Abstract
Nosema disease is one of the most important diseases of adult honey bees worldwide. It is known as silent killer because there are no characteristic symptoms. The aim of the present study was to determine prevalence of Nosema species in various towns of Kurdistan province in Iran. A multiplex polymerase chain reaction (multiplex-PCR) was performed for identification of Nosema species infecting European honeybee, Apis mellifera. A total of 100 samples were collected from apiaries (870 hives) in 10 counties of Kurdistan province, located in the west of Iran. Samples were examined using light microscope and PCR. The light microscope was used to determine the presence of Nosema spores in all of the collected samples. Multiplex-PCR based on 16S ribosomal RNA was used to differentiate N. apis from N. ceranae. Overall prevalence of the microscopic evaluation and PCR method were 29.00% and 32.00%, respectively. The analysis of Nosema isolates from interrogation of DNA databank entries of Kurdistan apiaries (based on rRNA sequence data) indicated that only N. ceranae was widespread in these apiaries, and it had already been found in high percentages (50.00%) in Marivan and Kamiaran counties of Kurdistan province. It was shown that only N. ceranae was found by PCR assay in the region.
Key Words: Iran, Kurdistan, Molecular identification, Nosema ceranae, Nosemosis
Introduction
Nosemosis (Nosema disease) is one of the most serious and prevalent adult honeybee diseases worldwide,1,2 which it caused by intracellular microsporidian parasites from a genus of Nosema. For decades, Nosema disease was particularly ascribed to a single species of Nosema, Nosema apis (N. apis), which was first identified in European honeybee, Apis mellifera.3 In 1996, another species of Nosema was first recognized in the Asian honey bee, Apis ceranae, thus named Nosema ceranae (N. cerana).4 In 2005, a natural infection of N. ceranae was reported in Apis mellifera colonies from Taiwan5 in a short period of time. Infection of Apis mellifera with N. ceranae was reported in Europe,6,7 the United States,8 China,9 Vietnam and worldwide.10 Nosema apis infection causes a fast acting and short duration syndrome; however, this has not been the case for N. ceranae, in which it has been noticed in relation to nonspecific symptoms such as gradual population decline, higher autumn/winter colony deaths or low honey production.11 Further, it has recently been shown that N. ceranae does not show the seasonality that is seen with N. apis.12 The different symptoms presented by these Nosema species in honey bees highlight the need to observe two different clinical types: Nosemosis type A caused by N. apis and Nosemosis type C caused by N. ceranae.12 Detection of Nosema species depends upon microscopic observation, molecular methods or transmission electron microscopy.13 The spores of the two Nosema species are very similar, and microscopic examination cannot differentiate between N. apis and N. ceranae.11 Several PCR protocols have been described, including PCR with specific primers,8 PCR-RFLP,10,14 Real-time PCR,15,16 or multiplex PCR.17 The aim of the present study was to determine prevalence of Nosema ceranae in various towns of Kurdistan province in Iran.
Materials and Methods
Study area. Kurdistan is located in the west of Iran with an area of 28,000 Km2. This province lies between the eastern longitude 45° 33′ 11″ and 51° 13′ 7″ and northern longitude 34° 24′ 16″ and 37° 52′ 12″. The weather conditions are similar to the Mediterranean area in which rainfall occurs in winter, moderate rain occurs in autumn and spring and no rainfall occurs during the summer season. With respect to the climate, the region is defined as having cold winters, hot summers, neutral springs and autumns with a wide range of temperature changes. According to the latest divisions of the country, this province has 10 counties.
Samples collection. A total of 100 apiaries were randomly sampled from November 2014 to September 2015 in 10 counties of Kurdistan province. The apiaries were selected randomly and according to the instructions of the Iranian Veterinary Organization (IVO). The required samples were taken from 5.00% of the colonies in each apiary18 and the samples were immediately transferred to the honey bee research department of Razi Vaccine and Serum Research Institute, Karaj, Iran.
Preparation of samples for microscopic examination. The abdomens of 20 adult dead honey bees were macerated in 10 mL distilled water and were crushed in a mortar. Then, the suspension was passed through a 100 µm mesh sieve to remove the debris and was centrifuged for 6 min at 800 g. Finally, the supernatant was discarded and the pellet was examined under the common light microscope at 400× magnification. This methodology was used to determine the presence of Nosema spores in all of the collected samples.13
Preparation of samples for PCR. The abdomens of 20 adult dead honey bees from each apiary were macerated in 10 mL distilled water (PCR grade), and the suspension was then filtered and centrifuged at 800 g for 6 min. Spore germination was induced with 200 μL freshly prepared germination buffer (0.50 M sodium chloride, 0.50 M sodium hydrogen carbonate, pH to 6.0 with ortho-phosphoric acid), and the mixture was incubated at 37 ˚C for 15 min.13, 17
DNA extraction. DNA was extracted using DNA extraction kit (Takapozist, Tehran, Iran) according to the manufacturer’s instructions.
Polymerase chain reaction. PCR amplification of 16S rRNA was performed using PCR kit (Sinaclon, Tehran, Iran) in an Eppendorf Mastercycler® gradient thermal cycler (Eppendorf, Hamburg, Germany) according to OIE terrestrial manual 2008 for N. ceranae, N. apis and N. bombi. For multiplex PCR amplification of partial 16S rRNA (= SSU rRNA) gene fragments, first 50 µL reaction mixture contains 5 ng genomic DNA, 3 mM MgCl2, 200 µM of each deoxyribonucleotide triphosphate, 100 ng of primers, 5 µL of 10X PCR buffer (100 mM Tris/HCl, pH 8.3; 15 mM MgCl2; 500 mM KCl) and 1 U of Taq polymerase. Conditions of amplification consist of an initial denaturation cycle at 94 ˚C for 15 sec followed by 25 cycles of denaturation (94 ˚C, 15 sec), primer annealing (61.80 ˚C, 30 sec), primer extension (72.00 ˚C, 45 sec) followed by additional extension step of 7 minutes at 72.00 ˚C.19 The PCR products were separated by electrophoresis on 1.00% agarose gel, stained with safe stain (Baiometra, Berlin, Germany) and visualized by UV transillumination. In this study, primers targeting small subunit 16S rRNA gene of N. apis, N. ceranae and N. bombi were used (Table 1). Positive controls for N. ceranae and N. apis were prepared from Department of Honeybee-Silkworm and Wildlife Diseases, Razi Vaccine and Serum Research Institute, Karaj, Iran. The PCR products were sent for both forward and reverse sequencing using Sanger method (Bioneer, Daejeon, South Korea), and revealed sequences were verified by Bioedit software (version 7.0.5; Ibis Therapeutics, Carlsbad, USA).20
Table 1.
Primers used for identification of Nosema species by multiplex PCR.
| Species | Primer sequence (5`-3`) | Fragment size | References |
|---|---|---|---|
| N. ceranae | 5’-CGGCGACGATGTGATATGAAAATATTAA-3’ 5’-CCCGGTCATTCTCAAACAAAAAACCG-3’ |
218-219 | 17 |
| N. apis | 5’-GGGGGCATGTCTTTGACGTACTATGTA-3’ 5’-GGGGGGCGTTTAAAATGTGAAACAACTATG-3’ |
321 | 17 |
| N. bombi | 5’-TTTATTTTATGTRYACMGCAG-3` 5`-GACTTAGTAGCCGTCTCTC-3` |
171 | 19 |
Results
The results of microscopic examination and PCR of all 100 samples are presented in Table 2. Microscopic examination showed 29.00% samples of apiaries were infected by Nosema spores (Fig. 1).
Table 2.
Distribution of erm genes in isolates.
| Counties | Apiary numbers | Microscopic positive (%) | PCR positive (%) |
|---|---|---|---|
| Bane | 10 | 10 | 10 |
| Bijar | 10 | 30 | 30 |
| Dehgolan | 10 | 30 | 30 |
| Divandarreh | 10 | 20 | 20 |
| Kamyaran | 10 | 50 | 60 |
| Marivan | 10 | 50 | 60 |
| Qorveh | 10 | 40 | 40 |
| Sanandaj | 10 | 40 | 50 |
| Saqqez | 10 | 20 | 20 |
| Sarvabad | 10 | 0 | 0 |
| Total (%) | 100 | 29 | 32 |
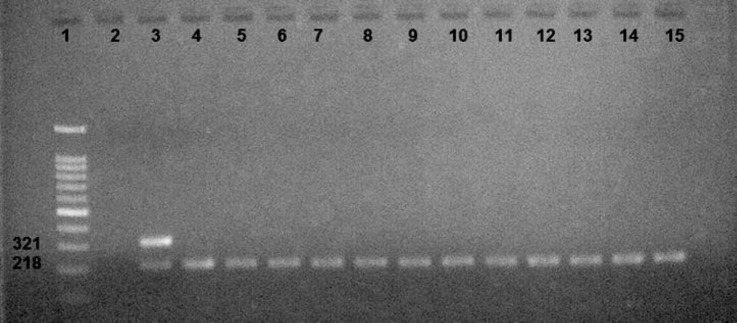
Fig. 1.
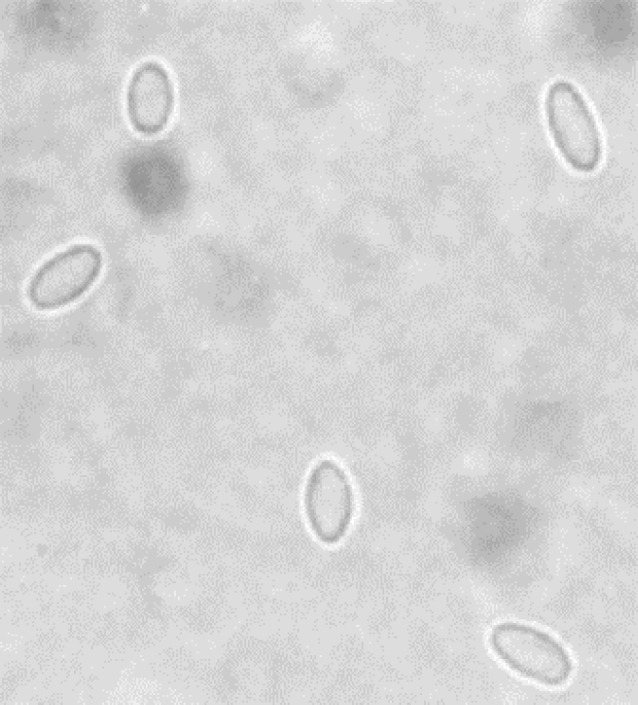
Spores of Nosema spices in light microscopy (400×).
In this study, PCR results showed that 32 samples (32.00%) were positive for N. ceranae, which showed a specific amplicon at 218bp (Fig. 2).
Fig 2.
Lane 1: Ladder 100 bp (Smobio, Hsinchu, Taiwan), Lane 2: Negative control, Lane 3: Positive control, Lanes 4 -15: N. ceranae.
In all of the positive samples, the presence of N. ceranae was detected. The analysis of Nosema isolates from interrogation of DNA databank entries of Kurdistan apiaries (based on rRNA sequence data) indicated that only N. ceranae was widespread in these apiaries, and it had already been found in high percentages (50%) in Marivan and Kamiaran counties of Kurdistan province.
Discussion
There is a little information about the epidemiological factors and clinical symptoms of this disease in different areas in Iran which have distinct beekeeping management and climatic conditions. The present study showed that N. ceranae was the only Nosema species found to infect honey bees from apiaries in Kurdistan province in the west of Iran. Lotfi et al. reported N. apis is widely distributed in East Azerbaijan province in northwest of Iran, being diagnosed in 85.00% of apiaries.21 Modirrousta et al. by multiplex PCR assay showed the samples were collected from five provinces of Iran (Alborz, East-Azerbaijan, Qazvin, Gilan and Tehran) in 2004-2013, were positive for N. ceranae.22 In Mazandaran province, north of Iran, only N. ceranae was found by PCR assay.23 Tavassoli et al. reported apiaries of Urmia, northwest of Iran, infected by N. apis.24 The prevalence of N. ceranae in East-Azerbaijan by microscopic and PCR methods was 58.10% and 67.10%, respectively.25 Aroee et al. reported that honey bee colony of Esfahan, Chaharmahal and Bakhtiari, and Fars was infected with N. ceranae.26 In Iran, there are reports showing that the frequency of Nosema infection has increased.25,27 During the last 10 years, an increase in infections by microsporidian parasite in honey bee (A. mellifera) has been detected in several European countries.28 Both in the North American continent29 and in Europe,17 the proportion of N. ceranae infections appears to dominate in warmer climates compared to more temperate regions, whereas N. apis may be presently more prevalent in cooler climates.1,7 Two types of Nosema that cause this disease are different from each other in terms of pathogenicity, symptoms and epidemiology, by which it is possible to diagnose the disease. Differences have been documented in Europe with regard to the epidemiological pattern of Nosema caused by N. ceranae,30 thus this disease has a prolonged incubation period without obvious clinical symptoms, which can lead to the death of colonies.6 The symptoms of N. ceranae include longer breeding period in the cold months, disproportion between the nurse bees and the larvae population in the hive in the warm months of the year, reduced honey production in the hive, wakened hive, reduced population of adult bees and destruction of hives over 1.5 to 2 years.31 There are two mechanisms involved in the loss of such contaminated hives: first, in the cold season more than 50.00% of the hive population is lost at one side of the hive, the mean number of spores per bee is over 10 million, and contamination is evident if the queen is found; second, the hive death occurs in early spring, the mean number of spores per bee is less, and the queen is not contaminated with disease. Further, in these two mechanisms the ratio of old/young bees is different in various seasons. In early spring, the ratio of contaminated/non-contaminated young bees is reduced, thereby postponing the queen contamination.12 A disease occurs due to a complex relationship between the triple sides of epidemiology, including disease cause, host, and environmental conditions. In domestic animals, the host is largely influenced by the maintenance and management conditions.32 The host is one of the triple sides involved in the occurrence of every disease. The hosting differences lead to differences in severity of the disease; for example, laboratory research using bees with different ages has been reported to yield different results indicating the effect of factors associated with the host on the results obtained from the research.33 Environmental conditions such as altitude effects, type of plants in the region, and management of apiary highly affect the parasitic relationships, as N. ceranae has been reported in Spain under the influence of temperature on the spread of N. ceranae. Currently, contamination of honey bee colonies with different species of Nosema around the world does not follow a different weather pattern.34 The studies conducted in regions where both types of Nosema have been prevalent have shown that N. ceranae is more prevalent in warmer areas, areas with Mediterranean climate, and prevalence of N. apis is higher in regions with moderate climate. This subject should be considered while the migratory bees enter such regions.1 Environmental factors have dramatically affected the competition between N. apis and N. ceranae, as in many areas around the world N. ceranae is the only species identified in honey bees.8-10,22,35 However, in some areas around the world, due to special weather conditions, N. apis is more prevalent.36,37 Recent studies at different levels (i.e. individual, colony, apiary, country, different races of honey bee) as well as analysis of the reasons involved in the prevalence of Nosema species around the world have indicated that no substitute has been found for N. apis and N. ceranae worldwide.34 Only N. ceranae is the dominant species during the year, and prevalence of N. apis depends on a specific epidemiological pattern (occurrence in spring and fall or more generally in colder seasons). This theory has been also proposed in the past,38,39 and recent studies in the U.S. have confirmed it.40 Many studies have been carried out on Nosema and its prevalence in spring, when the disease is severe.41,42 This is exactly when only N. apis is found as the cause of Nosemosis in the hives, an agent that is not active in tropical and subtropical areas;42 whereas, N. ceranae can be diagnosed all year in different geographical latitudes in colonies.17,34 There are a few studies on Nosema species in Iran, some of which have used microscopic observation technique. Given the many similarities of the spores of these two species, their differential diagnosis by microscopic observation is very difficult and at times impossible.11 Hence, molecular techniques must be used in this regard. Analysis of studies published in Iran demonstrates that studies that have used molecular techniques to investigate the causative agent of Nosema have reported microsporidian N. ceranae as the causative agent of this disease in Iran. On the other hand, studies using microscopic observation technique have introduced N. apis as the cause of this disease, and no report has ever been presented regarding the molecular diagnosis of N. apis in Iranian apiaries. Also, considering numerous reports from around the world about the replacement of N. ceranae with N. apis and changes in epidemiologic symptoms of the disease,6,7,10,17 N. ceranae is probably the cause of Nosema infection in Iran; however, further studies are required to confirm this issue.
Acknowledgments
This study was supported by grants received from Veterinary Office of Kurdistan (Grant No: 92/13245) and Razi Vaccine and Serum Research Institute, Karaj, Iran (Grant No: 4-53-18-93108).
Conflict of interest
The authors have no conflict of interest.
References
- 1.Fries I. Nosema ceranae in European honey bees (Apis mellifera) J Invertebr Pathol. 2010;103(S1):S73–79. doi: 10.1016/j.jip.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Matheson A. World bee health report. Bee world. 1993;74(4):176–212. [Google Scholar]
- 3.Zander E. Animal parasites as pathogens in the bee [German] Münchener Bienenzeitung. 1909;31:196–204. [Google Scholar]
- 4.Fries I, Feng F, da Silva A, et al. Nosema ceranae n sp (Microspora Nosematidae) morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis ceranae (Hymenoptera, Apidae) Eur J Protistol. 1996;32(3):356–365. [Google Scholar]
- 5.Huang W-F, Jiang J-H, Chen Y-W, et al. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie. 2007;38(1):30–37. [Google Scholar]
- 6.Paxton RJ, Klee J, Korpela S, et al. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie. 2007;38(6):558–565. [Google Scholar]
- 7.Chen Y, Evans JD, Smith IB, et al. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J Invertebr Pathol. 2008;97(2):186–188. doi: 10.1016/j.jip.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Chen YP, Huang ZY. Nosema ceranae, a newly identified pathogen of Apis mellifera in the USA and Asia. Apidologie. 2010;41(3):364–374. [Google Scholar]
- 9.Klee J, Besana AM, Genersch E, et al. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J Invertebr Pathol. 2007;96(1):1–10. doi: 10.1016/j.jip.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Fries I, Martín R, Meana A, et al. Natural infections of Nosema ceranae in European honey bees. J Apic Res. 2006;47(3):230–233. [Google Scholar]
- 11.Higes M, Martin-Hernandez R, Meana A. Nosema ceranae in Europe: An emergent type C nosemosis. Apidologie. 2010;41(3):375–392. [Google Scholar]
- 12.OIE. Nosemosis of honey bees. OIE terrestrial manual: OIE. 2013:1–6. [Google Scholar]
- 13.Tapaszti Z, Forgách P, Kövágó C, et al. First detection and dominance of Nosema ceranae in Hungarian honeybee colonies. Acta Vet Hung. 2009;57:383–388. doi: 10.1556/AVet.57.2009.3.4. [DOI] [PubMed] [Google Scholar]
- 14.Bourgeois LA, Rinderer TE, Beaman LD, et al. Genetic detection and quantification of Nosema apis and N ceranae in the honey bee. J Invertebr Pathol. 2010;103(1):53–58. doi: 10.1016/j.jip.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Cox-Foster DL, Conlan S, Holmes EC, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318(5848):283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 16.Martín-Hernández R, Meana A, Prieto L, et al. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl Environ Microbiol. 2007;73(20):6331–6338. doi: 10.1128/AEM.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bokaie S, Sharifi L, Mehrabadi M. Prevalence and epizootical aspects of varroasis in Golestan province, northern Iran. J Arthropod Borne Dis. 2014;8(1):102–107. [PMC free article] [PubMed] [Google Scholar]
- 18.Fries I, Chauzat MP, Chen YP, et al. Standard methods for Nosema research. J Apic Res. 2013;52(1):1–28. doi: 10.3896/IBRA.1.52.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall T. BioEdit: An important software for molecular biology. GERF Bull Biosci. 2011;2(1):60–61. [Google Scholar]
- 20.Lotfi AR, Jamshidi R, Aghdam Shahryar H, et al. The Prevalence of nosemosis in honey bee colonies in Arasbaran Region (Northwestern Iran) Am Eurasian J Agric Environ Sci. 2009;5(2):255–257. [Google Scholar]
- 21.Modirrousta H, Moharrami M, Mansouri MA. Retrospective study of the Nosema ceranae infection of honey bee colonies in Iran (2004-2013) Arc Razi Ins. 2014;69(2):197–200. [Google Scholar]
- 22.Nabian S, Ahmadi K, Nazem Shirazi M, et al. First detection of Nosema ceranae, a microsporidian protozoa of European honeybees (Apis mellifera) in Iran. Iran J Parasitol. 2011;6(3):89–95. [PMC free article] [PubMed] [Google Scholar]
- 23.Tavassoli M, Eiganinejad S, Alizadeh-Asl S. A survey on Nosema apis infection in apiaries of Urmia, North-West of Iran. Iran J Vet Sci Technol. 2010;1(1):35–40. [Google Scholar]
- 24.Razmaraii N, Karimi H. A survey of Nosema disease of honey bee (Apis mellifera) in East Azarbaijan province of Iran. J Anim Vet Adv. 2010;9(5):879–882. [Google Scholar]
- 25.Aroee F, Azizi H, Shiran B, et al. Molecular identification of Nosema species in provinces of Fars, Chaharmahal and Bakhtiari and Isfahan (Southwestern Iran) Asian Pac J Trop Biomed. 2017;7(1):10–13. [Google Scholar]
- 26.Davoudi J, Naderi A, Mohammadpour F, et al. Study of infection rate of suburb bee hives to parasites nosema apis, varroa spp and acarapis woodi in Miyaneh, Iran. J Now Agric Sci. 2009;4(13):39–43. [Google Scholar]
- 27.Botias C, Martin-Hernandez R, Garrido-Bailon E, et al. The growing prevalence of Nosema ceranae in honey bees in Spain, an emerging problem for the last decade. Res Vet Sci. 2012;93(1):150–155. doi: 10.1016/j.rvsc.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Williams GR, Shafer AB, Rogers RE, et al. First detection of Nosema ceranae, a microsporidian parasite of European honey bees (Apis mellifera), in Canada and central USA. J Invertebr Pathol. 2008;97(2):189–192. doi: 10.1016/j.jip.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 29.COLOSS. Nosema disease: lack of knowledge and work standardization. COST Action FA0803 - Prevention of honeybee colony losses. Guadalajava, Spain; 2009. pp. 1–39. [Google Scholar]
- 30.Higes M, Martín Hernández R, García Palencia P, et al. Horizontal transmission of Nosema ceranae (Micro-sporidia) from worker honeybees to queens (Apis mellifera) Environ Microbiol Rep. 2009;1(6):495–498. doi: 10.1111/j.1758-2229.2009.00052.x. [DOI] [PubMed] [Google Scholar]
- 31.Higes M, Meana A, Bartolomé C, et al. Nosema ceranae (Microsporidia), a controversial 21st century honey bee pathogen. Environ Microbiol Rep. 2013;5(1):17–29. doi: 10.1111/1758-2229.12024. [DOI] [PubMed] [Google Scholar]
- 32.Higes M, García-Palencia P, Martín-Hernández R, et al. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia) J Invertebr Pathol. 2007;94(3):211–217. doi: 10.1016/j.jip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Martín Hernández R, Botías C, Bailón EG, et al. Microsporidia infecting Apis mellifera: coexistence or competition Is Nosema ceranae replacing Nosema apis? Environ Microbiol. 2012;14(8):2127–2138. doi: 10.1111/j.1462-2920.2011.02645.x. [DOI] [PubMed] [Google Scholar]
- 34.Martínez J, Leal G, Conget P. Nosema ceranae an emergent pathogen of Apis mellifera in Chile. Parasitol Res. 2012;111(2):601–607. doi: 10.1007/s00436-012-2875-0. [DOI] [PubMed] [Google Scholar]
- 35.Budge G, Powell M, Roberts K, et al. What has Nosema got to do with losses? Monitoring both Nosema species in the UK. In: Kence M, editor. Ankara, Turkey : 4th European Conference of Apidology; 2010. p. 47. [Google Scholar]
- 36.Gisder S, Hedtke K, Mo¨ckel N, et al. Five-year cohort study of Nosema spp in Germany: Does climate shape virulence and assertiveness of Nosema ceranae? Appl Environ Microbiol. 2010;76(9):3032–3038. doi: 10.1128/AEM.03097-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bermejo FJO, Fernández PG. Nosema disease in the honey bee (Apis mellifera L) infested with varroa mites in southern Spain. Apidologie. 1997;28:105–112. [Google Scholar]
- 38.Pajuelo AG, Torres C, Bermejo FJO. Colony losses: a double blind trial on the influence of supplementary protein nutrition and preventative treatment with fumagillin against Nosema ceranae. J Apic Res. 2008;47:84–86. [Google Scholar]
- 39.Runckel C, Flenniken ML, Engel JC, et al. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One. 2011;6(6):e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey L. The epidemiology and control of nosema disease of the honeybee. Ann Appl Biol. 1955;43:379–389. [Google Scholar]
- 41.Martín-Hernández R, Meana A, García-Palencia P, et al. Effect of temperature on the biotic potential of honeybee microsporidia. Appl Environ Microbiol. 2009;75(8):2554–2557. doi: 10.1128/AEM.02908-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martín-Hernández R, Meana A, García-Palencia P, et al. Effect of temperature on the biotic potential of honeybee microsporidia. Appl Environ Microbiol. 2009;75(8):2554–2557. doi: 10.1128/AEM.02908-08. [DOI] [PMC free article] [PubMed] [Google Scholar]