Abstract
Sangju Ganmao tablet (SGT), a well-known Chinese patent medicine used to treat cold symptoms, is made from eight herbal medicines. In this study, an off-line hydrophilic interaction × reversed-phase two-dimensional liquid chromatography (HILIC × RP 2D-LC) method was developed to comprehensively separate the chemical constituents of SGT. Through optimization of the experimental conditions, a total of 465 peaks were finally detected in SGT, and the structures of 54 selected compounds were fully identified or tentatively characterized by quadrupole time-of-flight mass spectrometry (qTOF-MS) analysis. The established 2D-LC analysis showed high orthogonality (63.62%) and approximate 11-fold improvement in peak capacity (2399 and 1099, obtained by two calculation methods), in contrast to conventional one-dimensional RPLC separation. The eight component herbs of SGT were also respectively separated by using the 2D-LC system, and we found that a total of 12 peaks detected in SGT were not discovered in any component herbs. These newly generated chemical constituents would benefit better understanding of the compatibility mechanism of the component herbs. The strategy established in this study could be used for systematic chemical comparison of SGT and its component herbs, which contributes to exploration of herbal compatibility mechanism.
Keywords: two-dimensional liquid chromatography (2D-LC), hydrophilic interaction chromatography (HILIC), reverse-phase liquid chromatography (RPLC), Sangju Ganmao tablet (SGT), quadrupole time-of-flight mass spectrometry (qTOF-MS), compatibility mechanism
Introduction
Most of the Chinese patent medicines are composed of several or even 10s of herbal medicines, and they are very complicated chemical systems containing both hydrophilic and hydrophobic compounds (Li et al., 2010). While profiling the chemical constituents is critically important for modern investigations of a Chinese patent medicine, chromatographic separation of such a complex system is still a big challenge. On the other hand, the chemical profiling of a Chinese patent medicine is usually different from its component herbs, since chemical reactions and physical changes, such as oxidation and precipitation, sometimes occur during decocting together (Kim et al., 2014). Investigating the different chemical constituents between a Chinese patent medicine and its component herbs may be one of effective approaches to explore its compatibility mechanism.
Sangju Ganmao tablet (SGT), a well-known Chinese patent medicine, is currently used in clinical practice to treat cold symptoms, and has been officially recorded in China Pharmacopoeia (Chinese Pharmacopoeia Commission, 2015). It is composed of eight herbs, namely mulberry leaf (leaves of Morus alba L.), chrysanthemum (flowers of Chrysanthemum morifolium Ramat.), Fructus Forsythiae [fruits of Forsythia suspensa (Thunb.) Vahl], licorice (roots and rhizomes of Glycyrrhiza uralensis Fisch.), Semen Armeniacae Amarum (seeds of Prunus armeniaca L.), Platycodi Radix [roots of Platycodon grandiflorus (Jacq.) A.DC.], Phragmitis Rhizoma [roots and rhizomes of Phragmites australis (Cav.) Trin. ex Steud.], and mint (whole herbs of Mentha canadensis L.). The chemical constituents of SGT mainly include flavonoids (free flavonoids and flavonoid glycosides), triterpenoid saponins, phenylethanoid glycosides, and organic acids (Chang et al., 2014). Although several researches have studied the chemical profiling of these eight component herbs using various methods (Huang and Sheu, 2007; He et al., 2013; Chen et al., 2016; Song et al., 2017), systematic chemical analysis of SGT has never been reported so far. Recently, we developed a LC–MS method to profile the chemical constituents of SGT by optimizing different HPLC systems, and only less than 50 compounds were detected (Guo et al., 2017). Comprehensive profiling of chemical constituents in SGT is hindered due to its complex chemical composition, and the significantly different content renders those minor components difficult to be separated and detected. The chemical differences of SGT and its component herbs are also unknown.
In the past decade, two-dimensional liquid chromatography (2D-LC) has been proven to be a powerful tool in rapid separation and detection of chemical constituents in traditional Chinese medicines (TCM) (Shellie and Haddad, 2006; Guiochon et al., 2008; Marchetti et al., 2008). Among all the separation modes, combination of reversed-phase liquid chromatography (RPLC) and hydrophilic interaction chromatography (HILIC) is an effective method to separate complex mixtures with a wide range of polarities (Wang et al., 2008; Jandera and Hájek, 2018). A 2D-LC system can be operated in the on-line or off-line mode. Comparing with an on-line 2D-LC system with complex equipment settings, an off-line 2D-LC system can easily accomplish the flexible integration of different separation mechanisms without instrumental limitation (Li et al., 2014). Very recently, an off-line HILIC × RP 2D-LC method was established in our lab to achieve the comprehensive profiling of the chemical constituents of Ginkgo biloba extract (Ji et al., 2017).
In this study, a method based on off-line HILIC × RP 2D-LC coupled with qTOF-MS was established to comprehensively profile the chemical constituents of SGT, and the 2D-LC system was systematically optimized to achieve ideal orthogonality and peak capacity. The established method was used to investigate the chemical differences between SGT and its component herbs in order to explore the herbal compatibility mechanism.
Materials and Methods
Chemicals, Reagents, and Materials
The reference standards of isorhmnetin (3), diosmetin (6), isochlorogenic acid A (7), kaempferol (10), quercetin (14), isochlorogenic acid C (15), apigenin (19), forsythin (20), chlorogenic acid (23), liquiritin (25), luteolin-7-O-glucoside (26), astragalin (28), luteolin (29), buddleoside (31), quercetin-3-O-glucoside (33), amygdalin (37), caffeic acid (38), forsythoside A (39), glycyrrhizic acid (40), ferulic acid (42), rutin (44), hyperoside (50), and platycodin D (51) were purchased from Sichuan Weikeqi Biological Technology, Co., Ltd. (Chengdu, China). Mulberry leaf, chrysanthemum, Fructus Forsythiae, licorice, Semen Armeniacae Amarum, Platycodi Radix and Phragmitis Rhizoma were purchased from Guangdong Huiqun Chinese Traditional Medicine, Co., Ltd. (Shantou, China), and mint was provided by Jiangxi Jianmin Natural Perfume Factory (Ji’an, China). Their voucher specimens have been deposited at the Department of Pharmaceutical Analysis, Xuzhou Medical University, Xuzhou, China. All other reagents were of analytical grade and obtained commercially.
Sample Preparation
The simulative solution of SGT was prepared according to its record in China Pharmacopoeia (Chinese Pharmacopoeia Commission, 2015), and the detailed experimental process is described in Supporting information. A total of 23 reference standards were weighed accurately and dissolved in methanol to obtain their individual standard solutions at the concentration of 1 mg/mL, except for 26 (0.25 mg/mL). To optimize chromatographic condition of the 2D-LC system, the solutions of compounds 7, 15, 19, 20, 23, 25, 26, 28, 33, 37, 38, 39, 40, 42, 44, and 51 were mixed, dried under a gentle nitrogen flow, and reconstituted in methanol to produce mixed standard solutions. All the solutions were stored at 4 °C until use.
Off-Line Two-Dimensional Liquid Chromatography
The following HILIC columns were studied: Agilent Zorbax HILIC plus (2.1 mm × 150 mm, 3.5 μm, Agilent, Santa Clara, CA, United States) and Waters XBridge Amide (2.1 mm × 150 mm, 3.5 μm, Waters, Milford, MA, United States). For RPLC, Agilent Zorbax SB-C8 (4.6 mm × 150 mm, 5.0 μm), Agilent Zorbax SB-C18 (2.1 mm × 100 mm, 3.5 μm), Agilent Eclipse plus C18 (2.1 mm × 100 mm, 3.5 μm) and Agilent Eclipse plus C18 (4.6 mm × 100 mm, 3.5 μm) columns were studied.
The D1 LC (HILIC) was conducted on a Waters e2695 HPLC system, and the samples were separated on a Waters XBridge Amide column (2.1 mm × 150 mm, 3.5 μm). The mobile phase consisted of 0.1% formic acid (A) and acetonitrile (B), and a gradient elution program was used: 0 min, 90% B; 10 min, 60% B. The flow rate was 0.4 mL/min, and the column temperature was 35°C. The detection wavelength was 254 nm. From 1 to 10 min, a total of 10 fractions (Fraction 1 to Fraction 10) were collected with a one-min interval. The sample injection was repeated for five times, and the same fractions were combined, respectively. The resulting fractions (except for Fraction 1) was concentrated to dryness under a gentle flow of nitrogen gas at 37°C, and were then re-dissolved in 100 μL of 50% methanol for the next D2 LC analysis.
The D2 LC (RPLC) was performed on an Agilent 1290 UHPLC system. The samples were separated on an Agilent Eclipse Plus C18 column (4.6 mm × 100 mm, 3.5 μm), and a binary mobile phase composed of 0.1% formic acid (A) and methanol (B) was used following a gradient elution program: 0 min, 5% B; 30 min, 95% B. The flow rate was 1.0 mL/min, and the column temperature was 35°C. The detection wavelength was 254 nm.
Mass Spectrometry
An Agilent 6550 qTOF mass spectrometer equipped with an electrospray interface (ESI) was coupled to the D2 LC system, and the ion source was operated in the negative ion mode. The D2 LC eluent from the UV detector was introduced into the mass spectrometer by using a T-splitter, and the post-column splitting ratio was 8:1 (approximate 0.11 mL/min into the mass spectrometer). The MS parameters were set as follows: ionization mode, Dual AJS ESI (-); reference ion, 112.98 and 1033.98; capillary voltage, 3500 V; drying temperature, 280°C; dry gas flow, 12 L/min; atomizer pressure, 28 psig; sheath temperature, 350°C; sheath gas flow, 12 L/min; scan range, m/z 50–1500; cataclastic voltage, 300 V; collision energy, 15 and 45 eV.
Data Analysis
The raw DAD data were exported into OriginPro 9.1 software (OriginLab Corporation, United States) to construct the contour plots. Orthogonality (O) and peak capacity (n2D) were calculated based on literatures (Gilar et al., 2005; Rutan et al., 2012), as described in our previous publication (Ji et al., 2017).
Results and Discussion
Optimization of the Off-Line 2D-LC System
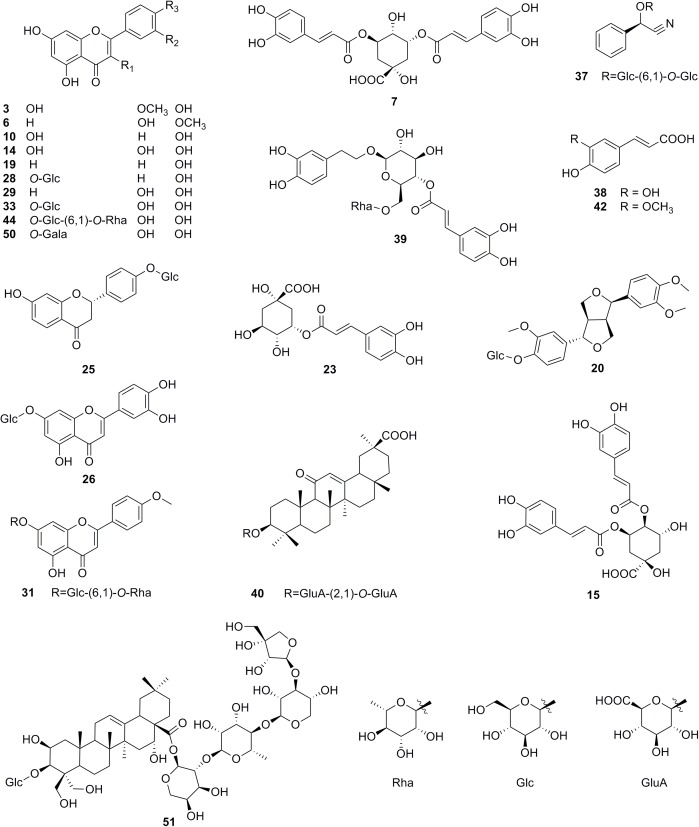
The separation modes of D1 and D2 in a 2D-LC system are determined based on the types of interactions between samples and stationary phases. The separation capacity of RPLC is mainly associated with non-polar selectivity, while HILIC is dominated by polar interaction. Therefore, the two different separation modes were combined to enhance orthogonality and peak capacity in this study, since the chemical constituents of SGT have a wide range of polarities. RPLC has been selected as the second dimension since it usually shows better peak shape of analytes and better compatibility with mass spectrometry, according to literatures (Jandera and Hájek, 2018). Among the 23 reference standards, 16 representative compounds (7, 15, 19, 20, 23, 25, 26, 28, 33, 37, 38, 39, 40, 42, 44, and 51), which are the major constituents of SGT with different structural types, were chosen and mixed to prepare standards solution (Figure 1). They were used to optimize the 2D-LC conditions.
FIGURE 1.
Chemical structures of 23 reference standards from SGT.
Optimization of HILIC
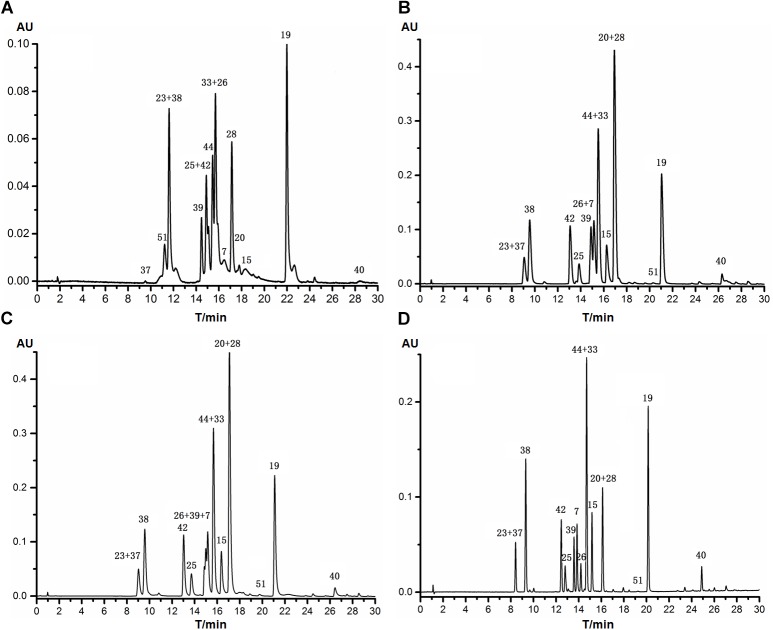
Two types of HILIC columns (Agilent Zorbax HILIC plus and Waters XBridge Amide) were evaluated by separating the above standard solution, with the gradient program described in Section “Off-Line Two-Dimensional Liquid Chromatography.” As shown in Figure 2, all the standards were co-eluted within 2 min with poor separation when they were separated on the Agilent Zorbax HILIC plus column. On the contrary, the Waters XBridge Amide column exhibited stronger retention capacity to the standards, and their identities were confirmed by mass spectrometry (Figure 2). A total of 14 peaks can be observed, and only four compounds were not fully separated. Therefore, we chose Amide as the stationary phase for the first dimension HILIC in this study, and gradient acetonitrile and 0.1% formic acid were chosen as the mobile phases.
FIGURE 2.
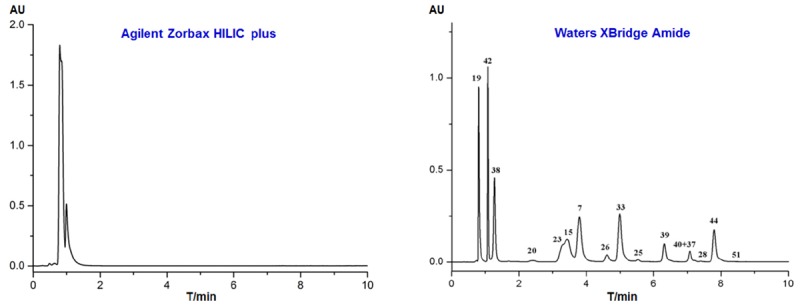
HPLC chromatograms of 16 reference standards separated with different HILIC columns with acetonitrile and 0.1% formic acid as the mobile phase (254 nm).
Optimization of RPLC
Four RPLC columns from Agilent Technologies with different packing materials or inner diameters were tested in respect of selectivity and resolution by using the gradient program described in Section “Off-Line Two-dimensional Liquid Chromatography.” The 16 reference standards were difficult to be fully separated in one single run, as shown in Figure 3. Apparently, the Eclipse plus C18 (4.6 mm, i.d.) column showed the highest resolution and best peak shape, and only six analytes were not fully separated. In particular, compounds 7, 26, and 39 could be well-resolved, though they were almost overlapped when separated on Zorbax SB-C8, Zorbax SB-C18 and Eclipse plus C18 (2.1 mm, i.d.). Therefore, Agilent Eclipse plus C18 (4.6 mm, i.d.) was tentatively chosen as the stationary phase for the second dimension RPLC. The mobile phases were then optimized, and detailed experimental process was described in Supporting information. As a result, 0.1% formic acid was used as the aqueous phase, and methanol was used as the organic phase.
FIGURE 3.
HPLC chromatograms of 16 reference standards separated with different Agilent RPLC columns with methanol and 0.1% formic acid as the mobile phase (254 nm). (A) Zorbax SB-C8 (4.6 mm × 150 mm, 5.0 μm); (B) Zorbax SB-C18 (2.1 mm × 100 mm, 3.5 μm); (C) Eclipse plus C18 (2.1 mm × 100 mm, 3.5 μm); (D) Eclipse plus C18 (4.6 mm × 100 mm, 3.5 μm).
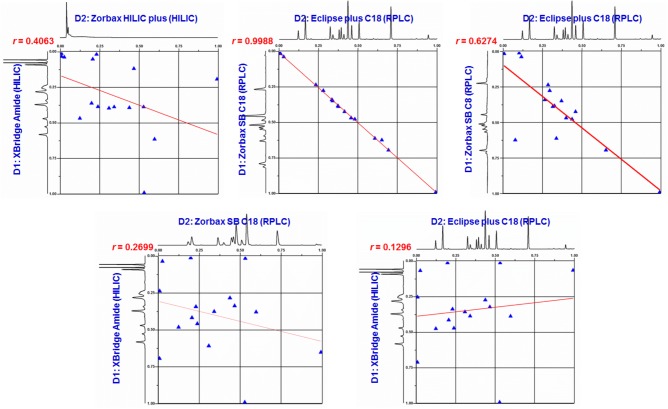
Subsequently, the orthogonality between Waters XBridge Amide and Agilent Eclipse plus C18 (HILIC × RPLC) was evaluated with Pearson linearity regression correlation coefficient (r) as the index. In addition, the orthogonality of several other combination modes, namely XBridge Amide × Zorbax HILIC plus, Zorbax SB C18 × Eclipse plus C18 (4.6 mm, i.d.), Zorbax SB C8 × Eclipse plus C18 (4.6 mm, i.d.) and XBridge Amide × Zorbax SB C18, was also determined. Retention times of each reference standard separated on these columns were firstly transformed into normalized retention times [, calculated according to our previous publication (Ji et al., 2017)], and the r-values of the two groups of RT(i)norm values were then calculated, respectively. As shown in Figure 4, the r-values of Amide × HILIC plus, SB C18 × plus C18 (4.6 mm, i.d.), SB C8 × plus C18 (4.6 mm, i.d.), Amide × SB C18 and Amide × plus C18 (4.6 mm, i.d.) were 0.4063, 0.9988, 0.6274, 0.2699, and 0.1296, respectively, suggesting that the combination modes of both HILIC × HILIC (Amide × HILIC plus) and RPLC × RPLC (SB C18 × plus C18 and SB C8 × plus C18) showed poor orthogonality with r > 0.40, due to the similar separation mechanisms between D1 and D2. As expected, the combination mode of HILIC × RPLC (Amide × SB C18 and Amide × plus C18) exhibited a higher orthogonality with r < 0.30. Then, the orthogonality and practical peak capacity of different combination modes were calculated according to our previous publication (Ji et al., 2017). As a result (Supplementary Table S1), Amide × plus C18 gave a significantly higher practical peak capacity (2303 and 1231) than Amide × SB C18 (1691 and 850), calculated by two different methods, though their orthogonality was the same (0.3969). Based on the above data, the choice of Waters XBridge Amide × Agilent Eclipse plus C18 for the chemical analysis of SGT was further confirmed in this study.
FIGURE 4.
Normalized retention time plots for HILIC × HILIC, RPLC × RPLC, and HILIC × RPLC systems.
Evaluation of the Off-Line 2D-LC System
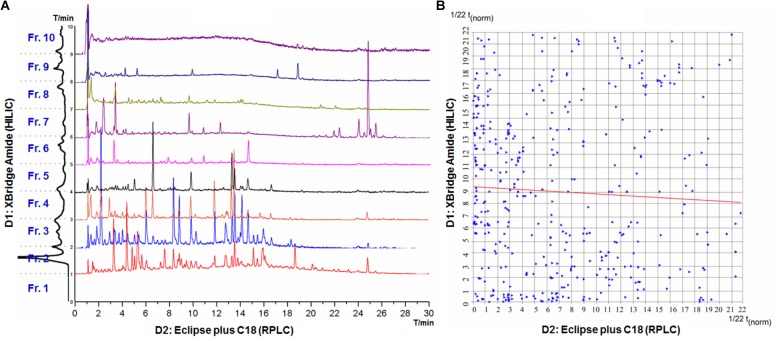
The optimized off-line 2D-LC system was used to analyze SGT, which mainly contains flavonoids, triterpenoid saponins, phenylethanoid glycosides, and organic acids. SGT was firstly separated on the XBridge Amide column in D1, and a total of 10 fractions (Fraction 1 to Fraction 10) were consecutively collected with 0.4 mL (1 min) per fraction (Supplementary Figure S1). The resulting fractions (except for Fraction 1) were further separated on the Zorbax Eclipse plus C18 column in D2, respectively, which was successively coupled with DAD and qTOF-MS as the detectors (Figure 5A). All the peaks in the nine chromatograms (Fraction 2 to Fraction 10) were assigned based on their retention time, UV spectra and MS data, and a total of 465 peaks were finally recognized in SGT. To evaluate the orthogonality and peak capacity of the 2D-LC system, the normalized retention times of all the peaks were calculated, and the separation space was divided into 22 × 22 rectangular bins, which (484) is close to Pmax (465). The rectangular bins were then superimposed with the data points, as shown in Figure 5B, and orthogonality and peak capacity were calculated according to literatures (Gilar et al., 2005; Rutan et al., 2012). Bins containing data plots (Σbins) covered about 50% of the separation space, and the orthogonality was calculated as 63.62%. In addition, the Pearson correlation coefficient (r) of the two groups of normalized retention time values was 0.0525, which suggested a good orthogonality.
FIGURE 5.
HPLC chromatograms and the 2D-LC plot of SGT. (A) HPLC chromatograms of nine fractions in the second dimension RPLC. (B) The normalized retention time plot for the chemical constituents of SGT separated by the 2D-LC system.
The average peak width of D1 and D2 was 0.738 and 0.145 min, respectively, and the effective gradient time for D1 and D2 was 10 and 30 min, respectively. Thus, the theoretical peak capacities for D1 and D2 were 14 and 207, respectively. The theoretical peak capacity of the 2D-LC system was 2898, and the practical peak capacity was 2399 and 1099, respectively, calculated following two methods described in our previous publication (Ji et al., 2017). The 2D-LC system increased the peak capacity by almost 11-fold comparing with the single-dimension D2. Improvement on peak capacity is beneficial to the exposure of more minor components, which could contribute to comprehensive separation and detection of chemical constituents in SGT and its component herbs.
Separation and Characterization of Chemical Constituents in SGT and Its Component Herbs by the Off-Line 2D-LC/qTOF-MS System
Chemical constituents of SGT were globally analyzed by the optimized off-line 2D-LC/qTOF-MS system, and a total of 465 peaks were detected. The eight component herbs were also respectively separated and detected by using the 2D-LC system, and we found that 12 peaks detected in SGT were not discovered in any component herbs. Their retention times and high-resolution mass spectral data were listed in Supplementary Table S2. The 12 compounds might be produced through chemical reactions of chemical constituents in component herbs during decocting together, such as hydrolysis, oxidation, and dissolution. They might cause the pharmacological activity differences between SGT and its eight component herbs, and contribute to exploration of the herbal compatibility mechanism. Further separation and purification are necessary to fully identify the chemical structures of the 12 compounds through nuclear magnetic resonance (NMR) spectroscopic analysis in future study.
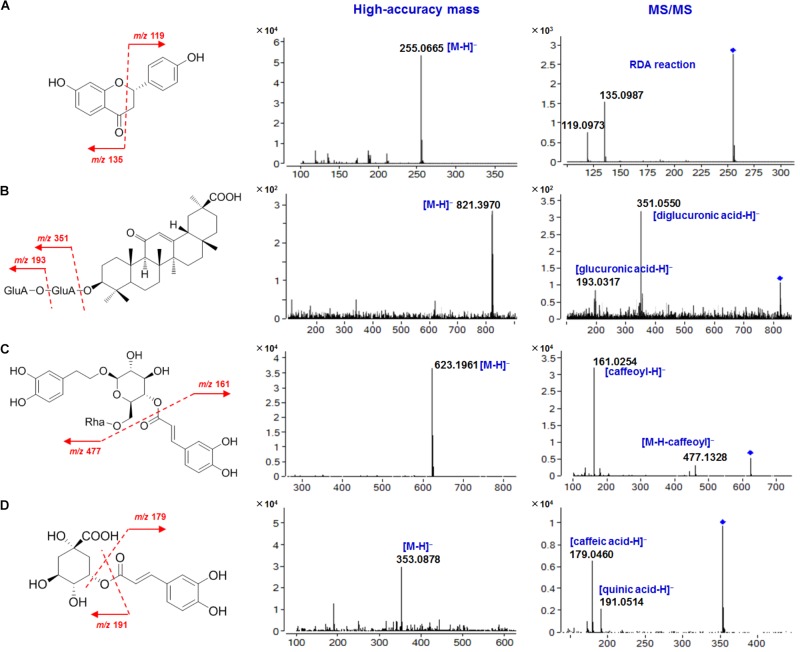
Among the 465 peaks, a selected group of constituents (54 compounds) were identified by comparing with reference standards (23 compounds), or tentatively characterized by comparing their high-resolution mass spectral data with previous literatures (31 compounds). These compounds included free flavonoids, flavonoid glycosides, triterpene saponins, phenylethanoid glycosides, organic acids, and others (Table 1). The qTOF-MS spectra of four representative compounds are illustrated in Figure 6.
Table 1.
Characterization of selected chemical constituents in SGT by qTOF-MS analysis.
| No. | Fr. | tR (min) | Formula | Predicted [M-H]- | Measured [M-H]- | Error (ppm) | Identification | Major fragment ions | Source |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 2.15 | C8H14O4 | 173.0819 | 173.0824 | 2.9 | Rengynic acid | 155.0187, 130.9664 | F |
| 2 | 2 | 7.00 | C20H30O12 | 461.1664 | 461.1665 | 0.2 | Forsythoside E | 315.1216, 108.0212 | F |
| 3∗ | 2 | 8.49 | C16H12O7 | 315.0510 | 315.0529 | 6.0 | Isorhmnetin | 246.9526, 207.0309 | M |
| 4 | 2 | 9.19 | C29H36O16 | 639.1931 | 639.1915 | -2.5 | Forsythoside C | 621.2144, 487.1683 | F |
| 5 | 2 | 11.53 | C15H12O4 | 255.0663 | 255.0665 | 0.8 | Liquiritigenin | 135.0987, 119.0973 | L |
| 6∗ | 2 | 11.99 | C16H12O6 | 299.0561 | 299.0547 | -4.7 | Diosmetin | 231.0859, 151.0047 | C |
| 7∗ | 2 | 12.08 | C25H24O12 | 515.1195 | 515.1182 | -2.5 | Isochlorogenic acid A | 353.1254, 179.1406 | C, M |
| 8 | 2 | 12.22 | C26H30O13 | 549.1614 | 549.1610 | -0.7 | Licuraside | 255.0691, 135.0090 | L |
| 9 | 2 | 12.35 | C21H22O9 | 417.1191 | 417.1179 | -2.9 | Isoliquiritin | 255.0681, 135.0085, | L |
| 10∗ | 2 | 13.75 | C15H10O6 | 285.0405 | 285.0392 | -4.6 | Kaempferol | 238.9991, 185.0123 | C, L |
| 11 | 2 | 14.00 | C21H22O10 | 433.1140 | 433.1124 | -3.7 | 5-Hydroxy-liquiritin | 271.0601, 151.0410, | L |
| 12 | 2 | 14.09 | C23H24O10 | 459.1297 | 459.1288 | -2.0 | Liquiritigenin-4′-O-(6-O-acetyl)-glucoside | 255.0661, 134.9757 | L |
| 13 | 2 | 14.21 | C16H18O9 | 353.0878 | 353.0885 | 2.0 | 1-Caffeoyl-quininic acid | 191.0684, 161.0344 | M |
| 14∗ | 2 | 14.30 | C15H10O7 | 301.0354 | 301.0325 | -9.6 | Quercetin | 178.9282, 151.0496 | F, C, L |
| 15∗ | 2 | 14.64 | C25H24O12 | 515.1195 | 515.1207 | 2.3 | Isochlorogenic acid C | 353.0944, 191.0433 | C, M |
| 16 | 2 | 15.38 | C26H30O13 | 549.1614 | 549.1639 | 4.5 | Isoliquiritin apioside | 417.1165, 255.0654 | L |
| 17 | 2 | 15.60 | C21H22O9 | 417.1191 | 417.1178 | -3.1 | Neoliquiritin | 255.0657, 135.0088 | L |
| 18 | 2 | 15.90 | C21H22O9 | 417.1191 | 417.1199 | 1.9 | Neoisoliquiritin | 255.0648, 135.0083 | L |
| 19∗ | 2 | 16.08 | C15H10O5 | 269.0455 | 269.0443 | -4.5 | Apigenin | 151.0977, 117.1972 | C |
| 20∗ | 2 | 17.09 | C27H34O11 | 533.2028 | 533.2019 | -1.7 | Forsythin | 371.0965 | F |
| 21 | 2 | 20.04 | C44H64O19 | 895.3969 | 895.3952 | -1.9 | 22β-Acetoxyl-licorice-saponin G2 | 832.9889, 351.0551 | L |
| 22 | 3 | 1.91 | C7H12O6 | 191.0561 | 191.0568 | 3.7 | Quinic acid | 173.0419, 110.0973 | S, L, C, F |
| 23∗ | 3 | 8.00 | C16H18O9 | 353.0878 | 353.0878 | 0.0 | Chlorogenic acid | 191.0514, 179.0460 | C, M |
| 24 | 3 | 10.54 | C27H32O13 | 563.1770 | 563.1798 | 5.0 | Isoliquiritoside | 473.0765, 327.0865 | L |
| 25∗ | 3 | 11.46 | C21H22O9 | 417.1191 | 417.1203 | 2.9 | Liquiritin | 255.0701, 134.9676 | L |
| 26∗ | 3 | 13.75 | C21H20O11 | 447.0933 | 447.0974 | 9.2 | Luteolin-7-O-glucoside | 285.1876, 175.1059 | C |
| 27 | 3 | 15.05 | C27H30O13 | 561.1614 | 561.1605 | -1.6 | Glycyroside | 267.0653, 146.9658 | L |
| 28∗ | 3 | 15.43 | C21H20O11 | 447.0933 | 447.0936 | 0.7 | Astragalin | 285.0438, 151.0036 | M, L, F |
| 29∗ | 3 | 18.28 | C15H10O6 | 285.0405 | 285.0401 | -1.4 | Luteolin | 151.0029, 133.0308 | C |
| 30 | 4 | 11.19 | C29H36O15 | 623.1981 | 623.1968 | -2.1 | Forsythoside I | 477.1656, 315.1085 | F |
| 31∗ | 4 | 12.10 | C28H32O14 | 591.1719 | 591.1711 | -1.3 | Buddleoside | 282.9350, 238.9306 | C |
| 32 | 4 | 12.82 | C29H36O15 | 623.1981 | 623.1979 | -0.3 | Forsythoside H | 477.0351, 161.0244 | F |
| 33∗ | 4 | 14.09 | C21H20O12 | 463.0882 | 463.0877 | -1.1 | Quercetin-3-O-glucoside | 300.0363, 151.0102 | C, M |
| 34 | 4 | 20.58 | C42H60O16 | 819.3809 | 819.3757 | -6.3 | Licorice-saponin E2 | 757.3807, 643.3443 | L |
| 35 | 4 | 21.17 | C44H64O18 | 879.4020 | 879.3969 | -5.8 | 22β-Acetoxyl-glycyrrhizic acid | 703.1909, 350.9911 | L |
| 36 | 4 | 21.58 | C42H62O17 | 837.3914 | 837.3924 | 1.2 | Licorice-saponin G2 | 661.3565, 485.2294 | L |
| 37∗ | 5 | 7.74 | C20H27NO11 | 456.1506 | 456.1501 | -1.1 | Amygdalin | 294.0426, 131.1087 | S |
| 38∗ | 5 | 9.08 | C9H8O4 | 179.0350 | 179.0359 | 5.0 | Caffeic acid | 161.1098, 135.0446 | R, F, C, S |
| 39∗ | 5 | 12.07 | C29H36O15 | 623.1981 | 623.1961 | -3.2 | Forsythoside A | 477.1328, 161.0254 | F |
| 40∗ | 5 | 24.16 | C42H62O16 | 821.3965 | 821.3970 | 0.6 | Glycyrrhizic acid | 351.0550, 193.0317 | L |
| 41 | 6 | 3.81 | C14H24O9 | 335.1348 | 335.1326 | -6.6 | Rengynic acid-1′-O-glucoside | 173.0578 | F |
| 42∗ | 6 | 4.22 | C10H10O4 | 193.0506 | 193.0502 | -2.1 | Ferulic acid | 149.0605, 134.0354 | C, R |
| 43 | 6 | 24.16 | C42H62O16 | 821.3965 | 821.3967 | 0.2 | Uralsaponin A | 645.1405, 469.1888 | L |
| 44∗ | 7 | 11.93 | C27H30O16 | 609.1461 | 609.1449 | -2.0 | Rutin | 301.0498, 285.1097 | C, M, F |
| 45 | 7 | 13.61 | C25H24O12 | 515.1195 | 515.1154 | -7.9 | Isochlorogenic acid B | 173.0481, 161.0267 | C |
| 46 | 7 | 20.19 | C59H94O29 | 1265.5808 | 1265.5902 | 7.4 | Platycodin A | 1133.512, 681.4071 | P |
| 47 | 7 | 23.91 | C48H72O21 | 983.4493 | 983.4523 | 3.0 | Licorice-saponin A3 | 645.1236, 351.0817 | L |
| 48 | 7 | 24.15 | C42H62O16 | 821.3965 | 821.3930 | -4.3 | Licorice-saponin H2 | 469.3447, 351.0641 | L |
| 49 | 7 | 24.83 | C42H62O16 | 821.3965 | 821.3973 | 1.0 | Licorice-saponin K2 | 645.2346, 351.0054 | L |
| 50∗ | 8 | 3.66 | C21H20O12 | 463.0882 | 463.0902 | 4.3 | Hyperoside | 300.9523 | F |
| 51∗ | 8 | 19.77 | C57H92O28 | 1223.5702 | 1223.5699 | -0.2 | Platycodin D | 959.4888, 681.3826 | P |
| 52 | 8 | 20.15 | C57H92O27 | 1207.5753 | 1207.5690 | -5.2 | Polygalacin D | 1075.4732, 665.3538 | P |
| 53 | 8 | 20.23 | C58H94O28 | 1237.5859 | 1237.5841 | -1.4 | Platycoside A | 1027.1965, 695.1736 | P |
| 54 | 9 | 16.60 | C69H112O38 | 1547.6759 | 1547.6749 | -0.6 | Platycoside E | 1415.6212, 989.0768 | P |
∗Identified by comparing with reference standards.
Fr., fractions collected from the first dimension HILIC; M, mulberry leaf; C, chrysanthemum; F, Fructus Forsythiae; L, licorice; S, Semen Armeniacae Amarum; P, Platycodi Radix; R, Phragmitis Rhizoma.
FIGURE 6.
High-accuracy mass and MS/MS spectra of four representative compounds in SGT. (A) Liquiritigenin (5); (B) Glycyrrhizic acid (40); (C) Forsythoside A (39); (D) Chlorogenic acid (23).
Characterization of Flavonoids
Flavonoids (free flavonoids and flavonoid glycosides) in SGT are mainly derived from mulberry leaf, chrysanthemum, Fructus Forsythiae and licorice. In the negative ion mode, free flavonoids in SGT could fragment on ring-C following the retro Diels-Alder (RDA) reaction at a relatively high collision energy, and they could also lose small molecules (like CO) or radicals (like CH3⋅) to produce diagnostic fragments (Fabre et al., 2001). For the flavonoid glycosides in SGT, flavonoid O-glycosides could undergo collision-induced dissociation (CID), and produce [M-H-162]- (-glucose), [M-H-146]- (-rhamnose) or [M-H-132]- (-apiose) ions by the neutral loss of sugar moieties. The resulting aglycones further fragment to produce the same fragments as the corresponding free flavonoids (Cuyckens et al., 2001). In contrast, flavonoid C-glycosides could fragment on the sugar ring to lose 60, 90, or 120 Da (0,4X, 0,3X, or 0,2X cleavage, respectively) at a relatively high collision energy (Lin et al., 2005). Based on these fragmentation behaviors, a selected group of flavonoids (7 free flavonoids and 16 flavonoid glycosides), including isoflavones, flavones, flavanones and chalcones, were identified or tentatively characterized in SGT. Here we choose liquiritigenin and its analogs, as well as their glycosides, as examples to describe their structural characterization.
Liquiritigenin (5) is the main free flavanone in licorice. Its [M-H]- ion at m/z 255.0665 fragmented to give m/z 135.0987 and m/z 119.0973 following the RDA reaction (Qiao et al., 2015). Liquiritin (liquiritigenin-4′-O-glucoside, 25) is a liquiritigenin O-glucoside with a [M-H]- ion at m/z 417.1203, and it eliminated a glucosyl residue to produce m/z 255.0701 corresponding to the aglycone. Like liquiritigenin, this fragment further fragmented to give m/z 134.9676 following the RDA reaction. Compound 9 (isoliquiritin) is a chalcone O-glucoside, and it showed the same fragmentation behavior as 25. However, its retention time on the Agilent Eclipse Plus C18 column was shorter than 25 (Qiao et al., 2015). Compound 11 (5-hydroxy-liquiritin) is also a flavanone O-glucoside, and its [M-H]- ion at m/z 433.1124 could easily lose a glucosyl residue to produce m/z 271.0601, which further fragmented to give m/z 151.0410 following the RDA reaction. This indicated that 11 was a hydroxylated derivative of 25 (Montero et al., 2016). Similarly, compounds 8, 12, 16, 17, 18, 24, and 27 were tentatively assigned to licuraside, liquiritigenin-4′-O-(6-O-acetyl)-glucoside, isoliquiritin apioside, neoliquiritin, neoisoliquiritin, isoliquiritoside, and glycyroside, respectively (Zhou et al., 2004; Qiao et al., 2015; Montero et al., 2016). The other selected flavonoids (3, 6, 10, 14, 19, 26, 28, 29, 31, 33, 44, and 50) were identified by comparing with reference standards.
Characterization of Triterpene Saponins
Triterpene saponins in SGT are mainly derived from Platycodi Radix and licorice, and most of them are oleanane-type triterpenes, which contain triterpene oleanolic acid or its analogs as the sapogenin and one or two oligosaccharide chains substituted at 3-OH or 28-OH. In the negative ion mode, they could lose sugar residues or sapogenins to produce corresponding diagnostic fragments. Based on this, the structures of 14 selected triterpene saponins were identified or tentatively characterized in this study.
Glycyrrhizic acid (40), with two connected glucuronic acid units as the saccharide chain, generated an [M-H]- ion at m/z 821.3970, indicating a molecular formula of C42H62O16. The [M-H]- ion fragmented into two characteristic fragments at m/z 351.0550 ([diglucuronic acid-H]-) and m/z 193.0317 ([glucuronic acid-H]-), respectively. Structural analogs of 40, including 22β-acetoxyl-licorice-saponin G2 (21), licorice saponin E2 (34), 22β-acetoxyl-glycyrrhizic acid (35), licorice-saponin G2 (36), uralsaponin A (43), licorice-saponin A3 (47), licorice-saponin H2 (48) and licorice-saponin K2 (49), were tentatively characterized (Zhou et al., 2004; Qiao et al., 2015; Montero et al., 2016). Platycodin D (51) contains two oligosaccharide chains, which are composed of five sugar residues, namely glucose, arabinose, rhamnose, xylose, and apiose. It had the molecular formula of C57H92O28 based on its [M-H]- ion at m/z 1223.5699, and its characteristic fragments were m/z 959.4888 ([M-apiose-xylose-H]-) and m/z 681.3826 ([M-apiose-xylose-rhamnose-arabinose-H]-), respectively. According to the similar fragmentation behaviors, the other selected saponins from Platycodi Radix (46, 52, 53, and 54) were tentatively characterized (Ha et al., 2010; Lee et al., 2015).
Characterization of Phenylethanoid Glycosides
Five selected phenylethanoid glycosides characterized in SGT (2, 4, 30, 32, and 39) were mainly derived from Fructus Forsythiae, and their chemical structures contained one or more aromatic rings, glucoses and caffeic acids. The [M-H]- ion of forsythoside A (39) at m/z 623.1961 suggested a molecular formula of C29H36O15, and the fragments at m/z 477.1328 and m/z 161.0254 were attributed to the loss of a caffeoyl group ([M-H-162]-) and the caffeoyl group ([caffeoyl-H]-), respectively. Similarly, forsythoside E (2), forsythoside C (4), forsythoside I (30), and forsythoside H (32) were tentatively characterized (Zhang et al., 2016).
Characterization of Phenolic Acids
Almost all of the component herbs contain phenolic acids, and a total of 10 selected phenolic acids in SGT were identified or tentatively characterized in this study. In the negative ion mode, phenolic acids could lose a carboxy group with the neutral loss of 44 Da (CO2) to generate diagnostic fragments, and the loss of small molecules (like H2O) or radicals (like CH3⋅) sometimes also occurred. For example, the [M-H]- ion of ferulic acid (42) at m/z 193.0502 produced characteristic fragments at m/z 149.0605 ([M-H-CO2]-) and m/z 134.0354 ([M-H-CH3⋅]-). For chlorogenic acid (23), where caffeic acid forms an ester bond with quinic acid, the ester bond fragmented to produce m/z 191.0514 and m/z 179.0460, representing a quinic acid unit and a caffeic acid unit, respectively. Based on these fragmentation patterns, the other selected phenolic acids (1, 7, 13, 15, 22, 38, 41, and 45) were tentatively characterized (Wang et al., 2014; Yan et al., 2014; Lee et al., 2015; Zhang et al., 2016; Zhu et al., 2017).
Conclusion
In summary, we developed an off-line HILIC × RP 2D-LC system to comprehensively separate the chemical constituents in SGT and its component herbs. The 2D-LC system showed high orthogonality (63.62%) and approximate 11-fold improvement in peak capacity in contrast to conventional one-dimensional RPLC separation. As a result, a total of 465 peaks were detected, and the structures of 54 selected compounds were fully identified or tentatively characterized by qTOF-MS analysis. In addition, 12 peaks detected in SGT were not discovered in any component herbs, and they might contribute partly to exploration of the compatibility mechanism of the component herbs. Integration of off-line HILIC × RP 2D-LC and high-resolution mass spectrometry is proven as a promising tool for chemical profiling and comparison of complicated Chinese patent medicines and their component herbs.
Author Contributions
SJ, M-ZG, and D-QT: participated in research design. SJ, Z-ZL, YD, and Z-YS: performed the experiments and data analysis. JW, T-YW, JH, and D-ZY: contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the scientific research funds of talents from Xuzhou Medical University (D2017016), Natural Science Foundation of Jiangsu Province (BK20170253), Jiangsu University Natural Science Foundation of China (16KJA350001 and 17KJB350013), Innovative and Entrepreneurial Talent Scheme of Jiangsu Province (2017), and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.01167/full#supplementary-material
References
- Chang Q., Wo S., Ngai K. L. K., Wang X., Fok B., Ngan T. M., et al. (2014). Bench to bed evidences for pharmacokinetic and pharmacodynamic interactions involving oseltamivir and Chinese medicine. Evid. Based Compl. Alt. 2014:354172. 10.1155/2014/354172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. X., Hu D. J., Lam S. C., Ge L., Wu D., Zhao J., et al. (2016). Comparison of antioxidant activities of different parts from snow Chrysanthemum (Coreopsis tinctoria Nutt.) and identification of their natural antioxidants using high performance liquid chromatography coupled with diode array detection and mass spectrometry and 2,2′-azinobis(3-ethylbenzthiazoline-sulfonic acid) diammonium salt-based assay. J. Chromatogr. A 1428 134–142. 10.1016/j.chroma.2015.10.037 [DOI] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Commission (2015). Pharmacopoeia of the People’s Republic of China. Beijing: Ministry of Health of the People’s Republic of China. [Google Scholar]
- Cuyckens F., Rozenberg R., de Hoffmann E., Claeys M. (2001). Structure characterization of flavonoid O-diglycosides by positive and negative nano-electrospray ionization ion trap mass spectrometry. J. Mass Spectrom. 36 1203–1210. 10.1002/jms.224 [DOI] [PubMed] [Google Scholar]
- Fabre N., Rustan I., de Hoffmann E., Quetin-Leclercq J. (2001). Determination of flavone, flavonol, and flavanone aglycones by negative ion liquid chromatography electrospray ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 12 707–715. 10.1016/S1044-0305(01)00226-4 [DOI] [PubMed] [Google Scholar]
- Gilar M., Olivova P., Daly A. E., Gebler J. C. (2005). Orthogonality of separation in two-dimensional liquid chromatography. Anal. Chem. 77 6426–6434. 10.1021/ac050923i [DOI] [PubMed] [Google Scholar]
- Guiochon G., Marchetti N., Mriziq K., Shalliker R. A. (2008). Implementations of two-dimensional liquid chromatography. J. Chromatogr. A 1189 109–168. 10.1016/j.chroma.2008.01.086 [DOI] [PubMed] [Google Scholar]
- Guo M., Han J., He D., Zou J., Li Z., Du Y., et al. (2017). Optimization and assessment of three different high performance liquid chromatographic systems for the combinative fingerprint analysis and multi-ingredients quantification of Sangju Ganmao TABLET. J. Chromatogr. Sci. 55 334–345. 10.1093/chromsci/bmw190 [DOI] [PubMed] [Google Scholar]
- Ha Y. W., Na Y. C., Ha I. J., Kim D. H., Kim Y. S. (2010). Liquid chromatography/mass spectrometry-based structural analysis of new platycoside metabolites transformed by human intestinal bacteria. J. Pharm. Biomed. Anal. 51 202–209. 10.1016/j.jpba.2009.08.002 [DOI] [PubMed] [Google Scholar]
- He J., Feng Y., Ouyang H., Yu B., Chang Y., Pan G., et al. (2013). A sensitive LC-MS/MS method for simultaneous determination of six flavonoids in rat plasma: application to a pharmacokinetic study of total flavonoids from mulberry leaves. J. Pharm. Biomed. Anal. 84 189–195. 10.1016/j.jpba.2013.06.019 [DOI] [PubMed] [Google Scholar]
- Huang W. Y., Sheu S. J. (2007). Separation and identification of the fifteen constituents in Forsythia Fructus. J. Food Drug Anal. 15 33–39. [Google Scholar]
- Jandera P., Hájek T. (2018). Mobile phase effects on the retention on polar columns with special attention to the dual hydrophilic interaction-reversed-phase liquid chromatography mechanism, a review. J. Sep. Sci. 41 145–162. 10.1002/jssc.201701010 [DOI] [PubMed] [Google Scholar]
- Ji S., He D., Wang T., Han J., Li Z., Du Y., et al. (2017). Separation and characterization of chemical constituents in Ginkgo biloba extract by off-line hydrophilic interaction × reversed-phase two-dimensional liquid chromatography coupled with quadrupole-time of flight mass spectrometry. J. Pharm. Biomed. Anal. 146 68–78. 10.1016/j.jpba.2017.07.057 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Shin H. K., Seo C. S. (2014). Chemical interaction between Paeonia lactiflora and Glycyrrhiza uralensis, the components of jakyakgamcho-tang, using a validated high-performance liquid chromatography method: herbal combination and chemical interaction in a decoction. J. Sep. Sci. 37 2704–2715. 10.1002/jssc.201400522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Ji S. H., Kim G. S., Song K. S., Um Y., Kim O. T., et al. (2015). Global profiling of various metabolites in Platycodon grandiflorum by UPLC-QTOF/MS. Int. J. Mol. Sci. 16 26786–26796. 10.3390/ijms161125993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Fu Q., Xin H., Ke Y., Jin Y., Liang X. (2014). Alkaloids analysis using off-line two-dimensional supercritical fluid chromatography × ultra-high performance liquid chromatography. Analyst 139 3577–3587. 10.1039/C4AN00438H [DOI] [PubMed] [Google Scholar]
- Li S., Zhang B., Jiang D., Wei Y., Zhang N. (2010). Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinformatics 11(Suppl. 11):S6. 10.1186/1471-2105-11-S11-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. C., Wu C. I., Sheu S. J. (2005). Determination of 12 pueraria components by high performance liquid chromatography-mass spectrometry. J. Sep. Sci. 28 1785–1795. 10.1002/jssc.200500126 [DOI] [PubMed] [Google Scholar]
- Marchetti N., Fairchild J. N., Guiochon G. (2008). Comprehensive off-line, two-dimensional liquid chromatography. application to the separation of peptide digests. Anal. Chem. 80 2756–2767. 10.1021/ac7022662 [DOI] [PubMed] [Google Scholar]
- Montero L., Ibáñez E., Russo M., di Sanzo R., Rastrelli L., Piccinelli A. L., et al. (2016). Metabolite profiling of licorice (Glycyrrhiza glabra) from different locations using comprehensive two-dimensional liquid chromatography coupled to diode array and tandem mass spectrometry detection. Anal. Chim. Acta. 913 145–159. 10.1016/j.aca.2016.01.040 [DOI] [PubMed] [Google Scholar]
- Qiao X., Song W., Ji S., Wang Q., Guo D., Ye M. (2015). Separation and characterization of phenolic compounds and triterpenoid saponins in licorice (Glycyrrhiza uralensis) using mobile phase-dependent reversed-phase × reversed-phase comprehensive two-dimensional liquid chromatography coupled with mass spectrometry. J. Chromatogr. A 1402 36–45. 10.1016/j.chroma.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Rutan S. C., Davis J. M., Carr P. W. (2012). Fractional coverage metrics based on ecological home range for calculation of the effective peak capacity in comprehensive two-dimensional separations. J. Chromatogr. A 1255 267–276. 10.1016/j.chroma.2011.12.061 [DOI] [PubMed] [Google Scholar]
- Shellie R. A., Haddad P. R. (2006). Comprehensive two-dimensional liquid chromatography. Anal. Bioanal. Chem. 386 405–415. 10.1007/s00216-006-0516-0 [DOI] [PubMed] [Google Scholar]
- Song W., Qiao X., Chen K., Wang Y., Ji S., Feng J., et al. (2017). Biosynthesis-based quantitative analysis of 151 secondary metabolites of licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal. Chem. 89 3146–3153. 10.1021/acs.analchem.6b04919 [DOI] [PubMed] [Google Scholar]
- Wang S., Hao L., Zhu J., Zhang Q., Wang Z., Zhang X., et al. (2014). Study on the effects of sulfur fumigation on chemical constituents and antioxidant activity of Chrysanthemum morifolium cv. Hang-ju. Phytomedicine 21 773–779. 10.1016/j.phymed.2013.10.019 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lu X., Xu G. (2008). Simultaneous separation of hydrophilic and hydrophobic compounds by using an online HILIC-RPLC system with two detectors. J. Sep. Sci. 31 1564–1572. 10.1002/jssc.200700663 [DOI] [PubMed] [Google Scholar]
- Yan X., Bai X., Liu Q., Liu S., Gao P., Li L., et al. (2014). Two new glycosides from the fruits of Forsythia suspense. J. Asian Nat. Prod. Res. 16 376–382. 10.1080/10286020.2014.884082 [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu S., Zhao Y., Zhang T., Liu D. (2016). Analysis on chemical constituents from Forsythiae Fructus by HPLC-Q-TOF-MS. Chin. Tradit. Herbal Drugs 47 2053–2060. [Google Scholar]
- Zhou Y., Wang M., Liao X., Zhu X., Peng S., Ding L. (2004). Rapid identification of compounds in Glycyrrhiza uralensis by liquid chromatography/tandem mass spectrometry. Chin. J. Anal. Chem. 32 174–178. 20443596 [Google Scholar]
- Zhu L., Peng Z., Zhang X., Yang J., Lai X., Yang G. (2017). Determination of polyphenols in Lycium barbarum leaves by high-performance liquid chromatography-tandem mass spectrometry. Anal. Lett. 50 761–776. 10.1080/00032719.2016.1202956 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.