Abstract
Background & Aims
Microvillus inclusion disease (MVID) is a congenital intestinal malabsorption disorder caused by defective apical vesicular transport. Existing cellular models do not fully recapitulate this heterogeneous pathology. The aim of this study was to characterize 3-dimensional intestinal organoids that continuously generate polarized absorptive cells as an accessible and relevant model to investigate MVID.
Methods
Intestinal organoids from Munc18-2/Stxbp2-null mice that are deficient for apical vesicular transport were subjected to enterocyte-specific differentiation protocols. Lentiviral rescue experiments were performed using human MUNC18-2 variants. Apical trafficking and microvillus formation were characterized by confocal and transmission electron microscopy. Spinning disc time-lapse microscopy was used to document the lifecycle of microvillus inclusions.
Results
Loss of Munc18-2/Stxbp2 recapitulated the pathologic features observed in patients with MUNC18-2 deficiency. The defects were fully restored by transgenic wild-type human MUNC18-2 protein, but not the patient variant (P477L). Importantly, we discovered that the MVID phenotype was correlated with the degree of enterocyte differentiation: secretory vesicles accumulated already in crypt progenitors, while differentiated enterocytes showed an apical tubulovesicular network and enlarged lysosomes. Upon prolonged enterocyte differentiation, cytoplasmic F-actin–positive foci were observed that further progressed into classic microvillus inclusions. Time-lapse microscopy showed their dynamic formation by intracellular maturation or invagination of the apical or basolateral plasma membrane.
Conclusions
We show that prolonged enterocyte-specific differentiation is required to recapitulate the entire spectrum of MVID. Primary organoids can provide a powerful model for this heterogeneous pathology. Formation of microvillus inclusions from multiple membrane sources showed an unexpected dynamic of the enterocyte brush border.
Keywords: Microvillus Atrophy, Disease Modeling, Brush Border Formation, Apical Vesicular Transport
Abbreviations used in this paper: 3D, 3-dimensional; DAPI, 4′,6-diamidino-2-phenylindole; EGFP, enhanced green fluorescent protein; FHL5, familial hemophagocytic lymphohistiocytosis type 5; IWP-2, inhibitor of WNT production-2; KO, knock-out; MVIs, microvillus inclusions; MVID, microvillus inclusion disease; PBS, phosphate-buffered saline; STXBP2, syntaxin binding protein 2; Stx3, syntaxin 3; TEM, transmission electron microscopy; VPA, valproic acid; WT, wild-type
Graphical abstract
See editorial on page 472.
Summary.
Using intestinal organoids from Munc18-2-/- mice we show that the phenotypic manifestation of microvillus inclusion disease is critically linked to the enterocyte differentiation state. Live-cell imaging showed that microvillus inclusions gradually form either intracellularly or by plasma membrane internalization.
Microvillus inclusion disease (MVID), also referred to as microvillus atrophy (OMIM 251850), is a congenital neonatal malabsorption disorder that manifests itself by protracted diarrhea. Total parenteral nutrition and small-bowel transplantation are the treatment options in severe cases. Histologic manifestations are villus atrophy, the absence of microvilli, the accumulation of apical secretory vesicles, and the sporadic formation of pathognomonic intracellular microvillus inclusions (MVIs).1, 2, 3 Genetic evidence that MVID is caused by defective apical vesicle trafficking was obtained first in Rab8 knock-out (KO) mice,4 and later via the identification of MYO5B mutations in patients.5 MYO5B mediates the tethering of RAB8a and other Rab guanosine triphosphatases for apical vesicle transport and membrane recycling.6, 7 Correct fusion of vesicles to the apical membrane also is critical; indeed, MVID can result from congenital mutations in STX3 gene coding for syntaxin 3, an apical vesicle receptor.8 Syntaxin binding protein 2 (STXBP2, also known as MUNC18-2) is a critical co-factor for syntaxin 3. Loss-of-function mutations in STXBP2 cause familial hemophagocytic lymphohistiocytosis type 5 (FHL5), a lymphocyte disorder in which cytotoxic granules fail to fuse with the plasma membrane.9 A broad spectrum of gastrointestinal symptoms has been described in FHL5,10 some of which resemble features of MVID.11, 12
The direct cellular consequences of disturbed apical trafficking are impaired microvillus formation and defective epithelial polarization.6, 13, 14 However, the spectrum of cellular defects in MVID is heterogeneous. Histologic accumulation of periodic acid–Schiff and CD10-positive intracytoplasmic bodies usually is found in the villus region, but also may affect lower crypt cells or be absent.15, 16, 17 Furthermore, the severity of villus blunting,18 the ectopic formation of basolateral microvilli, and the presence of ultrastructural features of MVIs may vary significantly.19 The cellular basis for the incomplete penetrance of MVIs is unknown and there is an open discussion as to whether the structures are formed by internalization of apical plasma membranes4, 7, 19, 20 and/or via intracellular microvillus nucleation.1, 21 A better understanding of the etiology should help us to design therapeutic strategies for improving absorptive functions.
In vitro cell-based models are required to address these questions. Both 2-dimensional and 3-dimensional (3D) culture models of transformed cell lines have been instrumental in identifying and characterizing the mechanisms that underlie vesicle transport and apicobasal polarity. However, the commonly used CaCo2 cell line does not recapitulate the entire phenotypic spectrum of MVID and the presence of mature MVIs rarely has been observed upon loss of MYO5B function.13, 22 Transformed cell lines have impaired differentiation potential and do not continuously self-renew; these are 2 important physiological characteristics of the intestinal epithelium that might have a strong impact in MVID. Mouse small intestinal organoids recapitulate the crypt–villus architecture of the gut via self-organization of stem cells in a 3D extracellular matrix.23 This results in a differentiation gradient with most differentiated cells located in the central villus-like domain. Wnt/R-spondin signaling are key players to maintain stem cells and proliferation in crypts and to prevent spontaneous villus differentiation.24 Hence, in the present study we used small intestinal organoids as a physiologic and accessible epithelial model of MVID biology.
Results
Murine Munc18-2 KO Organoids Recapitulate the Intestinal Features of FHL5 Patients
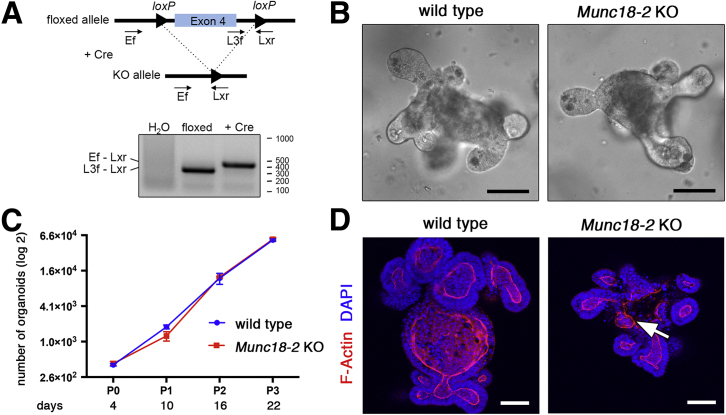
To analyze the molecular mechanism(s) underlying the enteropathy in MVID, we aimed to generate a Munc18-2/Stxbp2–deficient model. Using a constitutive mouse Stxbp2 KO allele, we were unable to obtain homozygous offspring from heterozygous crosses. indicating embryonic lethality. We therefore established small intestinal organoids from mice with a homozygous conditional Munc18-2–null allele. Ablation of exon 4 caused an early frame shift and was achieved by lentiviral Cre delivery, which resulted in the complete and stable absence of the nonrecombined allele (as confirmed by genomic polymerase chain reaction) (Figure 1A). Munc18-2 KO organoids showed a normal morphology, with branching crypt-like protrusions around a central villus-like domain (Figure 1B). Organoid expansion was not affected (Figure 1C) and the cultures could be maintained for many months, indicating that Munc18-2 is not required for cell proliferation or self-renewal. Despite loss of the trafficking regulator Munc18-2, the apicobasal epithelial polarity and crypt-villus architecture were unaffected, as evidenced by the position of the nucleus (DAPI staining) and F-actin staining (Figure 1D). We occasionally observed aberrant structures of the differentiated villus region of KO organoids (Figure 1D).
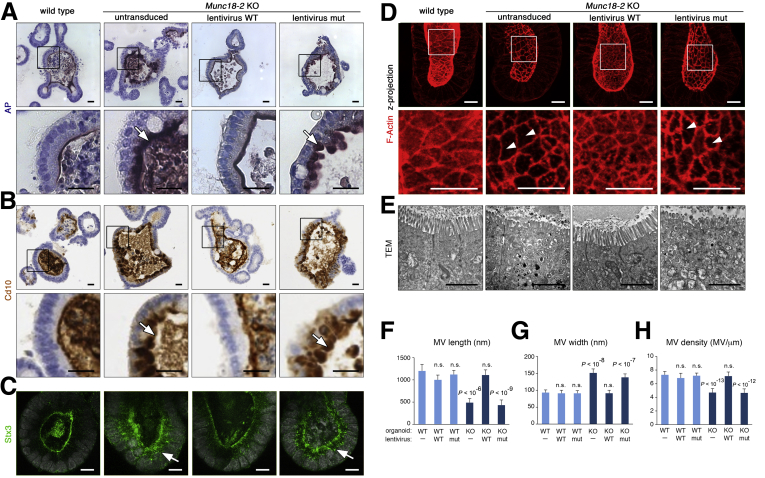
Figure 1.
Generation of a mouse organoid model for Munc18-2 deficiency. (A) Cre-mediated deletion of Munc18-2. Schematic genomic locus and genotyping polymerase chain reaction in organoids. (B) Light microscopy images of WT and Munc18-2 KO small intestinal organoids. (C) Growth curve of WT and Munc18-2 KO organoids (log2 scale). The mean organoid number (±SD) was determined in n = 3 independent wells during 22 days of culture (corresponding to 3 passages). (D) Z-projected confocal images of phalloidin staining and DAPI show maintenance of general morphology in Munc18-2 KO organoids. Irregular F-actin staining is seen in the villus region of KO organoids, indicating mild polarity defects (white arrow). Scale bars: 50 μm.
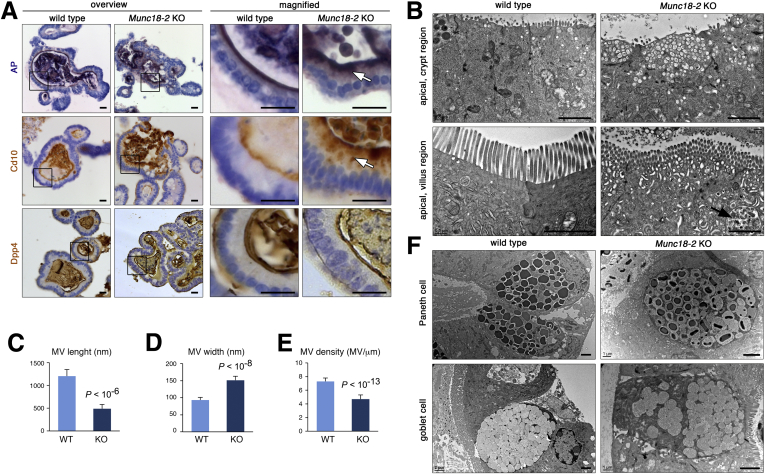
An immunohistochemical assessment showed subapical staining of alkaline phosphatase and Cd10, markers that are located only in the apical brush-border membrane in WT cells (Figure 2A). An abnormal subapical localization was more pronounced in the central villus regions of the organoids, which contain differentiated cells. In contrast, brush-border localization of Dpp4 was unaffected by loss of Munc18-2 (Figure 2A), as noted previously.12 TEM showed the following: (1) subapical vesicle accumulation throughout the epithelium, (2) more complex tubulovesicular structures in the differentiated regions, and (3) the presence of irregularly shaped endomembranes and (auto)lysosomes in the villus regions (Figure 2B). In Munc18-2 KO organoids, the microvilli were significantly shorter, wider, and less dense than in WT organoids (Figure 2C–E). No cytoplasmic vesicle accumulation was observed in the professional secretory cells (ie, goblet and Paneth cells), although the latter appeared to be less mature in the absence of Munc18-2 (Figure 2F). Importantly, we did not observe MVIs under these expansion conditions.
Figure 2.
Brush border and trafficking defects in Munc18-2 KO organoids. (A) Detection of alkaline phosphatase (AP) enzymatic activity (top), Cd10 (middle), and Dpp4 immunostaining (bottom) on paraffin sections of WT and Munc18-2 KO organoids. Overview (left) and magnified villus regions (right) are shown. White arrows show subapical intracellular accumulation of brush-border components. (B) TEM analysis of the apical region. Crypt regions (top) show accumulation of empty vesicles in the Munc18-2 KO. Villus regions (bottom) show the presence of tubulovesicular structures and (auto)lysosomes (arrow). No MVIs are found under expansion conditions. (C–E) Quantification of microvilli (MV) length, width, and density. In total, 70 MVs were measured in n = 7 organoids. Mean values ± SD are shown, P values were determined by t test. (F) Paneth cells (top) and goblet cells (bottom) show some immature aspects in Munc18-2 KO organoids, with reduced electron density of Paneth cell granules and decreased mucus content in goblet cells. Scalebars: (A) 10 μm, (B and F) 2 μm. All staining was confirmed in at least 2 independent experiments.
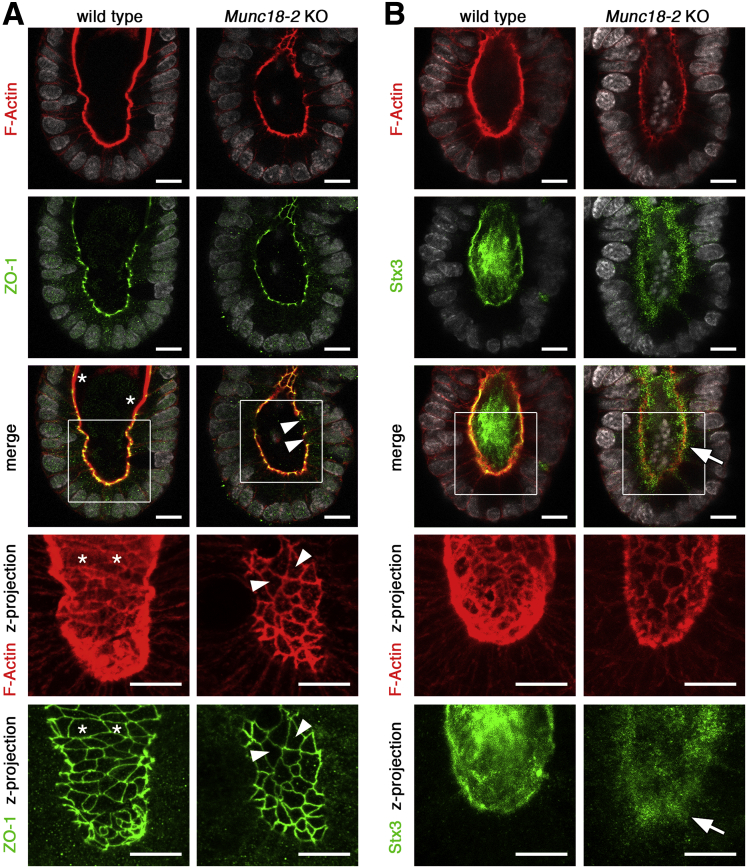
In organoids, self-renewal is driven by a continuous supply of cells from the crypt-like structures. We therefore sought to determine the onset of the observed trafficking defects in the undifferentiated crypt compartment. 3D confocal microscopy showed that brush-border levels of F-actin were very low in KO organoids, whereas the levels of tight-junction–associated F-actin (which co-localized with zonula occludens-1) were normal; this resulted in an interrupted apical staining pattern (Figure 3A). The accumulation of secretory vesicles was shown by immunostaining for syntaxin 3 (Stx3), which localized to apical membranes in WT cells but was dispersed in the apical cytoplasm in mutant cells (Figure 3B). Hence, mislocalization of this critical cargo receptor8 and Munc18-2 binding partner provides a mechanistic explanation for the sorting defects in Munc18-2 KO organoids. The defects observed in intestinal organoids from the Munc18-2 KO mouse thus recapitulate the subcellular intestinal phenotype described in FHL5 patients, in whom variable degrees of villus atrophy, apical vesicle accumulation, and apical trafficking defect were found.11, 12
Figure 3.
Confocal analysis of brush border and trafficking defects in Munc18-2 KO organoids. (A) Defective deposition of brush-border F-actin as shown by phalloidin staining (red). Zonula occludens-1 (ZO-1) co-staining (green) shows that tight junction–associated F-actin is not affected. Single confocal sections (with DAPI-stained nuclei in white). The magnified z-projected images (bottom) show a planar view onto the brush border. WT organoids show a continuous brush border (asterisks) that is interrupted in the Munc18-2 KO (white arrowheads). (B) Confocal analysis of F-actin (red) and Stx3 (green). Stx3 shows a diffuse subapical localization in the Munc18-2 KO (white arrow). Bottom: Magnified z-projected images. Scale bars: 10 μm. Staining was confirmed in 3 independent experiments.
Human MUNC18-2 Can Restore Apical Trafficking in Munc18-2 KO Organoids
We next wondered whether the mouse KO organoids could serve as a genetic model for testing the functionality of human MUNC18-2 variants. To this end, N-terminal GFP-fusions of the WT human protein and the variant from a MUNC18-2–deficient patient (P477L)9 were stably expressed in normal and KO organoids using lentiviral transduction. We found that expression of the WT protein rescued the brush-border localization of alkaline phosphatase, Cd10, Stx3, and F-actin, and the ultrastructural phenotype (Figure 4A–E). In contrast, P477L did not restore the KO organoid phenotype. Quantification of microvillus size and density confirmed that the WT’s rescue was complete, with no dominant-negative effects on control organoids (Figure 4F–H).
Figure 4.
Rescue of brush-border defect in Munc18-2 KO organoids by lentiviral complementation with the WT MUNC18-2 protein but not the P477L patient variant. (A and B) Characterization of apical protein trafficking: (A) alkaline phosphatase activity and (B) Cd10 immunostaining on paraffin sections. White arrows show subapical accumulation of brush-border components. Magnified images (boxes) are shown in the bottom rows. (C) Confocal analysis of Stx3 staining shows rescue of the apical location after expression of the WT MUNC18-2 protein. Diffuse cytoplasmic localization in the Munc18-2 KO (white arrow) after expression of the patient variant. (D) Confocal z-projection of F-actin staining shows a planar view onto the brush border in crypts. White arrowheads mark a disrupted brush border. Magnified images (boxes) are shown in the bottom row. (E) TEM images show restored ultrastructural phenotype by expression of the WT human protein. No MVIs were observed under expansion conditions. (F–H) Quantification of microvilli (MV) (F) length, (G) width, and (H) density. Mean values ±SD in n = 70 ±7, 40 ±2, 40 ±2, 70 ±7, 60 ±4, and 60 ±4 MVs each (with number of independent organoids in brackets). P values (t test) compared with the untransduced WT are shown. Scale bars: (A and B) 20 μm, (C and D) 10 μm, and (E) 2 μm. All staining was confirmed in at least 2 independent experiments.
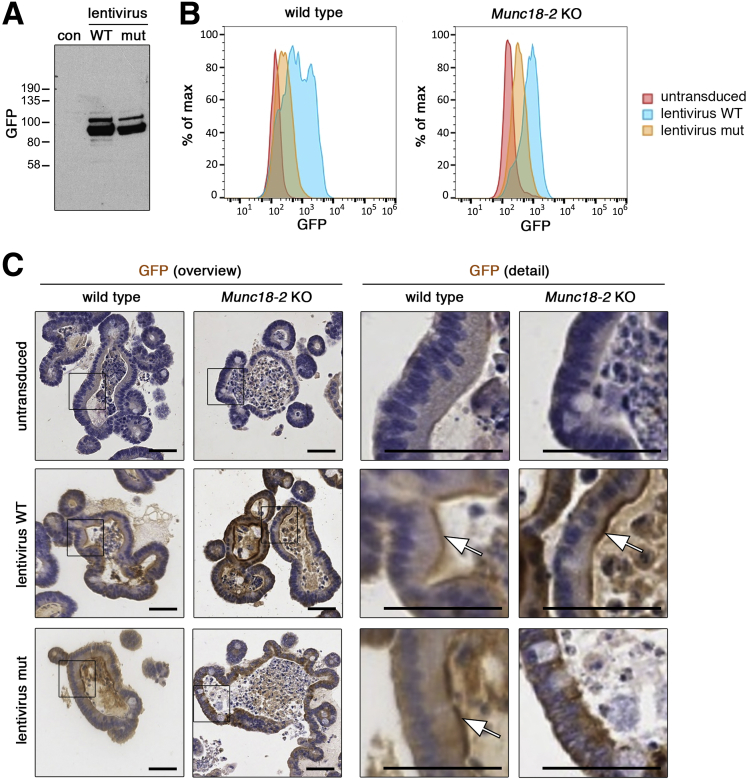
The integrity of both WT and mutant MUNC18-2 protein was confirmed by Western blot in HEK293T cells (Figure 5A). Flow cytometry of GFP expression showed strongly reduced expression of the P477L mutant protein in organoid cells (Figure 5B). Immunohistochemical assessment showed that both GFP fusion proteins were localized apically in WT organoids, however, the P477L mutant protein showed apical localization only in WT but not in KO organoids (Figure 5C). Taken as a whole, our data suggest that the P477L mutant carried by MUNC18-2–deficient patients is a null allele owing, at least in part, to low protein expression levels. These findings highlight the potential of organoids for functional screening of clinical variants in MVID.
Figure 5.
Reduced expression level of the P477L variant MUNC18-2 protein. (A) Western blot analysis of GFP–MUNC18-2 after transient transfection of HEK293T cells using the anti-GFP antibody. (B) Fluorescence-activated cell sorter analysis of GFP–MUNC18-2 expression in WT and Munc18-2 KO organoids. Continuous blasticidin antibiotic selection was performed to ensure homogenous expression. Histogram plot of GFP intensity shows reduced expression of the P477L point mutant protein compared with the WT. (C) Immunohistologic detection of GFP–MUNC18-2 expression in WT and Munc18-2 KO organoids using the anti-GFP antibody on paraffin sections. Specific apical brush-border signals could be detected (white arrows), but a more diffuse signal was detected with the point mutant protein in Munc18-2 KO organoids. Magnified images (boxes) are shown in the bottom rows. Scale bars: 50 μm. Staining was confirmed in 2 independent experiments. con, negative control; mut, mutant.
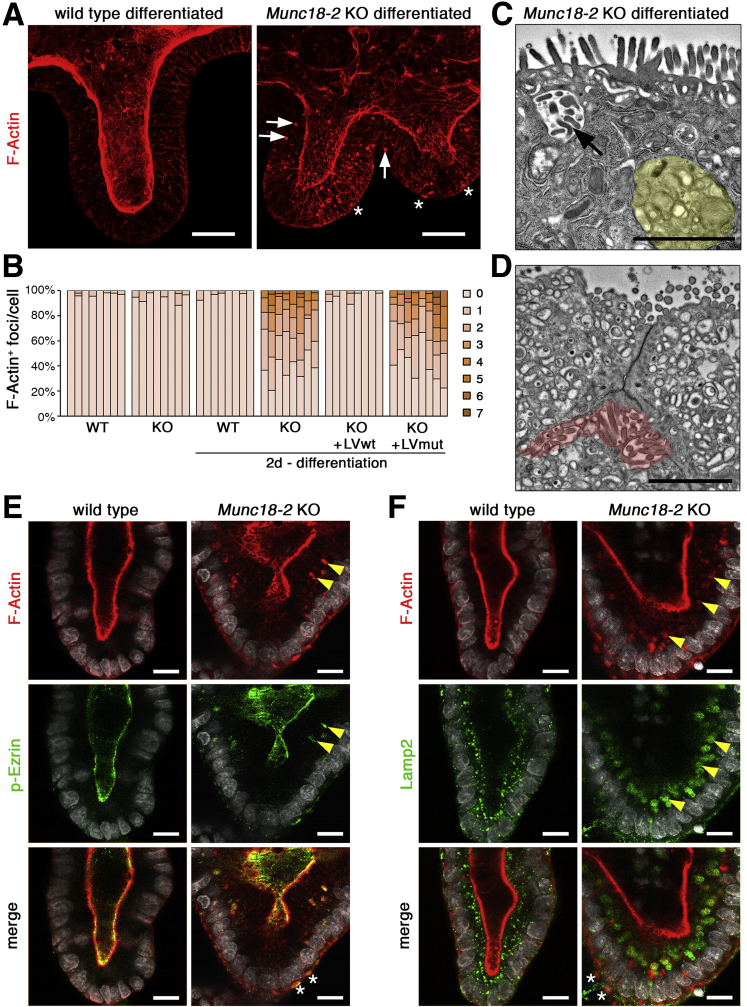
Munc18-2 Deletion Induces Nucleation of Cytoplasmic Microvilli Upon Enterocyte Differentiation
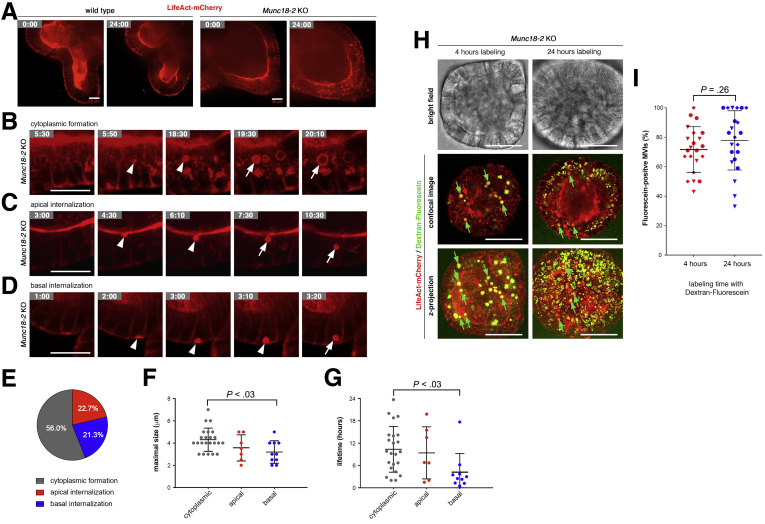
Given the observed association between phenotypic severity and enterocyte maturation, we wondered whether we could influence the cell phenotype by inducing villus differentiation. In the intestine, crypt maintenance and proliferation are critically dependent on Wnt signaling; withdrawal of the Wnt modulator R-spondin from the culture medium causes immediate differentiation in organoids.24 Two days after R-spondin withdrawal, we observed that Munc18-2 KO organoids, but not WT organoids, started to accumulate F-actin–positive (F-actin+) structures throughout the cytoplasm (Figure 6A and B). This phenotype could be fully rescued by lentiviral expression of the WT protein, and quantification on 3D confocal image stacks showed the presence of between 1 and 7 F-actin+ foci in more than 50% of all cells (Figure 6B). In TEM preparations, no classic MVIs were observed in the cytoplasm of Munc18-2 KO enterocytes. However, differentiation led to the formation of large lysosomes and transitional structures resembling the irregular inclusions described previously19 (Figure 6C). Basolateral microvillus-like structures also were observed (Figure 6D). To characterize these F-actin+ structures, we co-stained microvilli and lysosomes with p-Ezrin and Lamp2, respectively. Only in differentiated Munc18-2 KO organoids did we observe cytoplasmic p-Ezrin signals that partially co-localized with the F-actin signal (Figure 6E). Here, lysosomes strongly were increased in size and frequently surrounded the cytoplasmic F-actin+ foci (Figure 6F), indicating that lysosomal remodeling of the cytoplasmic vesicles is associated with the nucleation of presumptive microvilli in the mutant.
Figure 6.
Differentiation induces formation of F-actin+ cytosolic structures in Munc18-2 KO organoids. (A) Abundant presence of F-actin+ structures in Munc18-2 KO organoids (white arrowheads) 2 days after withdrawal of R-spondin. Z-projected confocal image stacks are shown. Asterisks mark basal F-actin. (B) Quantification of F-actin+ foci per cell. The fraction of cells with 0–7 aggregates was determined on consecutive confocal sections in n = 8 representative organoids per condition. (C and D) Ultrastructural characterization of differentiated Munc18-2 KO organoids. Note the irregular MVI (arrow), enlarged lysosome (yellow region), and basolateral MV-like structures (red region). (E and F) Confocal analysis of F-actin+ aggregates with p-Ezrin (E; microvilli) and Lamp2 (F; lysosomes). Yellow arrowheads mark regions of co-localization, asterisks mark basal F-actin. Scale bars: (A) 25 μm, (C and D) 2 μm, and (E and F) 10 μm. All staining was confirmed in at least 3 independent experiments. LV, lentivirus.
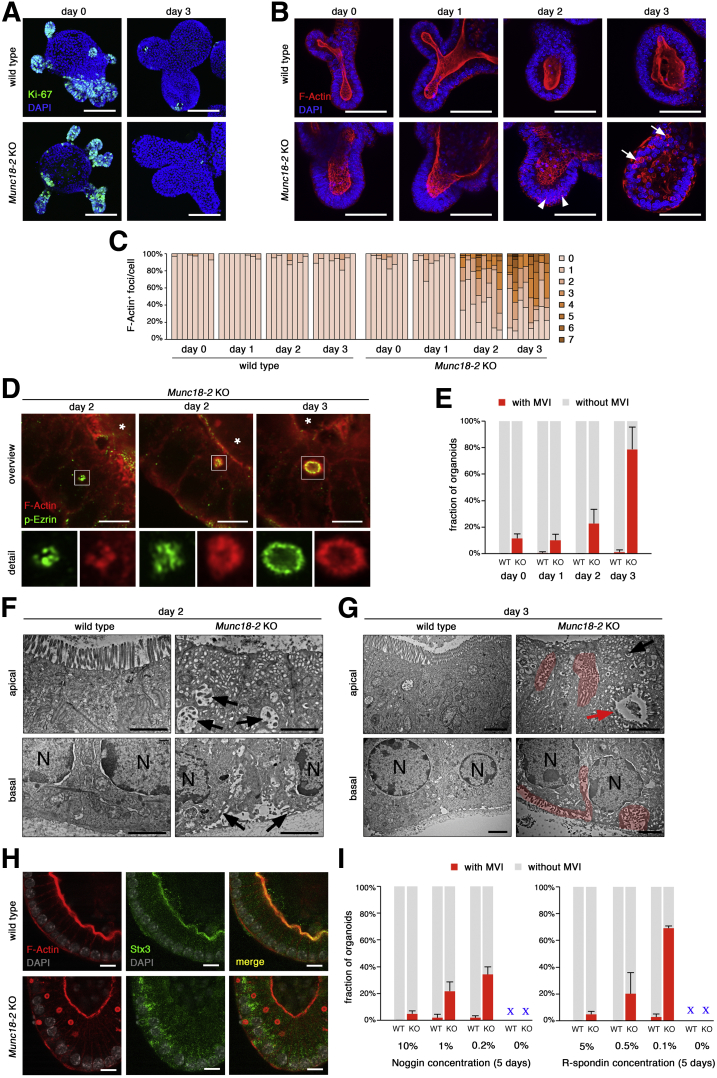
We next wondered whether prolonged culture of mature enterocytes would influence the severity of the phenotype. However, complete R-spondin withdrawal was not tolerated for longer than 2 days, given the progressive shedding of differentiated cells under this condition.25 As reported previously, treatment of mouse small intestinal organoids with a combination of the histone deacetylase inhibitor valproic acid (VPA) and the Wnt secretion inhibitor IWP-2 directs differentiation specifically toward the enterocyte lineage.26 Importantly, the resulting enterocytes can be kept for prolonged periods and are postmitotic, as observed by strongly reduced Ki-67 staining (Figure 7A), and thus allowed us to study phenotypic progression. By confocal microscopy, we observed small and irregular-shaped F-actin foci in the cytoplasm of KO organoids after 2 days of VPA/IWP-2 treatment (Figure 7B and C). After the third day of treatment, we detected larger, lumen-containing structures that showed a regular arrangement resembling classic MVIs. High-resolution confocal imaging of p-Ezrin and F-actin staining confirmed the presence of microvilli (Figure 7D). MVIs were observed in 76% ± 9% of all organoids (Figure 7E), and, on average, 24% ± 9% of all the cells were affected. TEM analysis confirmed the presence of differentiated enterocytes and the absence of secretory cells. On day 2, we observed a low brush-border density and an abundance of apical vesicles. These vesicles were heterogeneous with regard to their size, shape, and electron density, and some resembled irregular MVIs (Figure 7F), and on the basal side, MV-like structures were detected. On day 3, large portions of the apical surfaces were completely devoid of microvilli. In contrast, frequent MVIs (of various sizes and shapes) were detected in the cytoplasm (Figure 7G), and often were associated with the apical or basolateral membrane. Mature microvilli were observed on the basolateral membranes. We found that the MVIs were negative for Stx3 (Figure 7H). We tested alternative differentiation protocols and found that 5 days of culture under reduced levels of Noggin or R-spondin also resulted in MVIs, albeit with reduced efficiency and progressive organoid degeneration (Figure 7I). Overall, loss of Munc18-2 in mouse organoids resulted in a stepwise formation of intracytoplasmic actin+ foci and MV-like structures that increased in size, number, and maturation as the cells differentiated.
Figure 7.
Formation of microvillus inclusions in Munc18-2 KO organoids after prolonged enterocyte differentiation. (A) Confocal images of Ki-67 staining (green) after induced enterocyte differentiation (3 days after addition of IWP-2 + VPA). Z-projected image stacks are shown. (B) Confocal images of F-actin staining (red) after induced enterocyte differentiation. Appearance of F-actin+ foci at day 2 (arrowheads) and formation of lumen-containing F-actin+ MVIs at day 3 (arrows). Z-projected image stacks show a planar view on the brush border. (C) Quantification of F-actin+ aggregates per cell. The fraction of cells with 0–7 aggregates was determined on consecutive confocal sections in n = 8 representative organoids per condition. (D) High-resolution confocal analysis of F-actin (red) and p-Ezrin (green) staining after enterocyte differentiation. On day 2, irregular F-actin+/p-Ezrin+ foci were present in the cytoplasm. On day 3, lumen-containing F-actin+/p-E Ezrin+ MVIs were observed. White boxes show magnified regions and asterisks mark the central organoid lumen. (E) Quantification of mean organoid fraction (±SD) with MVIs (from n = 4 independent experiments). (F and G) TEM analysis of apical and basal regions. (F) On day 2 of differentiation, the apical brush border was reduced and intracellular and basal MV-like structures were found (black arrows). (G) On day 3, mature MVIs (red arrow) and abundant basolateral MVs (red regions) were found. N, nuclei. (H) Confocal analysis of Stx3 staining in differentiated organoids. Apical staining in WT and diffuse cytoplasmic localization in the Munc18-2 KO. MVIs were Stx3 negative. (I) Quantification of mean organoid fraction with MVIs (±SD) 5 days after reduction or withdrawal of Noggin (F, control was 10%) or R-spondin (G, control was 5%). Data from n = 3 independent experiments. Note that complete withdrawal of either factor resulted in organoid loss. Scale bars: (A) 100 μm, (B) 50 μm, (D) 5 μm, (F and G) 2 μm, and (H) 10 μm. All staining was confirmed in at least 2 independent experiments.
MVIs Form Dynamically Within the Cytoplasm or via Apical or Basolateral Membrane Internalization
To visualize the dynamic process of MVI formation, we used real-time confocal microscopy to track organoids after the introduction of a lentiviral F-actin reporter (LifeAct-mCherry) that faithfully recapitulated the phalloidin-staining pattern (Figure 8A). By high-resolution confocal microscopy, periodicity of the LifeAct signal was observed, confirming the detection of classic MVIs (Figure 8B). WT and KO organoids were tracked for the appearance of MVIs over a 24-hour period, 2 days after incubation in enterocyte differentiation medium. The WT cells showed a stable phenotype, with a dynamic cortical F-actin cytoskeleton but no intracellular LifeAct accumulation (Figure 9A and Supplementary Movie 1). In contrast, KO organoids initially contained many small LifeAct-positive foci—some of which increased in size and turned into lumen-containing MVIs (Figure 9A and B and Supplementary Movies 2 and 3). We also observed another mechanism of formation via internalization of apical membranes (Figure 9C and Supplementary Movie 4). This finding was reminiscent of previous observations in the Rab8 KO mice, in which luminal dye was taken up by a proportion of the MVIs, indicating the involvement of endocytosis or macropinocytosis.4 Furthermore, we detected a third route for MVI formation: the internalization of basolateral LifeAct-positive membranes (Figure 9D and Supplementary Movie 5). To distinguish the relative frequencies of these 3 mechanisms, we tracked 75 lumen-containing MVIs (Figure 9E). The majority was formed in the cytoplasm, followed by apical and basal membrane internalization. The MVI size and lifetime were heterogeneous, and basally formed MVIs were smaller and shorter-lived than MVIs formed in the cytoplasm (Figure 9F and G). To test the fraction of MVIs that formed by membrane internalization, we applied an extracellular dextran-fluorescein tracer. Labeling for 24 hours after 2 days of differentiation resulted in a large proportion of fluorescein-positive MVIs (78% ± 20%) (Figure 9H and I). However, upon incorporation for 4 hours after 3 days of differentiation, when MVIs had been formed, a similar fraction of tracer-positive MVIs was found (72% ± 15%). These results argue that MVIs are loaded with dye after they have been formed and that extracellular tracers, when administered with the medium, are not suitable to determine the fraction that is formed by membrane internalization.
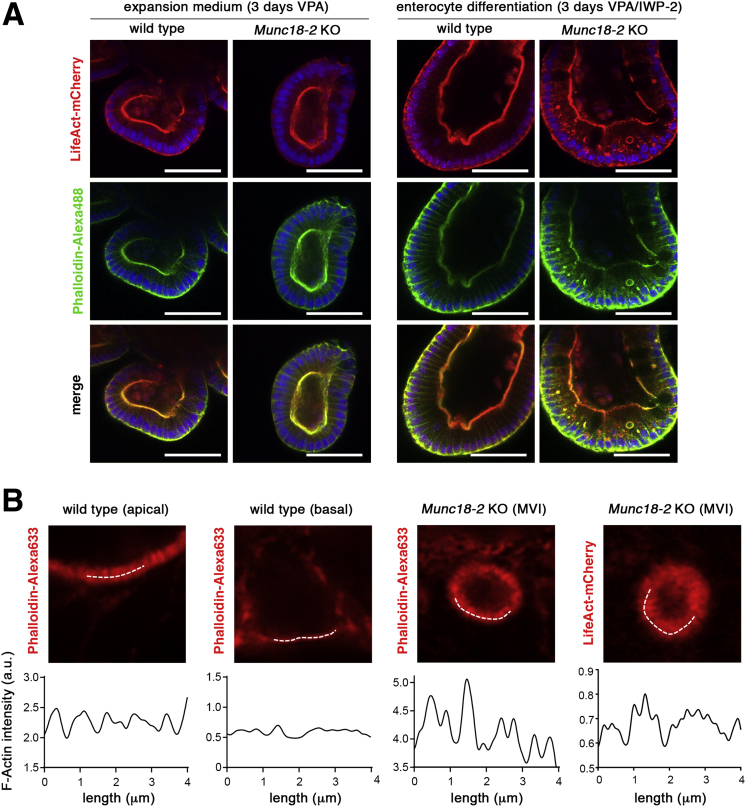
Figure 8.
Lentiviral expression of LifeAct reporter in organoids to monitor MVIs. (A) Confocal images of LifeAct reporter expression and phalloidin staining. LifeAct-mCherry (red signal, top) reporter transgenic WT and Munc18-2 KO organoids were fixed and stained by phalloidin-Alexa488 (green, middle). Nuclei were stained by DAPI. Organoids were kept in VPA-containing medium (expansion condition) or for 3 days in enterocyte differentiation medium (VPA + IWP-2) to induce MVIs. Both lumen-containing and noncontaining F-actin+ structures were co-stained in differentiated Munc18-2 KO organoids (yellow signal in merged image, bottom). Note that treatment with VPA alone reduced the presence of secretory cells (Paneth and goblet cells) in which spontaneous aggregation of the LifeAct reporter otherwise caused background signals (not shown). Scale bars: 50 μm. (B) High-resolution confocal analysis of LifeAct reporter expression and phalloidin staining. Organoids after 3 days in enterocyte differentiation medium. On the white lines (4 μm), scan of signal intensity was performed on apical microvilli (MV) in WT and on MVIs in Munc18-2 KO organoids; basal membrane in a WT organoid was used as a control, microvillus-free membrane.
Figure 9.
Dynamic characterization of MVI formation. (A–D) Still images from time-lapse, spinning-disc microscopy (see Supplementary Movie 1, Supplementary Movie 2, Supplementary Movie 3, Supplementary Movie 4, Supplementary Movie 5). LifeAct-mCherry–expressing organoids were tracked for 24 hours. Cells were differentiated for 2 days (IWP-2 + VPA) before start of imaging (t = 0). Time points are marked as hours:minutes. (A) Overview images and (B) detailed images of the formation of single MVIs in the cytoplasm, and by internalization of (C) apical or (D) basal membranes in Munc18-2 KO organoids. (B–D) The apical pole is orientated to the top. Arrowheads indicate precursor structures and arrows indicate mature MVIs that were tracked. Note that the basal LifeAct signal in WT cells marks cytoplasmic/junctional actin rather than MV. (A–D) Scalebars: 20 μm. All live-cell imaging experiments were performed at least 3 times independently. (E) Relative frequencies of MVI formation in the cytoplasm or by apical/basal internalization (determined in n = 75 MVIs). (F and G) Quantification of MVI size and lifetime from time-lapse data (n = 40 MVIs were tracked; means ± SD). (F) Analysis of the maximal size showed that structures formed in the cytoplasm were significantly bigger than basal MVIs. (G) Analysis of the lifetime showed that cytoplasmic formation was associated with a significantly longer lifetime than basal MVI formation. Statistical testing was performed by 1-way analysis of variance, only adjusted P values < .05 are shown. (H) Uptake of extracellular tracer in MVIs. Confocal images of LifeAct reporter (red) and dextran-fluorescein uptake (green) in Munc18-2 KO organoids. Organoids were kept for 3 days in enterocyte differentiation medium (VPA + IWP-2). Dextran-fluorescein was added to the medium during the last 4 or 24 hours of culture. Confocal sections (top) and z-projected confocal image stacks (bottom) are shown and fluorescein-positive and fluorescein-negative MVIs are labeled with green arrows and red arrows, respectively. Scale bars: 50 μm. (I) Quantification of the fraction of fluorescein-positive MVIs. Averages (±SD) from n = 22 organoids each are shown. Data were measured in 3 independent experiments as indicated by diamond, circle, and triangle symbols. No significant differences were found between 4 and 24 hours of labeling (P value from unpaired t test).
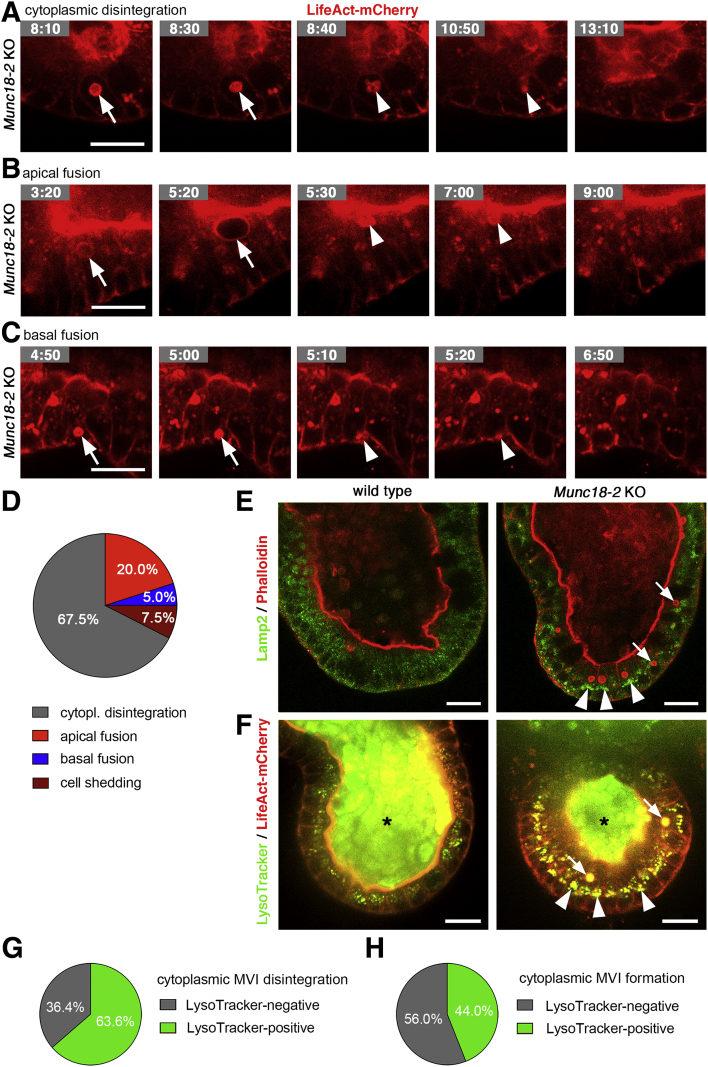
Furthermore, we observed distinct modes of disappearance (Figure 10A–D and Supplementary Movies 6, 7, and 8): cytoplasmic disintegration was most frequent, followed by apical fusion, and rarely basal fusion was observed. In total, we observed 3 of 75 instances in which MVI-containing cells were extruded from the epithelium (Supplementary Movie 9). Next, we investigated the link between MVI turnover and lysosomes. Live imaging using LysoTracker Green recapitulated the Lamp2 staining pattern (Figure 10E and F) and both markers showed enlarged lysosomes in differentiated Munc18-2 organoids. We noted that the apical lumen of organoids as well as the lumen of the majority of MVIs also was LysoTracker positive (87.5%; 35 of 40 MVIs analyzed), indicating acidic pH in both compartments. The remnants of 14 of 22 MVIs (63.6%) that disintegrated in the cytoplasm were LysoTracker positive (Figure 10G and Supplementary Movies 10 and 11), arguing for an involvement of lysosomal degradation. In addition, we observed that 11 of 25 (44.0%) LifeAct-positive foci that later developed into MVIs were associated with lysosomes (Figure 10H and Supplementary Movies 11 and 12), indicating that lysosomes also may play a role in cytoplasmic MVI formation. Taken together, our 3D time-lapse imaging in organoids has shown unexpected dynamics of the life cycle of MVIs.
Figure 10.
Dynamic characterization of MVI disappearance and association with lysosomes. (A–C) Still images from time-lapse microscopy of individual MVIs (as in Figure 9) show (A) cytoplasmic disintegration, and (B) apical or (C) basal fusion. LifeAct-mCherry–expressing Munc18-2 KO organoids (see Supplementary Movie 6, Supplementary Movie 7, Supplementary Movie 8). Arrows indicate MVIs that were tracked and arrowheads mark their remnants. (D) Relative frequencies of MVI disappearance in the cytoplasm, by fusion to apical/basal membranes or by cell shedding (determined in n = 40 MVIs). (E and F) Confocal imaging of lysosomes and MVIs in differentiated organoids (3 days VPA + IWP-2). (E) Lamp2 (green) and Phalloidin (red) staining on fixed cells. (F) Time-lapse imaging of LysoTracker (green) and LifeAct-mCherry reporter (red). Enlarged lysosomes were observed in differentiated Munc18-2 KO organoids (arrowheads). Note the LysoTracker signal in the organoid lumen (asterisks) and in MVIs (arrows). (G) Association of lysosomes with cytoplasmic MVI disintegration (see Supplementary Movies 10 and 11). MVIs (n = 22) were scored positive if LysoTracker stained their cytoplasmic remnants. (H) Association of lysosomes with cytoplasmic MVI formation (see Supplementary Movies 11 and 12). MVIs (n = 25) were scored as positive if LysoTracker stained their cytoplasmic precursors. Scale bars: 20 μm. All live-cell imaging experiments were performed at least 3 times independently.
Discussion
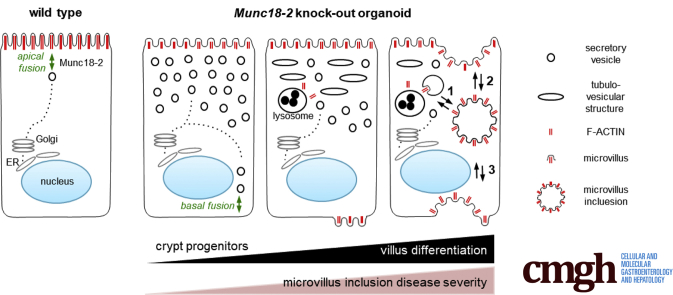
Diagnosis of MVID is based on the detection of structural and ultrastructural abnormalities in intestinal cells. The archetypal diagnostic feature is the presence of MVIs in the enterocyte’s cytoplasm. However, the intensity of the cellular abnormalities and the frequency of MVI formation are highly variable; hence, the origin and the pathogenic mechanism of MVI formation still are a subject of debate. Here, we have shown that the enterocyte differentiation state plays an important role for the cell phenotype in MVID. By using an appropriate murine Munc18-2–deficient organoid model, we evidenced a stepwise phenotypic progression of MVI formation along the crypt–villus axis. This study represents real-time visualization of MVIs. During enterocyte maturation, intracellular MVIs form progressively in the cytoplasm concomitantly with vesicular remodeling, but also via apical or ectopic basal internalization of microvilli (see graphic abstract). This gradual mechanism probably explains some of the reported variability between distinct gene defects (Supplementary Table 1) and among patients. We suggest that MVID should be considered as a phenotypic continuum.
MUNC18-2/STXBP2 catalyzes the fusion of vesicles with the plasma membrane,9 and thus influences the later stages in apical cargo trafficking. Loss of Munc18-2 in immature crypt cells results in a less severe phenotype, with undisturbed apicobasal polarity and residual microvilli. It is possible that vesicle fusion at the apical membrane still occurs (albeit at a low rate) in the absence of Munc18-2, thus allowing a limited amount of apical trafficking. In agreement with previous reports, we have observed Munc18-2–independent trafficking of Dpp4 in intestinal organoids.12 However, the poorly efficient, Munc18-2–deficient fusion machinery may not be able to cope with the progressive accumulation of vesicles during the differentiation process, leading to the MVI phenotype. This hypothesis is consistent with the following: (1) the predominant localization of secretory vesicle accumulation in the upper crypt regions,15 (2) the prevalence of MVIs mainly in villus/surface epithelial cells,19, 27 and (3) the formation of basolateral microvilli and MVIs in conditional mouse models only 3–4 days after deletion of Myo5b or Rab8/Rab11, respectively,21, 28 all of which further highlight the impact of the cell maturation status. When the function of other trafficking effectors (such as STX3 or MYO5B) is lost, an earlier phenotype might result from the more severe disruption of apical vesicle trafficking and thus more rapid vesicle accumulation. It is possible that MVI formation also is obscured by rapid cell loss upon differentiation and the appearance of a villus blunting phenotype, similar to our R-spondin withdrawal model. The fact that the mild phenotype caused by Munc18-2 loss in organoids can be transformed into a severe form of MVID argues that our IWP-2/VPA enterocyte differentiation protocol could be used for diagnosis, in particular for atypical or milder cases.
Previously described in vitro models (based on monolayers from biopsy-derived organoids or a 3D culture of STXBP2-deficient CaCo2 cells)12 did not result in the formation of mature MVIs. By controlled differentiation of murine small intestinal organoids, we documented a stepwise remodeling of secretory vesicles into a tubulovesicular network, before the formation of cytoplasmic/basolateral F-actin+ foci and MVIs. The presence of dot-like cytoplasmic F-actin+ foci has been noted previously.8 Basolateral microvilli most likely result from erroneous fusion of accumulated vesicles to the lateral membrane. The nucleation of ectopic microvilli probably occurs at a slow rate and/or might depend on the maturation stage or the size of the membrane compartments. Expanded lysosomes most likely reflect a cellular response to the vesicular accumulation and we observed their co-localization with F-actin+ foci. In addition, we have found that lysosomes are associated with disintegration of MVIs that occur frequently in the cytoplasm. Interestingly, the presence of mature MVIs only rarely induced cell death, indicating that these structures are not cytotoxic per se. In contrast, mature MVIs might have a cytoprotective role; the clearance of basolateral microvilli by invagination and fusion with the apical membrane may testify to a previously unrecognized plasticity in enterocyte membrane biology. Moreover, the rapid incorporation of dextran tracers into established MVIs indicates continuous (membrane) exchange with the extracellular milieu.
Mouse- or patient-derived organoids are useful tools for better understanding the pathophysiology of gastrointestinal disease and for functional testing of clinical variants. As a preclinical platform, rectal organoids are used currently for personalized testing of cystic fibrosis transmembrane conductance regulator–modulating drugs,29 and organoids have been instrumental in establishing a link between epithelial polarity defects and multiple intestinal atresia.30 In the future, organoid-based differentiation assays could improve the diagnosis of MVID and serve to test new therapeutic approaches for this group of disorders.
Materials and Methods
Mice
A L3 Stxbp2 mutant mouse line (containing a flipped Neo-LacZ cassette between exons 3 and 4, as well as a floxed exon 4 of mouse Stxbp2) was established at the Phenomin-Institut Clinique de la Souris (Illkirch, France; http://www.phenomic.fr) by using a targeting vector (PRPGS00057_B_A06) from the Knockout Mouse Project repository (www.komp.org). Before Flp-mediated recombination the L3 allele is constitutively null (so-called knockout-first allele). From heterozygous intercrosses we obtained 166 newborn pups, of which 35% were wild type (WT) and 65% heterozygous, indicating embryonic lethality of homozygous Stxbp2 loss. The conditional Stxbp2flox/flox mouse line containing a floxed exon 4 of mouse Stxbp2 was obtained by mating L3 Stxbp2 mice with a Flp recombinase transgenic mouse and maintained on a C57Bl/6 background. Mice were handled according to national and institutional guidelines.
Organoid Culture
Crypts were isolated from the small intestines of homozygous adult mice and cultured in BME-R1 (Amsbio, Abingdon, UK) as described.23 We have used 3-cm sections from the most proximal, middle, and distal third each, and the organoids were maintained as a polyclonal pool. A total of 5% R-spondin (vol/vol) and 10% Noggin IgG Fc-tag fusion (vol/vol) conditioned media were used and produced as described.31 For differentiation experiments cells were seeded at reduced density (1:8 split ratio) in normal medium and without puromycin selection. After 48 hours, R-spondin was removed from the medium followed by a second medium change after 10 minutes. For enterocyte-specific differentiation, cells were seeded in normal medium including 1 mmol/L valproic acid (Sigma-Aldrich, St. Louis, MO). After 48 hours, the medium was changed and additionally 2.5 μg/mL inhibitor of WNT production-2 (IWP-2) was added (Sigma-Aldrich), and after another 48 hours the medium was exchanged again. For differentiation in the presence of reduced levels of R-spondin and Noggin, the organoids were seeded in BME-R1 and cultured for 5 days using the indicated amounts of conditioned media.
Lentiviral Expression
Lentiviral expression was performed as described32 using a lentivirus containing an expression cassette encoding for nuclear localization sequence; Cre-recombinase; internal ribosome entry site; puromycin resistance (NLS-Cre-IRES-PuroR). Deletion was performed 3 weeks after establishment of the cultures and the cells were selected by addition of 1 μg/mL puromycin for 3 weeks before genotyping and cryopreservation. Experiments were performed within 2–3 months before a fresh cryovial was recovered. Genotyping was performed using the following 3 primers together: forward primer 1 5’-AGGGCATCACCAGTGAGTGGATG-3’, forward primer 2 5’-GTGGAAGACATCAACAAACGGAGAGA-3’, and reverse primer 5’-GCACATGGCTGAATATCGACGGT-3’. Stable maintenance of the recombined allele was confirmed repeatedly. For lentiviral expression of WT and P477L MUNC18-2 the human complementary DNAs were cloned as N-terminal enhanced green fluorescent protein (EGFP) fusion into the EcoRI and BamHI sites of pEGFP-C1 (Clontech), and then amplified by proofreading polymerase chain reaction (Phusion; NEB, Ipswich, MA) for subsequent infusion cloning (Clontech/Takara, Kyoto, Japan) into the XbaI and BamHI sites of lentivirus; CRISPR associated protein 9; blasticidin resistance (lentiCas9-Blast), which was a gift from Feng Zhang (plasmid 52962; Addgene, Cambridge, MA). The resulting vectors were confirmed by Sanger sequencing and Western Blot analysis using chicken anti-GFP antibody (Aves Labs, Tigard, OR). After transduction, organoids were cultured continuously in the presence of 2 μg/mL blasticidin to maintain transgene expression, which was confirmed by immunohistochemistry (see later) and fluorescene-cactivated cell sorter analysis on a BD (Franklin Lakes, NJ) Fortessa after organoid single-cell dispersal using TrypLE Express enzyme (Gibco, Waltham, MA). For lentiviral expression of LifeAct-mCherry, the open reading frame was amplified from pCMV-LifeAct-mCherry-Hyg-N1, which was a gift from Roland Wedlich-Söldner, and cloned into the lentivirus Pgk::EGFP–IRES-puro opened at XhoI and SalI sites to release the EGFP fragment. LifeAct-mCherry expression was maintained by continuous addition of 1 μg/mL puromycin.
Whole-Mount Organoid Immunofluorescence and Image Analysis
Organoids were collected in ice-cold medium, gently washed by sedimentation, and resuspended in 100 μL cold medium before addition of 10 mL ice-cold 2% paraformaldehyde/phosphate-buffered saline (PBS). Fixation was performed at 4°C overnight, except for p-Ezrin staining, which was performed in 4% PFA/PBS for 1 hour.
After permeabilization in PBS with 0.1% Tween 20 and 0.2% Triton X-100 (Sigma-Aldrich), cells were blocked in PBS with 0.1% Tween 20, 0.1% sodium azide, and 2% goat serum, and stored at 4°C until use. Antibodies were applied in blocking solution (as described earlier) in the following dilutions: rabbit anti–syntaxin 3 (ab133750, 1:50; Abcam, Cambridge, UK), rabbit anti–phospho-Ezrin (3726T, 1:50; Cell Signaling Technology, Danvers, MA), rat anti-Lysosome-associated membrane protein 2 (ab13524, clone GL2A7, 1:200; Abcam), and rabbit anti–zonula occludens-1 (61-7300, 1:200; Invitrogen, Waltham, MA). For phospho-Ezrin staining, phosphatase inhibitors (PhosSTOP; Roche, Basel, Switzerland) were added to the incubation buffer. After washes, suitable Alexa Fluor–coupled secondary antibodies (Invitrogen) were combined with 4′,6-diamidino-2-phenylindole (DAPI), rat anti-mouse Epithelial cell adhesion molecule - Allophycocyanin fusion (clone G8.8, 1:500; eBioscience, Waltham, MA), eFluor-660-conjugated rat anti-KI67 1:250 and/or fluorescent phalloidin-conjugates (Invitrogen) before washes and mounting in ProLong Gold Antifade (Invitrogen). Organoids were imaged on a Leica SP5 scanning confocal microscope using a 63× water immersion objective (room temperature; numeric aperture, 1.2) and a z-step size of 1 μm. High-resolution confocal microscopy was performed using a Zeiss (Oberkochen, Germany) LSM880 Airyscan and a 63× oil immersion objective (numeric aperture, 1.4); x-y resolution was approximately 140 nm. Line-scan analysis was performed using the Plot Profile function of Fiji software (https://fiji.sc/) on a 4-μm line (0.5-μm width for the apical plasma membrane and MVIs, 0.2-μm width for the basal plasma membrane).
For extracellular tracer experiments, differentiated LifeAct-mCherry transgenic organoids were incubated with 1 mg/mL dextran-fluorescein (70 kilodaltons, anionic, lysine fixable; Molecular Probes, Waltham, MA) for 4 or 24 hours before collection and washed using cold medium. Unfixed organoids were mounted as described earlier and fluorescein and mCherry signals were recorded immediately on a Leica SP5 scanning confocal microscope. Quantification was performed using Fiji software from 3D image stacks (z-step size, 1 μm). First, all lumen-containing MVIs were identified from the LifeAct signal and marked, and then classified as fluorescein-positive or fluorescein-negative. Six to 8 organoids were counted per condition and the experiment was repeated 3 times.
Z-projections were generated with Fiji software from image stacks of organoid hemispheres. F-actin+ aggregates and MVIs per cell were determined in 8 randomly selected organoids per condition. For quantification, 4 consecutive confocal z-sections (total z-range, 3 μm) were analyzed from midsagittal organoid images. Leica LAS software was used to determine the total number of cells (DAPI), and then all phalloidin+ cytoplasmic structures were counted per cell and plasma membrane staining (identified by the EPCAM signal) was excluded. MVIs were defined as lumen-containing cytoplasmic F-actin+ structures. The fraction of MVI+ organoids was determined by epifluorescence microscopy using the EVOS FL Imaging System (Thermo Fisher Scientific, Waltham, MA) in at least 25 organoids per condition and replicate.
Immunohistology and Histochemistry
Fixed organoids (as described earlier) were paraffin-embedded and 4-μm sections were rehydrated. For alkaline phosphatase staining, sections were incubated for 30 minutes at room temperature in 1× 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) substrate (ES006; Millipore, Burlington, MA, Vereinigte Staaten). Immunostainings were performed on a Leica Bond-Max, using the Bond Polymer Refine Detection System after epitope retrieval using EDTA, pH 9.0. Anti-GFP staining was performed using goat-anti GFP (ab6673, 1:200; Abcam) and mouse anti-human CD10 (clone 56C6, 1:50; AbD Serotech, Hercules, CA). For goat anti-mouse Dpp4 (AF954-SP, 1:500; R&D Systems, Minneapolis, MN) citrate, pH 6.0, epitope retrieval was performed. After 3,3′-diaminobenzidine tetra hydrochloride detection, the sections were counterstained with hematoxylin before embedding.
Transmission Electron Microscopy Analysis
Organoids were collected, washed, and resuspended as described earlier, and stored at 4°C in Trump’s fixative. Enhanced chemical fixation was performed in a mix of 4% paraformaldehyde with 2.5% glutaraldehyde in 0.1 mol/L cacodylate buffer overnight at 4°C. A 1.5-hour incubation in 1% OsO4 was followed by a 1.5-hour incubation with 2% uranyl acetate at ambient temperature. Organoids then were dehydrated through graded ethanol solutions, cleared in acetone, infiltrated, and embedded in Epon-Araldite mix (EMS hard formula; EMS hard formula, Hatfield, PA). We used adhesive frames (11560294 GENE-FRAME 65 μL; Thermo Fisher Scientific, Waltham, MA) for flat-embedding, which is more convenient for small samples such as organoids, as previously described.33 Polymerization was performed according to the manufacturer’s instructions. Ultrathin sections were cut on an ultramicrotome (UC7; Leica) and collected on formvar-coated slot grids (FCF2010-CU; EMS). Each organoid was sectioned in 5 different places (5 grids/organoid) with ≥10 μm between each grid to examine the sample over a large region. Each grid contained at least 10 consecutive sections of 70 nm. Transmission electron microscopy (TEM) grids were viewed using a JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan) operated at 120 kV, equipped with a Gatan Orius SC 1000 camera (Gatan, Pleasanton, CA), and piloted by the Digital Micrograph (Pleasanton, CA) program. Micrographs were analyzed using Fiji software.
Live-Cell Microscopy
Live-cell microscopy was performed after seeding organoids in BME-R1 on glass-bottom imaging plates (CG 1.0; Miltenyi Biotec, Bergisch Gladbach, Germany). A CQ1 spinning disc microscope (Yokogawa, Tokyo, Japan) was used and images were captured with a 40× air objective, 20% laser power, in 10-minute intervals, and at a z-step size of 1 μm. Imaging was performed for 24 hours at 37°C and 5% CO2. All experiments were repeated 3 times and in at least 5 organoids per genotype each. MVIs were defined as structures with a clearly discernable lumen, and 75 random MVIs that appeared during this period were selected and tracked in the 3D image data using the instrument’s software. Adjacent z-stacks were inspected to track the formation and disappearance of MVIs correctly, and to avoid misinterpretations caused by movement of (pre-existing) structures in or out of the focal plane. Forty of the 75 structures also disappeared during the imaging period and were used to determine lifetime and maximal size; 3 structures were excluded from the analysis because they could not be tracked. For co-labeling with lysosomes, LysoTracker Green DND-26 (1:20,000; Cell Signaling Technology) was added to the culture medium 3 hours before and during imaging. MVI disintegration or formation was classified as lysosome-positive if MVI remnants or precursors (marked by LifeAct), respectively, were adjacent (within ∼1 diameter) to small granular LysoTracker-positive structures. Twenty-five and 22 MVIs were tracked for formation and disintegration as described earlier, respectively. Time-lapse videos were generated using images from single z-steps that were exported and processed using Fiji software, and JPG (overview) or PNG (detail) video compression was applied.
Statistical Calculations
All error bars are expressed as SDs. Statistical analysis was performed using Prism 7 (GraphPad Software, La Jolla, CA). The number of experimental and biological replicates is indicated. An unpaired Student t test was used to assess significance and P values are shown in each figure. For multiple comparisons of life imaging data, 1-way analysis of variance was used (Tukey procedure) and adjusted P values are shown.
Acknowledgements
The authors would like to thank Stefan Stein and Petra Dinse for experimental assistance; Roland Wedlich-Söldner, Paul Ziegler, and Florian Greten for reagents; Suma Choorapoikayil for critical discussions; and Hans Clevers (Hubrecht Institute, NL) and Calvin Kuo (Stanford University, CA) for providing cell lines for the production of conditioned medium. The authors thank the electron microscopy unit of the MRic facility (Rennes, France).
Footnotes
Author contributions Ophélie Nicolle and Sophia Maschalidi contributed equally to this work; Mohammed H. Mosa, Ophélie Nicolle, Sophia Maschalidi, Aurelien Bidaud-Meynard, and Henner F. Farin performed the experiments; Fernando E. Sepulveda, Constantin Menche, and Birgitta E. Michels provided technical support; Mohammed H. Mosa, Ophélie Nicolle, Aurelien Bidaud-Meynard, Grégoire Michaux, and Henner F. Farin analyzed the data; Grégoire Michaux, Geneviève de Saint Basile, and Henner F. Farin conceived the study, supervised the experiments, and obtained funding; and Geneviève de Saint Basile and Henner F. Farin drafted the manuscript. All authors discussed the results and commented on the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the French National Institute of Health and Medical Research (INSERM), the Agence Nationale de la Recherche Hemophagocytic lymphohistiocytosis-Cytotox/ANR-12-BSV1-0020-01 (G.d.S.B.), the LOEWE Center for Cell and Gene Therapy Frankfurt, Hessisches Ministerium für Wissenschaft und Kunst (H.F.F.), and by the Centre national de la recherche scientifique and the Ligue Régionale Contre le Cancer (22/29/35/72) (G.M.).
Contributor Information
Grégoire Michaux, Email: gregoire.michaux@univ-rennes1.fr.
Geneviève de Saint Basile, Email: genevieve.de-saint-basile@inserm.fr.
Henner F. Farin, Email: farin@gsh.uni-frankfurt.de.
Supplementary Material
Supplementary Table 1.
Comparative Phenotypes in Different MVID Pathologies
| Normal intestine | MYO5B-deficient patients5, 21, 27 | STX3-deficient patients 8, 27 | MUNC18-2–deficient patients (FHL5)11, 12 | Munc18-2-/-mouse organoids | |
|---|---|---|---|---|---|
| Apical microvilli | Normal | Microvillus atrophy; severe loss | Microvillus atrophy; variable loss | Microvillus atrophy; variable loss | Microvillus atrophya |
| Microvillus inclusions | No | Frequent, observed in 10% of cells | Infrequent | Infrequent, not fully penetrant | Frequent, but only after differentiationa |
| Basolateral microvilli | No | Incomplete penetrance | Frequent | Frequent | Frequent, but only after differentiationa |
| Subapical vesicle accumulation | No | Vesicles and tubulovesicular; variable electron-dense and translucent | Vesicles and tubulovesicular; electron-dense and translucent (villus) | Electron-dense and translucent vesicles | Crypt: translucenta Villus: translucent tubulovesiculara |
| Lysosomal abnormalities | No | Enlarged (auto)lysosomes in villus | Enlarged (auto)lysosomes in villus | High number of lysosomes | Enlarged lysosomesa |
| STX3 localization | Apical membrane | Subapical vesicle and MVIs | Reduced/absent protein level | Subapical vesicle and MVIs | Diffuse in cytoplasm |
| F-actin | Strong in brush border | Reduced brush-border signal; MVIs positive | Presence of dot- and ring-like structures | Reduced brush border; MVIs stained; variable penetrance and severity | Reduced brush border, cytoplasmic foci, and MVIs staineda |
Dependence on the enterocyte differentiation status in Munc18-2-/- organoids.
Actin dynamics in a differentiated wild-type organoid. Overview of LifeAct-mCherry signal in WT (compare with Figure 6A). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). Images of a single z-plane are shown. Time points are indicated as hours:minutes, with 10-minute intervals (10 frames/s).
Dynamic MVI formation in a differentiated Munc18-2 KO organoid. Overview of LifeAct-mCherry signal in Munc18-2 KO (compare with Figure 6A). Cells were differentiated for 2 days in enterocyte differentiation medium before start of imaging (t = 0). Images of a single z-plane are shown. Time points are indicated as hours:minutes, with 10-minute intervals (10 frames/s).
Cytoplasmic MVI formation. Tracking of MVI formation by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 6B). Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). At t = 5:50, a cytoplasmic F-actin+ aggregate was formed (red arrow) that matured after t = 18:20 into a lumen-containing MVI (white arrow) and started to disintegrate at t = 21:20 (blue arrow). Note that a second MVI formed in the right half of the image. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that a pre-existing MVI had moved into the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
MVI formation by apical membrane internalization. Tracking of MVI formation by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 6C). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 4:00, the apical membrane invaginated (red arrow). At t = 7:10, a lumen-containing MVI pinched off (white arrow). A single z-plane is shown and apical is top. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
MVI formation by basal membrane internalization. Tracking of MVI formation by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 6D). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0:00). At t = 1:00, deposition of ectopic basal F-actin was observed (red arrow). At t = 3:00, membrane invagination and formation of a lumen-containing MVI were observed (white arrow). A single z-plane is shown and apical is top. Time points are indicated as hours:minutes, with 10-minute intervals (3 frames/s).
Disintegration of MVI in the cytoplasm. Tracking of MVI breakdown by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 7A). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 4:00, a central MVI was observed (red arrow) that started to disintegrate in the cytosol at t = 8:30 (white arrow) and disappeared at t = 13:10. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that the structure had moved out of the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Fusion of MVI to the apical plasma membrane. Tracking of MVI fusion to the apical membrane by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 7B). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 2:40, a central MVI was observed that increased in size (red arrow). At t = 5:30, it contacted the apical membrane and rapidly shrunk (white arrow). By t = 8:30 the structure had fully integrated into the apical membrane. A single z-plane is shown and apical is top. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Fusion of MVI to the basal plasma membrane. Tracking of MVI fusion to the basal membrane by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 7C). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 4:00, 1 MVI pinched off from the basal membrane (red arrow). After t = 5:00 the structure again fused to the basal membrane (white arrow) and the signal disappeared by t = 6:30. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks confirmed that the structure was not lost owing to z-movement. Time points are indicated as hours:minutes, with 10-minute intervals (3 frames/s).
Shedding of MVI-containing cell. Tracking of a cell shedding event by LifeAct-mCherry signal in Munc18-2 KO organoid. Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 0:30, 1 MVI was formed (red arrow) before the cell was extruded from the epithelium into the lumen at t = 2:30 (white arrow). A single z-plane is shown and apical is top. Inspection of adjacent z–stacks confirmed that the MVI was not lost owing to z-movement. Time points are indicated as hours:minutes, with 10-minute intervals (3 frames/s).
Lysosome-associated MVI disintegration in the cytoplasm. Tracking of MVI breakdown by LifeAct-mCherry and LysoTracker Green staining in Munc18-2 KO organoid. Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). One MVI was observed (white arrow) that had a LysoTracker-positive lumen that started to be lost from t = 10:20 (blue arrow). Note the proximity to lysosomes during this process. The MVI disappeared at t = 17:20. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that the structure had moved out of the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Lysosome nonassociated MVI formation and disintegration in the cytoplasm. Tracking of 1 MVI by LifeAct-mCherry and of lysosomes by LysoTracker Green staining in Munc18-2 KO. Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). At t = 3:10, a cytoplasmic F-actin+ aggregate was formed (red arrow). No apparent contact to lysosomes was observed. After t = 7:10, maturation into 1 MVI with LysoTracker-positive lumen was observed (white arrow) that started to disintegrate at t = 9:00 (blue arrow) without proximity to lysosomes. Note that the luminal dye of the MVI separated from the LifeAct-mCherry MVI remnants. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that the structure had moved into or out of the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Lysosome-associated MVI formation in the cytoplasm. Tracking of MVI formation by LifeAct-mCherry and LysoTracker Green staining in Munc18-2 KO organoid. Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). At t = 9:00, a cytoplasmic F-actin+ aggregate was formed (red arrow) that co-localized lysosomes (green) before maturation into a lumen-containing MVI at t = 9:50 (white arrow). A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that a pre-existing MVI had moved into the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
References
- 1.Cutz E., Rhoads J.M., Drumm B., Sherman P.M., Durie P.R., Forstner G.G. Microvillus inclusion disease: an inherited defect of brush-border assembly and differentiation. N Engl J Med. 1989;320:646–651. doi: 10.1056/NEJM198903093201006. [DOI] [PubMed] [Google Scholar]
- 2.Vogel G.F., Hess M.W., Pfaller K., Huber L.A., Janecke A.R., Müller T. Towards understanding microvillus inclusion disease. Mol Cell Pediatr. 2016;3:3. doi: 10.1186/s40348-016-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overeem A.W., Posovszky C., Rings E.H.M.M., Giepmans B.N.G., van IJzendoorn S.C.D. The role of enterocyte defects in the pathogenesis of congenital diarrheal disorders. Dis Model Mech. 2016;9:1–12. doi: 10.1242/dmm.022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T., Mushiake S., Kato Y., Sato K., Sato M., Takeda N., Ozono K., Miki K., Kubo Y., Tsuji A., Harada R., Harada A. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448:366–369. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- 5.Müller T., Hess M.W., Schiefermeier N., Pfaller K., Ebner H.L., Heinz-Erian P., Ponstingl H., Partsch J., Röllinghoff B., Köhler H., Berger T., Lenhartz H., Schlenck B., Houwen R.J., Taylor C.J., Zoller H., Lechner S., Goulet O., Utermann G., Ruemmele F.M., Huber L.A., Janecke A.R. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet. 2008;40:1163–1165. doi: 10.1038/ng.225. [DOI] [PubMed] [Google Scholar]
- 6.Roland J.T., Bryant D.M., Datta A., Itzen A., Mostov K.E., Goldenring J.R. Rab GTPase–Myo5B complexes control membrane recycling and epithelial polarization. Proc Natl Acad Sci. 2011;108:2789–2794. doi: 10.1073/pnas.1010754108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knowles B.C., Roland J.T., Krishnan M., Tyska M.J., Lapierre L.A., Dickman P.S., Goldenring J.R., Shub M.D. Myosin Vb uncoupling from RAB8A and RAB11A elicits microvillus inclusion disease. J Clin Invest. 2014;124:2947–2962. doi: 10.1172/JCI71651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiegerinck C.L., Janecke A.R., Schneeberger K., Vogel G.F., van Haaften-Visser D.Y., Escher J.C., Adam R., Thöni C.E., Pfaller K., Jordan A.J., Weis C.-A., Nijman I.J., Monroe G.R., van Hasselt P.M., Cutz E., Klumperman J., Clevers H., Nieuwenhuis E.E.S., Houwen R.H.J., van Haaften G., Hess M.W., Huber L.A., Stapelbroek J.M., Müller T., Middendorp S. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology. 2014;147:65–68.e10. doi: 10.1053/j.gastro.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Côte M., Ménager M.M., Burgess A., Mahlaoui N., Picard C., Schaffner C., Al-Manjomi F., Al-Harbi M., Alangari A., Le Deist F., Gennery A.R., Prince N., Cariou A., Nitschke P., Blank U., El-Ghazali G., Ménasché G., Latour S., Fischer A., de Saint Basile G. Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells. J Clin Invest. 2009;119:3765–3773. doi: 10.1172/JCI40732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagel J., Beutel K., Lehmberg K., Koch F., Maul-Pavicic A., Rohlfs A.-K., Al-Jefri A., Beier R., Bomme Ousager L., Ehlert K., Gross-Wieltsch U., Jorch N., Kremens B., Pekrun A., Sparber-Sauer M., Mejstrikova E., Wawer A., Ehl S., zur Stadt U., Janka G. Distinct mutations in STXBP2 are associated with variable clinical presentations in patients with familial hemophagocytic lymphohistiocytosis type 5 (FHL5) Blood. 2012;119:6016–6024. doi: 10.1182/blood-2011-12-398958. [DOI] [PubMed] [Google Scholar]
- 11.Stepensky P., Bartram J., Barth T.F., Lehmberg K., Walther P., Amann K., Philips A.D., Beringer O., Zur Stadt U., Schulz A., Amrolia P., Weintraub M., Debatin K.-M., Hoenig M., Posovszky C. Persistent defective membrane trafficking in epithelial cells of patients with familial hemophagocytic lymphohistiocytosis type 5 due to STXBP2/MUNC18-2 mutations. Pediatr Blood Cancer. 2013;60:1215–1222. doi: 10.1002/pbc.24475. [DOI] [PubMed] [Google Scholar]
- 12.Vogel G.F., van Rijn J.M., Krainer I.M., Janecke A.R., Posovzsky C., Cohen M., Searle C., Jantchou P., Escher J.C., Patey N., Cutz E., Müller T., Middendorp S., Hess M.W., Huber L.A. Disrupted apical exocytosis of cargo vesicles causes enteropathy in FHL5 patients with Munc18-2 mutations. JCI Insight. 2017 Jul 20;2(14):e94564. doi: 10.1172/jci.insight.94564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel G.F., Klee K.M.C., Janecke A.R., Müller T., Hess M.W., Huber L.A. Cargo-selective apical exocytosis in epithelial cells is conducted by Myo5B, Slp4a, Vamp7, and Syntaxin 3. J Cell Biol. 2015;211:587–604. doi: 10.1083/jcb.201506112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaux G., Massey-Harroche D., Nicolle O., Rabant M., Brousse N., Goulet O., Le Bivic A., Ruemmele F.M. The localisation of the apical Par/Cdc42 polarity module is specifically affected in microvillus inclusion disease. Biol Cell. 2016;108:19–28. doi: 10.1111/boc.201500034. [DOI] [PubMed] [Google Scholar]
- 15.Phillips A.D., Schmitz J. Familial microvillous atrophy: a clinicopathological survey of 23 cases. J Pediatr Gastroenterol Nutr. 1992;14:380–396. [PubMed] [Google Scholar]
- 16.Mierau G.W., Wills E.J., Wyatt-Ashmead J., Hoffenberg E.J., Cutz E. Microvillous inclusion disease: report of a case with atypical features. Ultrastruct Pathol. 2001;25:275–279. [PubMed] [Google Scholar]
- 17.Groisman G.M., Amar M., Livne E. CD10: a valuable tool for the light microscopic diagnosis of microvillous inclusion disease (familial microvillous atrophy) Am J Surg Pathol. 2002;26:902–907. doi: 10.1097/00000478-200207000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Phillips A.D., Szafranski M., Man L.-Y., Wall W.J. Periodic acid–Schiff staining abnormality in microvillous atrophy: photometric and ultrastructural studies. J Pediatr Gastroenterol Nutr. 2000;30:34–42. doi: 10.1097/00005176-200001000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Iancu T.C., Mahajnah M., Manov I., Shaoul R. Microvillous inclusion disease: ultrastructural variability. Ultrastruct Pathol. 2007;31:173–188. doi: 10.1080/01913120701350712. [DOI] [PubMed] [Google Scholar]
- 20.Weis V.G., Knowles B.C., Choi E., Goldstein A.E., Williams J.A., Manning E.H., Roland J.T., Lapierre L.A., Goldenring J.R. Loss of MYO5B in mice recapitulates microvillus inclusion disease and reveals an apical trafficking pathway distinct to neonatal duodenum. Cell Mol Gastroenterol Hepatol. 2016;2:131–157. doi: 10.1016/j.jcmgh.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneeberger K., Vogel G.F., Teunissen H., van Ommen D.D., Begthel H., El Bouazzaoui L., van Vugt A.H.M., Beekman J.M., Klumperman J., Müller T., Janecke A., Gerner P., Huber L.A., Hess M.W., Clevers H., van Es J.H., Nieuwenhuis E.E.S., Middendorp S. An inducible mouse model for microvillus inclusion disease reveals a role for myosin Vb in apical and basolateral trafficking. Proc Natl Acad Sci U S A. 2015;112:12408–12413. doi: 10.1073/pnas.1516672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruemmele F.M., Müller T., Schiefermeier N., Ebner H.L., Lechner S., Pfaller K., Thöni C.E., Goulet O., Lacaille F., Schmitz J., Colomb V., Sauvat F., Revillon Y., Canioni D., Brousse N., de Saint-Basile G., Lefebvre J., Heinz-Erian P., Enninger A., Utermann G., Hess M.W., Janecke A.R., Huber L.A. Loss-of-function of MYO5B is the main cause of microvillus inclusion disease: 15 novel mutations and a CaCo-2 RNAi cell model. Hum Mutat. 2010;31:544–551. doi: 10.1002/humu.21224. [DOI] [PubMed] [Google Scholar]
- 23.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 24.de Lau W., Barker N., Low T.Y., Koo B.-K., Li V.S.W., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., Stange D.E., van Es J.E., Guardavaccaro D., Schasfoort R.B.M., Mohri Y., Nishimori K., Mohammed S., Heck A.J.R., Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 25.Koo B.-K., Spit M., Jordens I., Low T.Y., Stange D.E., van de Wetering M., van Es J.H., Mohammed S., Heck A.J.R., Maurice M.M., Clevers H. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 26.Yin X., Farin H.F., van Es J.H., Clevers H., Langer R., Karp J.M. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel G.F., Janecke A.R., Krainer I.M., Gutleben K., Witting B., Mitton S.G., Mansour S., Ballauff A., Roland J.T., Engevik A.C., Cutz E., Müller T., Goldenring J.R., Huber L.A., Hess M.W. Abnormal Rab11-Rab8-vesicles cluster in enterocytes of patients with microvillus inclusion disease. Traffic. 2017;18:453–464. doi: 10.1111/tra.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Q., Bonder E.M., Engevik A.C., Zhang L., Tyska M.J., Goldenring J.R., Gao N. Disruption of Rab8a and Rab11a causes formation of basolateral microvilli in neonatal enteropathy. J Cell Sci. 2017;130:2491–2505. doi: 10.1242/jcs.201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., Graaf EA van de, Nieuwenhuis E.E.S., Houwen R.H.J., Vleggaar F.P., Escher J.C., Rijke YB de, Majoor C.J., Heijerman H.G.M., Groot KM. de W., Clevers H., Ent CK van der, Beekman J.M. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med. 2016;8:344ra84. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 30.Bigorgne A.E., Farin H.F., Lemoine R., Mahlaoui N., Lambert N., Gil M., Schulz A., Philippet P., Schlesser P., Abrahamsen T.G., Oymar K., Davies E.G., Ellingsen C.L., Leteurtre E., Moreau-Massart B., Berrebi D., Bole-Feysot C., Nischke P., Brousse N., Fischer A., Clevers H., Basile G. de S. TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J Clin Invest. 2014;124:328–337. doi: 10.1172/JCI71471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 32.Koo B.-K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., Clevers H. Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 2011;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 33.Kolotuev I. Positional correlative anatomy of invertebrate model organisms increases efficiency of TEM data production. Microsc Microanal. 2014;20:1392–1403. doi: 10.1017/S1431927614012999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Actin dynamics in a differentiated wild-type organoid. Overview of LifeAct-mCherry signal in WT (compare with Figure 6A). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). Images of a single z-plane are shown. Time points are indicated as hours:minutes, with 10-minute intervals (10 frames/s).
Dynamic MVI formation in a differentiated Munc18-2 KO organoid. Overview of LifeAct-mCherry signal in Munc18-2 KO (compare with Figure 6A). Cells were differentiated for 2 days in enterocyte differentiation medium before start of imaging (t = 0). Images of a single z-plane are shown. Time points are indicated as hours:minutes, with 10-minute intervals (10 frames/s).
Cytoplasmic MVI formation. Tracking of MVI formation by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 6B). Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). At t = 5:50, a cytoplasmic F-actin+ aggregate was formed (red arrow) that matured after t = 18:20 into a lumen-containing MVI (white arrow) and started to disintegrate at t = 21:20 (blue arrow). Note that a second MVI formed in the right half of the image. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that a pre-existing MVI had moved into the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
MVI formation by apical membrane internalization. Tracking of MVI formation by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 6C). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 4:00, the apical membrane invaginated (red arrow). At t = 7:10, a lumen-containing MVI pinched off (white arrow). A single z-plane is shown and apical is top. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
MVI formation by basal membrane internalization. Tracking of MVI formation by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 6D). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0:00). At t = 1:00, deposition of ectopic basal F-actin was observed (red arrow). At t = 3:00, membrane invagination and formation of a lumen-containing MVI were observed (white arrow). A single z-plane is shown and apical is top. Time points are indicated as hours:minutes, with 10-minute intervals (3 frames/s).
Disintegration of MVI in the cytoplasm. Tracking of MVI breakdown by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 7A). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 4:00, a central MVI was observed (red arrow) that started to disintegrate in the cytosol at t = 8:30 (white arrow) and disappeared at t = 13:10. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that the structure had moved out of the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Fusion of MVI to the apical plasma membrane. Tracking of MVI fusion to the apical membrane by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 7B). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 2:40, a central MVI was observed that increased in size (red arrow). At t = 5:30, it contacted the apical membrane and rapidly shrunk (white arrow). By t = 8:30 the structure had fully integrated into the apical membrane. A single z-plane is shown and apical is top. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Fusion of MVI to the basal plasma membrane. Tracking of MVI fusion to the basal membrane by LifeAct-mCherry signal in Munc18-2 KO organoid (compare with Figure 7C). Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 4:00, 1 MVI pinched off from the basal membrane (red arrow). After t = 5:00 the structure again fused to the basal membrane (white arrow) and the signal disappeared by t = 6:30. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks confirmed that the structure was not lost owing to z-movement. Time points are indicated as hours:minutes, with 10-minute intervals (3 frames/s).
Shedding of MVI-containing cell. Tracking of a cell shedding event by LifeAct-mCherry signal in Munc18-2 KO organoid. Cells were kept for 2 days in enterocyte differentiation medium before start of imaging (t = 0). At t = 0:30, 1 MVI was formed (red arrow) before the cell was extruded from the epithelium into the lumen at t = 2:30 (white arrow). A single z-plane is shown and apical is top. Inspection of adjacent z–stacks confirmed that the MVI was not lost owing to z-movement. Time points are indicated as hours:minutes, with 10-minute intervals (3 frames/s).
Lysosome-associated MVI disintegration in the cytoplasm. Tracking of MVI breakdown by LifeAct-mCherry and LysoTracker Green staining in Munc18-2 KO organoid. Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). One MVI was observed (white arrow) that had a LysoTracker-positive lumen that started to be lost from t = 10:20 (blue arrow). Note the proximity to lysosomes during this process. The MVI disappeared at t = 17:20. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that the structure had moved out of the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Lysosome nonassociated MVI formation and disintegration in the cytoplasm. Tracking of 1 MVI by LifeAct-mCherry and of lysosomes by LysoTracker Green staining in Munc18-2 KO. Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). At t = 3:10, a cytoplasmic F-actin+ aggregate was formed (red arrow). No apparent contact to lysosomes was observed. After t = 7:10, maturation into 1 MVI with LysoTracker-positive lumen was observed (white arrow) that started to disintegrate at t = 9:00 (blue arrow) without proximity to lysosomes. Note that the luminal dye of the MVI separated from the LifeAct-mCherry MVI remnants. A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that the structure had moved into or out of the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).
Lysosome-associated MVI formation in the cytoplasm. Tracking of MVI formation by LifeAct-mCherry and LysoTracker Green staining in Munc18-2 KO organoid. Cells were kept for 2 days in enterocyte differentiation medium before starting the imaging (t = 0). At t = 9:00, a cytoplasmic F-actin+ aggregate was formed (red arrow) that co-localized lysosomes (green) before maturation into a lumen-containing MVI at t = 9:50 (white arrow). A single z-plane is shown and apical is top. Inspection of adjacent z-stacks was performed to exclude that a pre-existing MVI had moved into the focus plane. Time points are indicated as hours:minutes, with 10-minute intervals (5 frames/s).