Genome-wide association studies have uncovered approximately 250 common loci with strong associations in inflammatory bowel disease (IBD).1 The current challenge is to understand the underlying molecular mechanisms whereby each locus predisposes to IBD.2 In this regard, transcriptomic profiling dissecting gene expression patterns in tissues where the chronic inflammation occurs, as well as studies on its genetic control (expression quantitative trail loci [eQTL]), are beginning to shed some light into IBD pathophysiology.3, 4, 5 However, data sets profiling patients to explore changes in gene expression across gastrointestinal locations relevant to the disease (eg, ileum vs colon) are lacking and no eQTL studies available are based on transcriptomic profiling of pediatric IBD.
We used genetic and transcriptomic data from the Risk Stratification and Identification of Immunogenetic and Microbial Markers of Rapid Disease Progression in Children with Crohn's Disease (RISK) study5 to explore the role of tissue (ileum vs rectum) and disease subtype (Crohn’s disease [CD] vs ulcerative colitis [UC]), and biopsy inflammation (present vs lacking) in shaping the transcriptome of pediatric IBD. Two main reasons motivate the study of these locations, namely each tissue is the most commonly affected for each IBD subtype (the ileum for CD and the rectum for UC), and both tissues are accessible on a routine basis during the diagnostic colonoscopy.
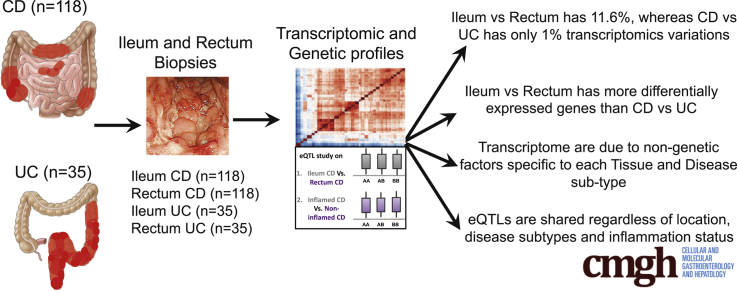
Our data set was constructed with 153 individuals stratified into 4 subgroups according to the nature of their disease and the location of the biopsy specimens collected in the intestine by histology: (1) ileum from CD patients (n = 118), and (2) rectum from the same individuals (n = 118), and the equivalent from UC patients; (3) ileum UC (n = 35); and (4) rectum UC (n = 35). Altogether, we performed 3 different analyses.
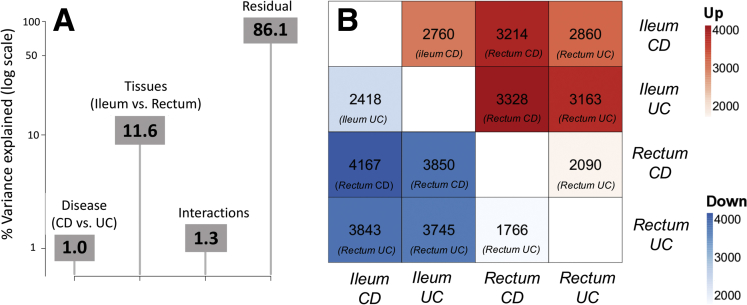
First, we used principal variance component analysis6 to assess the percentage of variance in transcriptomic profiles that is owing to tissue, disease subtype, inflammatory activity of the tissue, or interactions among them. As shown in Figure 1A, an analysis based on all available individuals showed that tissue (ileum vs rectum) accounted for the largest proportion in transcriptomic variation (11.6%), whereas disease subtype only explained a minor fraction (1%). Of note, the interaction between tissue and disease subtype played a slightly larger role than subtype alone (1.3%), implying that a fraction of the changes in patient transcriptomes are particular to each tissue/subtype combination. A second principal variance component analysis restricted to CD patients to evaluate the role of biopsy inflammation rendered a similar pattern, namely 13.5% for tissue vs 1.7% for inflammatory status. Indeed, the study of the total number of genes that were expressed differentially in pairwise comparisons between the 4 subgroups rendered similar patterns (Figure 1B), with the largest differences observed for comparisons across tissues. These results confirm that cell and tissue type are the key determinants in shaping the human transcriptome,7 although the significant variance explained by the interaction of tissue and subtype highlights the importance of profiling samples from patients of the IBD type under study.
Figure 1.
The transcriptome of pediatric IBD patients varies according to disease subtype and tissue profiled. (A) Barplot depicting (in log-scale) the amount of variance in gene expression explained by tissue, disease subtype, and the interaction among both along with the residual. (B) Heatmap showing the total number of differentially expressed genes in each pairwise comparison as detected by edgeR (see Supplementary Materials and Methods section). Top diagonal (in red colors) shows genes up-regulated and the bottom diagonal (in blue background) shows genes down-regulated, with the reference group included in parenthesis at the bottom of each cell.
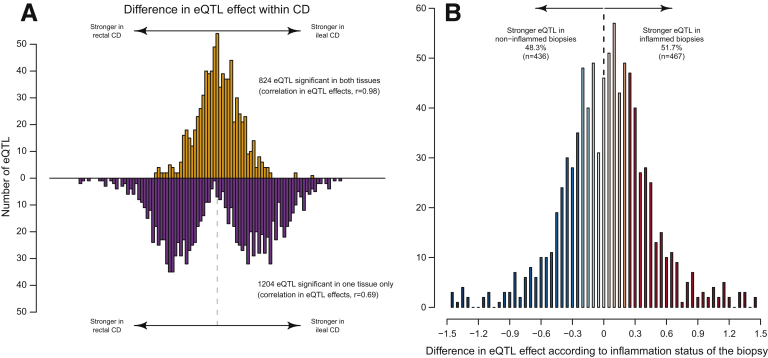
Next, we took advantage of the availability of single-nucleotide polymorphism data from the same patients to evaluate the degree to which the differences in transcriptome among subgroups are caused by variations in the local genetic control of gene expression. Sample size was the main determinant for the total number of genes that have at least 1 significant cis-eQTL (the genes whose expression levels have been associated with genetic variation at a specific genetic locus [eGenes]) identified in each genome-wide study (1026 and 1134 for ileum CD and rectum CD; 107 and 108 for ileum UC and rectum UC, respectively) (Supplementary Table 1). Nonetheless, as exemplified in the following exploration of ileum vs rectum in pediatric CD, there is an extensive sharing of eQTL effects (Figure 2). Specifically, Figure 2A shows the difference in effect size for top eQTLs for each eGene, classified according to whether they are detected as significant either in pediatric CD ileum or rectum only, or alternatively, in both. We can distinguish 3 behaviors from this analysis. First, when comparing the significant eQTL in both tissues (40.6%; n = 824) (Figure 2A, top), the effect sizes are similar and strongly correlated (r = 0.98), implying the same causal variant is acting in both tissues. Second, eQTL that putatively are tissue-specific (59.4%; n = 1204) (Figure 2A, bottom) have larger differences in effect size among the 2 tissues (r = 0.69 correlation in effect size). However, in turn, only a fraction of these present nominally significant (P < .05) tissue-by-subtype interactions, and hence harbor effects that are different between the 2 tissues (310 of 1204; 25.7%). Despite a proportion of effects that are particular to 1 of 2 tissues, overall these results imply that eQTLs are for the most part shared and therefore the major differences in the transcriptome are owing to nongenetic factors specific to each tissue and disease subtype. Interestingly, a fraction of eQTLs discovered in each tissue overlap with pathogenic variants discovered by genome-wide association studies1 (Supplementary Table 2).
Figure 2.
The genetic control of gene expression is shared regardless of disease subtype and tissue profiled. (A) Histogram depicting the difference in effect size (β, in SD units) of eQTLs detected in the ileum and rectum of CD patients. Top: For eGenes detected in both the ileum and rectum, the difference in effect size of the top eQTL in each tissue (n = 824). Bottom: The difference in effect size for eQTLs significant in only 1 of the 2 tissues (n = 1204). (B) Histogram of the difference in effect size between inflamed and noninflamed biopsy specimens for 903 eQTLs detected in ileum CD (see main text).
Finally, we aimed to determine whether tissue inflammation exacerbates the effect of eQTL variants that control gene expression. We split the ileal CD samples according to the inflammation status of the patient biopsy specimens, distinguishing between CD ileum inflamed (n = 83) and CD ileum noninflamed (n = 27). As shown in Figure 2B, the difference in effects at each eQTL are centered at zero, with only approximately half of the eQTL discovered in the full set of ileal CD samples showing a stronger effect in the inflamed subgroup (51.7%; n = 467 of 903; P = .32; binomial test). This balanced pattern fits the random expectation and indicates that eQTL effects are not preponderantly exacerbated in the inflamed subgroup. Supplementary Table 3 provides a full list of detected eQTLs along with the effect size in each of the subgroups discussed in our analyses.
Taken together, we show that transcriptomic signatures in pediatric IBD cluster mainly by location in the gastrointestinal tract. This result agrees with recent evidence based on genetic risk scores,8 overall enhancing the important role of disease location in shaping disease variability in IBD. On the other hand, we observed that eQTLs are shared regardless of location, disease subtypes, and inflammation status, which have implications for study design involving molecular profiling of IBD patients.
Acknowledgment
The authors are thankful to H. Somineni, K. Mondal, and colleagues for their valuable suggestions, support, and helpful comments.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Supplementary Materials and Methods
Cohort
The RISK study was started in 2008 as an observational prospective cohort study that aimed to develop risk models for predicting the complicated course in children with CD. This study ended up in 2012, with more than 1800 treatment-naive children and adolescents younger than 17 years, including newly diagnosed with CD and UC, from 28 pediatric gastroenterology centers in North America.1, 2, 3 Before diagnosis and treatment, all patients underwent a baseline colonoscopy and confirmed typical chronic active colitis/ileitis by histology. We restricted to 153 patients (118 with CD and 35 with UC) from our RISK study, who had both ileum and rectum biopsy specimen transcriptomic data and the genotypes from peripheral blood.
Transcriptomic Data Processing
Isolation of RNA from ileum biopsy specimens from colonoscopy at diagnosis, and the extraction of RNA sequencing gene expression profiles have been described in our previously studies.1, 2, 3 We followed the same protocol to extract the rectal RNA sequencing gene expression profiles. TopHat 2.0.13 (Baltimore, MA)4 with default parameters was used to map the reads with human genome 19, SAMtools (Cambridge, MA)5 was used to transform the aligned reads, and HTSeq-0.6.1 (Heidelberg, Germany)6 with the default union mode was used to quantify the number of reads at the gene level.
Genotyping
We started with 192,523 variants from peripheral blood genotyped by immunochip array. All the variants were mapped to Genome Reference Consortium Human Build 37 (GRCh37) using the SNPlocs.Hsapiens.dbSNP.20120608 package.7 We removed any variants with nonbiallelic variants, Hardy-Weinberg equilibrium (P < 10-3), Minor allele frequency <5%, >1% missing data, and >0.10% genotype missing rates using PLINK (Boston, MA). To check relatedness among samples, we calculated pairwise IBD based on 27,728 single-nucleotide polymorphisms obtained after LD pruning using the PLINK routine -indep 50 5 2, confirming no pairs were first-degree relatives (PI_HAT > 0.25). After all quality control steps, we retained 91,809 single-nucleotide polymorphisms available for all 153 samples.
Differential Gene Expression Analysis Between Disease and Tissue Subtypes
We profiled the differential gene expression between (1) tissue types (ileum and rectum), (2) disease types (CD and UC), and (3) inflamed and noninflamed regions within CD samples. Raw counts were compiled and processed with edgeR8 to obtain normalized counts through trimmed mean of M values normalization. An in-house R script then was used to inverse rank transform expression estimates for each gene into a standard normal distribution with a mean of 0 and a variance of 1. The trimmed mean of M values were transformed further into the reads per kilobase per million mapped reads metric and the genes with reads per kilobase per million mapped reads greater than 1 and greater than 6 read counts in at least 10 individuals were retained. Finally, we looked for differentially expressed genes between these groups by using the differential expression analysis method edgeR and obtained the fold change differences as indicated.
eQTL Study and Statistical Analysis
We used unsupervised surrogate variable analysis9 and supervised normalization of the microarray10 procedure to identify and remove any hidden confounding factors in our gene expression data sets. The known variables such as sex and disease status were protected in this process and we included them as covariates in the eQTL mapping. The eQTL mapping was performed with a linear mixed model implemented in GEMMA.11 We adjusted for population structure and relatedness among individuals as a random effect through a genetic relationship matrix from a LD-pruned single-nucleotide polymorphism data set. Finally, the association was performed between each gene (normalized read counts) and the single-nucleotide polymorphism located within ±1 Mb region of the gene transcription start site.
Comparison With Existing Studies
Various eQTL studies have been performed in human intestine in general, across the population, and different tissues such as ileum and colon showed the divergence of gene regulations.12, 13, 14, 15 We calculated the percentage of replicability with the Kabakchiev et al15 study, which showed only 655 and 553 eQTLs are common in ICD and RCD, respectively (Supplementary Table 3, ICD, RCD - tables). The comparisons on common eQTLs were not performed because of the absence of effect size from the Kabakchiev et al15 study.
Supplemental Graphical Summary.
Supplementary Table 1.
The Total Number of Significant eQTLs, eGenes, and the genetic variants associated with transcript expression levels (eSNPs) Detected in Disease, Tissues, Tissue-Relevant Disease-Specific, Inflamed, and Noninflamed Data Sets
| Data sets | Total samples | Total eQTLs | eQTLs (FDR P < .05) | eGenes (FDR P < .05) | eSNPs (FDR P < .05) |
|---|---|---|---|---|---|
| Ileum CD | 118 | 1,603,615 | 13,471 | 1026 | 9790 |
| Ileum UC | 35 | 1,592,326 | 1103 | 107 | 1061 |
| Rectum CD | 118 | 1,585,034 | 16,173 | 1134 | 11,333 |
| Rectum UC | 35 | 1,573,929 | 1125 | 108 | 1026 |
| Ileum CD inflamed | 83 | 885,592 | 10,257 | 801 | 7927 |
| Ileum CD non-inflamed | 25 | 880,584 | 639 | 60 | 554 |
| Rectum CD inflamed | 69 | 862,451 | 7485 | 662 | 5964 |
| Rectum CD non-inflamed | 37 | 861,664 | 3083 | 311 | 2708 |
Supplementary Material
References
- 1.Liu J.Z. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGovern D.P. Gastroenterology. 2015;149:1163–1176 e1162. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granlund A. PLoS One. 2013;8 e56818. [Google Scholar]
- 4.Haberman Y. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kugathasan S. Lancet. 2017;389:1710–1718. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boedigheimer M.J. BMC Genomics. 2008;9:285. doi: 10.1186/1471-2164-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mele M. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleynen I. Lancet. 2016;387:156–167. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Haberman Y. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kugathasan S. Lancet. 2017;389:1710–1718. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marigorta U.M. Nat Genet. 2017;49:1517–1521. doi: 10.1038/ng.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H. Bioinformatics. 2009;25.16:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anders S. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pages H. SNPlocs.Hsapiens.dbSNP.20120608; R package version 0.99.11. 2017
- 8.Dai Z. F1000Res. 2014;3:95. doi: 10.12688/f1000research.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leek J.T. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mecham B.H. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulur I. BMC Genomics. 2015;16:138. doi: 10.1186/s12864-015-1292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh T. Inflamm Bowel Dis. 2015;21:251–256. doi: 10.1097/MIB.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo C.C. J Dig Dis. 2016;17:600–609. doi: 10.1111/1751-2980.12399. [DOI] [PubMed] [Google Scholar]
- 15.Kabakchiev B. Gastroenterology. 2013;144:1488–1496. doi: 10.1053/j.gastro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.