Abstract Abstract
To date, only one mitochondrial genome (mitogenome) in the Eumeninae has been reported in the world and this is the first report in China. The mitogenome of O.a.aterrimus is 17 972 bp long, and contains 38 genes, including 13 protein coding genes (PCGs), 23 tRNA genes, two rRNA genes, a long non-coding region (NCR), and a control region (CR). The mitogenome has 79.43% A + T content, its 13 PCGs use ATN as the initiation codon except for cox1 using TTG, and nine genes used complete translation termination TAA and four genes have incomplete stop codon T (cox2, cox3, nad4, and cytb). Twenty-two of 23 tRNAs can form the typical cloverleaf secondary structure except for trnS1. The CR is 1 078 bp long with 84.69% A+T content, comprising 28 bp tandem repeat sequences and 13 bp T-strech. There are two gene rearrangements which are an extra trnM2 located between trnQ and nad2 and the trnL2 in the upstream of nad1. Within all rearrangements of these mitogenomes reported in the family Vespidae, the translocation between trnS1 and trnE genes only appears in Vespinae, and the translocation of trnY in Polistinae and Vespinae. The absent codons of 13 PCGs in Polistinae are more than those both in Vespinae and Eumeninae in the family Vespidae. The study reports the complete mitogenome of O.a.aterrimus, compares the characteristics and construct phylogenetic relationships of the mitogenomes in the family Vespidae.
Keywords: Eumeninae , mitochondrial genomes, Orancistrocerus aterrimus aterrimus , phylogenetic analysis, Vespidae
Introduction
Animal mitochondrial genomes (mitogenomes) have been widely used in studies of molecular evolution, population genetic structure, and phylogeny because of their stable gene content, rapid evolutionary rate, relatively conserved gene arrangement, maternal inheritance, and infrequent recombination (Wolstenholme 1992; Saccone et al. 1999; Oliveira et al. 2008; Li et al. 2017). The family Vespidae has more than 5000 known species worldwide, which are divided into six subfamilies, Euparagiinae, Masarinae, Eumeninae, Stenogastrinae, Polistinae, and Vespinae (Carpenter 1993), but their phylogenetics have not been settled. There have been ten mitogenomes sequences reported in the Vespidae (seven in the subfamily Vespinae, three in Polistinae, and one in Eumeninae) (Table 1). Among these six subfamilies, there are more than 3600 species in the subfamily Eumeninae worldwide, more than half of the known species of Vespidae. The species in Eumeninae, also known as potter wasps, are solitary, and mostly catch caterpillars as food for their next generation in the environment of farmlands, forests, and orchards, which can directly control caterpillar pests. To date, there is only one species (Abispaephippium) with its mitogenome published (Cameron et al. 2008). Orancistrocerusaterrimusaterrimus, the species under study in this work, belongs to the Eumeninae, and is widely distributed in China (Jiangsu, Anhui, Fujian, Jiangxi, Hunan, Guangxi, Chongqing, Sichuan, Yunnan provinces), and Laos, Vietnam (Li 1985; Selis 2018).
Table 1.
The information of Vespidae mitogenomes used in the phylogenetic analysis in the present study.
| Subfamily | Species | Migenome size (bp) | Gene number | GenBank Accession | Reference |
| Ingroup (Vespidae) | |||||
| Eumeninae | Orancistrocerus aterrimus aterrimus | 17972 | 38 | KY941926 | This study |
| Eumeninae | Abispa ephippium | 16953 | 41 | EU302588 | Cameron et al. (2008) |
| Polistinae | Polistes jokahamae | 16616 | 34 | KR052468 | Song et al. (2016) |
| Polistinae | Polistes humilis synoecus | 14741 | 34 | EU024653 | Cameron et al. (2008) |
| Polistinae | Parapolybia crocea | 16619 | 37 | KY679828 | Peng et al. (2017) |
| Vespinae | Vespula germanica | 16342 | 33 | KR703583 | Zhou et al. (2016) |
| Vespinae | Vespa ducalis | 15779 | 37 | KX950825 | Kim et al. (2017a) |
| Vespinae | Vespa mandarinia | 15902 | 37 | KR059904 | Chen et al. (2015) |
| Vespinae | Vespa bicolor | 16937 | 35 | KJ735511 | Wei et al. (2014) |
| Vespinae | Vespa velutina nigrithorax | 16475 | 37 | KY091645 | Kim et al. (2017b) |
| Vespinae | Vespa orientalis | 16101 | 37 | KY563657 | Nizar et al (2017) |
| Vespinae | Dolichovespula panda | 17137 | 37 | KY293679 | Fan et al (2017) |
| Outgroup (Formicidae) | |||||
| Formicinae | Formica selysi | 16752 | 37 | KP670862 | Yang et al. (2015) |
In the present study, the complete mitogenome of O.a.aterrimus was sequenced using Illumina sequencing technique, and its characteristics analyzed, including gene rearrangements, nucleotide composition, codon usage, etc. More importantly, the phylogenetic relationships of 12 species of mitogenomes in Vespidae are constructed and discussed based on nucleotide sequences of 13 PCGs using both Maximum Likelihood (ML) and Bayesian Inference (BI) methods. The study updates phylogenetic research based on the mitogenomes, and provides basic information framework of mitogenomes in Vespidae for further research on the phylogenetic relationships of both genera and subfamilies in this family.
Materials and methods
Sample collection and DNA preparation
The specimens of O.a.aterrimus were collected from Yangshuo county of Guangxi province, preserved in the 100% ethanol, and stored at -20 °C. Total DNA of a single adult specimen was extracted from the muscle tissues using the DNeasy DNA Extraction Kit (QIAGEN) in accordance with the manufacturer’s instructions. The concentration of genomic DNA in extraction product was assayed on a Qubit fluorometer using a dsDNA High-sensitivity Kit (Invitrogen).
Mitogenomes sequencing and assembling
The Illumina TruSeq library was constructed from the gDNA with the average length of the inserted fragment of 480 bp. The library was sequenced on a full run of Illumina Hiseq 2500 with 500 cycles and paired-end sequencing (250 bp reads). High-quality reads were used in de novo assembly with IDBA-UD after removing adapters, unpaired, short and low quality reads (Peng et al. 2012). With IDBA-UD, these parameters have a similarity threshold of 98% and minimum and maximum k values of 80 and 240 bp, respectively. To identify the mitogenome assemblies from the pooled sequencing files, two different fragments of mtDNA (cox1 and rrnS) were amplified as bait sequences by standard PCR reactions using primers designed with reference of Simon et al. (2006). Using BLASTN search against the reference of bait sequences, matching rate of 100% was confirmed as the mitogenome of O.a.aterrimus. The identical or near-identical overlapping terminal regions of mitogenome sequences were examined and circularized by Geneious (http://www.geneious.com/).
Sequence annotations and analysis
PCGs and rRNA genes were aligned with other published Vespidae insect mitogenomes by Clustal X (Thompson et al. 1997). The majority of the tRNA gene locations and secondary structures were identified by tRNAscan-SE Search Server v.1.21 (Lowe and Eddy 1997), and the remaining tRNA were identified in comparison with other known species of tRNAs in Vespidae (Cameron et al. 2008; Song et al. 2016). The CRand the tandem repeat sequence were analyzed with Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html) (Benson 1999). Base composition and codon usage in all 12 mitogenomes of Vespidae were calculated by MEGA v 6. 0 (Tamura et al. 2013). In addition, the AT skew = [A - T] / [A + T] and GC skew = [G - C] / [G + C] were computed (Perna and Kocher 1995).
Phylogenetic analysis
Eleven known mitogenome sequences in the family Vespidae and the mitogenome sequence of Formicaselysi (KP670862) in the family Formicidae were downloaded from GeneBank, and that of O.a.aterrimus was produced in the present study (Table 1). The phylogenetic tree of 12 mitogenomes sequences in the family Vespidae was constructed using ML and BI methods with MEGA 6.0 (Tamura et al. 2013) and MrBayes 3.1.1 (Huelsenbeck and Ronquist 2001), and the Formicaselysi (KP670862) was used as outgroup. The nucleotide sequences of 13 PCGs were applied in the phylogenetic inference, and the best fitting substitution model was detected using Mrmodeltest 2.3 (Nylander 2004). The bootstrap values were calculated based on 1000 replications, and the confidence values of the topology is high.
Results and discussion
Genomic organization
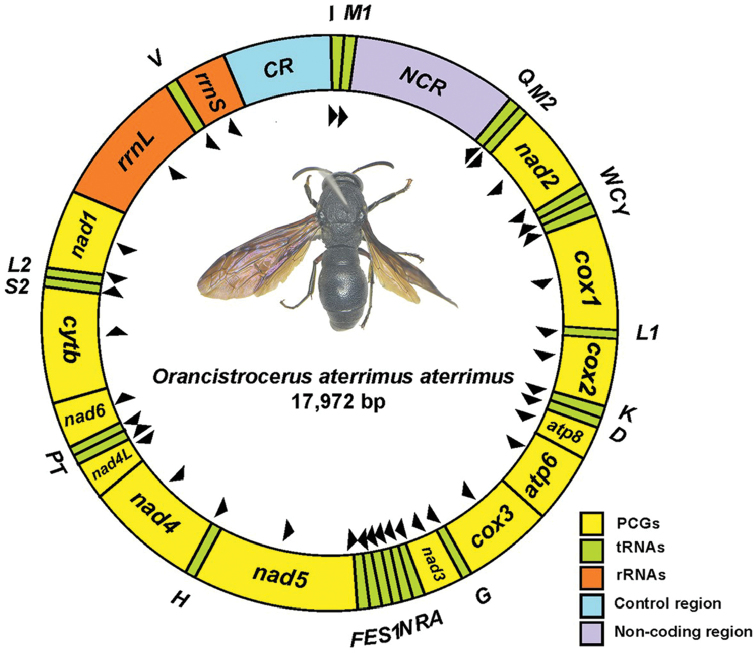
The complete mitogenome of O.a.aterrimus is a double-strand of circular molecular DNA and 17,972 bp. It contains 38 genes: 13 PCGs, 23 tRNAs, two rRNAs, a control region (CR), and a long non-coding region (NCR) (Figure 1), of which 24 genes are situated in the majority strand (J-strand) and the other 14 genes are located in the minority strand (N-strand) (Table 2). An extra trnM2 and a long NCR were found in the mitogenome. The gene trnM2 is 67 bp and located in 2 142-2 208 between trnQ and nad2. The NCR is 1 946 bp long, located in 128-2 073 between trnM1 and trnQ. With the exception of the NCR (1 946 bp), 14 intergenic spacers exist and sum to 174 bp, of which the longest spacer is 48 bp long, located between nad4l and trnT. In addition, a total of 24 bp overlaps was identified in 12 genes, with the overlap length of each gene ranging from 1 to 8 bp.
Figure 1.
The mitochondrial genome of O.a.aterrimus. Arrows indicate the direction of genes. Abbreviations of the gene name are as follows: nad1-4 and nad4L act as nicotinamide adenine dinucleotide hydrogen dehydrogenase subunits 1-6 and 4L; cox1, cox2, and cox3 act as the cytochrome C oxidase subunits; cytb act as cytochrome b; atp8 and atp6 act as adenosine triphosphate synthase subunits 6 and 8; rrnL and rrnS act as large and small rRNA subunits; In addition, CR indicates control region and NCR indicates non-coding region.
Table 2.
Mitochondrial genome annotation of O.a.aterrimus.
| Gene | Direction | Location | Size (bp) | Anticodon | Codon | Intergenic nucleotides | |
| Start | Stop | ||||||
| trnI | F | 1–63 | 63 | 30–32 GAT | |||
| trnM1 | F | 63–127 | 65 | 93–95 CAT | -1 | ||
| non-coding region | 1946 | ||||||
| trnQ | R | 2074–2138 | 65 | 2108–2110 TTG | 0 | ||
| trnM2 | F | 2142–2208 | 67 | 2173–2175 CAT | 3 | ||
| nad2 | F | 2209–3234 | 1026 | ATC | TAA | 0 | |
| trnW | F | 3249–3315 | 67 | 3280–3282 TCA | 14 | ||
| trnC | R | 3308–3374 | 67 | 3342–3344 GCA | -8 | ||
| trnY | R | 3383–3447 | 65 | 3416–3418 GTA | 8 | ||
| cox1 | F | 3446–4981 | 1536 | TTG | TAA | -2 | |
| trn L1 | F | 5006–5073 | 68 | 5035–5037 TAA | 24 | ||
| cox2 | F | 5074–5752 | 679 | ATC | T- | 0 | |
| trnK | F | 5753–5824 | 72 | 5785–5787 CTT | 0 | ||
| trnD | F | 5824–5893 | 70 | 5858–5860 GTC | -1 | ||
| atp8 | F | 5894–6049 | 156 | ATC | TAA | 0 | |
| atp6 | F | 6049–6720 | 672 | ATG | TAA | -1 | |
| cox3 | F | 6742–7525 | 784 | ATG | T- | 21 | |
| trnG | F | 7526–7593 | 68 | 7556–7558 TCC | 0 | ||
| nad3 | F | 7594–7947 | 354 | ATT | TAA | 0 | |
| trnA | F | 7947–8011 | 65 | 7977–7979 TGC | -1 | ||
| trnR | F | 8011–8074 | 64 | 8038–8040 TCG | -1 | ||
| trnN | F | 8078–8147 | 70 | 8108–8110 GTT | 3 | ||
| trn S1 | F | 8147–8206 | 60 | 8168–8170TCT | -1 | ||
| trnE | F | 8214–8277 | 64 | 8244–8246 TTC | 7 | ||
| trnF | R | 8277–8342 | 66 | 8307–8309 GAA | -1 | ||
| nad5 | R | 8344–10032 | 1689 | ATT | TAA | 1 | |
| trnH | R | 10033–10096 | 64 | 10065–10067 GTG | 0 | ||
| nad4 | R | 10097–11402 | 1306 | ATA | T- | 0 | |
| nad4l | R | 11399–11677 | 279 | ATT | TAA | -4 | |
| trnT | F | 11726–11789 | 64 | 11756–11758 TGT | 48 | ||
| trnP | R | 11789–11858 | 70 | 11823–11825 TGG | -1 | ||
| nad6 | F | 11860–12399 | 540 | ATG | TAA | 1 | |
| cytb | F | 12403–13534 | 1132 | ATG | T- | 3 | |
| trnS2 | F | 13544–13612 | 69 | 13572–13574 TGA | 9 | ||
| trnL2 | R | 13640–13707 | 68 | 13676–13678 TAG | 27 | ||
| nad1 | R | 13708–14676 | 969 | ATA | TAA | 0 | |
| rrnL | R | 14682–16044 | 1363 | 5 | |||
| trnV | R | 16043–16106 | 64 | 16074–16076 TAC | -2 | ||
| rrnS | R | 16107–16894 | 788 | 0 | |||
| Control region | 16895–17972 | 1078 | 0 | ||||
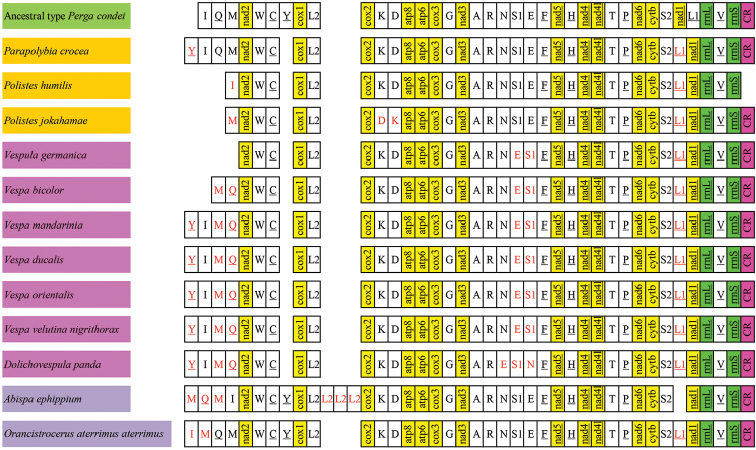
Gene rearrangements
The gene order of 13 PCGs and two rRNAs in O.a.aterrimus mitogenome is consistent with the putative hymenopteran ancestor: the sawfly Pergacondei (Hymenoptera: Symphyta: Pergidae:) (Castro and Dowton 2005). However, there are two rearrangements of tRNAs in the mitogenome (Figure 2), namely, an extra trnM2 and trnL2 in the upstream of nad1, contributing to the novel gene order: trnL2 - nad1 - rrnL - trnV - rrnS - CR - trnI - trnM1 - trnQ - trnM2 - nad2 (Figure 2). In the mitogenome of Abispaephippium, another species in the subfamily Eumeninae, the gene order of rearrangements is trnL2 - trnM1 - trnQ - trnM2 - trnI, trnL1 - trnL1 - trnL1 - trnL1 and trnS2 - nad1 (Figure 2) (Cameron et al. 2008). In the subfamily Polistinae, the translocation between nad1 and trnL1 is present in three reported species. In addition, the translocation of trnY in Parapolybiacrocea occurs, trnQ, trnM and trnY genes are lost in Polisteshumilis mitogenome, and in Polistesjokahamae mitogenome, not only trnD is in the upstream of trnK but also trnI, trnQ and trnY are missing (Figure 2) (Cameron et al. 2008; Song et al. 2016; Peng et al. 2017). In the subfamily Vespinae, except for the incomplete mitogenomes of Vespulagermanica and Vespabicolor, there is the same rearrangements in other four reported species, such as the translocation of trnY, the translocation between trnQ and trnM genes, between trnS1 and trnE genes, and between nad1 and trnL2(CUN) genes, respectively and Dolichovespulapanda is different from other four species: the translocation between trnS1 and trnE genes in exchange for shuffling of trnN and trnE (Figure 2) (Chen et al. 2016; Fan et al. 2017; Kim et al. 2017a; Kim et al. 2017b; Nizar et al. 2017). In general, the rearrangement frequency in Eumeninae is lower than those of both Vespinae and Polistinae. The rearrangement of tRNAs is a typical event in the mitogenomes of Hymenoptera (Dowton and Austin 1999; Dowton et al. 2009; Chen et al. 2016).
Figure 2.
Mitochondrial gene arrangement of 12 species of Vespidae. The red fonts indicate the rearrangement of the genes.
Nucleotide composition
To date, the nucleotide compositions of ten complete mitogenomes have been reported in the family Vespidae. In the subfamily Eumeninae, the overall A + T content of O.a.aterrimus and Abispaephippium mitogenomes is 79.43% and 80.61%, respectively (Table 3). Among all Vespidae mitogenomes, there are no significant differences of the A + T content of Polistinae, i.e., P.humilis being 84.73%, P.jokahamae 83.41%, and Parapolybiacrocea 82.94%, respectively. In the subfamily Vespinae, there are a little differences of the A + T content from Vespamandarinia 79.39% to Dolichovespulapanda being 84.61%. Generally speaking, the A + T content of Eumeninae is lower than those of both Vespinae and Polistinae. According to these different regions of O.a.aterrimus mitogenome, the A + T content of 13 PCGs is 78.27% near to A.ephippium (78.67%). In tRNAs, rRNAs, and CR, the A + T content is 83.41%, 84.29% and 84.69%, respectively. From the A + T content of all known Vespidae complete mitogenomes (Table 3), a universal feature is presumed that A + T content of tRNAs and rRNAs higher than that of PCGs.
Table 3.
Nucleotide composition of different regions in all complete Vespidae mitogenomes.
| Species | Regions | Size(bp) | A% | T% | G% | C% | (A+T)% | AT-skew | GC-skew |
|---|---|---|---|---|---|---|---|---|---|
| Orancistrocerus aterrimus aterrimus | Whole genome | 17972 | 39.53 | 39.9 | 8.06 | 12.51 | 79.43 | -0.005 | -0.216 |
| Protein coding genes | 11122 | 33.15 | 45.12 | 10.02 | 11.72 | 78.27 | -0.153 | -0.078 | |
| tRNA genes | 1525 | 42.69 | 40.72 | 9.25 | 7.34 | 83.41 | 0.024 | 0.115 | |
| rRNA genes | 2151 | 41.89 | 42.4 | 10.79 | 4.93 | 84.29 | -0.006 | 0.373 | |
| Control region | 1078 | 39.8 | 44.9 | 6.49 | 8.81 | 84.69 | -0.06 | -0.152 | |
| Abispa ephippium | Whole genome | 16953 | 39.55 | 41.05 | 6.02 | 13.38 | 80.61 | -0.019 | -0.38 |
| Protein coding genes | 11305 | 35.2 | 43.48 | 10.12 | 11.21 | 78.67 | -0.105 | -0.051 | |
| tRNA genes | 1787 | 44.66 | 38.84 | 8.95 | 7.55 | 83.49 | 0.07 | 0.085 | |
| rRNA genes | 2180 | 43.62 | 38.35 | 5.14 | 12.89 | 81.97 | 0.064 | 0.43 | |
| Control region | 308 | 43.83 | 46.1 | 1.3 | 8.77 | 89.94 | -0.025 | -0.742 | |
| Polistes jokahamae | Whole genome | 16616 | 41.97 | 41.45 | 5.8 | 10.79 | 83.41 | 0.006 | -0.301 |
| Protein coding genes | 10852 | 36.77 | 46.61 | 8.11 | 8.51 | 83.38 | -0.118 | -0.024 | |
| tRNA genes | 1318 | 44.76 | 42.64 | 6.98 | 5.61 | 87.4 | 0.024 | 0.108 | |
| rRNA genes | 2257 | 43.95 | 41.25 | 4.3 | 10.5 | 85.2 | 0.032 | 0.419 | |
| Control region | 1096 | 39.05 | 46.53 | 6.84 | 7.57 | 85.58 | -0.087 | -0.051 | |
| Polistes humilis | Whole genome | 14741 | 43.09 | 41.65 | 5.32 | 9.95 | 84.73 | 0.017 | -0.303 |
| Protein coding genes | 10852 | 36.77 | 46.61 | 8.11 | 8.51 | 83.38 | -0.118 | -0.024 | |
| tRNA genes | 1258 | 47.22 | 41.02 | 6.52 | 5.25 | 88.24 | 0.07 | 0.108 | |
| rRNA genes | 1932 | 43.27 | 43.22 | 9.16 | 4.35 | 86.49 | 0.001 | 0.356 | |
| Control region | * | * | * | * | * | * | * | * | |
| Parapolybia crocea | Whole genome | 16619 | 43.39 | 39.55 | 5.91 | 11.15 | 82.94 | 0.046 | -0.307 |
| Protein coding genes | 11022 | 35.48 | 45.16 | 9.54 | 9.82 | 80.65 | -0.12 | -0.015 | |
| tRNA genes | 1486 | 44.01 | 42.13 | 7.67 | 6.19 | 86.14 | 0.022 | 0.107 | |
| rRNA genes | 2176 | 40.3 | 45.96 | 9.38 | 4.37 | 86.26 | -0.066 | 0.365 | |
| Control region | 1316 | 42.25 | 46.05 | 5.17 | 6.53 | 88.3 | -0.043 | -0.117 | |
| Vespa ducalis | Whole genome | 15779 | 40.32 | 39.8 | 5.8 | 14.08 | 80.12 | 0.006 | -0.417 |
| Protein coding genes | 11159 | 34.32 | 43.46 | 10.36 | 11.86 | 77.78 | -0.118 | -0.067 | |
| tRNA genes | 1487 | 45.46 | 40.15 | 8.14 | 6.25 | 85.61 | 0.062 | 0.131 | |
| rRNA genes | 2299 | 44.58 | 39.58 | 11.44 | 4.39 | 84.17 | 0.059 | 0.445 | |
| Control region | 166 | 46.99 | 45.78 | 0 | 7.23 | 92.77 | 0.013 | -1 | |
| Vespa mandarinia | Whole genome | 15902 | 38.88 | 40.51 | 6.07 | 14.53 | 79.39 | -0.021 | -0.41 |
| Protein coding genes | 11119 | 33.73 | 43.37 | 10.56 | 12.35 | 77.09 | -0.125 | -0.078 | |
| tRNA genes | 1505 | 45.12 | 40.47 | 8.37 | 6.05 | 85.58 | 0.054 | 0.161 | |
| rRNA genes | 1569 | 43.91 | 39.64 | 12.11 | 4.33 | 83.56 | 0.051 | 0.473 | |
| Control region | 200 | 49 | 39.5 | 0.5 | 11 | 88.5 | 0.107 | -0.913 | |
| Vespa velutina nigrithorax | Whole genome | 16475 | 40.3 | 41.44 | 5.43 | 12.83 | 81.74 | -0.014 | -0.406 |
| Protein coding genes | 11197 | 34.99 | 44.75 | 9.42 | 10.83 | 79.74 | -0.122 | -0.07 | |
| tRNA genes | 1514 | 44.58 | 41.35 | 8.12 | 5.94 | 85.93 | 0.038 | 0.155 | |
| rRNA genes | 2319 | 45.11 | 40.06 | 10.52 | 4.31 | 85.17 | 0.059 | 0.419 | |
| Control region | 132 | 50.76 | 41.67 | 0 | 7.58 | 92.42 | 0.098 | -1 | |
| Vespa orientalis | Whole genome | 16101 | 40.65 | 40.3 | 5.86 | 13.19 | 80.95 | 0.004 | -0.384 |
| Protein coding genes | 10653 | 34.5 | 44.08 | 9.74 | 11.68 | 78.58 | -0.122 | -0.09 | |
| tRNA genes | 1481 | 45.51 | 40.51 | 7.97 | 6.01 | 86.02 | 0.058 | 0.14 | |
| rRNA genes | 2079 | 43.67 | 39.15 | 11.5 | 5.68 | 82.83 | 0.055 | 0.339 | |
| Control region | 60 | 48.33 | 41.67 | 8.33 | 1.67 | 90 | 0.074 | 0.667 | |
| Dolichovespula panda | Whole genome | 17136 | 42.8 | 41.81 | 5.39 | 10 | 84.61 | 0.012 | -0.3 |
| Protein coding genes | 11276 | 35.82 | 46.78 | 8.78 | 8.62 | 82.6 | -0.133 | 0.009 | |
| tRNA genes | 1506 | 45.88 | 40.44 | 7.9 | 5.78 | 86.32 | 0.063 | 0.155 | |
| rRNA genes | 2126 | 43.7 | 40.87 | 10.68 | 4.75 | 84.57 | 0.033 | 0.384 | |
| Control region | 586 | 67.24 | 32.42 | 0 | 0.34 | 99.66 | 0.349 | -1 |
* P.humilis (EU024653), not sequenced for the control region
Two other parameters, AT-skew and GC-skew, have been widely used to measure the nucleotide compositional behaviors of mitogenome in addition to the A + T content (Enrico et al. 2011). The AT skew of O.a.aterrimus mitogenome is -0.005 near to 0, and the GC skew (-0.216) is negative. The base composition bias plays an important role in researching the mechanism of replication and transcription of mitogenomes (Wei et al. 2010).
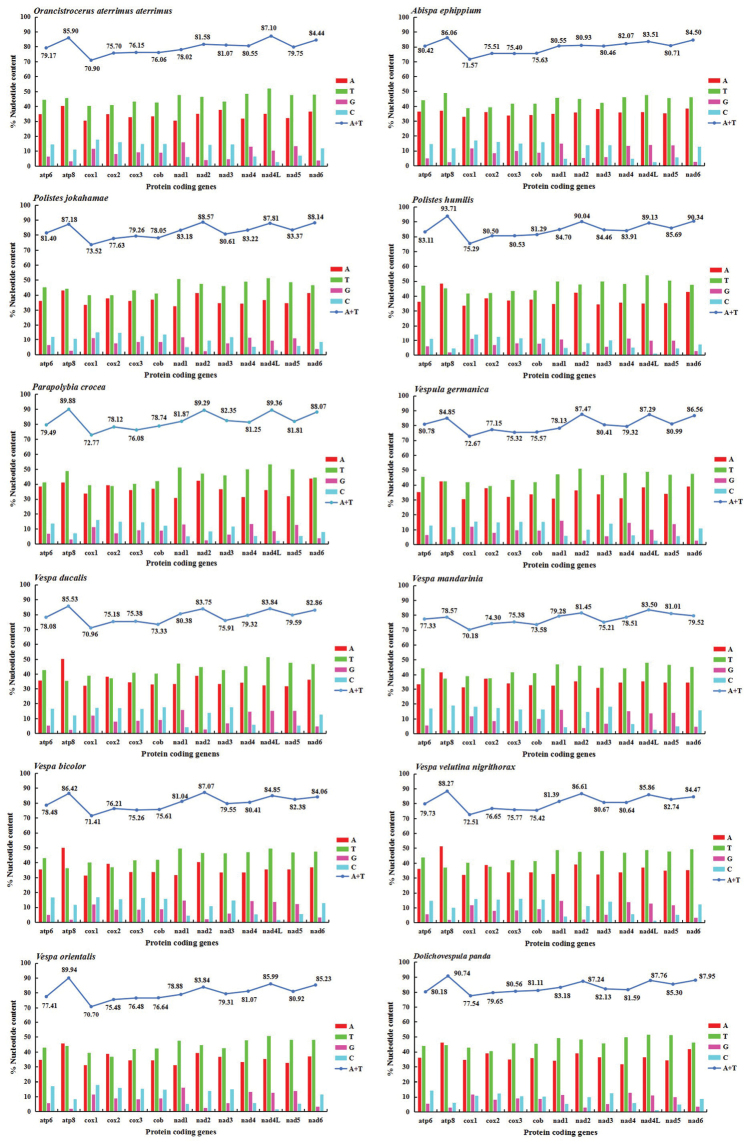
Among the PCGs of 12 Vespidae species (containing two incomplete mitogenomes), the A + T content of cox1 is the lowest in 13 PCGs, ranging from 70.18% (Vespamandarinia) to 75.29% (P.humilis) (Figure 3). The A + T content of atp8, nad2, and nad4L is highest (Figure 3). This result ascertains cox1 is conserved relatively again, which is the reason for former abundant phylogenetic analysis in other insects (Rivera and Currie 2009; Santos et al. 2015). In addition, it is a common phenomenon that T content is more than A, and C content is slightly more than G (Figure 3).
Figure 3.
Nucleotide composition of all 13 PCGs of eleven species of Vespidae.
Protein coding genes
In the 13 PCGs of the O.a.aterrimus mitogenome, nine PCGs are encoded in the J-strand, and the other four PCGs are located in the N-strand. The total length of PCGs is 11 122 bp. All PCGs use the conventional start codons ATN except for cox1 using TTG which was also employed as the initiation codon in other insects (Sheffield et al. 2008; Li et al. 2012a). The termination codons of nine PCGs in O.a.aterrimus mitogenome use complete TAA (nad2, cox1, atp8, atp6, nad3, nad5, nad4l, nad6 and nad1), and other four genes have incomplete stop codons T (cox2, cox3, nad4 and cytb). In general, the termination codons of insect mitogenomes PCGs were the TAA or incomplete T (Ojala et al. 1981; Li et al. 2012a).
There is a total of 3697 codons in O.a.aterrimus mitogenome, excluding termination codons, which is within the range of the common insect mitogenomes codon number (3585-3746) (Cha et al. 2007). According to the relative synonymous codon usage (RSCU), all of these 12 Vespidae species frequently used UUU, UUA, AUU and AUA (Figure 4), leading to the high A + T content in the PCGs of the family Vespidae mitogenomes. CUG is absent in O.a.aterrimus mitogenome and CGC and AGC are absent in A.ephippium. Some codons are also lacking in other species of Vespidae. For example, CGC and AGC in Vespaorientalis, CUG, GCG, CGC in V.bicolor and CCG, ACC, ACG, GCG, UGC, and CGC in Dolichovespulapanda are absent, respectively. There are several codons missing in Polistesjokahamae, namely, CUG, GUC, ACG, GCG, CGC, CGG, AGC; and CUG, GUC, GCG, CGC, and GGC are also lacked in P.humilis (Figure 4). Thus, the amount of absent codons in Vespinae and Polistinae is more than in Eumeninae.
Figure 4.
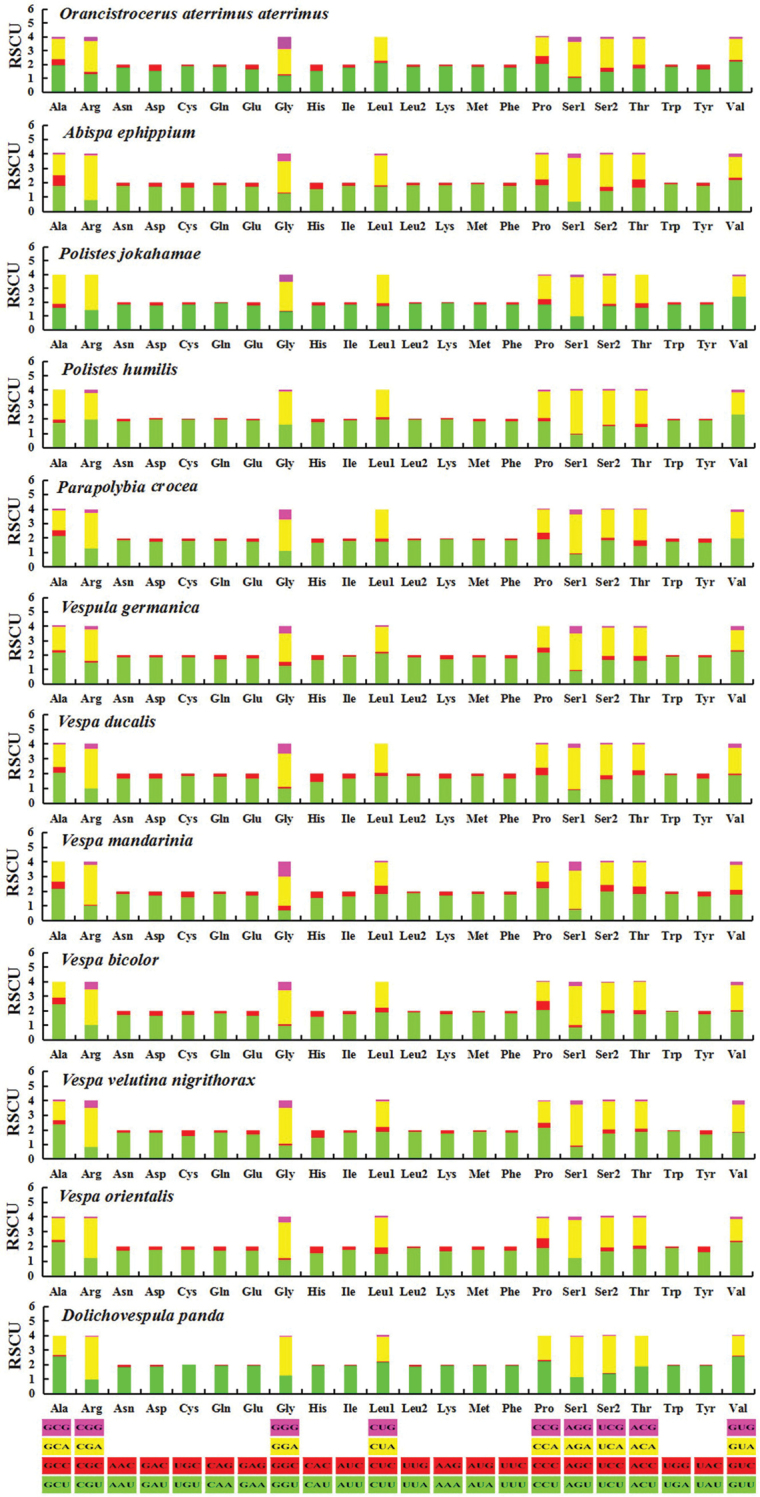
Relative synonymous codon usage (RSCU) in Vespidae. Codon families are displayed along the x-axis.
Transfer RNA and ribosomal RNA genes
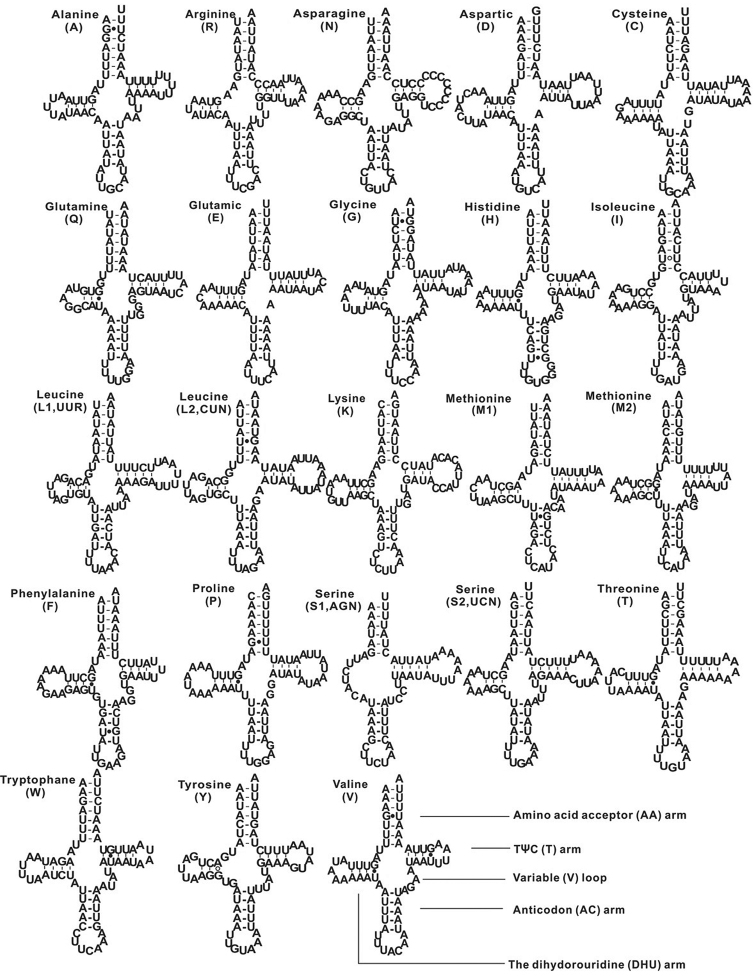
There are 23 tRNAs found in O.a.aterrimus mitogenome and their lengths range from 60 bp (trnS1) to 72 bp (trnK) including an extra trnM2, whereas usually there are 22 tRNAs in other insects (Boore 1999; Chen et al. 2015). Among 23 anticodons of these tRNAs, 21 are coincident with the majority of insects mitogenomes (Lee et al. 2008; Hua et al. 2016), but trnI and trnS1 change from CCT to GAT, and GCT to TCT, respectively. Except for trnS1, the other 22 tRNAs have the capability of folding into typical clover-leaf secondary structures. The secondary structure of trnS1 lacks the dihydrouridine DHU arm and reduces its shape to a simple loop (Figure 5), which is a common phenomenon in metazoan mitogenomes (Wolstenholme 1992; Li et al. 2012b). There are 20 mismatches in 13 tRNAs, including 18 unmatched GU base pairs, an unmatched AG, and an unmatched UU (Figure 5).
Figure 5.
Secondary structures of 23 tRNAs of O.a.aterrimus mitochondrial genome. Watson-Crick bonds are showed by dashes, GU pairs by filled dots, and AG and UU by open dots.
The length of rrnL is 1 363 bp long, located between nad1 and trnV, and rrnS 788 bp long in minority strand between trnV and CR. The A + T content of two genes is 84.29% (rrnL and rrnS) (Table 3).
A control region and a non-coding region
The CR plays an important role in regulating of replication and transcription of mitogenomes (Taanman 1999; Saito et al. 2005). The CR of O.a.aterrimus mitogenome is 1078 bp long, located between rrnS and trnI. The A + T content of this region (84.69%) is higher than other region of the O.a.aterrimus mitogenome. There is a tandem repeat model of 28 bp (TATTCCATTTAAGTTCGTAAAAACTAAT) which occurs more than eight times in the O.a.aterrimus mitogenome. Tandem repeat structures in the CR are different in different species (Peng et al. 2017). There is also a poly-T stretch of 13 bp, which may be as recognition site for the initiation of replication in the mitogenomes (Andrews et al. 1999). In the O.a.aterrimus mitogenome, a NCR is situated in position 128 - 2 073 (1 946 bp) between trnM1 and trnQ, which is reported in most insect mitogenomes (Saito et al. 2005; Cameron et al. 2008; Jiang et al. 2016). The A + T content of NCR is 73.69%, among which there is 97 bp (close to trnQ gene) with obviously high A + T content 90.72%. In addition, two tandem repetitive sequences are found in the NCR, which repeated 17 and 18 times, respectively.
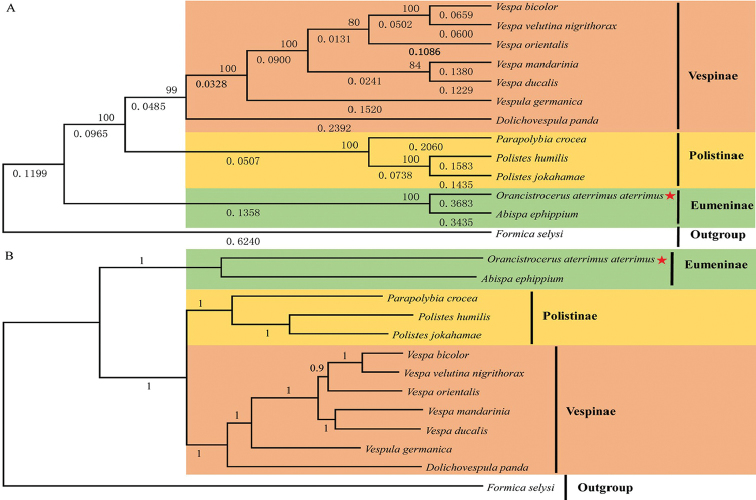
Phylogenetic relationships
The best fitting model GTR + G + I was selected for ML analysis. The phylogeny of mitogenomes in Vespidae was constructed based on the nucleotide sequences of 13 PCGs of 13 species using ML and BI methods (Figure 6). The phylogenetic relationships between 12 species in the family Vespidae are (((((Vespabicolor + Vespavelutinanigrithorax) + Vespaorientalis) + (Vespamandarinia + Vespaducalis)) + Vespulagermanica+ Dolichovespulapanda) + (Parapolybiacrocea+ (Polisteshumilissynoecus + Polistesjokahamae))) + (Orancistrocerusaterrimusaterrimus + Abispaephippium) (Figure 6). O.a.aterrimus and A.ephippium belong to the subfamily Eumeninae, which is concordant with morphological classification. In the present study, Eumeninae is the sister group with (Polistinae + Vespinae), which is different from morphological classification “(Eumeninae + (Stenogastriinae + (Vespinae + Polistinae))) ” (Carpenter 1982, 2003). So far, there is no reported mitogenome in the subfamily Stenogastriinae, so the relationships among the four subfamilies Eumeninae, Stenogastriinae, Vespinae and Polistinae based on mitogenomes need to be further explored in our follow-up studies.
Figure 6.
The phylogenetic relationships were established by the 13 PCGs using ML (A) and BI (B) methods. Numbers abutting branches were bootstrap percentages with 1000 replicates (A) and Bayesian posterior probabilities (B). Red pentagram refers to the mitogenome sequences of O.a.aterrimus.
Conclusions
According to nine complete mitogenomes reported in the family Vespidae, gene numbers of two species (38 and 41 genes) of the subfamily Eumeninae are more than those of the other seven species (34 - 37 genes) of both Polistinae and Vespinae. The rearrangements of tRNAs are common in Vespidae, but rearrangement rules are different in different subfamilies. The translocation between trnS1 and trnE only happens in the subfamily Vespinae, and there are the same rearrangements in these four complete mitogenomes of Vespamandarinia, V.ducalis, V.orientalis, and V.velutinanigrithorax. The translocation of trnY occurs in both Vespinae and Polistinae, whereas trnY location in Eumeninae is consistent with that of the sawfly Pergacondei. The number of absent codons in Eumeninae is less than Vespinae and Polistinae. The phylogenic results of mitogenomes show that O.a.aterrimus and Abispaephippium belong to Eumeninae and (Polistinae + Vespinae) and Eumeninae constitute a sister group. Lastly, these results of this study might suggest that Eumeninae derived earlier than both Polistinae and Vespinae, which is consistent with reported research based on morphology.
Acknowledgements
We are very grateful to James M. Carpenter and Rogério Lopes for their critical comments. This study was funded by the National Natural Science Foundation of China (Nos: 31772490, 31372247, 31000976, 31372265), Young Talent Incubation Program of Chongqing Normal University (14CSDG07), and the Par-Eu Scholars Program.
Citation
Zhang Q-H, Huang P, Chen B, Li T-J (2018) The complete mitochondrial genome of Orancistrocerusaterrimusaterrimus and comparative analysis in the family Vespidae (Hymenoptera, Vespidae, Eumeninae). ZooKeys 790: 127–144. https://doi.org/10.3897/zookeys.790.25356
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. (1999) Reanalysis and revision of Cambridge reference sequence for human mitochondrial DNA. Nature Genetics 23(2): 147. 10.1038/13779 [DOI] [PubMed]
- Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. (1999) Animal mitochondrial genomes. Nucleic Acids Research 27(8): 1767–1780. 10.1093/nar/27.8.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SL, Dowton M, Castro LR, Ruberu K, Whiting MF, Austin AD, Diement K, Stevens J. (2008) Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 51(10): 800–808. 10.1139/G08-066 [DOI] [PubMed] [Google Scholar]
- Carpenter JM. (1982) The phylogenetic relationships and natural classification of the Vespoidea (Hymenoptera). Systematic Entomology 7: 11–38. 10.1111/j.1365-3113.1982.tb00124.x [DOI] [Google Scholar]
- Carpenter JM. (1993) Biogeographic patterns in the Vespidae (Hymenoptera): Two views of Africa and South America. Biological relationships between Africa and South America. Yale University Press, New Haven, 139–155. 10.2307/j.ctt22726mc.11 [DOI]
- Carpenter JM. (2003) On “Molecular phylogeny of Vespidae (Hymenoptera) and the evolution of sociality in wasps”. American Museum Novitates 3389: 1–20. 10.1206/0003-0082(2003)389%3C0001:OMPOVH%3E2.0.CO;2 [DOI] [Google Scholar]
- Castro LR, Dowton M. (2005) The position of the Hymenoptera within the Holometabola as inferred from the mitochondrial genome of Pergacondei (Hymenoptera: Symphyta: Pergidae). Molecular Phylogenetics & Evolution 34(3): 469–479. 10.1016/j.ympev.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim I. (2007) The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombusignitus (Hymenoptera: Apidae). Gene 392(1–2): 206–220. 10.1016/j.gene.2006.12.031 [DOI] [PubMed] [Google Scholar]
- Chen PY, Wei SJ, Liu JX. (2015) The mitochondrial genome of the Vespamandarinia Smith (Hymenoptera: Vespidae: Vespinae) and a phylogenetic analysis of the Vespoidea. Mitochondrial DNA 27(6): 4414–4415. 10.3109/19401736.2015.1089550 [DOI] [PubMed] [Google Scholar]
- Chen PY, Zheng BY, Liu JX, Wei SJ. (2016) Next-Generation sequencing of two mitochondrial genomes, from Family Pompilidae (Hymenoptera: Vespoidea) Reveal Novel Patterns of Gene Arrangement. International Journal of Molecular Sciences 17: 1641. 10.3390/ijms17101641 [DOI] [PMC free article] [PubMed]
- Dowton M, Austin AD. (1999) Evolutionary dynamics of a mitochondrial rearrangement “hot spot” in the Hymenoptera. Molecular Biology and Evolution 16(2): 298–309. 10.1093/oxfordjournals.molbev.a026111 [DOI] [PubMed] [Google Scholar]
- Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF. (2009) Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Molecular Biology and Evolution 26: 1607–1617. 10.1093/molbev/msp072 [DOI] [PubMed] [Google Scholar]
- Enrico N, Massimiliano B, and Tomaso P. (2011) The mitochondrial genome of the ascalaphid owlfly Libelloidesmacaronious and comparative evolutionary mitochondriomics of neuropterid insects. BMC Genomics 12(5): 221. [DOI] [PMC free article] [PubMed]
- Fan XL, Gong YJ, Chen PY, Tan QQ, Tan JL, Wei SJ. (2017) Next-generation sequencing of the mitochondrial genome of Dolichovespulapanda (Hymenoptera: Vespidae) with a phylogenetic analysis of Vespidae. Journal of Asia-Pacific Entomology 20(3): 971–976. 10.1016/j.aspen.2017.07.009 [DOI] [Google Scholar]
- Hua YQ, Din YR, Yan ZT, Si FL, Luo QC, Chen B. (2016) The complete mitochondrial genome of Anophelesminimus (Diptera: Culicidae) and the phylogenetics of known Anopheles mitogenomes. Insect Science 23(3): 353–365. 10.1111/1744-7917.12326 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jiang P, Li H, Song F, Cai Y, Wang JY, Liu JP, Cai WZ. (2016) Duplication and remolding of tRNA genes in the mitochondrial genome of Reduviustenebrosus (Hemiptera: Reduviidae). International Journal of Molecular Sciences 17(6): 951. 10.3390/ijms17060951 [DOI] [PMC free article] [PubMed]
- Kim JS, Jeong JS, Jeong SY, Kim MJ, Kim I. (2017a) Complete mitochondrial genome of the black-tailed hornet, Vespaducalis (Hymenoptera: Vespidae): Genomic comparisons in Vespoidea. Entomological Research 47: 129–136. 10.1111/1748-5967.12218 [DOI] [Google Scholar]
- Kim JS, Jeong JS, Kim I. (2017b) Complete mitochondrial genome of the yellow-legged Asian hornet, Vespavelutinanigrithorax (Hymenoptera: Vespidae). Mitochondrial DNA Part B: Resources 2(1): 82–84. 10.1080/23802359.2017.1285211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Oh J, Kim YU, Kim N, Yang S, Hwang UW. (2008) Mitome: dynamic and interactive database for comparative mitochondrial genomics in metazoan animals. Nucleic Acids Research 36: 938–942. 10.1093/nar/gkm763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Leavengood Jr JM, Chapman EG, Burkhardt D, Song F, Jiang P, Liu JP, Zhou XG, Cai WZ. (2017) Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proceedings of the Royal Society B: Biological Sciences 284: 1–10. 10.1098/rspb.2017.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu H, Shi AM, Štys P, Zhou XG, Cai WZ. (2012a) The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp.. (Hemiptera: Enicocephalidae). PLoS ONE 7: e29419. 10.1371/journal.pone.0029419 [DOI] [PMC free article] [PubMed]
- Li H, Liu HY, Song F, Shi AM, Zhou XG, Cai WZ. (2012b) Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera). PLoS ONE 7: e45925. 10.1371/journal.pone.0045925 [DOI] [PMC free article] [PubMed]
- Li TS. (1985) Economic insect faund of China, Hymenoptera: Vespoidea. Science Press, Beijing, 120–121.
- Lowe TM, Eddy SR. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizar JH, Kosai AN, Bent P, Love D, Nikolaj B, Thomas SP. (2017) Complete mitochondrial genome of the Oriental hornet, Vespaorientalis F. (Hymenoptera: Vespidae). Mitochondrial DNA Part B: Resources 2(1): 139–140. 10.1080/23802359.2017.1292480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Uppsala University: Evolutionary Biology Centre.
- Ojala D, Montoya J, Attardi G. (1981) tRNA punctuation model of RNA procession in human mitochondria. Nature 290: 470–474. 10.1038/290470a0 [DOI] [PubMed] [Google Scholar]
- Oliveira DCSG, Rhitoban R, Lavrov DV, Werren JH. (2008) Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Molecular Biology and Evolution 25: 2167–2180. 10.1093/molbev/msn159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Chen B, Li TJ. (2017) Complete sequence and analysis of Parapolybiacrocea mitochondrial genome (Hymenoptera: Vespidae). Acta Entomologica Sinica 60(4): 464–474. [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL. (2012) IBDA-UD: a de novo assembler for single-cell and metagenomic sequencing data woth highly uneven depth. Bioinformatics 28: 1420–1428. 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- Perna NT, Kocher TD. (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution 41: 353–358. 10.1007/BF01215182 [DOI] [PubMed] [Google Scholar]
- Rivera J, Currie DC. (2009) Identification of Nearctic black flies using DNA barcodes (Diptera: Simuliidae). Molecular Ecology Resources 9: 224–236. 10.1111/j.1755-0998.2009.02648.x [DOI] [PubMed] [Google Scholar]
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. (1999) Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238: 195–209. 10.1016/S0378-1119(99)00270-X [DOI] [PubMed] [Google Scholar]
- Saito S, Tamura K, Aotsuka T. (2005) Replication origin of mitochondrial DNA in insects. Genetics 171(4): 1695–1705. 10.1534/genetics.105.046243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos BF, Payne A, Pickett KM, Carpenter JM. (2015) Phylogeny and historical biogeography of the paper wasp genus Polistes (Hymenoptera: Vespidae): implications for the overwintering hypothesis of social evolution. Cladistics 2015 31(5): 535–549. 10.1111/cla.12103 [DOI] [PubMed]
- Selis M. (2018) Additions to the knowledge of solitary wasps (Hymenoptera: Vespidae: Eumeninae), with description of eight new species. Zootaxa 4403(3): 441–468. 10.11646/zootaxa.4403.3.2 [DOI] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF. (2008) A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Molecular Biology and Evolution 25: 2499–2509. 10.1093/molbev/msn198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. (2006) Incorporating Molecular Evolution into Phylogenetic Analysis, and a New Compilation of Conserved Polymerase Chain Reaction Primers for Animal Mitochondrial DNA. Annual Review of Ecology Evolution & Systematics 37(1–2): 545–579. 10.1146/annurev.ecolsys.37.091305.110018 [DOI] [Google Scholar]
- Song SN, Chen PY, Wei SJ, Chen XX. (2016) The mitochondrial genome of Polistesjokahamae and a phylogenetic analysis of the Vespoidea (Insecta: Hymenoptera). Mitochondrial DNA 27(4): 1–2. [DOI] [PubMed] [Google Scholar]
- Taanman JW. (1999) The mitochondrial: structure, transcription, translation and replication. Biochimica et Biophysica Acta 1410(2): 103–123. 10.1016/S0005-2728(98)00161-3 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6. 0. Molecular Biology and Evolution 30(12): 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Niu FF, Tan JL. (2014) The mitochondrial genome of the Vespabicolor Fabricius (Hymenoptera: Vespidae: Vespinae). Mitochondrial DNA 27(2): 1–2. [DOI] [PubMed] [Google Scholar]
- Wei SJ, Shi M, Chen XX, Sharkey MJ, Achterberg C van, Ye GY, He JH. (2010) New views on strand asymmetry in insect mitochondrial genomes. PLoS ONE 5(9): e12708. 10.1371/journal.pone.0012708 [DOI] [PMC free article] [PubMed]
- Wolstenholme DR. (1992) Animal mitochondrial DNA: structure and evolution. International Review of Cytology 141(6): 173–216. 10.1016/S0074-7696(08)62066-5 [DOI] [PubMed] [Google Scholar]
- Yang S, Li X, Cai LG, Qian ZQ. (2015) Characterization of the complete mitochondrial genome of Formicaselysi (Insecta: Hymenoptera: Formicidae: Formicinae). Mitochondrial DNA 27: 3378–3380. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Hu YL, Xu ZF, Wei SJ. (2016) The mitochondrial genome of the German wasp Vespulagermanica (Fabricius, 1793) (Hymenoptera: Vespoidea: Vespidae). Mitochondrial DNA 27(4): 1–2. [DOI] [PubMed] [Google Scholar]