such as “antibiotics”, “IV antibiotics”, and “antibiosis” were also documented but are not included in Table 6.
Table 6.
SSI-Related Antibiotics
| Generic Name (if used) |
Trade Name (if used) |
Common Abbreviations |
|---|---|---|
| amoxicillin- clavulanate |
Augmentin | - |
| cefazolin | Ancef | - |
| piperacillin- tazobactam |
Zosyn | pip-tazo |
| levofloxacin | Levaquin | - |
| metronidazole | Flagyl | - |
| ciprofloxacin | - | cipro |
| vancomycin | - | vanco, vanc |
| tigecycline | - | - |
| clindamycin | Cleocin | clinda |
| linezolid | Zyvox | - |
| mupirocin | Bactroban | - |
| ertapenem | Invanz | Erta |
| meropenem | Merrem | mero |
| cephalexin | Keflex | - |
| nafcillin | - | - |
| ampicillin- sulbactam |
Unasyn | - |
| trimethoprim- sulfamexazole |
Bactrim | TMP-SMX |
| ceftriaxone | Rocephin | CTX |
| micafungin | - | mica |
| fluconazole | Diflucan | - |
| minocycline | - | mino |
| doxycycline | - | doxy |
Clinically significant organisms were documented in the clinical notes. These were recorded when associated with a wound or abscess culture (Table 7). Organisms comprised 111 (7%) of the total SSI-related terms. An organism isolated from a wound or abscess culture is sufficient on its own to diagnose an SSI [5]. Terms such as “gram positive cocci”, “lactose fermenting rods”, and “coagulase negative staphylococcus” were also recorded but not included in the table due to their generality in describing many infections.
Table 7.
SSI Related Organisms
| Documented Organisms | |
|---|---|
| klebsiella | proteus |
| escherichia coli | staphylococcus |
| prevotella | pseudomonas |
| corynebacterium | pasteurella |
| streptococcus | anerococcus |
| achromobacter | veillonella |
| enterococcus | peptostreptococcus |
| bacteroides | clostridium |
Wound care items were commonly encountered terms. Types of dressings including: gauze, Kerlix, NuGauze, Aquacel, wet to dry, and xeroform were frequently documented as SSI-related terms. The most commonly encountered wound care item was “wound vac”.
SSI-associated abbreviations and acronyms were relatively uncommon, but repeated frequently. The recorded abbreviations are included in Table 8 They are grouped according to type of abbreviation.
Table 8.
SSI Related Abbreviations
| Category | Abbreviation |
|---|---|
| Anatomic Location | IT, LLQ, LUQ, RUQ, RLQ, EC, abd, RP |
| Microbiology | cx, abx, GM+, GPC, GNR, GPR, UTI, CoNS, ifx, MSSA, MRSA, VRE, VSE, GNB, SSI |
| Imaging | CT, A/P, US, IR |
| Exam | c/d/i, CDI, Tmax, TTP |
| Laboratory | WBC |
| Frequency | BID, TID, QID |
| Miscellaneous | POD, op, IV, PO, s/s, I&D, ID, vac, JP |
Misspellings in the clinical notes relating to surgical site infections were infrequent. There were only 12 instances of misspelling found in SSI related terms. The most commonly misspelled word was “dehiscence”. Inaccuracies of documentation associated with SSIs also appeared to be rare. Although it is difficult to assess inaccuracies by solely a retrospective search of the clinical notes, only one obvious inaccuracy was discovered. A pelvic abscess was incorrectly documented in a telephone note.
SSI Expressions in Notes Versus Original Expert Terms
As demonstrated in Table 1, there were 43 unique phrases determined by expert consensus that were included in the original set of “strong features”. These original phrases can be broken down into 22 base terms (observations and actions) and 24 modifiers.
Overall, extraction of SSI signals from clinical notes resulted in an overlap of 17 base observations and actions (77%) with the original set of expert phrases with 5 terms from the original list not found in our corpus. There were 24 modifiers identified from the original features, with an overlap of 21 modifiers (88%). Only 3 modifiers were not found in our corpus. Combined, there was 83% overlap when accounting for both base terms and modifiers. One term in the original set, “antibiotics”, was categorized in our other SSI-related features. In addition, we identified 62 new terms from the corpus: 47 new base observations and 15 new actions. Eleven of these terms were in the top 25% of frequency in the 54 cases reviewed. All of our antibiotics, organisms, and abbreviations/acronyms were new compared with the originial set.
Modifiers of Base Terms & Validation of Modifiers
Modifiers of the main SSI-related base terms (observations and actions) were extracted in analysis of each base term. Modifiers were also classified as evidence for an SSI diagnosis (positive) and against an SSI diagnosis (negative). Overall, there were 383 modifiers among all of the base terms. Only unique modifiers were recorded for each base term. Repeated modifers were recorded only if used for different base terms. There was a wide range of modifiers per base term, with a median of 2 (range 0-49).
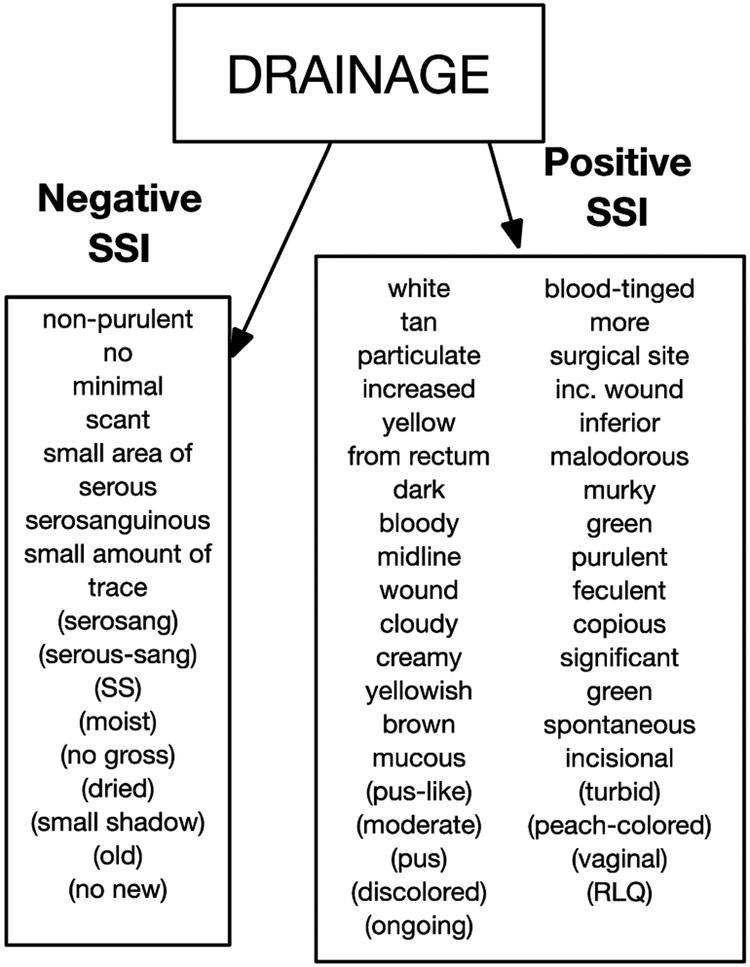
Three terms: “fluid collection”, “drainage”, and “infection” were tested with the NLP-PIER search engine on a new set of 25 patients with an SSI and 25 patients without an SSI in 2015 to validate the utility of the previously identified modifiers and to determine if there were additional modifiers associated with these base terms. From chart review, “fluid collection”, “drainage”, and “infection” had the most modifiers, with 32, 49, and 41 respectively. Only 49 new modifiers were encountered in this clinical note query of 50 patients (13 for “fluid collection”, 18 for “drainage”, and 18 for “infection”). Figure 1 is an example of a base term with its modifiers.
Figure 1.
Base Term Example and Modifiers. Additional modifiers identified by NLP-PIER are included in parentheses
Discussion
Automated or semi-automated SSI detection has the potential to decrease present day manual abstraction required in most cases. While quality improvement registries such as NSQIP demonstrate tangible benefits to patient outcomes in hospitals in the private and public sectors [7;11], increased automation around outcome extraction for post-operative complications like SSIs could reduce cost barriers creating wider adoption. Our study demonstrated a number of important types of signals in clinical texts which we did not recognize previously with the assistance of expert consensus. This speaks to the variability of language used in our documentation of patient care, and it is likely that these findings can be leveraged to improve the performance of these algorithms.
The method used in this study was a two-step approach by first empirically analyzing the content of clinical notes in positive cases of SSI and then performing a validation of the base terms and associated modifiers to ensure good coverage of the identified modifiers. A previous study reported defining SSI patterns using two conceptual groups of terms in text-mining: bacteriology and surgery [12]. In our study, we discovered additional groups of SSI-related terms. We opted to classify our concepts into observations, actions, and plans related to SSIs. Our SSI-related observations were used to describe exam, laboratory, and imaging findings. SSI-related actions were used for procedures and tasks performed by clinicans. The clinical plan includes future tasks to be performed and next steps in SSI management. Other information related to SSIs included antibiotics, organisms, and wound care. We also observed abbreviations for many of these concepts.
We observed that antibiotics and organisms comprised 23% of total terms. These terms appear to have a relatively high sensitivity in detecting SSIs since only certain organisms and antibiotics tend to be associated with post-operative infections. Unfortunately, these same antibiotics and organisms can often be found in infections not relevant to SSIs, resulting in a low specificity for SSI detection.
The original set of “strong features” identified by expert opinion included a portion of terms that could be used for SSI identification. Compared to our original set, 62 new base terms and 362 new modifiers were identified in chart review of patients with an SSI or a high probability score on the SSI algorithm. In addition to the base terms, there were six clinical plan categories as well as antibiotics, organisms, and abbrevations/acronyms that could be leveraged for better SSI detection. While most modifiers were already discovered in the initial review of SSI cases, 49 additional new modifiers were identified through NLP-PIER searches which added to the robustness of the associated set of signals.
As expected, key data for the determination of SSIs is stored in the clinical notes. While structured data is useful for detecting SSIs, clinician judgement and physical examination remain key; significant, unique details about SSIs are only found within clinical notes. NLP and text-mining can be used to detect adverse events in clinical notes with better performance than manual review and other methods utilizing structured data for automated detection [13]. Recently, a text-mining approach using two categories was used to detect SSIs in a neurosurgery department [12]. Our findings demonstrate, however, that a constellation of terms is needed to determine the presence of an SSI. It is likely that improved discrimination for SSIs can be achieved by accounting for more complex phrases and/or base terms and their modifiers to capture more complex SSI semantics. Further organization including analysis of descriptor groupings and locations may be useful in classifying terms for the identification of SSIs.
This project did not assess pre-operative risk factors and predictors of SSIs in surgical patients. By studying post-operative outcomes and improving outcome abstraction in surgical patients, we are building our knowledge of these risk factors and predictors. These risk factors have even more complexity, ranging from patient physical characteristics, past medical and surgical history, additional laboratory data, to operation-specific signals. With more data analysis, perhaps a future project could assess pre-operative clinical notes to develop a separate algorithm to predict SSI risk prior to or immediately following an operation.
Our study has several limitations including its relatively small sample size and the use of data from a single institution. Future work to validate these findings on a separate dataset including any regional variability of SSI language and additional variability associated with specialities with lower rates of SSI (e.g. neurosurgery or otolaryngology) is needed.
Conclusions
The language behind SSIs is complex. There are many categories of terms that may contribute to an SSI determination, including observations, actions, clinical plans, antibiotics, and specific organisms. Empiric analysis of SSI cases was an effective method for uncovering the complexity of SSI-related expressions in clinical texts. These findings may provide value in improving the performance of SSI detection algorithms.
Acknowledgements
This research was supported by the University of Minnesota Academic Health Center Faculty Development Award (GS, GM), Agency for Healthcare Research and Quality (R01HS24532), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program (UL1TR000114), Fairview Health Services, and University of Minnesota Physicians.
References
- [1].Anderson DJ, Podgorny K, Berríos-Torres SI, Bratzler DW, Dellinger EP, Greene L, … Kaye KS (2014). Strategies to prevent surgical site infections in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol, 35 Suppl 2, S66–88. [DOI] [PubMed] [Google Scholar]
- [2].Ban KA, Minei JP, Laronga C, Harbrecht BG, Jensen EH, Fry DE, … Duane TM (2016). American College of Surgeons and Surgical Infection Society: Surgical Site Infection Guidelines, 2016 Update. J Am Coll Surg doi: 10.1016/j.jamcollsurg.2016.10.029 [DOI] [PubMed] [Google Scholar]
- [3].Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, … Team, E. I. P. H.-A. I. a. A. U. P. S. (2014). Multistate point-prevalence survey of health care-associated infections. N Engl J Med, 370(13), 1198–1208. doi: 10.1056/NEJMoa1306801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wick EC, Vogel JD, Church JM, Remzi F, & Fazio VW (2009). Surgical site infections in a "high outlier" institution: are colorectal surgeons to blame? Dis Colon Rectum, 52(3), 374–379. [DOI] [PubMed] [Google Scholar]
- [5].National NHS. Surgical Site Infection (SSI) Event. (2013) Atlanta, GA: Center for Disease Control and Prevention. [Google Scholar]
- [6].Ingraham AM, Richards KE, Hall BL, & Ko CY (2010). Quality improvement in surgery: the American College of Surgeons National Surgical Quality Improvement Program approach. Adv Surg, 44, 251–267. [DOI] [PubMed] [Google Scholar]
- [7].Hollenbeak CS, Boltz MM, Wang L, Schubart J, Ortenzi G, Zhu J, & Dillon PW (2011). Cost-effectiveness of the National Surgical Quality Improvement Program. Ann Surg, 254(4), 619–624. [DOI] [PubMed] [Google Scholar]
- [8].Hu Z, Melton GB, Moeller ND, Arsoniadis EG, Wang Y, Kwaan MR, Jensen EH, Simon GJ. Accelerating Chart Review Using Automated Methods on Electronic Health Record Data for Postoperative Complications. Proceedings of the American Medical Informatics Association Symposium. 2016: 1822–31. [PMC free article] [PubMed] [Google Scholar]
- [9].Hu Z, Simon GJ, Arsoniadis EG, Wang Y, Kwaan MR, & Melton GB (2015). Automated Detection of Postoperative Surgical Site Infections Using Supervised Methods with Electronic Health Record Data. Stud Health Technol Inform, 216, 706–710. [PMC free article] [PubMed] [Google Scholar]
- [10].McEwan R, Melton GB, Knoll BC, Wang Y, Hultman G, Dale JL, … Pakhomov SV (2016). NLP-PIER: A Scalable Natural Language Processing, Indexing, and Searching [PMC free article] [PubMed]
- [11].Dimick JB, Chen SL, Taheri PA, Henderson WG, Khuri SF, & Campbell DA (2004). Hospital costs associated with surgical complications: a report from the private-sector National Surgical Quality Improvement Program. J Am Coll Surg, 199(4), 531–537. [DOI] [PubMed] [Google Scholar]
- [12].Campillo-Gimenez B, Garcelon N, Jarno P, Chapplain JM, & Cuggia M (2013). Full-text automated detection of surgical site infections secondary to neurosurgery in Rennes, France. Stud Health Technol Inform, 192, 572–575. doi: 10.1016/j.jamcollsurg.2004.05.276 [DOI] [PubMed] [Google Scholar]
- [13].Melton GB, & Hripcsak G (2005). Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc, 12(4), 448–457. doi: 10.1197/jamia.M1794 [DOI] [PMC free article] [PubMed] [Google Scholar]