Abstract
Hepatic recurrence of gastric cancer (GC) is uncontrollable. Discovery of causative oncogenes and the development of sensitive biomarkers to predict hepatic recurrence are required to improve patients’ outcomes. In this study, recurrence pattern‐specific transcriptome analysis of 57 749 genes was conducted to identify mRNAs specifically associated with hepatic metastasis of patients with stage III GC who underwent curative resection. GC cell lines were subjected to mRNA expression analysis, PCR array analysis, and siRNA‐mediated knockdown. The expression levels of primary cancer tissues from 154 patients with resectable GC were determined and correlated with clinicopathological variables. Among 21 genes significantly overexpressed specifically in patients with hepatic recurrence, Sushi domain containing 2 (SUSD2) was selected as a promising target. PCR array analysis revealed that SUSD2 mRNA levels positively correlated with those of FZD7, CDH2, TGFB1, SPARC, ITGA5, and ZEB1. Functional analysis revealed that knockdown of SUSD2 significantly reduced the proliferation, migration, and invasiveness GC cell lines. Patients with high SUSD2 expression were more likely to experience shorter disease‐free and overall survival. Analysis of the relation between disease recurrence pattern and SUSD2 levels revealed that significantly more patients with hepatic metastases expressed higher levels of SUSD2 mRNA. The cumulative incidence of hepatic recurrence was greater in patients with high SUSD2 expression. In conclusion, SUSD2 likely contributes to the malignant potential of GC and may serve as a novel biomarker that predicts hepatic recurrence after curative resection.
Keywords: expression, gastric cancer, hepatic recurrence, prognosis, SUSD2
1. INTRODUCTION
Despite an overall decline in incidence over the last several decades,1 gastric cancer (GC) is the third and fifth leading cause of cancer‐related deaths in males and females, respectively, worldwide.2 Regardless of improvements in multimodal management strategies, approximately 40%‐80% of patients relapse,3 and despite the precision of curative resection, micrometastases remain outside the stomach and cause recurrence. Particularly, hepatic relapse contributes to the high incidence of GC‐related fatalities and represents a frequent and crucial problem for oncologists.4, 5 To address this serious problem, we require novel, sensitive biomarkers that predict hepatic recurrence and serve as targets to prevent or treat hepatic recurrence. A better understanding of the molecular mechanisms of GC progression is essential for developing such clinical tools. For example, advances in basic molecular oncological research led to the development of trastuzumab (anti‐HER‐2/neu antibody), which benefits many patients suffering from GC.6
To select candidate genes associated with the progression and metastasis of GC, we conducted a recurrence, pattern‐specific transcriptome analysis of 57 749 genes of patients with stage III GC who underwent curative resection. We identified sushi domain containing 2 (SUSD2) as a candidate oncogene that was associated with hepatic recurrence. To assess the significance of SUSD2 in recurrent GC, we used GC cell lines and clinical samples to conduct detailed studies of SUSD2 expression, function, as well as the clinical significance of SUSD2 overexpression.
2. MATERIALS AND METHODS
2.1. Ethics
This study conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki, Ethical Principles for Medical Research Involving Human Subjects and was approved by the Institutional Review Board of Nagoya University, Japan.
2.2. Transcriptome analysis
We conducted a recurrence, pattern‐specific transcriptome analysis of 57 749 genes to identify candidates specific to patients with stage III GC with hematogenous metastasis who underwent curative resection followed by S‐1 adjuvant therapy. For this purpose, we used the HiSeq platform (Illumina, San Diego, CA) to analyze primary GC tissues and their respective corresponding noncancerous adjacent gastric mucosa.7
2.3. Sample collection
The GC cell lines MKN1, MKN7, MKN45, MKN74, NUGC2, NUGC3, NUGC4, IM95, OCUM1, and SC‐6‐JCK cell lines were obtained from the Japanese Collection of Research Bio Resources Cell Bank (JCRB; Osaka, Japan). The AGS, KATOIII, and N87 cell lines were obtained from the American Type Culture Collection (Manassas, VA), and the GCIY cell line was obtained from Tohoku University (Miyagi, Japan). Cells were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM; Sigma‐Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum in an atmosphere containing 5% CO2. Cell lines were analyzed using the short tandem repeat‐polymerase chain reaction (PCR) method and authenticated by the JCRB Cell Bank during June 2015.8
Primary GC tissues and the corresponding noncancerous adjacent tissues were collected from 154 patients who underwent gastric resection for GC at the Department of Gastroenterological Surgery, Nagoya University Hospital between 2001 and 2014. Written informed consent for the use of clinical samples and data, as required by the institutional review board, was obtained from all patients. The tissue samples were immediately frozen in liquid nitrogen and stored at −80°C. Since 2010, specimens have been histologically classified according to the 7th edition of the Union for International Cancer Control (UICC) classification system. Patients recruited before 2010 were reclassified accordingly. Since 2006, adjuvant chemotherapy using S‐1 (a fluorinated pyrimidine)9 has been orally administered to all patients with UICC stages II/III GC, unless contraindicated by a patient's condition.10
2.4. Analysis of SUSD2 mRNA levels
SUSD2 mRNA levels in cell lines and clinical samples were determined using a quantitative real‐time reverse‐transcription PCR (qRT‐PCR) assay. Total RNAs (10 µg per sample) were used to generate cDNAs that were amplified with primers specific for SUSD2 (Table S1) as follows: initial denaturation at 95°C for 10 minutes, 40 cycles at 95°C for 10 seconds, and 60°C for 30 seconds. Samples were tested in triplicate, and samples without template were included in each PCR plate as negative controls. The ABI StepOnePlus Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA) was used for real‐time detection of the emission intensity of SYBR‐Green fluorescence. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) mRNA served as an internal standard, and the expression level of each sample was calculated as the value of SUSD2 mRNA divided by that of GAPDH mRNA.11
2.5. PCR array analysis
To identify genes coordinately expressed with SUSD2 in GC cell lines, we used the Human Epithelial to Mesenchymal Transition (EMT) RT2 Profiler PCR Array (Qiagen, Hilden, Germany). This array includes 84 “key” genes that encode proteins with the functions as follows: transcription factor, extracellular matrix protein as well as proteins involved in the epithelial‐mesenchymal transition (EMT), cell differentiation, morphogenesis, growth, proliferation, migration, cytoskeleton, and signaling pathways.7
2.6. Small interfering RNA (siRNA)‐mediated knockdown of SUSD2
We designed four siRNAs specific for SUSD2 (SUSD2‐siRNA). MKN1 and AGS cells were cultured in 24‐well plates (5 × 104 cells/mL). Cells were transiently transfected the next day with 100 nmol/L siRNAs specific for SUSD2 (SUSD2‐siRNA; Table S1) or a control siRNA (siControl). A NEON electroporation system (Invitrogen, Massachusetts, USA) was used to introduce the siRNAs into cells. MKN1 cells were subjected to a pulse voltage = 1400 V, pulse width 20 ms (two pulses), and AGS cells were subjected to a pulse voltage = 1500 V, pulse width 10 ms (three pulses). Knockdown efficiency was determined using qRT‐PCR 24 hours after transfection and Western blotting analysis 72 hours after transfection. Western blotting analysis using a mouse anti‐SUSD2 polyclonal antibody (ab168162; Abcam, Cambridge, UK) diluted 1:100 were performed as previously described.12 Cells were cultured in RPMI medium without antibody for 72 hours and then used for functional assays.
2.7. Assays of cell proliferation, migration, and invasion
Briefly, cell proliferation was evaluated using the Cell Counting Kit‐8 (Dojindo Molecular Technologies, Inc, Kumamoto, Japan). Cells (5 × 103 cells per well) were incubated, and the optical density of the solution in each well was measured on days 1, 3, 5, 7 after the addition of 10 µL of Cell Counting Kit‐8 solution. The ability of GC cells to invade Matrigel was determined using BioCoat Matrigel invasion chambers (BD Biosciences, Bedford, MA) according to the manufacturer's protocol. Cells (2.5 × 104) in serum‐free DMEM were added to each upper well of the chamber. After 44 hours, cells on the lower surface of the membrane were fixed, stained, and a microscope (200 × magnification) was used to count the cells in eight randomly selected fields. Cell migration was evaluated using wound‐healing assays as previously described. The width of the wound was measured at 100‐mm intervals measurements per well, 40 × magnification.
2.8. Evaluation of the clinical significance of SUSD2 expression
SUSD2 mRNA levels were determined in 154 matched pairs of resected gastric tissues from patients with stage I, II, or III GC to determine the risk of recurrence after curative resection. Patients were stratified into high or low expression groups (greater or lower than the median SUSD2 value of GC tissues, respectively). For external validation of the survival data, we accessed a public‐domain integrated dataset comprising 1065 patients with GC from three major cancer research centers (Berlin, Bethesda, and Melbourne; https://kmplot.com/analysis/).
2.9. Statistical analysis
Differences in the values of qualitative variables were compared between groups using the chi‐square test, and quantitative variables were compared using the Mann‐Whitney test. The significance of the difference between two variables was assessed using Spearman's rank correlation coefficient. Overall and disease‐free survival rates were calculated using the Kaplan‐Meier method, and the difference between survival curves was analyzed using the log‐rank test. Risk factors for recurrence and survival were assessed using the Cox hazards ratio model. P < 0.05 was considered statistically significant. Statistical analyses were performed using JMP 13 software (SAS Institute Inc, Cary, NC, USA).
3. RESULTS
3.1. Identification of candidate markers
We identified 21 candidate markers that were specifically expressed at significantly higher levels in hepatic metastatic GC tissues (Table 1). We chose to pursue a study of SUSD2 for the reasons as follows: (a) association with gastric cancer progression (SUSD2 is a membrane protein that may mediate interactions between cells and between cells and cell‐matrix adhesion molecules.), (b) no report of an association of SUSD2 expression with GC, (c) results of a pilot study of GC cell lines (Shinichi Umeda (US), Mitsuro Kanda (MK), Takashi Miwa (TM), Haruyoshi Tanaka (HT), Chie Tanaka (CT), Daisuke Kobayashi (DK), Masaya Suenaga (MS), Norifumi Hattori (NH), Masamichi Hayashi (MH), Suguru Yamada (SY), Goro Nakayama (GN), Michitaka Fujiwara (MF), Yasuhiro Kodera (YK)).
Table 1.
List of candidate genes upregulated in gastric cancer tissues from patients with hepatic recurrence not with peritoneal and lymph node recurrence
| Symbol | H‐rec/Non‐rec | Full name | Location | Function | P‐rec/Non‐rec | N‐rec/Non‐rec | |||
|---|---|---|---|---|---|---|---|---|---|
| Log2 | P value | Log2 | P value | Log2 | P value | ||||
| SUSD2 | 2.976 | <0.001 | Sushi domain containing 2 | 22q11.23 | Cytokine receptor | 0.464 | 0.4 | 0.302 | 0.583 |
| GAL | 4.278 | <0.001 | Galanin and GMAP prepropeptide | 11q13.2 | Endocrine hormone of nervous systems | 2.076 | 0.11 | 1.34 | 0.192 |
| COMP | 4.187 | <0.001 | Cartilage oligomeric matrix protein | 19p13.11 | Extracellular matrix protein | 0.76 | 0.452 | 1.173 | 0.075 |
| IGSF1 | 3.546 | <0.001 | Immunoglobulin superfamily member 1 | Xq26.2 | Immunoglobulin | −0.899 | 1 | 0.601 | 1 |
| BCAM | 2.123 | <0.001 | Basal cell adhesion molecule | 19q13.32 | Laminin receptor | −0.554 | 0.269 | 0.042 | 0.933 |
| ASGR2 | 3.56 | <0.001 | Asialoglycoprotein 2 | 17p13.1 | Mediator of endocytosis of glycoproteins | −0.124 | 1 | 0.452 | 1 |
| RNF182 | 5.362 | <0.001 | Ring finger protein 182 | 6p23 | Mediator of MHC‐I antigen | −0.124 | 1 | 2.317 | 1 |
| CYP2W1 | 6.809 | <0.001 | Cytochrome P450 family 2 subfamily W member 1 | 7p22.3 | Metabolic enzyme | 1.45 | 0.138 | 1.667 | 0.12 |
| FABP3 | 3.774 | <0.001 | Fatty acid binding protein 3 | 1p35.2 | Metabolic enzyme | 0.051 | 0.953 | 0.992 | 0.229 |
| TKTL1 | 6.109 | <0.001 | Transketolase like 1 | Xq28 | Metabolic enzyme | −2.08 | 1 | −2.758 | 1 |
| GPC3 | 2.99 | <0.001 | Glypican 3 | Xq26.2 | Multifunction membrane protein | −0.997 | 0.151 | 0.465 | 0.491 |
| TCF7L1 | 2.288 | <0.001 | Transcription factor 7 like 1 | 2p11.2 | Regulator of cell cycle | −0.26 | 0.651 | −0.386 | 0.497 |
| HIF3A | 4.168 | <0.001 | Hypoxia‐inducible factor 3 alpha subunit | 19q13.32 | Regulator of hypoxia‐inducible genes | 0.29 | 0.717 | −0.018 | 0.979 |
| MYO18B | 4.731 | <0.001 | Myosin XVIIIB | 22q12.1 | Regulator of muscle structure | 4.325 | 0.094 | −0.66 | 1 |
| TNNT1 | 3.316 | <0.001 | Troponin T1, slow skeletal type | 19q13.42 | Regulator of muscle structure | 1.675 | 0.067 | −0.637 | 0.355 |
| RBP4 | 3.549 | <0.001 | Retinol binding protein 4 | 10q23.33 | Retinol carrier | −1.186 | 0.196 | 0.834 | 0.253 |
| PRSS1 | 4.203 | <0.001 | Protease, serine 1 | 7q34 | Serine protease | 0.122 | 0.906 | 0.952 | 0.361 |
| GATA5 | 2.944 | <0.001 | GATA binding protein 5 | 20q13.33 | Transcriptional factor | −1.401 | 0.096 | −0.398 | 0.554 |
| HIC2 | 3.434 | <0.001 | HIC ZBTB transcriptional repressor 2 | 22q11.21 | Transcriptional factor | 0.523 | 0.352 | 0.843 | 0.135 |
| HMGA2 | 3.291 | <0.001 | High mobility group AT‐hook 2 | 12q14.3 | Transcriptional factor | 0.421 | 0.535 | 0.612 | 0.343 |
| SMTNL2 | 4.739 | <0.001 | Smoothelin like 2 | 17p13.2 | Unknown | −0.879 | 0.338 | 1.108 | 0.227 |
H‐rec, hepatic recurrence; Non‐rec, no recurrence; P‐rec. Peritoneal recurrence; L‐rec, lymph node recurrence.
3.2. Expression of SUSD2 in GC cell lines
SUSD2 mRNA levels varied among GC cell lines. Differentiated GC cell lines expressed higher levels of SUSD2 mRNA compared with those of undifferentiated GC cell lines. Cell lines established from metastatic sites in the liver, such as MKN1 and MKN45 cells, expressed the highest levels of SUSD2 mRNA (Figure 1A).
Figure 1.
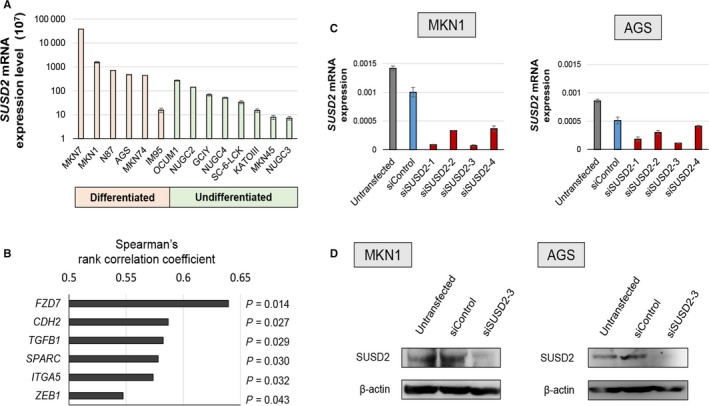
Analysis of SUSD2 mRNA levels of gastric cancer (GC) cell lines, cancer‐related genes expressed cooperatively with SUSD2, and knockdown efficacy. A, SUSD2 mRNA levels in GC cell lines. B, SUSD2 mRNA and mRNAs of genes expressed at similar differential levels were identified using PCR array analysis. Spearman's rank correlation coefficient between the mRNA expression levels of SUSD2 and those of FZD7, CDH2, TGFB1, SPARC, ITGA5, and ZEB1 are shown. C, siRNA‐mediated SUSD2‐knockdown efficacy in MKN1 and AGS cell lines was determined using qRT‐PCR analysis. D, SUSD2‐knockdown efficacy was also determined using Western blotting analysis
3.3. PCR array analysis
There is little evidence that SUSD2 contributes to the mechanism that regulates hematogenous metastasis associated with the epithelial‐mesenchymal transition (EMT). Therefore, we conducted a PCR array analysis to identify cancer‐related genes expressed coordinately with SUSD2 with the aim of acquiring evidence to implicate SUSD2 in cancer progression. We found that mRNAs encoding FZD7, CDH2, TGFB1, SPARC, ITGA5, and ZEB1 were expressed at levels corresponding to those of SUSD2 (Figure 1B).
3.4. Effect of SUSD2 knockdown on the malignant phenotype of GC cells
We selected MKN1 and AGS cells for subsequent analyses, because they expressed the second and fourth highest levels of SUSD2 mRNA SUSD2 mRNA expression levels in MKN7 and N87 cells were high; however, these cells were not analyzed, because they were unsuitable for the invasion assay based on our pilot experiments (unpublished data). qRT‐PCR analysis revealed that siSUSD2‐3 yielded the highest level of inhibition (Figure 1C). The knockdown efficacy of siSUSD2‐3 was confirmed by Western blotting analysis (Figure 1D).
We next determined the effects of siSUSD2‐3 on cell proliferation, invasion, and migration. Inhibition of SUSD2 expression significantly decreased the proliferation of MKN1 cells (40% and 37% decreases on days 3 and 5, respectively) and slightly decreased the proliferation of AGS cells (Figure 2A). Further, siSUSD2 inhibited the invasion of Matrigel by MKN1 and AGS cells by 44% 46%, respectively, compared with untransfected cells (Figure 2B). siSUSD2 inhibited the migration of MKN1 and AGS cell by 10% and 57%, respectively, compared with untransfected cells 12 hours after transfection (Figure 3).
Figure 2.
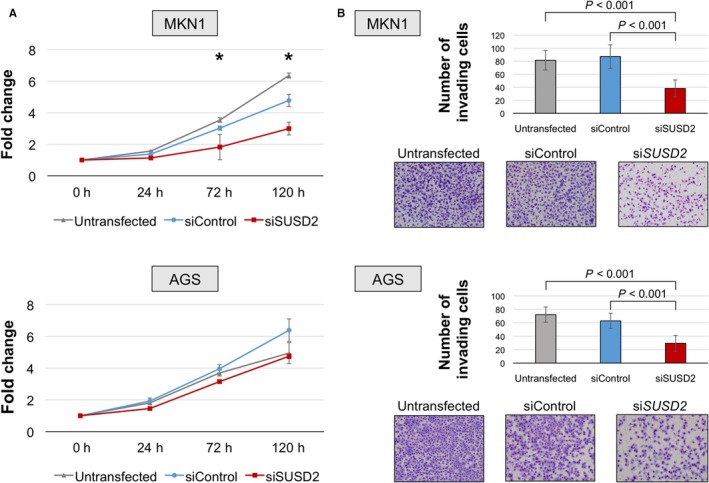
Cell proliferation and invasion assays. A, Cell proliferation assay. Inhibition of SUSD2 expression significantly decreased the proliferation of MKN1and AGS cells. *P < 0.05. B, Cell invasion assays. The number of invading cells was significantly lower in cells transfected with the SUSD2‐siRNA
Figure 3.
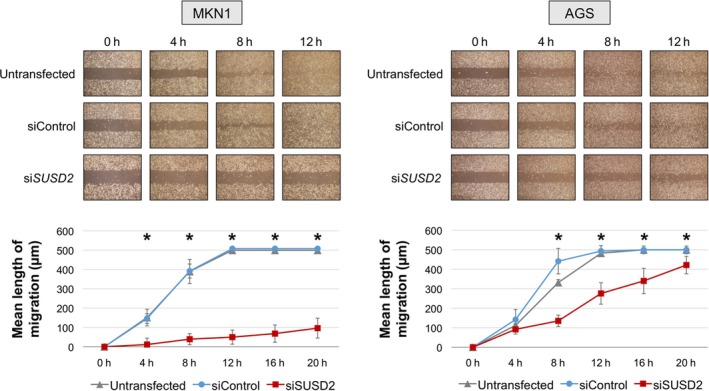
Cell migration assay. The migration of MKN1 and AGS cells transfected with the SUSD2‐sRNA was significantly decreased vs control cells. *P < 0.05
3.5. Clinical significance of SUSD2 expression
The patient population included 114 males and 40 females aged 26‐96 years (65.7 ± 10.6 years, mean ± SD), and 83 and 71 patients were diagnosed with undifferentiated or differentiated GC, respectively. According to the 7th edition of the UICC classification, 46, 40, and 68 patients were in stages I, II, and III, respectively. High SUSD2 expression significantly associated with differentiation but not with tumor depth, invasive growth, lymph node metastasis, lymphatic involvement, vessel invasion, or tumor stage (Table S2). Patients in the high group were more likely to experience shorter disease‐free survival and overall survival (Figure 4A). Similar results were acquired using the external‐validation cohort (Figure 4B). Multivariable analysis identified high SUSD2 mRNA levels as an independent prognostic factor for recurrence of patients with resected GC (hazard ratio, 2.89; 95% confidence interval, 1.52‐5.82; P = 0.001; Table 2). Further, SUSD2 mRNA levels were an independent prognostic factor of overall survival (hazard ratio, 3.13; 95% confidence interval, 1.53‐6.82; P = 0.002; Table S3).
Figure 4.
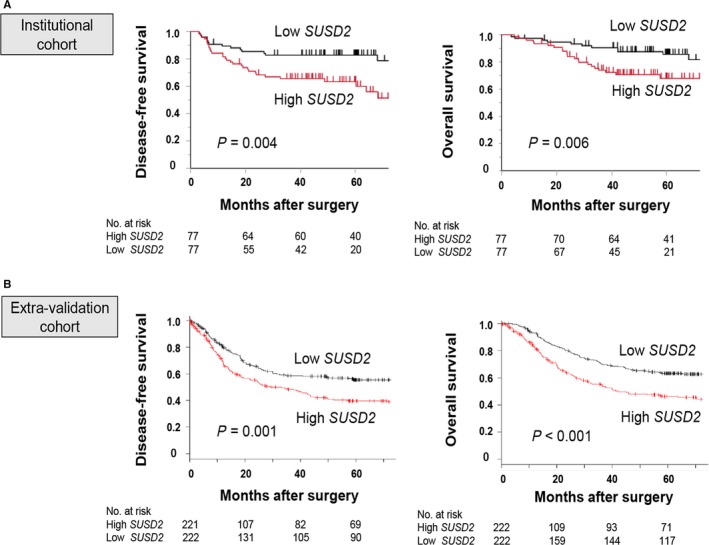
Prognostic implications of SUSD2 mRNA expression in patients with GC after curative resection. A, Kaplan‐Meyer analysis of disease‐free survival and overall survival. B, Kaplan‐Meyer analysis of disease‐free survival and overall survival of patients in the external‐validation cohort
Table 2.
Prognostic factors for disease‐free survival of 154 patients
| Univariate | Multivariable | |||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | |
| Age (≥65) | 1.13 | 0.62‐2.12 | 0.679 | |||
| Gender (male) | 0.84 | 0.45‐1.66 | 0.606 | |||
| Tumor location (Lower) | 0.84 | 0.44‐1.55 | 0.584 | |||
| Tumor multiplicity | 0.47 | 0.08‐1.54 | 0.247 | |||
| Tumor size (≥60 mm) | 2.45 | 1.35‐4.44 | 0.004 | 1.87 | 1.02‐3.44 | 0.044* |
| Carcinoembryonic antigen (>5 ng/mL) | 1.67 | 0.75‐3.33 | 0.194 | |||
| Carbohydrate antigen 19‐9 (>37 IU/mL) | 2.73 | 1.34‐5.17 | 0.007 | 1.75 | 0.84‐3.46 | 0.132 |
| Tumor depth (pT4) | 2.98 | 1.64‐5.45 | <0.001 | 1.92 | 1.03‐3.64 | 0.041* |
| Lymph node metastasis | 14.6 | 5.28‐60.3 | <0.001 | 7.26 | 2.42‐32.3 | <0.001* |
| Tumor differentiation (undifferentiated) | 1.51 | 0.83‐2.86 | 0.179 | |||
| Lymphatic involvement | 2.73 | 1.35‐5.17 | 0.007 | 1.72 | 0.27‐33.6 | 0.607 |
| Vascular invasion | 4.32 | 2.17‐9.58 | <0.001 | 2.27 | 1.08‐5.45 | 0.029* |
| Postoperative adjuvant chemotherapy | 1.65 | 0.91‐2.99 | 0.10 | |||
| High SUSD2 expression | 2.47 | 1.33‐4.81 | 0.004 | 2.89 | 1.52‐5.82 | 0.001* |
Statistically significant in multivariable analysis. CI, confidence interval; UICC, Union for International Cancer Control.
Of all 154 patients, 44 patients (28.6%) experienced recurrence and there were a total of 50 initial relapse sites. Analysis of the association of initial recurrence patterns and SUSD2 mRNA levels revealed that significantly more patients were included in the high group (P = 0.037; Figure 5A). The cumulative occurrence of hepatic recurrence was greater in the high group, in contrast to patients with peritoneal recurrence (Figure 5B).
Figure 5.
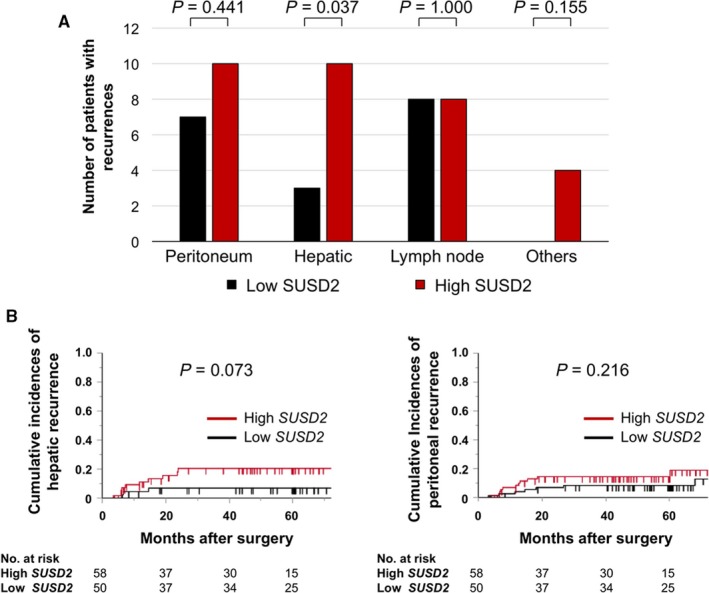
Analysis of recurrence patterns. A, Numbers of sites of initial recurrence in the high and low expression groups. B, Cumulative incidence of hepatic and peritoneal recurrence
4. DISCUSSION
Here we used transcriptome analysis to identify SUSD2 mRNA levels as a candidate marker of hepatic recurrence and survival of patients with GC who underwent curative resection. Human SUSD2 was identified in a cDNA library enriched for genes that encode membrane and secreted proteins that are highly expressed in cancer cells, but at low levels in normal tissues.13 SUSD2 resides on chromosome 22q11.23, comprises fifteen exons, and encodes a type I membrane protein (822 amino acid residues). The predicted SUSD2 amino acid sequence comprises a somatomedin B and adhesion‐associated domains present in MUC4 and other proteins (AMOP) as well as von Willebrand factor type D and sushi domains. The latter play significant roles in mediating intercellular and cell‐to‐matrix adhesion.14 As predicted by its transmembrane domain, SUSD2 localizes to the plasma membrane in vivo.15
Evidence indicates that SUSD2 contributes to oncogenesis. For example, SUSD2 increases the invasiveness of breast cancer cells and may act as a component of the mechanism of immune evasion through induction of apoptosis of the Jurkat T cell line.14 SUSD2 recruits macrophages into the tumor microenvironment, and promotes M2 polarization, indicating that inhibiting the function of SUSD2 may serve as an effective therapy for patients with breast cancer.16
In contrast, SUSD2 may act as a tumor suppressor. For example, SUSD2 exhibits tumor suppressor activity in high‐grade serous ovarian carcinomas17 and may function as a tumor suppressor in renal cell carcinoma and lung cancer.18 Low levels of SUSD2 expression correlate with the aggressive behavior of non‐small cell lung cancer cells.19 SUSD2 is expressed in endometrial carcinoma cells, and suppressing its expression following treatment with TGFβ or a specific siRNA increases apoptosis and senescence. The present study presents the first evidence, to our knowledge, that SUSD2 acts as an oncogene in GC.20
We show here that inhibiting SUSD2 mRNA expression using a specific siRNA inhibits cell proliferation, invasion, and migration, indicating that SUSD2 is required to establish the malignant phenotype of GC cells. To address this issue in more detail, we performed PCR array analysis and found that FZD7, CDH2, TGFB1, SPARC, ITGA5, and ZWB1 mRNAs were overexpressed in concert with SUSD2. These findings implicate the contribution of the WNT signaling pathway to the malignant phenotype of GC. Specifically, the evolutionarily conserved WNT signaling pathway controls intercellular interactions during embryogenesis, and dysfunction of this pathway is implicated in a spectrum of human diseases, particularly solid and hematologic malignancies.21 Thus, WNT signaling may serve as a target of cancer therapy.22 Fzd7 is the WNT receptor most commonly upregulated in diverse cancers and plays a significant role in stem cell biology and cancer development and progression. Small molecules that act as classic GPCR modulators targeting Fzd7 to regulate WNT/β‐catenin signaling may therefore represent potential cancer therapeutics.23
The EMT involves the conversion of epithelial cells to migratory and invasive cells, and the activation of the EMT is closely associated with the motility and invasiveness of GC cells.24, 25 FZD7, CDH2, TGFB1, SPARC, ITGA5, and ZWB1 are associated with the EMT and malignant cell function, indicating that SUSD2 may play a role in the WNT pathway and the EMT.23, 26, 27, 28, 29, 30 Therefore, SUSD2 may serve as a therapeutic target for inhibiting inappropriate WNT signaling or induction of the EMT.
Although the levels of SUSD2 mRNA were not significantly associated with clinicopathological variables that influence the malignant phenotype of GC (tumor size, lymph nodes metastasis, and UICC stage), SUSD2 mRNA levels were closely associated with recurrence and survival after curative surgery. These results are consistent with those of our analysis of a validation cohort. Therefore, SUSD2 mRNA level may reflect the malignant potential of GC independent of clinicopathological markers. Moreover, multivariable analysis revealed that SUSD2 mRNA levels served as an independent risk factor of recurrence and death after complete resection of GC, indicating the utility of SUSD2 expression as a novel predictor of prognosis.
To translate our findings into clinical practice, SUSD2 mRNA levels, determined using biopsy or surgical specimens, might be useful to predict patients who are at high risk of relapse. Stronger adjuvant chemotherapy such as cisplatin or taxane, or neoadjuvant chemotherapy, may prevent recurrence.31, 32 Frequent follow‐up may facilitate an earlier diagnosis of recurrence, allowing immediate administration of chemotherapy and curative resection of the site of recurrence, which will likely improve prognosis.
The present study shows that SUSD2 expression predicted recurrence and survival, as well as the recurrence pattern after curative resection of GC. It is important to note that high levels of SUSD2 expression are closely associated with hematogenous metastasis vs lymphatic and peritoneal recurrence. Therefore, we recommend that patients with high SUSD2 mRNA levels, which indicate hematogenous recurrence, undergo intensive preoperative and postoperative surveillance, such as with Gd‐EOB‐DTPA, enhanced magnetic resonance imaging of the liver,33 bone scintigraphy, and positron emission tomography for early detection.34 It is interesting to note an increase in the incidence of western‐type GC, which develops on the esophagogastric junction or upper stomach in the absence of Helicobacter pylori infection, and tends to recur via the hematogenous route.31, 35, 36 The prediction and control of hepatic recurrence are more important for these patients, lending weight to the potentially important clinical implications of our findings.
There are some limitations to this study. First, PCR array analyses identified mRNAs encoding proteins associated with the EMT or the WNT signaling pathway, although there is no evidence that SUSD2 contributes to the activities of these pathways. Therefore, pathway analyses should be conducted to further understand the biological functions of SUSD2 in GC. Second, this was a retrospective study of a small number patients treated at a single center. External validation using large cohorts from multiple institutions as well as bioinformatics analysis of large datasets are required to validate our present findings. In summary, our results indicate that SUSD2 expression reflects the malignant potential of GC and will serve as novel biomarker that predicts recurrence and prognosis after curative resection of GC SUSD2 may serve as a target of therapy and therefore will facilitate the development of effective therapeutic strategies.
CONFLICTS OF INTEREST
None declared.
Supporting information
Umeda S, Kanda M, Miwa T, et al. Expression of sushi domain containing two reflects the malignant potential of gastric cancer. Cancer Med. 2018;7:5194–5204. 10.1002/cam4.1793
REFERENCES
- 1. Carcas LP. Gastric cancer review. J Carcinog. 2014;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 3. Gallo A, Cha C. Updates on esophageal and gastric cancers. World J Gastroenterol. 2006;12:3237‐3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kanda M, Tanaka H, Shimizu D, et al. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018. in press. 10.1038/s41388-018-0335-8 [DOI] [PubMed] [Google Scholar]
- 5. Kinoshita T, Kinoshita T, Saiura A, Esaki M, Sakamoto H, Yamanaka T. Multicentre analysis of long‐term outcome after surgical resection for gastric cancer liver metastases. Br J Surg. 2015;102:102‐107. [DOI] [PubMed] [Google Scholar]
- 6. Gong SJ, Jin CJ, Rha SY, Chung HC. Growth inhibitory effects of trastuzumab and chemotherapeutic drugs in gastric cancer cell lines. Cancer Lett. 2004;214:215‐224. [DOI] [PubMed] [Google Scholar]
- 7. Tanaka H, Kanda M, Shimizu D, et al. FAM46C serves as a predictor of hepatic recurrence in patients with resectable gastric cancer. Ann Surg Oncol. 2017;24:3438‐3445. [DOI] [PubMed] [Google Scholar]
- 8. Miwa T, Kanda M, Tanaka H, et al. FBXO50 enhances the malignant behavior of gastric cancer cells. Ann Surg Oncol. 2017;24:3771‐3779. [DOI] [PubMed] [Google Scholar]
- 9. Sasako M, Sakuramoto S, Katai H, et al. Five‐year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S‐1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29:4387‐4393. [DOI] [PubMed] [Google Scholar]
- 10. Kanda M, Kobayashi D, Tanaka C, et al. Adverse prognostic impact of perioperative allogeneic transfusion on patients with stage II/III gastric cancer. Gastric Cancer. 2016;19:255‐263. [DOI] [PubMed] [Google Scholar]
- 11. Kanda M, Shimizu D, Tanaka H, et al. Metastatic pathway‐specific transcriptome analysis identifies MFSD4 as a putative tumor suppressor and biomarker for hepatic metastasis in patients with gastric cancer. Oncotarget. 2016;7:13667‐13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanda M, Shimizu D, Tanaka H, et al. Significance of SYT8 For the Detection, Prediction, and Treatment of Peritoneal Metastasis From Gastric Cancer. Ann Surg. 2018;267:495‐503. [DOI] [PubMed] [Google Scholar]
- 13. Egland KA, Vincent JJ, Strausberg R, Lee B, Pastan I. Discovery of the breast cancer gene BASE using a molecular approach to enrich for genes encoding membrane and secreted proteins. Proc Natl Acad Sci U S A. 2003;100:1099‐1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watson AP, Evans RL, Egland KA. Multiple functions of sushi domain containing 2 (SUSD2) in breast tumorigenesis. Mol Cancer Res. 2013;11:74‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sugahara T, Yamashita Y, Shinomi M, et al. Isolation of a novel mouse gene, mSVS‐1/SUSD2, reversing tumorigenic phenotypes of cancer cells in vitro. Cancer Sci. 2007;98:900‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hultgren EM, Patrick ME, Evans RL, Stoos CT, Egland KA. SUSD2 promotes tumor‐associated macrophage recruitment by increasing levels of MCP‐1 in breast cancer. PLoS One. 2017;12:e0177089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sheets JN, Iwanicki M, Liu JF, et al. SUSD2 expression in high‐grade serous ovarian cancer correlates with increased patient survival and defective mesothelial clearance. Oncogenesis. 2016;5:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng Y, Wang X, Wang P, et al. SUSD2 is frequently downregulated and functions as a tumor suppressor in RCC and lung cancer. Tumour Biol. 2016;37:9919‐9930. [DOI] [PubMed] [Google Scholar]
- 19. Cai C, Shi R, Gao Y, et al. Reduced expression of sushi domain containing 2 is associated with progression of non‐small cell lung cancer. Oncol Lett. 2015;10:3619‐3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang S, Zeng N, Alowayed N, et al. Downregulation of endometrial mesenchymal marker SUSD2 causes cell senescence and cell death in endometrial carcinoma cells. PLoS One. 2017;12:e0183681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwab R, Amin N, Flanagan DJ, Johanson TM, Phesse TJ, Vincan E. Wnt is necessary for mesenchymal to epithelial transition in colorectal cancer cells. Dev Dyn. 2017;247:521‐530. [DOI] [PubMed] [Google Scholar]
- 22. Tabatabai R, Linhares Y, Bolos D, Mita M, Mita A. Targeting the Wnt pathway in cancer: a review of novel therapeutics. Target Oncol. 2017;12:623‐641. [DOI] [PubMed] [Google Scholar]
- 23. King TD, Zhang W, Suto MJ, Li Y. Frizzled7 as an emerging target for cancer therapy. Cell Signal. 2012;24:846‐851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319‐8326. [DOI] [PubMed] [Google Scholar]
- 25. Song Y, Li ZX, Liu X, Wang R, Li LW, Zhang Q. The Wnt/beta‐catenin and PI3K/Akt signaling pathways promote EMT in gastric cancer by epigenetic regulation via H3 lysine 27 acetylation. Tumour Biol. 2017;39:1010428317712617. [DOI] [PubMed] [Google Scholar]
- 26. Ma T, Zhao Y, Wei K, et al. MicroRNA‐124 functions as a tumor suppressor by regulating CDH2 and epithelial‐mesenchymal transition in non‐small cell lung cancer. Cell Physiol Biochem. 2016;38:1563‐1574. [DOI] [PubMed] [Google Scholar]
- 27. Zong W, Yu C, Wang P, Dong L. Overexpression of SASH1 Inhibits TGF‐beta1‐Induced EMT in Gastric Cancer Cells. Oncol Res. 2016;24:17‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fenouille N, Tichet M, Dufies M, et al. The epithelial‐mesenchymal transition (EMT) regulatory factor SLUG (SNAI2) is a downstream target of SPARC and AKT in promoting melanoma cell invasion. PLoS One. 2012;7:e40378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morandi EM, Verstappen R, Zwierzina ME, Geley S, Pierer G, Ploner C. ITGAV and ITGA5 diversely regulate proliferation and adipogenic differentiation of human adipose derived stem cells. Sci Rep. 2016;6:28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT‐activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518‐529. [DOI] [PubMed] [Google Scholar]
- 31. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. The Lancet. 2016;388:2654‐2664. [DOI] [PubMed] [Google Scholar]
- 32. Kodera Y. Neoadjuvant chemotherapy for gastric adenocarcinoma in Japan. Surg Today. 2017;47:899‐907. [DOI] [PubMed] [Google Scholar]
- 33. Fukumoto W, Nakamura Y, Higaki T, Tatsugami F, Iida M, Awai K. Additional value of diffusion‐weighted MRI to Gd‐EOB‐DTPA‐enhanced hepatic MRI for the detection of liver metastasis: the difference depending on the experience of the radiologists. Hiroshima J Med Sci. 2015;64:15‐21. [PubMed] [Google Scholar]
- 34. Polat E, Bostanci EB, Aksoy E, et al. The impact of PET/CT on the management of hepatic and extra hepatic metastases from gastrointestinal cancers. Eur J Radiol. 2015;84:1165‐1170. [DOI] [PubMed] [Google Scholar]
- 35. Kodera Y, Fujitani K, Fukushima N, et al. Surgical resection of hepatic metastasis from gastric cancer: a review and new recommendation in the Japanese gastric cancer treatment guidelines. Gastric Cancer. 2014;17:206‐212. [DOI] [PubMed] [Google Scholar]
- 36. Hwang EC, Hwang I, Jung SI, et al. Prognostic factors for recurrence‐free and overall survival after adrenalectomy for metastatic carcinoma: a retrospective cohort pilot study. BMC Urol. 2014;14:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


