Abstract
Production of lipid-derived inositol phosphates including IP4 and IP5 is an evolutionarily conserved process essential for cellular adaptive responses that is dependent on both phospholipase C and the inositol phosphate multikinase Ipk2 (also known as Arg82 and IPMK). Studies of Ipk2, along with Arg82 prior to demonstrating its IP kinase activity, have provided an important link between control of gene expression and IP metabolism as both kinase dependent and independent functions are required for proper transcriptional complex function that enables cellular adaptation in response to extracellular queues such as nutrient availability. Here we define a promoter sequence cis-element, 5’-CCCTAAAAGG-3’, that mediates both kinase-dependent and independent functions of Ipk2. Using a synthetic biological strategy, we show that proper gene expression in cells lacking Ipk2 may be restored through add-back of two components: IP4/IP5 production and overproduction of the MADS box DNA binding protein, Mcm1. Our results are consistent with a mechanism by which Ipk2 harbors a dual functionality that stabilizes transcription factor levels and enzymatically produces a small molecule code, which together coordinate control of biological processes and gene expression.
1. Introduction
Inositol phosphates (IPs) are conserved eukaryotic signaling molecules implicated in the regulation of many biological processes, including the export of mRNA from the nucleus, embryonic development, signaling, and responses to environmental stress (Wilson et al., 2013; Lee et al., 2012; Folkmann et al., 2011; Chakraborty et al., 2011; Hatch and York, 2010; Michell, 2008; Bhandari et al., 2007; Irvine and Schell, 2001; Majerus, 1992). In the yeast Saccharomyces cerevisiae, the generation of IPs begins with a single phospholipase C enzyme (Plc1) that hydrolyzes the lipid phosphatidylinositol 4,5-bisphosphate (PIP2) to release the soluble head group inositol 1,4,5-trisphosphate (IP3) from diacylglycerol (DAG) (Flick and Thorner, 1993; York et al., 1999). Inositol phosphate multikinase, Ipk2 (also known as IPMK and Arg82), converts IP3 to inositol 1,4,5,6-tetrakisphosphate (IP4) and inositol 1,3,4,5,6-pentakisphosphate (IP5) by sequential phosphorylation at the 6- and 3-positions, respectively (York et al., 1999; Odom et al., 2000; Saiardi et al., 1999). IP5 is converted to inositol 1,2,3,4,5,6-hexakisphosphate (IP6), the most abundant species in wild-type S. cerevisiae, by the activity of Ipk1 (York et al., 1999; Ives et al., 2000). The abridged yeast lipid-dependent inositol phosphate pathway (Fig. 1A) is evolutionarily conserved (Hatch and York, 2010); however in some eukaryotes Ipk2 exhibits distinct “multikinase” activities that lead to more than one metabolic pathway leading to formation of IP6 (Hatch and York, 2010; Otto et al., 2007; Verbsky et al., 2005; Stevenson-Paulik et al., 2005; Seeds et al., 2004).
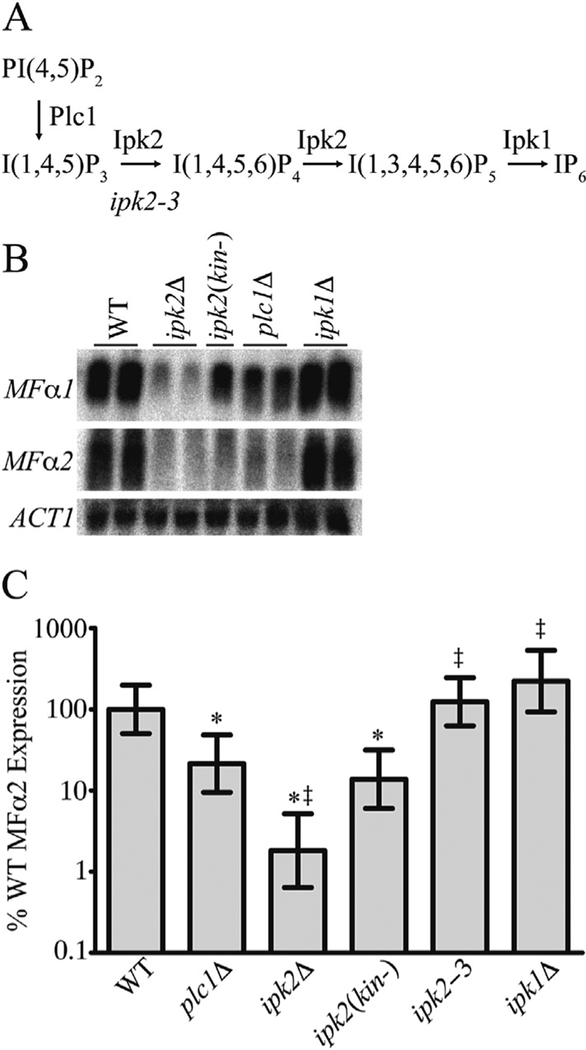
Fig. 1. Ipk2 and IPs are required for efficient MFα2 expression.
A) Diagram outlining the IP production pathway in S. cerevisiae. Diphosphate species and phosphatases have been omitted for simplicity. B) Northern blot analysis of MFα1 and MFα2 expression in WT and IP pathway mutant strains. ACT1 is shown as a loading control. C) Real-time PCR quantification of MFα2 expression levels in WT and IP pathway mutant strains. Values are the mean ± SD of at least three independent cultures, * indicates significant difference from WT expression (p < 0.05), and z indicates significant difference from WT expression (p < 0.05), and ‡ indicates significant difference from plc1Δ (p < 0.05).
The discovery that Ipk2 encodes an inositol phosphate kinase isogenic to the transcriptional regulator Arg82 (ArgRIII complementation group) provided an unanticipated link between inositol phosphate signaling and the control of gene expression (Odom et al., 2000). Prior to defining its enzymatic function, studies of Arg82 defined its role as a transcriptional regulator in response to nutrient conditions of arginine as the sole nitrogen source (El Bakkoury et al., 2000; Dubois and Messenguy, 1994; Messenguy and Dubois, 1993; Messenguy et al., 1991; Qiu et al., 1990; Dubois et al., 1987; Bercy et al., 1987; Delforge et al., 1975; Bechet et al., 1970). Arg82 is a member of the ArgR-Mcm1 transcription complex that regulates the transcription of genes in the arginine metabolic pathway. The ArgR-Mcm1 complex consists of four members, Mcm1, Arg80, Arg81 and Ipk2 (Messenguy and Dubois, 1993). Mcm1 is a founding member of the family of MADS (Mcm1, Agamous, Deficients, Serum Response Factor) box transcription factors, and it is essential for cell viability (Messenguy and Dubois, 2003; Treisman and Ammerer, 1992). Arg80 is also a MADS box transcription factor, but its function appears to be limited to the regulation of arginine metabolic genes (Messenguy and Dubois, 2003). Together, Mcm1 and Arg80 are able to bind elements in the promoters of arginine-responsive genes, but when excess arginine is present in the medium, Arg81, the arginine sensor, and Arg82 also assemble on DNA to form the complete complex (Yoon et al., 2004). In addition, studies of metazoan Ipk2/IPMK protein suggest an evolutionarily conserved function as a partner with a variety of nuclear proteins to regulate nutrient responses and gene expression (Kim et al., 2016; Xu and Snyder, 2013; Xu et al., 2013a,b; Kim et al., 2013; Wu et al., 2011; Kim and Snyder, 2011; Kim et al., 2011).
In the past decade studies have documented Ipk2’s role in regulating gene expression occurs through both kinase-dependent and independent effects. Kinase-dependence and production of IP products has been shown to be required for a number of transcriptional responses to nutrient changes, including amino acids, phosphate and nitrogen (Odom et al., 2000; Guzinska et al., 2009; Steger et al., 2003; El Alami et al., 2003). Additionally, conditional-phenotypes observed in ipk2 null yeast, including temperature sensitivity and growth on arginine as the sole nitrogen source, are complemented by heterologous expression of either plant, fly or metazoan Ipk2 in a kinase-dependent fashion, consistent with an essential role for its enzymatic function and therefore the IP products (Endo-Streeter et al., 2012; Seeds et al., 2005; Xia et al., 2003; Stevenson-Paulik et al., 2002). Kinase-independent functions of Ipk2 have also been reported for assembly of the ArgR-Mcm1 complex onto promoter elements (Odom et al., 2000), stabilization of Mcm1 (El Alami et al., 2003); and the expression of certain transcripts in response to nutrient changes (El Alami et al., 2003; Bosch and Saiardi, 2012; Dubois et al., 2000).
To gain further mechanistic insights into the role of Ipk2 and its inositol phosphate products in transcriptional control, we define a cis-acting promoter element required for mediating both kinase-dependent and kinase-independent regulation of gene expression. This element confers two activities of Ipk2, one as a protein scaffold and the other as a source for enzymatic production of a small-molecule inositol phosphate code, both of which are necessary to provide unique features required for proper promoter function.
2. Results
2.1. Transcription of MFα2 requires Ipk2
Ipk2 has been reported to play a role in the transcription of several Mcm1-dependent genes, including mating type-specific genes (Dubois and Messenguy, 1994; Guzinska et al., 2009). However, the mechanisms by which Ipk2 and its IP products control these processes remain unclear. To gain further insights, we set out to identify specific genes requiring Ipk2 IP4/IP5 kinase activity using whole-genome microarray analysis on a variety of mutant strains including plc1 null (plc1Δ), ipk2Δ, and ipk1Δ as well as ipk2(kin−) (kinase dead). Transcripts whose expression fluctuated in a manner dependent on IP4/IP5 production were defined as similarly altered in plc1Δ, ipk2Δ and ipk2(kin−), but not ipk1Δ mutant strains as compared to wild-type. From this analysis we identified MFα2, encoding one of the mating pheromones secreted by MATα haploids, requires Ipk2. Array analysis results were first validated using northern blot analysis (Fig. 1B) and indeed expression of MFα2 requires the lipid-derived production of IP4/IP5 but not IP6. It is important to note that plc1Δ strains harbor a fully functional Ipk2 protein and therefore possess the kinase-independent functions of Ipk2, yet are unable to convert the lipid PIP2 into the IP3 substrate required for conversion to IP4/IP5 (York et al., 1999), thus represent a critical control alongside the ipk2Δ and ipk2(kin−) mutants. Since no alterations of MFα2 transcript levels were observed in ipk1Δ cells we conclude that IP6 production is not required; however, we cannot exclude an alternative hypothesis that the metabolic build-up of IP4/IP5 reported in these cells is somehow able to compensate for the loss of IP6 (York et al., 1999). Additionally, we performed northern blot analysis on the other mating pheromone expressed in MATα haploids, MFα1, and consistent with other reports we found this to be altered although the dependence on IP4/IP5 production appeared to be less robust (Fig. 1B) (Dubois and Messenguy, 1994; Guzinska et al., 2009).
Using a third methodology that allows for precise quantification, we performed quantitative real-time PCR on MFα2 transcript levels in the various strains (Fig. 1C). Loss of soluble production of IPs, either by deletion of Plc1 (plc1Δ) or mutation in the catalytic site of Ipk2, ipk2kin- (kinase-dead), resulted in approximately a 5-fold reduction in expression. Strikingly, in cells completely lacking Ipk2 (ipk2Δ) this effect was further enhanced such that the transcription of MFα2 was reduced to approximately 1% of the wild-type. These data highlight the dual nature of the Ipk2-mediated effects on MFα2 transcription, both kinase-dependent and independent.
Deletion of Ipk1, the kinase downstream of Ipk2 in the IP metabolic pathway that is responsible for generating IP6, had no effect on MFα2 transcription (Fig. 1C). Therefore, although depletion of all IPs by deletion of Plc1 impaired MFα2 expression, eliminating IP6 production had no effect on MFα2 expression. These data indicate that an IP species upstream of IP6 is important for this process.
The ipk2–3 strain carries a hypomorphic allele of IPK2 that at 30 °C is able to catalyze the addition of a phosphate group to the 6-position of IP3 to generate IP4, but is impaired in its ability to catalyze the addition of a phosphate to the 3-position of IP4 to generate IP5 (York et al., 1999; Odom et al., 2000). MFα2 expression is unaffected in this strain (Fig. 1C). This indicates that either the ability to produce at least IP4 is important for MFα2 expression or that the impaired IP4-kinase activity of the ipk2–3 mutant is sufficient to support MFα2 transcription.
These data highlight the dual nature of the Ipk2 dependence of MFα2 transcription. Ipk2 has a kinase-dependent role in the transcription of MFα2, as shown by the impaired expression levels in the plc1Δ and ipk2(kin-) strains. Ipk2 must also play a kinase-independent function in this process because the ipk2Δ strain has a more profound impairment of MFα2 transcription than the plc1Δ or ipk2(kin-) strains. This indicates that the yeast Ipk2 protein alone can provide some kinase-independent function that is not compromised in the plc1Δ or the ipk2(kin-) strains.
There are conflicting data regarding the kinase-dependence of the transcription of MFα1 (Guzinska et al., 2009; El Alami et al., 2003). The data presented here indicate that MFα1 expression is reduced in the absence of IPs or Ipk2 kinase activity (Fig. 1B). However, MFα2 expression is more severely affected in the absence of IPs so MFα2 transcription was further characterized as a means of investigating the roles of IPs in transcriptional activation.
2.2. The kinase-dependent and kinase-independent functions of Ipk2 can be separated
The role of Ipk2 in the ArgR-Mcm1 complex has been proposed to be primarily independent of its IP kinase activities (El Alami et al., 2003). This assertion appears to be supported by the observation that plc1Δ and ipk2(kin-) strains grow on media containing ornithine as the nitrogen source (Fig. 2A). Previous reports have implicated the poly-D loop, a specific acidic region of the yeast Ipk2 protein, as being important for mediating the kinase-independent function of Ipk2 in the ArgR-Mcm1 complex (El Alami et al., 2003). Expression of Ipk2 from the plant Arabidopsis thaliana (AtIpk2) rescued the activity of the ArgR-Mcm1 complex in ipk2Δ cells, as assayed by the ability to grow on media containing ornithine (Fig. 2A). AtIpk2 lacks the poly-D loop found in the yeast protein (Stevenson-Paulik et al., 2002), and its ability to rescue the ornithine growth phenotype of ipk2Δ cells was dependent on IP kinase activity (Fig. 2A). While Ipk2 has a kinase-independent role in the ArgRMcm1 transcription complex, its kinase activity alone is sufficient to maintain the function of the complex.
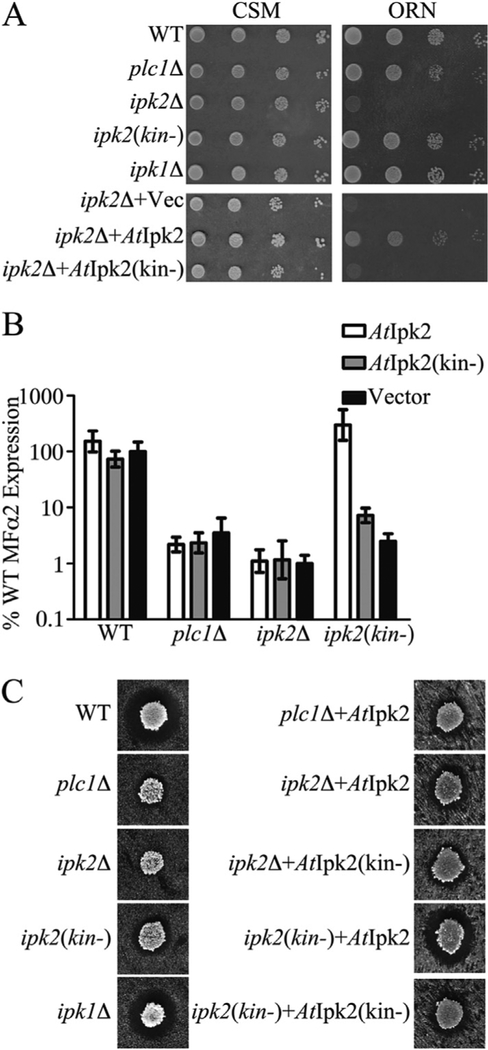
Fig. 2. Ipk2-dependent processes have different IP requirements.
A) Ten-fold serial dilutions of the indicated strains grown on either CSM (left panels) or Orn media (right panels). B) Real-time PCR quantification of MFα2 expression levels in the indicated strains complemented with AtIpk2, the kinase-dead mutant (AtIpk2(kin-)) or the vector control (Vector). Values are the mean ± SD of at least 3 independent cultures. C) Halo assay of pheromone production in WT and IP pathway mutants. Representative images are shown.
To test whether Ipk2 homologues are generally able to rescue the transcription phenotypes associated with deletion of IPK2, we assayed MFα2 transcription levels in yeast transformed with various Ipk2 constructs. Expression of AtIpk2 had no effect on the WT or plc1Δ strains and failed to rescue the expression of MFα2 in the ipk2Δ strain (Fig. 2B) indicating that Ipk2-kinase activity alone is not sufficient to support MFα2 expression. These data differ from those previously presented (Fig. 1B) because the strains were grown in minimal media to maintain selection for the plasmids carrying the indicated AtIpk2 constructs. This difference in media magnifies the differences in MFα2 expression observed between the WT and the plc1Δ and ipk2(kin-) strains. Expression of AtIpk2 rescued MFα2 expression in the ipk2(kin-) strain in a kinase-dependent fashion (Fig. 2B) confirming that MFα2 expression requires both a kinase-independent function provided by the yeast Ipk2 protein and its IP-kinase activity.
The amount of pheromone produced in IP pathway mutants was also examined using a halo assay. When exposed to pheromones of the opposite mating type, haploid yeast arrest their cell-cycle in preparation for mating. MATα haploids were plated on a lawn of MATa cells unable to recover from pheromone exposure. The amount of clearing around each MATα spot is a rough measure of the total pheromone produced by each strain. Deletion of IPK2 resulted in a complete inability to form a cleared halo on the MATa lawn (Fig. 2C). The plc1Δ and ipk2(kin-) strains form much smaller halos than the WT strain and deletion of IPK1 has no effect on halo production. Adding back Ipk2 kinase activity by expressing AtIpk2 constructs only rescues the ability to form a cleared halo in the ipk2(kin-) background (Fig. 2C). This assay further confirms that both the yeast Ipk2 protein and its kinase activity are required for efficient pheromone production (Fig. 2C). These data are in close agreement with the real-time PCR quantification of MFα2 expression (Fig. 2B), but because this assay is a measure of both MFα1 and MFα2 production it is a less specific measure of IP-dependent transcription than real-time PCR.
These data confirm that MFα2 expression requires both the yeast Ipk2 protein and its IP kinase activity and shows that the kinase-dependent and kinase-independent functions of Ipk2 can be provided on separate proteins. In the absence of Ipk2, its kinase activity alone is not sufficient to support MFα2 transcription. This indicates that the kinase-dependent function of Ipk2 requires the kinase-independent function. This data also presents another distinction between MFα2 transcription and the activity of the ArgR-Mcm1 complex in which kinase activity alone is sufficient to coordinate transcriptional activity.
2.3. IP4 and/or IP5 production is required for MFα2 transcription
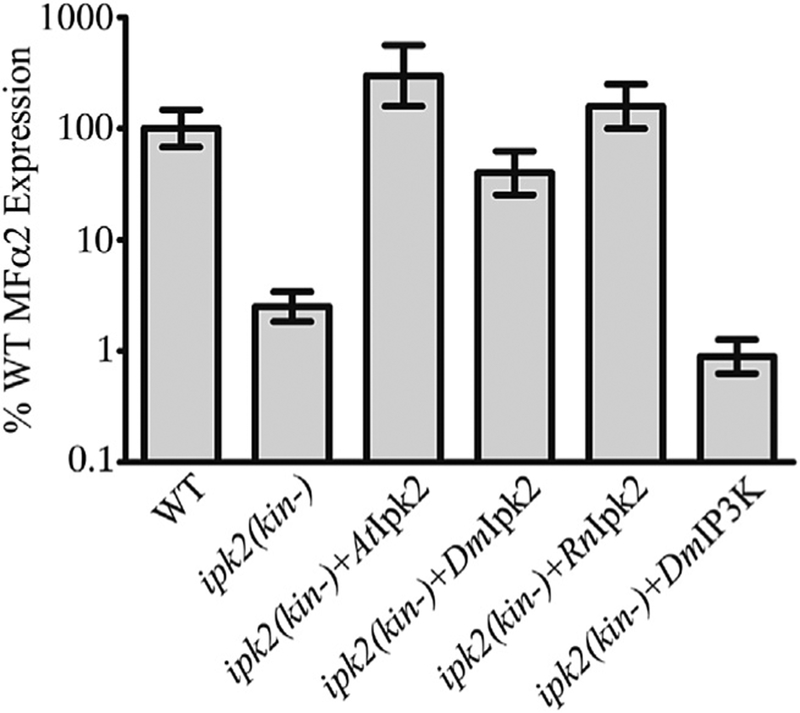
The kinase-dependent function of Ipk2 was probed by testing the abilities of additional IP kinase constructs to rescue MFα2 transcription in the ipk2(kin-) strain. The ipk2(kin-) strain provides a background in which the kinase-independent function of Ipk2 is fulfilled. This allows for the direct examination of the IP species that provide the kinase-dependent function of Ipk2 in MFα2 expression. The Ipk2 homolog from plants has the same 6-/3-kinase activities as the yeast enzyme (Stevenson-Paulik et al., 2002) and expression of AtIpk2 in the ipk2(kin-) background provides robust rescue of MFα2 (Figs. 2B and3). The Ipk2 homolog from the fruit fly Drosophila melanogaster (DmIpk2) is either a 6-/3-kinase or a 3-/6-kinase in vitro depending on the conditions (Seeds et al., 2004) and it is also able to rescue MFα2 transcription in the ipk2(kin-) strain (Fig. 3). The Ipk2 homolog from the rat Rattus norvegicus (RnIpk2) is a 3-/6-kinase (Fujii and York, 2005) and is also capable of rescuing MFα2 transcription in the ipk2(kin-) strain (Fig. 3). This indicates that there may be multiple IP isomers capable of supporting MFα2 transcription because the rescuing constructs catalyze the production of I(1,4,5,6)P4, I(1,3,4,5)P4 and I(1,3,4,5,6)P5. I(1,3,4,5)P4 is unlikely to be capable of supporting MFα2 transcription for several reasons. This species can be produced in vivo by an alternative Kcs1-dependent pathway in cells lacking Ipk2 kinase activity (Seeds et al., 2005), but the ipk2(kin-) strain is deficient in MFα2 expression. Also, expressing an IP3 3-kinase from Drosophila melanogaster (DmIP3K) in the ipk2(kin-) strain does not rescue its ability to express MFα2 (Fig. 3). While I(1,3,4,5)P4 production does not provide the kinase-dependent function of Ipk2, we cannot formally exclude I(1,4,5,6)P4 or I(1,3,4,5,6)P5 as the IP species that provides the kinase-dependent function of Ipk2 in MFα2 transcription in vivo.
Fig. 3. MFα2 expression requires a specific IP isomer.
Real-time PCR quantification of MFα2 expression in WT and ipk2(kin-) strains carrying the indicated constructs. Values are the mean ± SD of at least three independent cultures.
2.4. MFα2 transcription requires a unique regulatory sequence
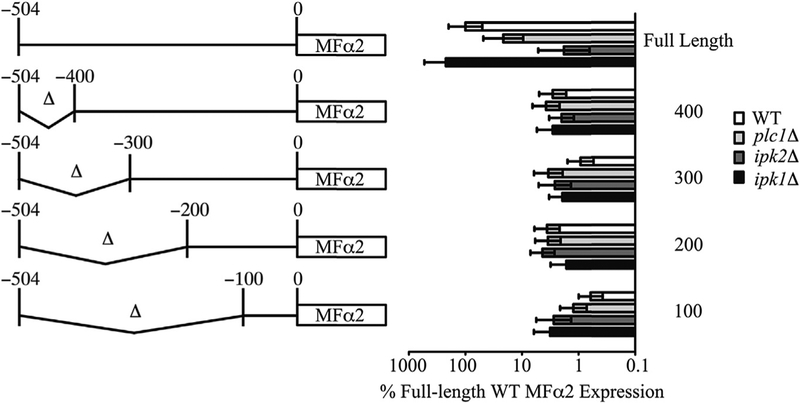
To characterize the sequence elements within the MFα2 promoter that mediate Ipk2 dependent expression, we devised a truncation strategy. Beginning approximately 500 bp upstream of the translation start site, fragments of 100 bp were deleted from the genomic DNA (Fig. 4). This strategy generated strains in which the promoter at the genomic locus was modified, and the effects of mutations could be assayed in the context of the normal chromosome environment. By assaying MFα2 mRNA levels in these strains, it was possible to determine the contributions of elements within the promoter to the transcription of MFα2. Assays of MFα2 expression levels in strains carrying truncated promoters showed that deleting the region between approximately 400 bp and 500 bp upstream of the MFα2 translation start site reduced expression to a level similar to deletion of IPK2 (Fig. 4). Further truncations of the promoter did not further impair expression.
Fig. 4. Truncation of the MFα2 promoter identifies an essential regulatory region.
At left is a schematic showing promoter truncation mutants and the corresponding naming convention. The coordinates are indicated relative to the start codon of MFα2. The graph on the right shows real-time PCR quantification of MFα2 expression levels in the promoter truncation mutants. Values are the mean ± SD of at least 3 independent cultures.
2.5. A putative Mcm1-binding site is required for transcriptional activation
Expression of mating type-specific genes is a process requiring Mcm1 and mating type-specific transcription factors. The sequences sufficient to confer a mating type-specific transcription consist of a bipartite binding sequence called the PQ box (Fig. 4A) (Jarvis et al., 1988). Mcm1 binds to these sites, which are generally understood to be the sequence elements that regulate α mating type-specific transcription, in conjunction with mating type-specific factors. Deletion of the PQ sequence, which spans the region between 292 bp and 317 bp upstream of the translation start site (Jarvis et al., 1988), from the MFα2 promoter did not reduce MFα2 expression levels to the same extent as deletion of IPK2 (Fig. 4B). This result indicates that there may be a distinct regulatory sequence in the MFα2 promoter that mediates its dependence on Ipk2.
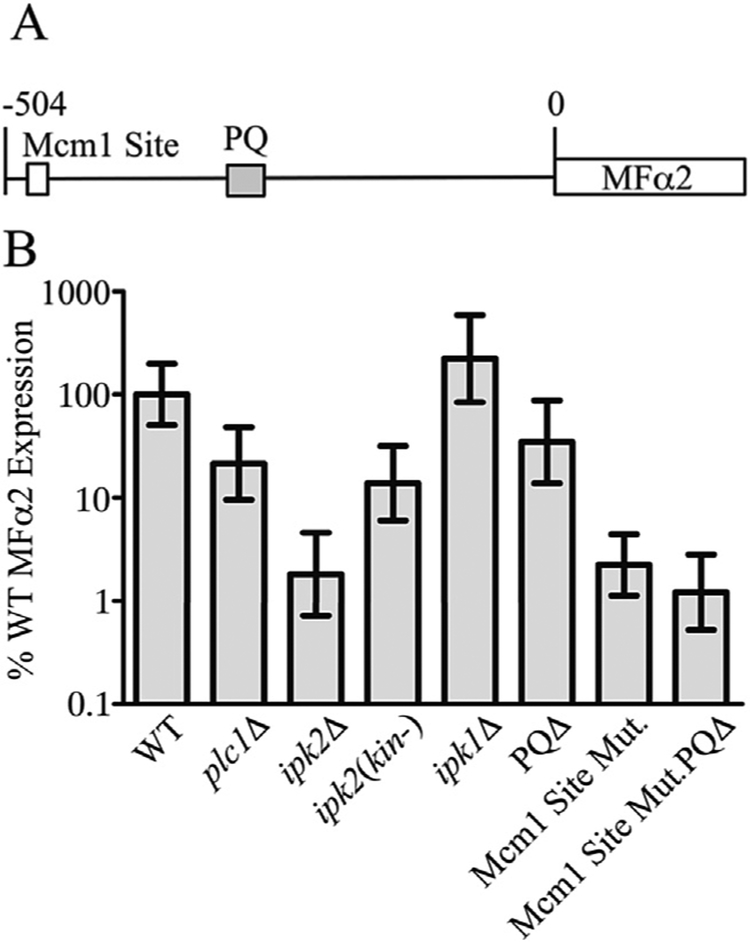
Truncation of the MFα2 promoter showed that the sequence approximately 400 bp to 500 bp upstream of the translation start site is required for efficient MFα2 transcription. Examination of the sequence between 400 bp to 500 bp upstream of MFα2 showed that it contains a consensus Mcm1-binding sequence (Fig. 5A, Fig. 6A) (Wu et al., 2011). Mutation of three nucleotides within this consensus sequence in the context of the genomic MFα2 promoter resulted in significantly inhibited transcription to a level equivalent to deletion of IPK2 (Fig. 5B, Mcm1 site Mut.). This result was not an effect of the genetic manipulations used because generating a WT strain carrying the full-length promoter using the same technique did not perturb MFα2 expression. The double mutation of the PQ and Mcm1 sites did not appreciably reduce expression beyond the Mcm1 site mutant alone (Fig. 4B). The presence of additional Mcm1-binding sites in the promoters of α-specific genes has previously been reported (Jarvis et al., 1988), but this is the first description of any function for this putative Mcm1-binding sequence in the transcription of MFα2.
Fig. 5. Summary of sequence elements required for MFα2 transcription.
A) Schematic of the MFα2 promoter. The white box indicates the putative Mcm1-binding site. The grey box indicates the PQ sequence. Features are not drawn to scale. B) Real-time PCR quantification of MFα2 expression in WT, IP pathway mutant strains, and strains with mutations in the genomic MFα2 promoter. Values are the mean ± SD of at least three independent cultures.
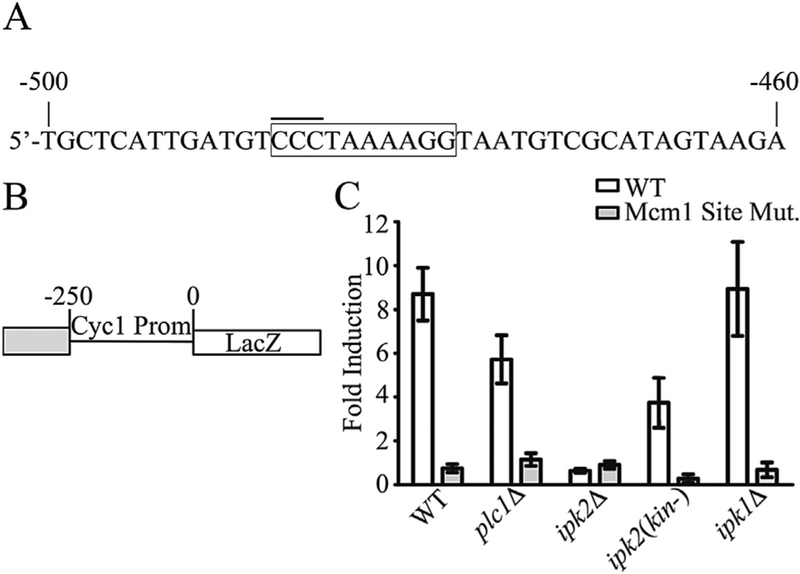
Fig. 6. A putative Mcm1-binding site is a transcriptional activator.
A) The sequence of the MFα2 promoter containing a putative Mcm1-binding site. The coordinates are indicated relative to the start codon of MFα2. The box encloses the putative Mcm1-binding site. The overlined nucleotides were mutated to adenosines to generate the Mcm1-binding site mutant (Mcm1 Site Mut.). B) Schematic of the construct used for studies of transcriptional activation. The grey box indicates the location of the MFα2 sequence that was cloned into pCM64 upstream of the minimal Cyc1 promoter and bacterial β-galactosidase (LacZ) gene. C) Assays of transcriptional activation in WT and IP pathway mutant strains. LacZ activity was measured in Miller Units as previously described (Ausubel, 1987). Data are presented as the fold induction over the values obtained from the empty vector in each strain. Values are the mean ± SEM of at least 6 independent cultures.
To further study this region of the MFα2 promoter, a plasmid-based assay of transcriptional activation was used to determine whether this region was sufficient to activate Ipk2-dependent transcription. The sequence containing the putative Mcm1-binding site and approximately 15 nucleotides on either side was cloned into a reporter plasmid upstream of the minimal promoter of the CYC1 gene fused to the bacterial LacZ reading frame (Fig. 6B). Assays of LacZ activity in strains transformed with this reporter showed that the sequence between 450 bp and 504 bp upstream of the MFα2 translation start site was sufficient to activate transcription from a minimal promoter in a WT strain (Fig. 6C). Mutation of the putative Mcm1-binding site eliminates the ability of this sequence to activate transcription (Fig. 6C). Deletion of IPK2 resulted in a complete inability to activate transcription from this sequence. We therefore propose that this region contains an Ipk2-responsive transcriptional activation element that is dependent on an intact Mcm1-binding sequence. To investigate whether IPs are required for the ability to activate transcription IP-pathway mutants were tested in this assay. The ipk1Δ showed transcriptional activation similar to the WT strain (Fig. 6C) in this assay. This is in agreement with previous results showing that deletion of IPK1 does not affect MFα2 expression (Fig. 1C). The plc1Δ strain is impaired in its ability to activate transcription from this sequence (Fig. 6C). The ipk2(kin-) strain also exhibits an impaired ability to activate transcription from the sequence containing the putative Mcm1-binding site (Fig. 6C). The integrity of the putative Mcm1-binding site was critical for transcriptional activation in all of the strains tested. The transcriptional activation assay indicates that IPs are required for the full transcriptional activation from this sequence. We can conclude that both Ipk2 and the integrity of the putative Mcm1-bindings site are required for the ability of this short sequence from the MFα2 promoter to activate transcription from a minimal promoter.
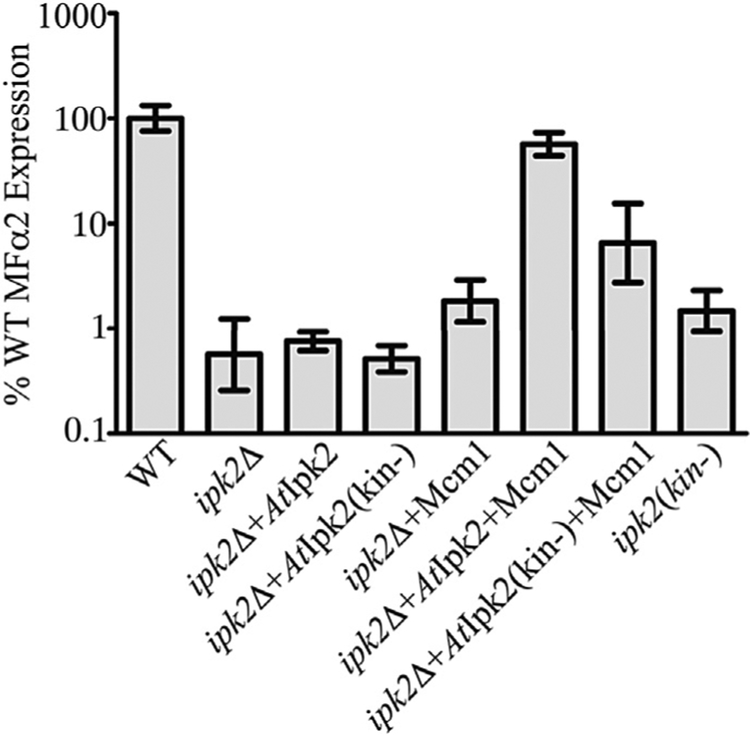
2.6. MFα2 expression requires both IP production and sufficient Mcm1 levels
Deletion of IPK2 causes a defect in MFα2 expression that is not rescued by supplementing IP production by expressing the homologous kinase AtIpk2. MFα2 expression is therefore dependent on some unique kinase-independent function provided by the yeast Ipk2 protein. Ipk2 is required to maintain the stability of Mcm1, and overexpressing Mcm1 has been reported to rescue the ornithine growth phenotype of an ipk2Δ strain (Dubois and Messenguy, 1994). Because MFα2 expression is also known to require Mcm1 and a putative Mcm1-binding site we sought to test whether the MFα2 expression defect observed in ipk2Δ yeast may also be related to Mcm1 levels. Mcm1 was expressed under the control of its own promoter from a multi-copy plasmid. Expression of AtIpk2 or Mcm1 alone was not sufficient to rescue MFα2 expression in the ipk2Δ line (Fig. 7). Expressing AtIpk2 and Mcm1 together rescued MFα2 expression in the ipk2Δ strain, and this rescue was dependent on IP-kinase activity (Fig. 7). Because the requirement for the yeast Ipk2 protein can be bypassed by supplementing Mcm1 expression these results indicate that the kinase-independent function of Ipk2 in MFα2 transcription may be to provide a sufficient amount of Mcm1 to facilitate efficient expression. These data also confirm that while Mcm1 is required for MFα2 expression, IPs produced by the kinase activity of Ipk2 are also essential for full transcriptional activity.
Fig. 7. MFα2 expression requires both IP production and sufficient Mcm1.
Real-time PCR quantification of MFα2 expression in WT, IP pathway mutant strains, and strains complemented with plasmids carrying AtIpk2 constructs and Mcm1. Values are the mean ± SD of at least three independent cultures.
3. Discussion
Deletion of IPK2 results in several significant phenotypes in yeast and is fatal in metazoans. The multiple roles of Ipk2 make it an intriguing focus of study but also complicate the interpretations of experiments aimed at understanding the mechanisms of its functions. Differentiating between the IP kinase-dependent and -independent functions of Ipk2 has been an area of particular biological interest.
This study shows that Ipk2 plays both kinase-dependent and kinase-independent roles in the transcription of MFα2 and identifies elements required for MFα2 transcription. Previous reports have shown that the ability to produce IPs is associated with several Mcm1-dependent transcription processes, including the expression of mating type specific genes (Guzinska et al., 2009). IP production is also sufficient to rescue the activity of the ArgR-Mcm1 transcription complex as assayed by the ability to utilize ornithine as the sole source of organic nitrogen (Fig. 2A) (Seeds et al., 2005). Ipk2 kinase activity is also important for regulating the activities of the SWI/SNF and Ino80 chromatin remodeling complexes affecting the expression of Pho5 in response to environmental phosphate (Steger et al., 2003), and for rescuing the temperature sensitivity of the ipk2Δ strain (Seeds et al., 2005; Stevenson-Paulik et al., 2002). These findings establish that Ipk2 kinase activity mediates important functions within the cell, including several transcriptional processes.
The ability to produce either I(1,4,5,6)P4 or I(1,3,4,5,6)P5 via Ipk2 is critical for efficient MFα2 expression. The finding that RnIpk2, which produces I(1,3,4,5,6)P5 via the I(1,3,4,5)P4 isomer, is capable of rescuing MFα2 transcription in the ipk2(kin-) background is consistent with a model in which I(1,3,4,5,6)P5 provides the kinase-dependent function of Ipk2. However, we cannot exclude I(1,4,5,6)P4 as playing a role in MFα2 expression because we are currently unable to produce I(1,4,5,6)P4 in vivo without also producing some amount of I(1,3,4,5,6)P5. It is also possible that multiple different IP isomers are able to provide functional complementation in the expression of MFα2. Our data also indicate the effects observed are not due to potential lipid-kinase activities attributed to Ipk2 as we observed 1) that deletion of Plc1 has a similar transcriptional defect to the ipk2(kin-), and 2) full complementation with AtIpk2, which does not harbor lipid kinase activity (Maag et al., 2011; Blind et al., 2012). Further study is required to establish which IP isomer supports MFα2 production in WT cells. An alternative model is that IP6 is the preferred IP species in WT cells and either IP4 or IP5 is able to provide the kinase-dependent function of Ipk2. Formal assignment of the kinase-dependent function of Ipk2 to a specific IP isomer will require the development of new methods to produce individual IP species in vivo.
Expression of MFα2 also requires a kinase-independent function provided by the yeast Ipk2 protein. Ipk2 has been reported to stabilize the general transcription factor Mcm1 (El Bakkoury et al., 2000). This stabilization has been interpreted as being significant for the transcription of Mcm1-dependent genes (El Alami et al., 2003). The kinase-independent function of yeast Ipk2 in MFα2 expression may be to regulate Mcm1 levels because the need for Ipk2 can be bypassed by supplementing Mcm1 expression in conjunction with IP-kinase activity. Future investigations of the kinase-dependent function of Ipk2 will determine whether IPs regulate Mcm1 directly or whether additional factors mediate the IP-dependence of MFα2 transcription. In either case, MFα2 expression is regulated by IPs and requires sufficient amounts of the general transcription factor Mcm1.
Mcm1 binds to sequences in the promoters of α-specific genes called PQ boxes, and these sequences are sufficient to bestow α-specific transcription to a minimal promoter (Jarvis et al., 1988; Keleher et al., 1989). Expression of MFα2 requires an Ipk2-responsive element containing a putative Mcm1-binding site upstream of the canonical PQ box. The relationship between the upstream Mcm1-binding site and the PQ box is an interesting area for future investigation. The putative Mcm1-binding site described here is essential for MFα2 transcription, but it is possible that there are additional elements in the MFα2 promoter that are also required. There must be some differences between the upstream putative Mcm1-binding site and the PQ site in the regulation of MFα2 transcription. Disruption of the upstream Mcm1-binding site inhibits MFα2 transcription to a greater extent than deletion of the PQ site. Whether this difference is an effect of the sequences of the specific Mcm1-binding sites or the relative locations of these sites remains a question requiring further examination. Mcm1-binding sites have been implicated in transcriptional activation through the recruitment of Gcn4 and the SWI/SNF chromatin-remodeling complex (Hong and Yoon, 2011; Yoon and Hinnebusch, 2009). It is possible that the upstream Mcm1 site, which is critical for MFα2 expression, could be acting through a similar mechanism.
Expression of the mating pheromone gene MFα2 requires both the production of IPs downstream of IP3, a putative Mcm1-binding site in the promoter sequence and a sufficient amount of Mcm1 that is not otherwise present in ipk2Δ cells. This study identifies multiple elements required for the transcription of an Ipk2-dependent gene and provides multiple avenues of further investigation of biological processes regulated by IPs.
Acknowledgements
We thank the members of the York lab for helpful discussions. This work was supported by funds from the Howard Hughes Medical Institute and the National Institutes of Health, RO1 HL-55672 (both to J.D.Y.).
Appendix
Materials and methods
Strains, media and manipulations –
All yeast strains were grown at 30 °C in either rich medium (yeast extract-peptonedextrose, YPD) or synthetic complete medium (CSM) lacking nutrients to maintain plasmids carrying markers and were handled according to standard protocols. Ornithine plates were made as previously described (Odom et al., 2000; Gimeno et al., 1992). Yeast strains were transformed using the standard lithium-acetate procedure (Ausubel, 1987). Expression of Ipk2 homologues from plasmids was induced by including 100 μM CuSO4 in the growth medium.
Strain construction –
All strains were MATα haploids generated from the w303 (leu2–3112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15) background. The single exception is the MATa strain used in the halo assays which is a bar1 variant of the BF264–15Du background (ade1 his2 leu2–3112 trp1–1 ura3Δns) (McMillan et al., 1998). Strains carrying mutations in the IP metabolic pathway were previously described (York et al., 1999; Odom et al., 2000). Ipk2 constructs and DmIP3K were expressed as in-frame fusions with GFP and three copies of the c-myc epitope tag from copper inducible plasmids that were constructed as previously described (Stevenson-Paulik et al., 2002). Mcm1, including 553 bp upstream of the translation start site, was cloned between the NotI and XhoI sites of plasmid pRS426. A c-myc epitope tag was added to the C-terminus of Mcm1 during cloning. The sense primer 5’- CAT GCG GCG GCG GCC GCC ATG CGA GAG TAA GAG ATG CC and antisense primer 5’-TAC CTC GAG TCA CAG ATC TTC TTC AGA AAT AAG TTT TTG TTC GTA TTG GCC TTG TTG CGG were used to amplify myc-tagged Mcm1 and its promoter from genomic DNA. Strains carrying plasmids were freshly transformed before use in experiments. Strains in which the genomic MFα2 promoter was mutated were constructed using the method of transplacement. The MFα2 coding region and 1000 bp upstream of the ATG translation start site were cloned between the KpnI and SacI sites of the yeast integrating vector pRS306. Site-directed mutagenesis was used to insert a BglII site approximately 500 bp upstream of the translation start site and an XhoI site immediately upstream of the translation start site. These sites were used for the insertion of modified promoter constructs. Site-directed mutagenesis was also used to modify the upstream putative Mcm1-binding site and to delete the PQ box sequence. Plasmids containing constructs with the desired mutations were linearized using XhoI and transformed into the yeast strains to be modified, and transformants were selected using agar plates containing synthetic complete medium without uracil (CSM-Ura). Colonies that grew on CSM-Ura were subsequently grown overnight in YPD liquid culture and plated to obtain single colonies on YPD agar plates. Recombinants were then selected by replica plating on media containing 5-fluoroorotic acid (FOA). Strains in which the genomic MFα2 promoter had been modified were identified using diagnostic PCR on genomic DNA extracted from FOA-resistant colonies.
Northern blot analysis –
Total RNA was prepared using the RNeasy Midi kit according to the manufacturer’s protocol (Qiagen). Probes were generated by PCR amplification of specific ORFs from genomic DNA, except for the ACT1 gene fragment, which was kindly provided by Susan Wente (Vanderbilt University), and labeled using the Random Primed DNA Labeling kit (Roche) according to the manufacturer’s protocol.
Real-time PCR –
Single colonies were grown to early log phase (OD600 0.5–0.8 for YPD cultures or OD600 0.3–0.6 for cultures in selective media) in liquid media, and RNA was extracted using hot trizol as previously described (Schmitt et al., 1990). RNA was treated with RQ1 DNAse (Promega) according to the manufacturer’s instructions, and 1 μg was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) according to the supplier’s protocol. Real-time PCR data collection was performed using an iQ5 multicolor real-time PCR detection system. Reaction mixes contained 7.5 μl 2× SYBR Green master mix (Bio-Rad), 3 pmol each of the forward and reverse primers, and 2 μl cDNA diluted 1:10 in a total reaction volume of 15 ml. Relative expression levels were quantified using the comparative ΔΔCt method with Act1 as the reference gene. The primers used for amplification of Act1 were sense 5’-GCCTTCTACGTTTCCATCCA and antisense 5’-GGCCAAATCGATTCTCAAA. The primers used for amplification of MFα2 were sense 5’-ACGCTACCGCCAGTGGGCTAT and antisense 5’-GTGCCAAGCGTCGGCAACAG.
Halo assays –
Halo assays of mating factor production were performed as previously described (Guthrie and Fink, 1991).
LacZ assays –
The plasmids used in assays of transcriptional activation were generated by ligating annealed oligonucleotides into the BglII and XhoI sites of plasmid pCM64 (kindly provided by Dennis Thiele, Duke University) The sequences of the oligonucleotides used to generate construct carrying the WT sequence were sense 5’-GAT CTCACTTGCTCATTGATGTCCC TAAAAGGTAATGTCGCATAGTAAGATGTATGTCCCC and antisense 5’-TCGAGGGGACATACATC TTACTATGCGACATTACCTT TTAGGGACATCAATGAGCAAGTGA. The mutations made to generate the defective Mcm1-binding site are indicated in the text. LacZ assays were performed as previously described (Ausubel, 1987).
Ornithine growth –
Media containing ornithine as the sole nitrogen source were prepared as previously described (Odom et al., 2000; Gimeno et al., 1992). Single colonies of strains to be grown on ornithine plates were first grown overnight in 1 ml CSM liquid cultures. An equivalent number of cells of each strain to be tested were collected from the liquid cultures based on the OD600 and washed twice with 0.5 ml sterile water. Washed cells were diluted in serial 10-fold increments using sterile water and immediately plated on either CSM or ornithine media.
Statistical analysis –
Analysis of real-time PCR data was performed using GraphPad Prism 5 software (GraphPad Software). Differences were examined using a one-way analysis of variance (ANOVA) test followed by Tukey’s test and were considered significant if the calculated p value was <0.05. LacZ reporter data were analyzed using a two-way ANOVA and differences were considered significant if the calculated p value was <0.05.
References
- Ausubel FM, 1987. Current Protocols in Molecular Biology. J. Wiley, Media, Pa. [Google Scholar]
- Bechet J, Greenson M, Wiame JM, 1970. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem./FEBS 12 (1), 31–39. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Bercy J, Dubois E, Messenguy F, 1987. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene 55 (2–3), 277–285. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Bhandari R, Chakraborty A, Snyder SH, 2007. Inositol pyrophosphate pyrotechnics. Cell metab. 5 (5), 321–323. 10.1016/j.cmet.2007.04.008. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Blind RD, Suzawa M, Ingraham HA, 2012. Direct modification and activation of a nuclear receptor-PIP(2) complex by the inositol lipid kinase IPMK. Sci.Signal. 5 (229), ra44 10.1126/scisignal.2003111. PubMed PMID: ; PMCID: PMC3395721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch D, Saiardi A, 2012. Arginine transcriptional response does not require inositol phosphate synthesis. J. Biol. Chem 287 (45), 38347–38355. 10.1074/jbc.M112.384255. PubMed PMID: ; PMCID: 3488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Kim S, Snyder SH, 2011. Inositol pyrophosphates as mammalian cell signals. Sci. Signal 4 (188), re1 10.1126/scisignal.2001958. PubMed PMID: ; PMCID: 3667551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delforge J, Messenguy F, Wiame JM, 1975. The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR- mutations and the general control of amino-acid biosynthesis. Eur. J. Biochem./FEBS 57 (1), 231–239. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Dubois E, Messenguy F, 1994. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol. Gen. Genet 243 (3), 315–324. Epub 1994/05/10. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Dubois E, Bercy J, Messenguy F, 1987. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast.Mol. Gen. Genet 207 (1), 142–148. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Dubois E, Dewaste V, Erneux C, Messenguy F, 2000. Inositol polyphosphate kinase activity of Arg82/ArgRIII is not required for the regulation of the arginine metabolism in yeast. FEBS Lett. 486 (3), 300–304. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- El Alami M, Messenguy F, Scherens B, Dubois E, 2003. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol. Microbiol 49 (2), 457–468. Epub 2003/06/28. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- El Bakkoury M, Dubois E, Messenguy F, 2000. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1 by the pleiotropic factor ArgRIII is required for their stability. Mol. Microbiol 35 (1), 15–31. Epub 2000/01/13. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD, 2012. Structural studies and protein engineering of inositol phosphate multikinase. J. Biol. Chem 287 (42), 35360–35369. 10.1074/jbc.M112.365031. PubMed PMID: ; PMCID: 3471723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick JS, Thorner J, 1993. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol.Cell Biol 13 (9), 5861–5876. Epub 1993/09/01. PubMed PMID: ; PMCID: 360334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Noble KN, Cole CN, Wente SR, 2011. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus 2 (6), 540–548. 10.4161/nucl.2.6.17881. PubMed PMID: ; PMCID: 3324343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, York JD, 2005. A role for rat inositol polyphosphate kinases rIPK2 and rIPK1 in inositol pentakisphosphate and inositol hexakisphosphate production in rat-1 cells. Epub 2004/11/06 J. Biol. Chem 280 (2), 1156–1164. 10.1074/jbc.M412006200. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR, 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68 (6), 1077–1090. Epub 1992/03/20. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego: xxxvii, 933 p. p. [Google Scholar]
- Guzinska K, Varghese R, Vancura A, 2009. Role of Plc1p in regulation of Mcm1p-dependent genes. Epub 2009/05/23 FEMS Microbiol. Lett 295 (2),245–250. 10.1111/j.1574-6968.2009.01602.x. PubMed PMID: ; PMCID: 2761037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch AJ, York JD, 2010. SnapShot: inositol phosphates, 1030–e1 Cell 143 (6). 10.1016/j.cell.2010.11.045. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Yoon S, 2011. Mcm1p binding sites in the ARG1 promoter positively regulate ARG1 transcription and S. cerevisiae growth in the absence of arginine and Gcn4p. Epub 2010/07/14 Amino acids 40 (2), 623–631. 10.1007/s00726-010-0687-z. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ, 2001. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. cell Biol 2 (5), 327–338. 10.1038/35073015. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Ives EB, Nichols J, Wente SR, York JD, 2000. Biochemical and functional characterization of inositol 1,3,4,5, 6-pentakisphosphate 2-kinases. Epub 2000/08/29 J. Biol. Chem 275 (47), 36575–36583. 10.1074/jbc.M007586200. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Jarvis EE, Hagen DC, Sprague GF Jr., 1988. Identification of a DNA segment that is necessary and sufficient for alpha-specific gene control in Saccharomyces cerevisiae: implications for regulation of alpha-specific and a-specific genes. Mol. Cell Biol 8 (1), 309–320. Epub 1988/01/01. PubMed PMID: ; PMCID: 363126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher CA, Passmore S, Johnson AD, 1989. Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell Biol 9 (11),5228–5230. Epub 1989/11/01. PubMed PMID: ; PMCID: 363677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Snyder SH, 2011. Nutrient amino acids signal to mTOR via inositol polyphosphate multikinase. Cell cycle 10 (11), 1708–1710. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, Chakraborty A, Koldobskiy MA, Cha SH, Barrow R, Snowman AM, Snyder SH, 2011. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell metab. 13 (2), 215–221. 10.1016/j.cmet.2011.01.007. PubMed PMID: ; PMCID: 3042716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Tyagi R, Lee JY, Park J, Kim YR, Beon J, Chen PY, Cha JY, Snyder SH, Kim S, 2013. Inositol polyphosphate multikinase is a coactivator for serum response factor-dependent induction of immediate early genes. Proc. Natl. Acad. Sci. U. S. A 110 (49), 19938–19943. 10.1073/pnas.1320171110. PubMed PMID: ; PMCID: 3856792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Beon J, Lee S, Park J, Kim SIPMK, 2016. A versatile regulator of nuclear signaling events. Adv. Biol. Regul 61, 25–32. 10.1016/j.jbior.2015.11.005. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim YR, Park J, Kim S, 2012. Inositol polyphosphate multikinase signaling in the regulation of metabolism. Ann. N. Y. Acad. Sci 1271, 68–74. 10.1111/j.1749-6632.2012.06725.x. PubMed PMID: ; PMCID: PMC3499638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Maxwell MJ, Hardesty DA, Boucher KL, Choudhari N, Hanno AG, Ma JF, Snowman AS, Pietropaoli JW, Xu R, Storm PB, Saiardi A, Snyder SH, Resnick AC, 2011. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Natl. Acad. Sci. U. S. A 108(4), 1391–1396. 10.1073/pnas.1017831108. PubMed PMID: ; PMCID: PMC3029688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, 1992. Inositol phosphate biochemistry. Annu. Rev. Biochem 61, 225–250. 10.1146/annurev.bi.61.070192.001301. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- McMillan JN, Sia RA, Lew DJ, 1998. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol 142 (6), 1487–1499. Epub 1998/09/23. PubMed PMID: ; PMCID: 2141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E, 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell Biol 13(4), 2586–2592. Epub 1993/04/01. PubMed PMID: ; PMCID: 359592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E, 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316,1–21. Epub 2003/10/18. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Messenguy F, Dubois E, Boonchird C, 1991. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters.Mol. Cell Biol 11 (5), 2852–2863. PubMed PMID: ; PMCID: 360071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell RH, 2008. Inositol derivatives: evolution and functions. Nat. Rev. Mol. cell Biol 9 (2), 151–161. 10.1038/nrm2334. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD, 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287 (5460), 2026–2029. Epub 2000/03/17. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Otto JC, Kelly P, Chiou ST, York JD, 2007. Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc. Natl. Acad. Sci. U. S. A 104 (40), 15653–15658. 10.1073/pnas.0705729104. PubMed PMID: ; PMCID: 1994134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu HF, Dubois E, Broen P, Messenguy F, 1990. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Gen. Genet 222 (2–3), 192–200. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH, 1999. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol 9 (22), 1323–1326. Epub 1999/11/27. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL, 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18 (10), 3091–3092. Epub 1990/05/25. PubMed PMID: ; PMCID: 330876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds AM, Sandquist JC, Spana EP, York JD, 2004. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. Epub 2004/08/24 J. Biol. Chem 279 (45), 47222–47232. 10.1074/jbc.M408295200. PubMed PMID: 15322119. [DOI] [PubMed] [Google Scholar]
- Seeds AM, Bastidas RJ, York JD, 2005. Molecular definition of a novel inositol polyphosphate metabolic pathway initiated by inositol 1,4,5-trisphosphate 3-kinase activity in Saccharomyces cerevisiae. Epub 2005/06/10 J. Biol. Chem 280 (30), 27654–27661. 10.1074/jbc.M505089200. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK, 2003. Regulation of chromatin remodeling by inositol polyphosphates. Epub 2002/11/16 Science 299 (5603), 114–116. 10.1126/science.1078062. PubMed PMID: ; PMCID: 1458531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Odom AR, York JD, 2002. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. Epub 2002/09/13 J. Biol. Chem 277 (45), 42711–42718. 10.1074/jbc.M209112200. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD, 2005. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. U. S. A 102 (35), 12612–12617. 10.1073/pnas.0504172102. PubMed PMID: ; PMCID: 1194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R, Ammerer G, 1992. The SRF and MCM1 transcription factors. Curr. Opin. Genet. Dev 2 (2), 221–226. Epub 1992/04/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW, 2005. The pathway for the production of inositol hexakisphosphate in human cells. J.Biol. Chem 280 (3), 1911–1920. 10.1074/jbc.M411528200. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Wilson MS, Livermore TM, Saiardi A, 2013. Inositol pyrophosphates: between signalling and metabolism. Biochem. J 452 (3), 369–379. 10.1042/BJ20130118. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Wu W, Huang X, Cheng J, Li Z, de Folter S, Huang Z, Jiang X, Pang H, Tao S, 2011. Conservation and evolution in and among SRF- and MEF2-type MADS domains and their binding sites. Epub 2010/08/21 Mol. Biol. Evol 28 (1), 501–511. 10.1093/molbev/msq214. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- Xia HJ, Brearley C, Elge S, Kaplan B, Fromm H, Mueller-Roeber B, 2003. Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant cell 15 (2), 449–463. PubMed PMID: ; PMCID: 141213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Snyder SH, 2013. Gene transcription by p53 requires inositol polyphosphate multikinase as a co-activator. Cell cycle 12 (12), 1819–1820. 10.4161/cc.25119. PubMed PMID: ; PMCID: 3735690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Sen N, Paul BD, Snowman AM, Rao F, Vandiver MS, Xu J, Snyder SH, 2013a. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci. Signal 6 (269), ra22 10.1126/scisignal.2003405. PubMed PMID: ; PMCID: 4108196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Paul BD, Smith DR, Tyagi R, Rao F, Khan AB, Blech DJ, Vandiver MS, Harraz MM, Guha P, Ahmed I, Sen N, Gallagher M, Snyder SH, 2013b. Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc. Natl. Acad. Sci. U. S. A 110 (40), 16181–16186. 10.1073/pnas.1315551110. PubMed PMID: ; PMCID: 3791727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Hinnebusch AG, 2009. Mcm1p binding sites in ARG1 positively regulate Gcn4p binding and SWI/SNF recruitment. Epub 2009/02/24 Biochem. biophysical Res. Commun 381 (1), 123–128. 10.1016/j.bbrc.2009.02.045. PubMed PMID: ; PMCID: 2683423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Govind CK, Qiu H, Kim SJ, Dong J, Hinnebusch AG, 2004. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Epub 2004/08/04 Proc. Natl. Acad. Sci. U. S. A 101 (32), 11713–11718. 10.1073/pnas.0404652101. PubMed PMID: ; PMCID: 511042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR, 1999. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285 (5424), 96–100. Epub 1999/07/03. PubMed PMID: . [DOI] [PubMed] [Google Scholar]