Abstract
Skeletal muscle plays a pivotal role in regulating systemic glucose homeostasis in part through the conserved cellular energy sensor AMPK. AMPK activation increases glucose uptake, lipid oxidation, and mitochondrial biogenesis, leading to enhanced muscle insulin sensitivity and whole-body energy metabolism. Here we show that the muscle-enriched H19 long noncoding RNA (lncRNA) acts to enhance muscle insulin sensitivity, at least in part, by activating AMPK. We identify the atypical dual-specificity phosphatase DUSP27/DUPD1 as a potentially important downstream effector of H19. We show that DUSP27, which is highly expressed in muscle with previously unknown physiological function, interacts with and activates AMPK in muscle cells. Consistent with decreased H19 expression in the muscle of insulin-resistant human subjects and rodents, mice with genetic H19 ablation exhibit muscle insulin resistance. Furthermore, a high-fat diet downregulates muscle H19 via both posttranscriptional and epigenetic mechanisms. Our results uncover an evolutionarily conserved, highly expressed lncRNA as an important regulator of muscle insulin sensitivity.
Introduction
Skeletal muscle, the largest insulin-sensitive organ, accounting for up to 60% of whole-body glucose disposal, plays an essential role in maintaining systemic glucose homeostasis. Muscle insulin resistance contributes critically to the pathogenesis of major metabolic disorders, including type 2 diabetes (T2D) and cardiovascular diseases. The evolutionarily conserved AMPK is a key sensor of the cellular energy balance that exerts a fundamental role in metabolic regulation. Expressed in major metabolic organs such as muscle, liver, fat, and hypothalamus, AMPK is activated by energy deprivation, exercise, and hormones that affect cellular metabolism. In muscle, AMPK activation enhances glucose uptake, fatty acid oxidation, and mitochondrial biogenesis, leading to increased muscle insulin sensitivity and whole-body energy metabolism. Thus, AMPK has emerged as a promising therapeutic target for metabolic diseases (1).
AMPK is a heterotrimeric Ser/Thr kinase consisting of one catalytic α subunit and two regulatory subunits, β and γ. AMPK is activated by upstream kinases that phosphorylate a conserved threonine (Thr172) in the α subunit. The primary kinases that phosphorylate Thr172 are LKB1 and CaMKKβ. Binding of AMP to the γ subunit leads to allosteric activation of AMPK and protection of Thr172 from dephosphorylation. AMPK can also be inactivated by protein phosphatases and by inhibitory phosphorylation (1–3). AMPK modulates many downstream targets, including acetyl-CoA carboxylase (ACC) and peroxisome proliferator–activated receptor-γ coactivator 1α (PGC-1α). ACC is a rate-limiting enzyme for the synthesis of malonyl-CoA, an essential substrate for fatty acid biosynthesis and a potent inhibitor of lipid oxidation. Phosphorylation by AMPK inactivates ACC, resulting in inhibition of lipogenesis while increasing lipid oxidation (1). PGC-1α is a master regulator of mitochondrial biogenesis and lipid oxidation (4,5). AMPK activates PGC-1α through multiple mechanisms, including direct phosphorylation. As a transcriptional coactivator, PGC-1α activates different transcription factors to drive transcription of vast gene networks involved in all aspects of energy homeostasis, including glucose utilization and fatty acid oxidation, as well as mitochondrial biogenesis (including mitochondrial DNA replication) and function. Among the numerous genes activated by PGC-1α are Pgc-1α (autoregulation), carnitine palmitoyltransferase 1b (Cpt1b), pyruvate dehydrogenase kinase 4 (Pdk4), and mitofusin-2 (Mfn2). As PGC-1α positively regulates its own transcription, activation by AMPK can also increase PGC-1α protein levels.
The evolutionarily conserved H19 long noncoding RNA (lncRNA) is abundantly expressed in skeletal muscle, but its expression is significantly decreased in patients with T2D and insulin-resistant rodents (6). Whether H19 is involved in regulation of muscle insulin sensitivity and how H19 is downregulated in the diabetic muscle are not clear.
Research Design and Methods
Animals
All animal work was approved by the Yale University Institutional Animal Care and Use Committee. All mice used in this report were male. The wild-type (WT) and knockout (KO) mice on a background of C57BL/6J were gifts from Dr. Luisa Dandolo (Institut Cochin, Paris, France). Mice were housed at 22–24°C with a 12-h light/12-h dark cycle with regular chow (RC) (Teklad no. 2018, 18% calories from fat; Harlan) or high-fat diets (HFDs) (D12451, 45% calories from fat; Research Diets) and water provided ad libitum. For HFD studies, 7-week-old WT mice were exposed to HFD or RC for 11 days or 13 weeks, followed by the indicated experiments.
Body Composition and Glucose and Insulin Tolerance Tests
Body composition was measured in vivo by MRI (EcoMRI; Echo Medical Systems). Glucose tolerance tests (GTTs) were performed in 16 h–fasted mice as previously described (7). Each animal received an i.p. injection of 2 g/kg body weight glucose (DeltaSelect) in sterile saline. Blood glucose levels were measured at 0, 15, 30, 45, 60, 90, and 120 min after the injection. Insulin tolerance tests (ITTs) were performed in ad libitum fed mice (7). Each animal received an i.p. injection of insulin, 1 unit/kg (Humulin R; Eli Lilly and Company). Blood glucose levels were monitored at 0, 15, 30, 45, 60, 90, and 120 min after the injection.
Hyperinsulinemic-Euglycemic Clamp and Measurement of Muscle 2-[14C]deoxy-d-glucose Uptake
The experiments were performed on 11-week-old WT and KO mice as previously described with minor modification (8). In brief, mice were anesthetized and cannulated. After recovery for 7 days, mice were fasted overnight (14 h) followed by infusion of d-[3-3H]glucose to assess the basal rate of whole-body glucose turnover. After the basal period, a 2-h hyperinsulinemic-euglycemic clamp was conducted with a fixed amount of insulin (4 mU/[kg ⋅ min]) and a variable amount of 20% dextrose to maintain euglycemia. A 10-μCi bolus of 2-[14C]deoxy-d-glucose was injected into mice at 90 min to determine tissue-specific glucose uptake. After collection of the final blood sample, the mice were anesthetized and tissues were harvested, frozen in liquid nitrogen, and stored at −80°C until later use.
Plasmid Construction
Plasmids pAV-PGK-DUSP27 (wt-DUSP27) and pAV-PGK-DUSP27C146S (mt-DUSP27), which express WT and mutant dual-specificity protein phosphatase 27 (DUSP27), respectively, were constructed by cloning the open reading frames of WT and mutant mouse DUSP27 (NM_001013826.2) into the pAV-PGK expression vector (Vigene Biosciences Inc., Rockville, MD). The expression of both proteins was driven by a phosphoglycerate kinase 1 (PGK-1) promoter. The accuracy of the clones was confirmed by sequencing.
RNA Extraction and Real-time Quantitative PCR
Total RNAs were extracted from frozen tissue samples or from cultured myotubes using the PureLink RNA Mini Kit (12183018A; Ambion). cDNA was synthesized using the PrimeScript RT Reagent Kit (TAKARA, RR037A; Invitrogen, Grand Island, NY) in a 20-μL reaction containing 0.2–0.5 μg of total RNA. Real-time quantitative PCR (RT-qPCR) was performed in a 15-μL reaction containing 0.5–1 μL of cDNA using iQSYBRGreen (Bio-Rad) in a Bio-Rad iCycler. PCR was performed by initial denaturation at 95°C for 5 min, followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis. Gene expression levels were normalized against the following housekeeping genes: β-tubulin for muscle tissues, β-actin for myotubes, and U6 for let-7. Real-time PCR primers are listed in Supplementary Table 2.
Western Blot Analysis
Myotubes were detached from plates by trypsin digestion and collected in culture medium. After gentle centrifugation, the cell pellet was quickly lysed in 2× SDS sample buffer (100 μL per well of a 24-well plate) by heating at 100°C for 5 min with occasional vortexing. For muscle tissue samples, 5 mg of frozen tissue was quickly homogenized on ice in 200 μL of 2× SDS sample buffer in the presence of phosphatase inhibitors (Cocktail 2 [P5726] and Cocktail 3 [P0044]; Sigma-Aldrich) at a final concentration of 1×. Samples were then heated at 100°C for 5 min with occasional vortexing. Additional phosphatase inhibitors were added at a final concentration of 1× before loading the samples onto a 10% SDS-PAGE gel, followed by Western blot analysis. Detailed information on antibodies and conditions is listed in Supplementary Table 3. Western blot gels were quantified using ImageJ (https://imagej.nih.gov/ij). β-Actin was used for normalization. For DUSP27 Western blot analysis, mouse IgG κ binding protein (m-IgGk BP, sc-516102; Santa Cruz Biotechnology) conjugated to horseradish peroxidase was used instead of anti-mouse IgM secondary antibody to increase specificity and minimize background.
Myotube Culture and Transfection
Undifferentiated mouse C2C12 myoblasts (91031101-iVL; Sigma-Aldrich) were maintained in growth medium (GM) (DMEM, 11965-092, supplemented with 10% FBS, heat inactivated, 1% penicillin/streptomycin, 1% l-glutamine, and 1 mmol/L sodium pyruvate; Gibco). To prepare for differentiation, cells were seeded at a density of 1.6 × 105 cells/well or 2.0 × 104 cells/well in GM in 6-well or 24-well plates, respectively. Differentiation was initiated 2 days later when cells became confluent by replacing GM with differentiation medium (DM) containing 2% horse serum in place of 10% FBS. The medium was changed every other day until transfection, which was performed on day 4–5 after initiation of differentiation.
For small interfering RNA (siRNA) or let-7 transfection, culture medium was gently removed, followed by the addition of transfection cocktails to cover the myotubes. To prepare transfection cocktails for each well of 24-well plates, 125 pmol of siRNA (with a stock solution of 5 μmol/L) was mixed with 150 μL of OPTI-MEM by gentle pipetting. For iLet7 rescue experiments, 125 pmol of H19-specific siRNA (siH19) and 75 pmol of iLet7 were mixed together with 150 μL of OPTI-MEM. In parallel, 6.25 μL of Lipofectamine 2000 was diluted in 150 μL of OPTI-MEM by gentle pipetting. After 5 min incubation at room temperature, the two were combined by gentle pipetting. After incubation at room temperature for 25 min, the resulting cocktail (300 μL) was gently added to the myotubes. After 8–10 h incubation in a tissue culture incubator, the cocktail was gently replaced with fresh DM. Forty-eight hours after the transfection, RNA and protein were extracted for analysis.
Plasmid DNA (Vec; wt-DUSP27 or mt-DUSP27) transfections were performed in a 24-well plate scale. To prepare plasmid DNA mix per well of myotubes, 0.4 μg of DNA was mixed with 1 μL of P3000 (L3000-008; Invitrogen) and 40 μL of OPTI-MEM by gentle pipetting. In a separate tube, 0.75 μL of Lipofectamine 3000 was mixed with 40 μL of OPTI-MEM. After 5 min incubation at room temperature, the two were mixed and incubated at room temperature for 10 min. The resulting 80 μL of transfection cocktail was gently added to myotubes. After 6 h of incubation in a tissue culture incubator, 800 μL of DM was added and incubation was continued for an additional 48 h, followed by protein extraction and Western blot analysis.
Glucose Uptake Assay
These were performed in a 96-well plate scale using the Glucose Uptake Cell-Based Assay Kit (600470; Cayman Chemical) according to the manufacturer’s instructions with minor modifications. To prepare the siRNA mix per well of myotubes, 25 pmol of siRNA (control siRNA [siCon] or siH19) was mixed with 30 μL of OPTI-MEM by gentle pipetting. In a separate tube, 2.5 μL of Lipofectamine 3000 was mixed with 30 μL of OPTI-MEM. After 5 min incubation at room temperature, the two were mixed and incubated at room temperature for 10 min. To prepare the plasmid DNA mix per well of myotubes, 0.1 μg of Vec or wt-DUSP27 was mixed with 0.3 μL of P3000 and 10 μL of OPTI-MEM by gentle pipetting. In a separate tube, 0.2 μL of Lipofectamine 3000 was mixed with 10 μL of OPTI-MEM. After 5 min incubation at room temperature, the two were mixed and incubated at room temperature for 10 min. The resulting 60 μL of siRNA mix and 20 μL of plasmid DNA mix were combined to make a final transfection cocktail of 80 μL, which was gently added to one well of myotubes. After 6 h of incubation in a tissue culture incubator, 200 μL of DM was added and incubation was continued for an additional 48 h. On the day of the glucose uptake assay, DM was replaced with 200 μL of glucose-free DMEM (11966-025; Gibco) and incubation was carried out for 2 h. Then, the medium was replaced with 100 μL of new glucose-free DMEM in the presence or absence of 100 nmol/L of insulin for 15–20 min. Subsequently, 100 μL of new glucose-free DMEM containing fluorescent 2-NBDG at a final concentration of 150 μg/mL was added. Incubation was performed in the dark for an additional 15 min in the tissue culture incubator. The medium was then removed, and the myotubes were washed once with 200 μL of ice-cold PBS. After adding 100 μL of new ice-cold PBS to the myotubes, fluorescent intensity was immediately determined using the fluorescent plate reader (FilterMax F3 and F5 Multi-Mode Microplate Reader; Molecular Devices). Results are presented with myotubes without insulin stimulation arbitrarily set as 1.
RNA-Seq and Data Analysis
RNAs were extracted from WT and KO soleus muscle tissue samples using the Purelink RNA Mini Kit (12183018A). Genome-wide transcriptome analysis (RNA-Seq) libraries were prepared using the Illumina TruSeq Stranded Total RNA LT Kit with Ribo-Zero Human/Mouse/Rat, set A (rs-122-2201) according to the sample preparation protocol. In brief, 1 μg of total RNA was subjected to Ribo-Zero depletion to remove rRNAs. The remaining RNA was purified, fragmented, and primed with random hexamers for cDNA synthesis. After first and second cDNA synthesis, cDNA fragments were adenylated and then ligated to indexing adapters. The cDNA fragments were enriched by PCR, purified, and then sequenced on an Illumina NextSeq500 using paired-end chemistry and 76–base pair cycles. Sequences are available from the GEO with accession number GSE103202.
Illumina BaseSpace (https://basespace.illumina.com/) embedding tools were used to analyze the RNA-Seq data. RNA-Seq Alignment version 1.0.0 was used to map sequencing reads to mm10 genome and quantify reads of genes. DESeq2 version 1.0.0 was applied to calculate differential expression of genes.
Immunoprecipitation
To prepare antibodies, 8 μL of packed beads (protein L agarose beads [sc-2336; Santa Cruz Biotechnology] for anti-DUSP27 and rabbit preimmune IgM; protein A sepharose beads for anti-AMPKα [2032; Cell Signaling Technology] and rabbit preimmune IgG) was incubated with 16 μg of anti-DUSP27, anti-AMPKα, preimmune IgM, or preimmune IgG in 250 μL immunoprecipitation (IP) buffer (1% Triton X-100, 300 mmol/L NaCl, 10 mmol/L Tris-HCl at pH 7.5, and 10 mmol/L EDTA) at 4°C overnight. The next day, the beads were washed three times with IP buffer and kept on ice until used. To prepare tissue lysates, 35 mg of frozen muscle tissue was homogenized on ice in 100 μL of freshly prepared gentle lysis buffer (GLB; 1% Triton X-100, 10 mmol/L NaCl, 10 mmol/L Tris-HCl at pH 7.5, 10 mmol/L EDTA, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L dithiothreitol, and 1× protease inhibitor cocktail [Calbiochem]), followed by the addition of 900 μL of GLB and 10 s centrifugation in a benchtop microcentrifuge. After transferring the supernatant (∼900 μL) to a new tube on ice, the remaining tissue pellet was homogenized again in the residual ∼100 μL of GLB. Then, the supernatant was added back to the tube containing the tissue pellet and the tube was incubated on ice for 10 min. After removing insoluble materials by centrifugation at 12,000 rpm at 4°C for 15 min, 4 mol/L NaCl was added to the lysate at a final concentration of 300 mmol/L. The lysate was then split into two tubes (∼500 μL per tube) and incubated with 10 μL of packed protein L beads (for anti-DUSP27 IP) or protein A beads (for anti-AMPKα IP) at 4°C for 1 h to minimize nonspecific binding. The precleared lysates were then transferred to tubes containing the respective antibody or preimmune IgM/IgG-coated beads (each tube contained ∼250 μL of precleared lysate), and IP was carried out by rotating the tubes at 4°C for 3 h. After IP, the beads were quickly washed with 1 mL of cold IP buffer twice and then washed an additional three times by rotating the tube at 4°C for 3 min each time. After the final wash, residual liquid was completely removed using a long flat pipette tip. Beads were eluted with 20 μL of 2× SDS at 100°C for 3 min. Ten microliters of eluant was loaded onto each well of 12% SDS-PAGE gels.
Immunofluorescence
For DUSP27 and AMPKα double immunostaining, myotubes grown in a 1-μ Slide eight-well ibiTreat microscopy chamber (80826; Research Products International Corp.) were washed briefly with TBS (20 mmol/L Tris-HCl, pH 7.4, 225 mmol/L NaCl), fixed with 3% paraformaldehyde/TBS for 20 min, permeabilized with 1% SDS/TBS for 5 min, and blocked with 10% BSA/0.1% goat serum/TBS for 1 h. Cells were then incubated with anti-DUSP27 and anti-AMPKα antibody at a dilution of 1:50 at 4°C overnight, washed with TBS, and then incubated with Alexa Fluor 555 (for DUSP27) and Alexa Fluor 488 (for AMPKα) conjugated secondary antibodies (1:500 dilution; Invitrogen) for 1 h. After washing with TBS, cells were mounted with mounting medium containing DAPI (Vector Laboratories, Burlingame, CA). Images were obtained under confocal microscopy (Leica SP5) at a magnification of 60×.
In Vitro Pulldown Assays
For glutathione S-transferase (GST) pulldown, GST-AMPKα (4 μg, H00005563-P01; R&D Systems) or GST (202039; GenScript) was incubated with 8 μL of glutathione beads (16100; ThermoFisher) in 200 μL of binding buffer (BB; 25 mmol/L HEPES [pH 7.3], 100 mmol/L NaCl, 10% glycerol, 1% Triton X-100, 1 mmol/L dithiothreitol, 0.25 mmol/L phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [Calbiochem]) in the presence of 0.1% BSA at 4°C for 6 h. The beads were then washed with cold BB once, followed by incubation with 2 μg of FL-DUSP27 (TP314361; OriGene) in 200 μL of BB (without BSA) at 4°C overnight. The next day, beads were subjected to quick wash with BB once, slow wash (by rotating the tube at 4°C for 3 min) with medium-salt BB (final NaCl concentration to 200 mmol/L), and then high-salt BB (final NaCl concentration to 300 mmol/L) once each. Bound (pellet) and unbound (Sup) fractions were resolved on SDS-PAGE, followed by Western blot analysis.
For FLAG-DUSP27 pulldown, FL-DUSP27 (2.5 μg) or FLAG peptide (F3290-4MG; Sigma-Aldrich) was incubated with 8 μL of anti-FLAG magnetic agarose beads (M8823; Sigma-Aldrich) in 200 μL of BB in the presence of 0.1% BSA at 4°C for 6 h. The beads were then washed with cold BB once, followed by incubation with 1 μg of GST-AMPKα in 200 μL of BB (without BSA) at 4°C overnight. The next day, beads were subjected to quick wash with BB once and slow wash with medium-salt BB and then high-salt BB once each. Bound (pellet) and unbound (Sup) fractions were resolved on SDS-PAGE, followed by Western blot analysis.
Quantitative Methylation-Specific PCR
Genomic DNA was extracted from soleus muscle tissues from WT mice after exposure to RC or HFD using Quick-gDNA MicroPrep (D3021; Zymo Research). Fifteen microliters of elution buffer was used to elute DNA from each column. For bisulfite treatment, 200 ng of DNA was used for each column using the EZ DNA Methylation-Gold Kit (D5006; Zymo Research). Elution buffer (100 μL) was used to elute DNA from each column. RT-qPCR was performed in a 15-μL reaction containing 5 μL of the eluant using iQSYBRGreen (Bio-Rad) in a Bio-Rad iCycler. The PCR primers (Supplementary Table 2) for methylated NCTC1 were used at a final concentration of 0.6 μmol/L in each PCR reaction. PCR was performed by initial denaturation at 95°C for 5 min, followed by 40 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis. Albumin DNA was used as loading controls for all quantitative methylation-specific PCR (QMSP) normalizations.
Statistical Analysis
Statistical analyses and figure construction were performed using GraphPad Prism version 7.01 for Mac (www.graphpad.com; GraphPad Software, La Jolla, CA). A value of P < 0.05 was considered statistically significant. Statistical significance was determined by unpaired, two-tailed Student t test between two groups. One-way ANOVA, followed by Tukey multiple comparisons test, was used for comparisons between three groups. Nonparametric Spearman correlations were performed for gene correlation analyses. One asterisk represents P < 0.05, and two asterisks represent P < 0.01. Error bars are shown as mean ± SEM.
Results
Genetic Ablation of H19 Leads to Muscle Insulin Resistance
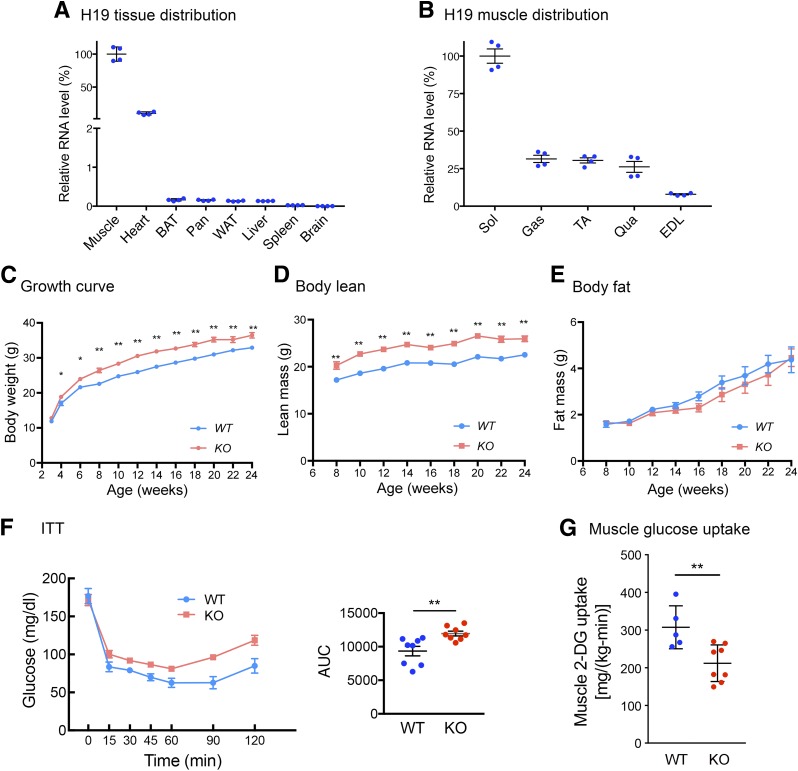
H19 is highly expressed in skeletal muscle (Fig. 1A), confirming previous reports (9). We also observed that its expression is the highest in the oxidative fiber-enriched soleus muscle (Fig. 1B). As siRNA-mediated H19 knockdown impaired insulin-stimulated glucose uptake in cultured mouse myotubes (6), we tested whether H19 regulates muscle insulin sensitivity in vivo. To this end, we used a whole-body H19 KO mouse model that carries a targeted deletion of the entire H19 transcription unit (10). The KO mice exhibited an overgrowth phenotype (Fig. 1C), consistent with previous observations (10,11). Overgrowth could be attributed to an increase in the lean mass (Fig. 1D and E), which was not surprising because the KO mice exhibit skeletal muscle hyperplasia and hypertrophy (11). Yet, ITTs performed in ad libitum fed mice unexpectedly revealed decreased whole-body insulin sensitivity in the KO mice (Fig. 1F). To assess sites where insulin action was affected, hyperinsulinemic-euglycemic clamp studies after overnight fasting were carried out. Results showed that insulin-stimulated glucose uptake in muscles was significantly lower in the KO compared with the WT mice (Fig. 1G and Supplementary Fig. 1). Taken together with our previous observation that H19 knockdown by siRNA in myotubes impaired insulin-stimulated glucose uptake (6), we suggest that H19 is important for appropriate physiological regulation of insulin action in muscle.
Figure 1.
H19 deletion impairs muscle insulin sensitivity. A: Results of RT-qPCR analysis of H19 expression in WT mice (n = 4). The H19 level in gastrocnemius muscle is arbitrarily set as 100%. BAT, brown adipose tissue; Pan, pancreas, WAT, white adipose tissue. B: H19 expression in various muscle types from WT mice (n = 4). EDL, estensor digitorum; Gas, gastrocnemius; Qua, quadriceps; Sol, soleus; TA, tibialis anterior. C: Growth curve of WT and KO mice. Each growth point represents n = 15–20 mice. D and E: Body composition as assessed by EcoMRI. Each point represents n = 15–20 mice. F: ITT in ad libitum fed WT (n = 8) and KO (n = 8) mice, with quantification on the right. AUC, area under curve. G: Insulin-stimulated glucose uptake in gastrocnemius and soleus muscles during hyperinsulinemic-euglycemic clamp studies. n = 5–8 animals in each genotype. All data represent mean ± SEM. C–G: *P < 0.05 and **P < 0.01, compared with WT mice.
AMPK Activity Is Decreased in KO Muscle
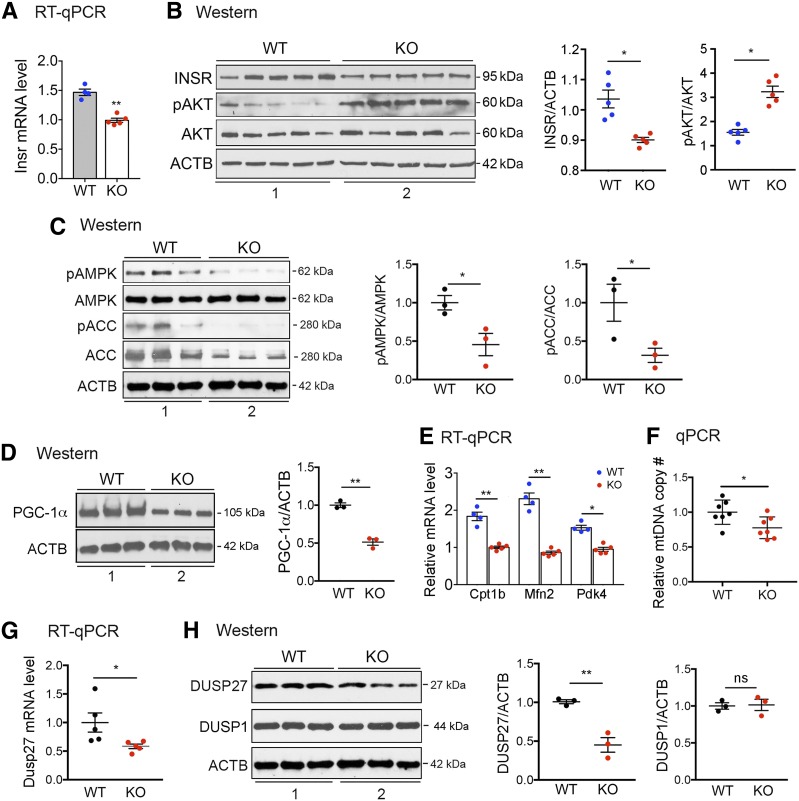
To begin to elucidate the molecular mechanism underlying the insulin resistance observed in the H19 KO muscle, we first tested whether the insulin signaling pathway is affected. Binding of insulin to insulin receptor (INSR) initiates a cascade of phosphorylation events, leading to phosphorylation and activation of AKT, which mediates most of insulin’s metabolic effects, including increasing glucose uptake in muscle (reviewed in Boucher et al. [12]). Thus, INSR expression and AKT phosphorylation at Ser473 (phosphorylation at this site is required for full activation of AKT [12]) were examined in WT and KO muscles. We observed decreased INSR expression at both mRNA (Fig. 2A) and protein levels (Fig. 2B, top blot in left panel [compare lane 2 to lane 1], and middle panel) in KO as compared with WT muscles. This is consistent with the previously reported mechanism of H19-dependent regulation of INSR expression in both human and rodent muscles (6). Unexpectedly, we observed an increase in AKT phosphorylation in KO muscle (Fig. 2B, middle two blots in left panel [compare lane 2 to lane 1], and right panel). Given that an increase in the basal level of AKT-dependent insulin signaling was similarly reported in HFD-induced insulin-resistant mouse muscle (13), we speculate that the basal AKT phosphorylation increase in insulin-resistant muscle might serve as a compensatory mechanism that warrants future investigation.
Figure 2.
H19 deletion leads to decreased AMPK activity and DUSP27 expression in muscle. A: RT-qPCR results of INSR mRNA from WT and KO muscle. n = 4–5 animals in each genotype. B: Representative Western blots (left) and densitometry (middle and right) results for INSR, phosphorylation of AKT at Ser473 (pAKT), total AKT (AKT), and β-actin (ACTB, as a loading control) in muscle samples from WT and KO mice fed ad libitum. Muscle extracts from five mice from each genotype were loaded. C: Representative Western blots (left) and densitometry (middle and right) results for phosphorylation of AMPKα at Thr172 (pAMPK), total AMPKα (AMPK), phosphorylation of ACC at Ser79 (pACC), total ACC (ACC), and ACTB in muscle samples from WT and KO mice fed ad libitum. Muscle extracts from three mice from each genotype were loaded. D: Representative Western blots (left) and densitometry (right) results of PGC-1α protein. Muscle extracts from three mice from each genotype were loaded. E: RT-qPCR results of indicated mRNAs from WT and KO muscle. n = 4–5 animals in each genotype. F: Relative mitochondrial DNA (mtDNA) copy numbers from WT and KO muscle. n = 7 animals in each genotype. G: RT-qPCR results of DUSP27 mRNA from WT and KO muscle. n = 5 animals in each genotype. H: Representative Western blots (left) and densitometry (middle and right) results of DUSP27 and DUSP1 proteins. All data are shown as mean ± SEM. *P < 0.05 and **P < 0.01, compared with WT mice.
Next, we tested whether AMPK activity is altered in KO muscle, as chronic muscle-specific AMPK inactivation exacerbates the development of diet-induced muscle insulin resistance (14) and constitutive muscle-specific AMPK activation enhances insulin sensitivity (15). Thus, we performed Western blot analysis to assess phosphorylation of AMPKα at Thr172 in WT and KO muscles. The phosphorylation was significantly decreased in KO as compared with WT muscle (Fig. 2C, top two blots in left panel [compare lane 2 to lane 1], and middle panel). As expected, phosphorylation of ACC at Ser79 was also decreased (Fig. 2C, left and right panels). The apparent decrease in ACC total protein level (Fig. 2C, second blot from bottom in left panel, compare lane 2 to lane 1) was not surprising because an increase in both ACC protein abundance and Ser79 phosphorylation was reported in mice with chronic muscle-specific AMPK activation (15). As decreased (or increased) ACC phosphorylation would inhibit (or enhance) lipid oxidation, the total ACC protein level decrease (or increase) may reflect a failed compensatory mechanism.
It was previously shown that mice with phosphodefective ACC1 and/or ACC2 (the mutated enzymes hence were not sensitive to AMPK-dependent phosphorylation) had decreased fatty acid oxidation, as indicated by increased levels of triacylglycerol (TAG) and diacylglycerol in muscle (16,17). To assess whether altered ACC phosphorylation/expression in H19 KO muscle affects lipid accumulation, muscle TAG contents were measured, which showed no significant difference between the WT and KO animals (Supplementary Fig. 2). Our results highlight the complexity of an ACC-mediated mechanism of lipid metabolism regulation. Indeed, one study reported that global ACC2 KO mice had increased fatty acid oxidation and muscle insulin sensitivity in addition to reduced fat mass (18), whereas another study showed no change in fatty acid oxidation and little effect on body weight and fat mass in muscle-specific ACC2 KO mice (19). Further, the latest research findings have demonstrated that not all muscle insulin resistance shows increases in total intramuscular lipids and/or lipid intermediates (20–22). Future studies are certainly needed to determine how decreased AMPK activity in H19 KO muscle affects lipid metabolism–mediated ACC.
As an additional readout for AMPK activity, Western blot analysis was performed on PGC-1α and showed decreased protein level in KO muscle (Fig. 2D). This is in agreement with the well-established role of AMPK in promoting PGC-1α expression via the PGC-1α autoregulatory pathway (4,5). Further, a decrease in expression of PGC-1α targets, Cpt1b, Mfn2, and Pdk4 (Fig. 2E), as well as in mitochondrial DNA contents (Fig. 2F), was evident in the KO muscle, consistent with PGC-1α’s role in stimulating mitochondrial biogenesis and fat oxidation. Based on the finding that H19 KO muscle exhibits both decreased ACC phosphorylation and decreased expression of PGC-1α, we conclude that H19 positively affects AMPK activity in muscle.
DUSP27 Expression Is Downregulated in KO Muscle
To delineate the molecular mechanism linking H19 to AMPK activation, we analyzed gene expression profiles of muscles from WT and KO mice by RNA-Seq. We noted a recent report that in human cardiac fibroblast cells, H19 activates ERK by regulating expression of DUSP5 (although the mechanism by which H19 regulates DUSP5 was not defined) (23). This was intriguing because ERK inactivates AMPK by phosphorylating AMPKα at Ser487/491 (3). Therefore, we first examined expression and phosphorylation of ERK in WT and KO muscle but did not find any significant difference between the two groups (data not shown), suggesting that ERK is likely not involved in AMPK inactivation in H19 KO muscle. Next, we turned our attention to DUSP27 (also called DUPD1) (24), as our RNA-Seq analysis revealed a significant decrease in its expression in KO versus WT muscles, whereas no significant difference was detected in other DUSPs, including DUSP1 (Supplementary Table 1 and GEO accession no. GSE103202). The DUSP27 decrease was confirmed by RT-qPCR (Fig. 2G) and Western blotting (Fig. 2H).
DUSP27 is evolutionarily conserved; the human DUSP27 protein shares 87%, 86%, and 81% identity with mouse, rat, and dog, respectively (24). DUSP27 is a member of a heterogeneous group of protein phosphatases capable of dephosphorylating both phosphotyrosine and phosphoserine/phosphothreonine residues within the same substrate (25). DUSPs are divided into six subgroups that include mitogen-activated protein kinase phosphatases (MKPs) and atypical DUSPs (25). DUSP27 falls in the atypical category (24) and has been shown to act as a phosphatase to deactivate ERK and p38 MAPKs in ovarian cells (26). Notably, in mice, DUSP27 is expressed highly in skeletal muscle, fat, and liver (24), whereas in humans, its mRNA is detected only in skeletal muscle (http://www.proteinatlas.org/ENSG00000188716-DUPD1/tissue). Although the physiological significance of this striking tissue-specific expression is unclear, its decreased expression in H19 KO muscle suggests a regulation of DUSP27 by H19.
H19 Regulates DUSP27 Expression at the Posttranscriptional Level
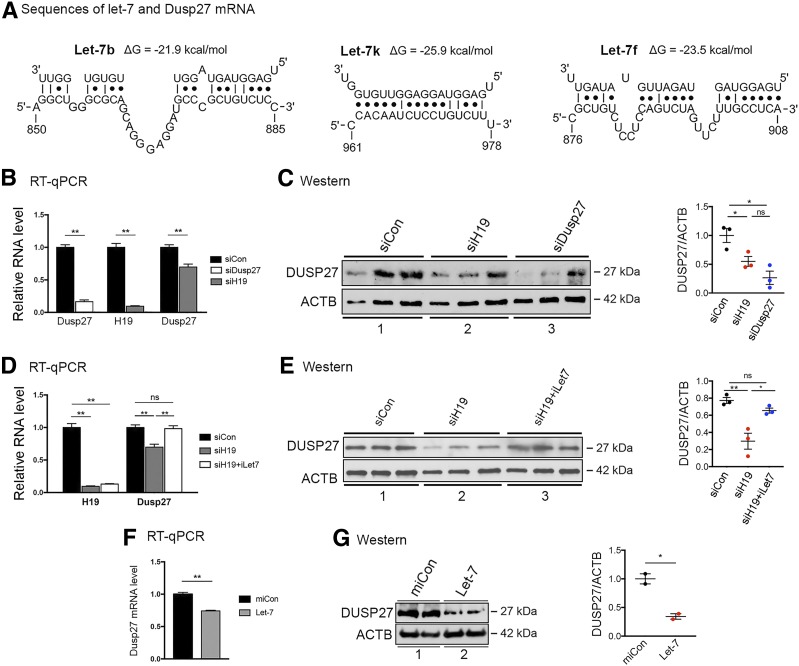
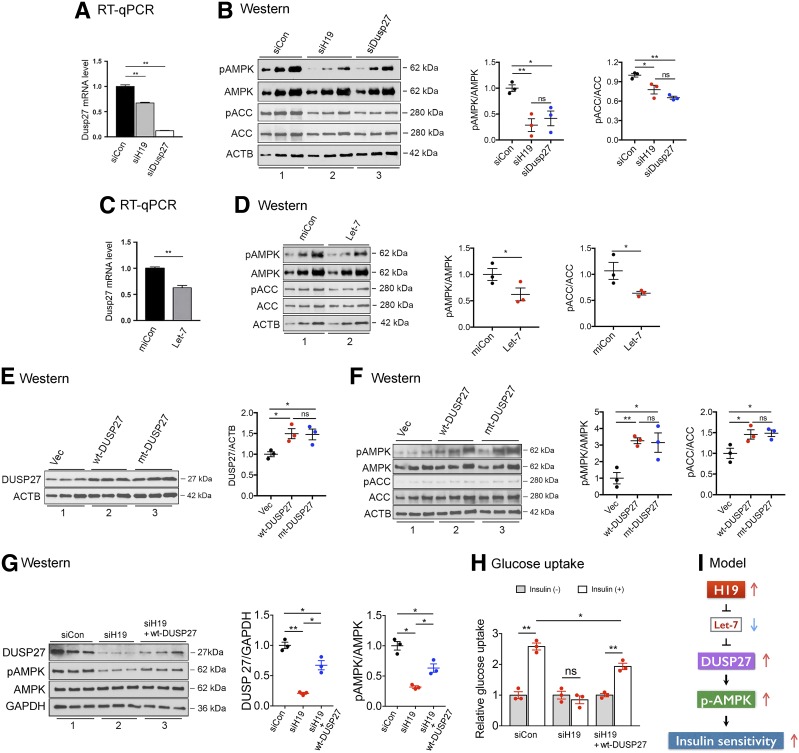
We have previously reported the H19/let-7 axis where H19 functions to reduce the bioavailability of microRNA let-7 by acting as a molecular sponge. H19 contains six let-7–binding sites that sequester let-7 and prevent it from binding to target mRNAs (27). Thus, it is not the absolute amounts of let-7 but rather the relative abundance between H19 and let-7 that determines the expression level of genes regulated by let-7. Let-7 inhibits target gene expression by binding to complementary sequences in the mRNA, leading to translational repression and/or mRNA degradation. Therefore, we performed bioinformatics analysis and predicted three let-7–binding sites in the 3′-UTR of DUSP27 mRNA (Fig. 3A). To determine whether H19 regulates DUSP27 via the H19/let-7 axis, we first tested the effect of H19 siRNA knockdown on DUSP27 expression in mouse C2C12 myotubes (herein called myotubes). While knocking down DUSP27 (Fig. 3B, left column, compare white bar to black bar) expectedly reduced its protein level (Fig. 3C, top blots in left panel, compare lane 3 to lane 1; right panel, compare blue dots to black dots), downregulation of H19 (Fig. 3B, middle column, compare gray bar to black bar) also decreased DUSP27 expression at both the mRNA (Fig. 3B, right column, compare gray bar to black bar) and protein levels (Fig. 3C, top blots in left panel, compare lane 2 to lane 1; right panel, compare red dots to black dots), suggesting that H19 positively regulates DUSP27 expression. Next, we tested whether this regulation is mediated by let-7. We have previously documented in other cell types that H19 knockdown increases the bioavailability of let-7, but in the presence of iLet7 (a let-7–specific inhibitor), the effects of H19 downregulation (i.e., let-7 release and target gene inhibition) were attenuated (6,28–30). As expected, in myotutes, H19 knockdown (Fig. 3D, left column, compare gray bar to black bar) led to a decrease in DUSP27 expression at the levels of both mRNA (Fig. 3D, right column, compare gray bar to black bar) and protein (Fig. 3E, top blots in left panel, compare lane 2 to lane 1; right panel, compare red dots to black dots). However, when H19 was knocked down in the presence of iLet7 (Fig. 3D, left column, compare white bar to black bar), there was no longer a decrease in DUSP27 mRNA (Fig. 3D, right column, compare white bar to black bar) or protein (Fig. 3E). Further, when let-7 was transfected, it suppressed DUSP27 expression at both the mRNA (Fig. 3F) and protein levels (Fig. 3G). Taken together, these results suggest that in muscle cells, H19 posttranscriptionally regulates DUSP27 expression by reducing the bioavailability of let-7.
Figure 3.
H19 regulates DUSP27 expression via the H19/let-7 axis. A: Bioinformatics predicted three let-7 binding sites at the 3′-UTR of mouse DUSP27 mRNA. Sequences of three let-7 subtypes (top strands) and partial sequences of DUSP27 mRNA (bottom strands) are shown. Numbers are in nucleotides relative to the transcriptional start site of DUSP27. B: RT-qPCR results of DUSP27 and H19 expression in myotubes 48 h after transfection of siCon, siH19, or DUSP27-specific siRNA (siDusp27). C: Western blots (left) and densitometry results (right) of DUSP27 and ACTB proteins from experiments shown in B. Lanes were loaded in increasing amounts of cell lysates. D: RT-qPCR results of H19 and DUSP27 expression in myotubes 48 h after transfection of siCon, siH19, and siH19 plus iLet-7. E: Western blots (left) and densitometry (right) results of DUSP27 and ACTB protein levels from experiments shown in D. F: RT-qPCR results of DUSP27 mRNA levels in myotubes 24 h after transfection of miCon (negative control miRNA) or let-7. Lanes were loaded in duplicate. G: Western blots (left) and densitometry (right) results of DUSP27 and ACTB protein levels from experiments shown in F. All data are shown as mean ± SEM (n = 3). *P < 0.05; **P < 0.01. ns, no statistical difference.
DUSP27 Interacts With AMPKα
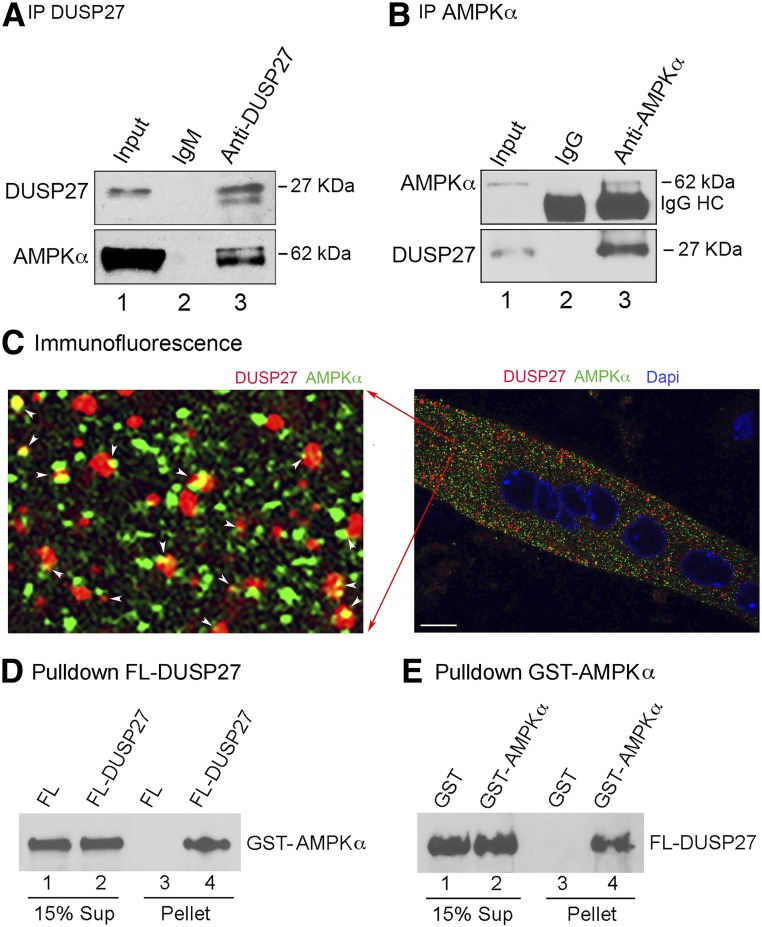
To determine whether DUSP27 mediates, at least in part, the H19-dependent regulation of AMPK, coimmunoprecipitation (co-IP) experiments were performed using extracts from WT muscle. The DUSP27 antibody (anti-DUSP27) was able to pull down DUSP27 (Fig. 4A, top blot, lane 3), together with AMPKα (bottom blot, lane 3), whereas the preimmune IgM control antibody did not (lane 2). In reciprocal experiments, co-IP using an AMPKα-specific antibody (31) pulled down AMPKα (Fig. 4B, top blot, lane 3), together with DUSP27 (bottom blot, lane 3). Next, immunofluorescence was performed on myotubes using the same anti-AMPKα antibody whose specificity had been previously validated for both IP (31) and immunofluorescence (32). Results showed that a small fraction of DUSP27 colocalizes with AMPKα in the cytoplasm (Fig. 4C, left panel, yellow dots indicated by white arrowheads). The observation that not all DUSP27 and AMPKα colocalize with each other is consistent with AMPKα being a multifunctional protein known to have many interacting partners. Our results indicate that an appreciable fraction of DUSP27 is in complex with AMPKα. To determine whether a direct protein-protein interaction is involved, in vitro pulldown experiments using purified recombinant proteins were conducted. As seen in Fig. 4D, FLAG-tagged DUSP27 (FL-DUSP27) was able to pull down GST-AMPKα (lane 4), whereas FLAG peptide alone did not (lane 3). On the other hand, when GST-AMPKα (but not GST alone) was used as a bait, it pulled down FL-DUSP27 (Fig. 4E, compare lane 4 to lane 3). Collectively, our results identify DUSP27 as a new interacting partner of AMPKα.
Figure 4.
DUSP27 interacts with AMPKα. A: Mouse gastrocnemius muscle extract was immunoprecipitated with rabbit IgM DUSP27 antibody (preimmune rabbit IgM as a negative control). Representative Western blot results are shown; 3% of input was loaded. B: Mouse soleus muscle extract was immunoprecipitated with rabbit AMPKα antibody (preimmune rabbit IgG as a negative control). Representative Western blot results are shown. HC, IgG heavy chain. C: Representative immunofluorescence image of a myotube with DUSP27 stained red, AMPKα green, and nuclei blue. Arrowheads indicate colocalization of DUSP27 and AMPKa. Original magnification ×60. Scale bar, 10 μm. D: Purified recombinant protein FL-DUSP27 (or FLAG peptide as a negative control) prebound on anti-FLAG beads was incubated with purified recombinant protein GST-AMPKα. Bound (pellet) and unbound (Sup) fractions were resolved on SDS-PAGE, followed by Western blot analysis using the anti-AMPKα antibody. E: Purified GST-AMPKα (or GST as a negative control) prebound on glutathione beads was incubated with FL-DUSP27. Bound and unbound fractions were resolved on SDS-PAGE, followed by Western blot analysis using the anti-DUSP27 antibody.
DUSP27 Contributes to H19-Dependent AMPK Activation
To determine whether DUSP27 affects AMPK activity, DUSP27 was knocked down in myotubes, followed by Western blot analysis. Downregulation of DUSP27 (Fig. 5A, compare white bar to black bar) decreased AMPK phosphorylation (Fig. 5B, top blots in left panel, compare lane 3 to lane 1; middle panel, compare blue dots to black dots), as well as phosphorylation of ACC (Fig. 5B, left and right panels), suggesting that DUSP27 positively affects AMPK activity. Next, when DUSP27 expression was reduced as a result of H19 downregulation (Fig. 5A, compare gray bar to black bar), decreased AMPK phosphorylation (Fig. 5B, left and middle panels) accompanied by decreased phosphorylation of ACC (Fig. 5B, left and right panels) was observed. Further, when DUSP27 expression was inhibited by let-7 (Fig. 5C), there was a decrease in phosphorylation of both AMPK (Fig. 5D, left and middle panels) and ACC (Fig. 5D, left and right panels). To determine whether increasing DUSP27 affects AMPK activity, plasmids expressing WT DUSP27 (wt-DUSP27) or mutant DUSP27 (mt-DUSP27, in which the catalytically critical Cys-146 was replaced by Ser) were transfected into myotubes. Western blot analysis showed that increasing either wt-DUSP27 (Fig. 5F, top blot in left panel, compare lane 2 to lane 1; right panel, compare red dots to black dots) or mt-DUSP27 (Fig. 5E, top blot in left panel, compare lane 3 to lane 1; right panel, compare blue dots to black dots) was able to increase phosphorylation of both AMPK and ACC (Fig. 5F). These results suggest that DUSP27 likely acts as a cofactor for AMPK and that the catalytic activity of DUSP27 does not appear to be required for this regulation. To determine whether DUSP27 contributes to H19-mediated AMPK activation, H19 knockdown in combination with exogeneous DUSP27 expression experiments were conducted. As expected, H19 knockdown decreased DUSP27 expression (Fig. 5G, left panel, top blot, compare lane 2 to lane 1; middle panel, compare red dots to black dots) and AMPK phosphorylation (left and right panels). Exogeneous expression of DUSP27 with H19 knockdown (left panel, top blot, compare lane 3 to lanes 2 and 1, and middle panel) partially restored AMPK phosphorylation to the control level (left and right panels). Glucose uptake assays showed that H19 knockdown impaired insulin-stimulated glucose uptake (Fig. 5H, compare middle column to left column), whereas exogeneous DUSP27 expression partially restored insulin-stimulated glucose uptake to the control level (right column). The lack of full restoration of AMPK phosphorylation and glucose uptake to control levels likely reflected the lack of full restoration of DUSP27 expression, as the balance between H19 siRNA knockdown efficiency and DUSP27 overexpression without compromising cell viability had limited the amount of DUSP27 to be transfected. Taken together, these results support the notion that the H19/DUSP27/AMPK axis regulates muscle insulin sensitivity in a cell-autonomous manner (Fig. 5I).
Figure 5.
DUSP27 contributes to H19-dependent modulation of AMPK activity. A: RT-qPCR results of DUSP27 expression in myotubes 48 h after transfection of siCon, siH19, or DUSP27-specific siRNA (siDusp27). B: Western blots and densitometry results of phosphorylation of AMPK (pAMPK), AMPK, pACC, ACC, and ACTB proteins from experiments shown in A. Lanes were loaded in increasing amounts of proteins. C: RT-qPCR results of DUSP27 expression in myotubes 24 h after transfection of miCon (negative control miRNA) or let-7. D: Western blots and densitometry results of pAMPK, AMPK, pACC, ACC, and ACTB proteins from experiments shown in C. E and F: Myotubes were transfected with empty vector (Vec), wt-DUSP27, or mt-DUSP27, followed by Western blot analysis 48 h later. Western blots and densitometry results of DUSP27, pAMPK, AMPK, pACC, ACC, and ACTB proteins are shown. G: Myotubes were transfected with siCon, siH19, or siH19 plus wt-DUSP27, followed by Western blot analysis 48 h later. Western blots and densitometry results of DUSP27, pAMPK, AMPK, and GAPDH proteins are shown. H: Glucose uptake of myotubes from experiments shown in G. Results are presented as relative glucose uptake, with values in absence of insulin stimulation set as 1. I: A proposed model of H19-dependent regulation of AMPK activity. All data are shown as mean ± SEM (n = 3). *P < 0.05; **P < 0.01. ns, no statistical difference.
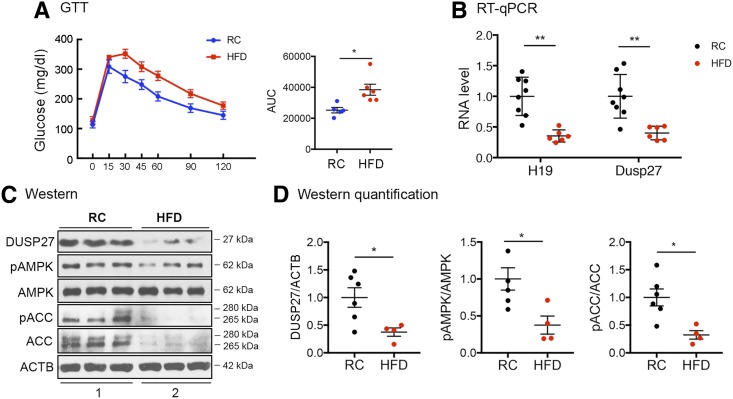
DUSP27 Is Downregulated in Muscle of HFD-Induced Glucose-Intolerant Mice
Our previous studies showed that H19 is downregulated in muscle of insulin-resistant patients and rodents (6). As DUSP27 is regulated by H19 (Fig. 3) and as its expression is decreased in KO muscle (Fig. 2G and H), we asked whether DUSP27 expression is altered in the muscle of HFD-induced glucose-intolerant mice (Fig. 6A). The expression of H19 was expectedly decreased in the HFD as compared with RC muscle (Fig. 6B, left column), and the expression of DUSP27 was also reduced at levels of both mRNA (Fig. 6B, right column) and protein (Fig. 6C, top blot, compare lane 2 to lane 1, and Fig. 6D, left panel). This was accompanied by a decrease in phosphorylation of both AMPK (Fig. 6C and middle panel in Fig. 6D) and ACC (Fig. 6C and right panel in Fig. 6D). The decreased ACC total protein level in the HFD muscle is consistent with our observation in H19 KO muscle (Fig. 2C), possibly reflecting a failed compensatory mechanism. Collectively, our results further support that the H19/DUSP27/AMPK signaling pathway contributes to muscle insulin sensitivity regulation.
Figure 6.
HFD-induced downregulation of H19, DUSP27, and AMPK signaling in muscle. A: Results of GTTs of WT mice after 13-week exposure to RC or HFD. B: RT-qPCR results of H19 and DUSP27 expression from RC and HFD muscles. C: Representative Western blots of DUSP27, phosphorylation of AMPK (pAMPK), AMPK, pACC, ACC, and ACTB proteins from experiments shown in B. Extracts from three mice were loaded in each group. D: Quantification results of Western blots. All data are shown as mean ± SEM of four to five animals per group. *P < 0.05 and **P < 0.01, compared with the RC group. AUC, area under the curve.
H19 Downregulation Involves Both Posttranscriptional and Epigenetic Mechanisms
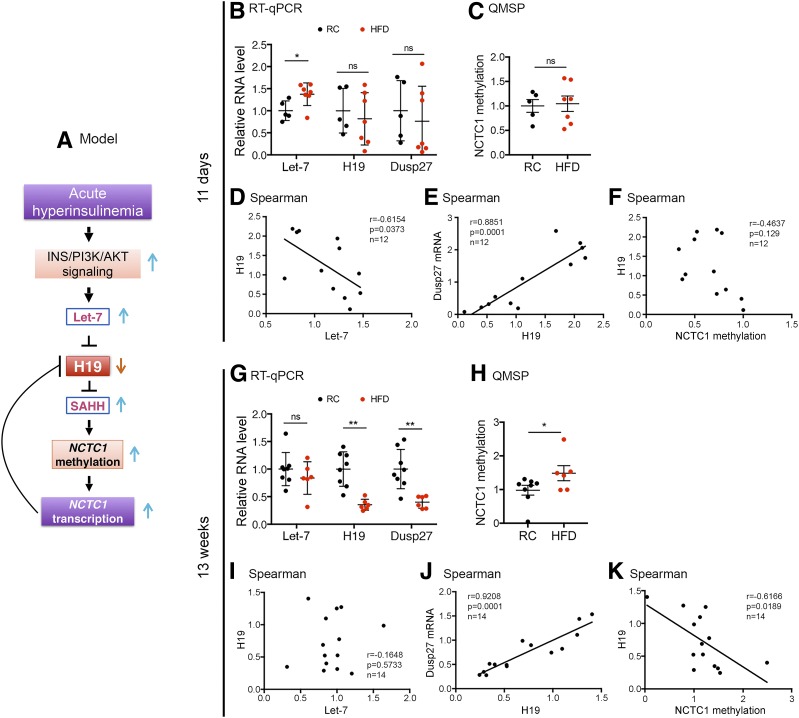
So far, our results strongly suggest that H19 plays an important role in muscle insulin sensitivity regulation, at least in part, by activating AMPK via the putative cofactor DUSP27. An important question remains: what induces and maintains H19 downregulation in the muscle of humans and rodents with diabetes? Addressing this question has an important clinical implication as it will help to identify potential molecular targets for the prevention and treatment of insulin resistance. Our previous work showed that in nondiabetic mice, acute hyperinsulinemia upregulates let-7 via activation of the insulin-PI3K-AKT pathway in skeletal muscle; let-7 binds to H19 and induces H19 degradation, thereby temporally reducing H19 levels (6) (Fig. 7A). The system then corrects itself to restore H19 to normal levels at later time points after acute hyperinsulinemia. This regulation thus appears to serve as a protective mechanism in individuals without diabetes to prevent muscle from overusing circulating glucose, which otherwise would be toxic to the muscle (6). Importantly, the high-dose insulin–induced let-7 increase and H19 decrease are dependent on intact insulin signaling, as such effects were no longer observed in HFD muscle (6). We also showed that H19 functions to modulate DNA methylation by inhibiting S-adenosylhomocysteine hydrolase (SAHH) (the H19/SAHH axis) (33,34). Specifically, in mouse myotubes, H19 siRNA knockdown activates SAHH (H19 normally binds to SAHH and inhibits its enzymatic activity), leading to increases in gene body methylation and subsequently increased transcription of NCTC1, a lncRNA-encoding gene located 20 kb downstream of H19 (33) (Fig. 7A). This NCTC1 transcription increase leads to inhibition of H19 expression via a mechanism involving promoter competition (the NCTC1 promoter recruits RNA polymerase II for its own transcription at the expense of H19 transcription) (33,35). Taken together, these previous findings led us to hypothesize that the pathway illustrated in Fig. 7A may contribute to the underlying mechanism of H19 repression in diabetic muscles.
Figure 7.
Mechanisms of H19 downregulation in diabetic muscle. A: A proposed model. Short-term (B–F) and long-term (G–K) effects of HFD on gene expression and NCTC1 methylation. B and G: RT-qPCR results of let-7, H19, and DUSP27 RNA from RC and HFD muscles. C and H: QMSP results of NCTC1 gene body methylation of RC and HFD muscles. D–F and I–K: Spearman correlations with P values and total animal numbers (RC plus HFD) marked. B, C, G, and H: Data are shown as mean ± SEM of five to eight animals per group. *P < 0.05 and **P < 0.01, compared with the RC group. ns, no statistical significance.
To further explore this model, we examined the expression of let-7, H19, and DUSP27 as well as NCTC1 gene body methylation in muscles of mice after 11-day (prediabetes) and 13-week (chronic diabetes) exposure to RC or HFD, respectively. These time points were chosen based on a previous study showing that HFD-fed mice started to exhibit glucose intolerance (as determined by GTTs) at day 3, developed hepatic insulin resistance (as determined by hyperinsulinemic-euglycemic clamp studies) at day 7, and developed muscle insulin resistance at day 21 (36). After 11 days of HFD feeding, the mice started to show increased let-7 expression (Fig. 7B, left column), likely as a result of hyperinsulinemia-induced let-7 upregulation. Importantly, there was a statistically significant negative correlation between let-7 and H19 levels (Fig. 7D). A significantly positive correlation between H19 and DUSP27 was also observed (Fig. 7E), underscoring an in vivo regulation of DUSP27 by H19. The apparent heterogeneity in the levels of let-7, H19, and DUSP27 among the individual mice highlights their differential responses to HFD exposure during early stages of diabetic development. The lack of difference in the average levels of H19 (Fig. 7B, middle column) and DUSP27 (Fig. 7B, right column) between the two groups likely reflected self-correction after hyperinsulinemia assaults at early stages of diabetes (6). When NCTC1 methylation was assessed using QMSP (33), no significant differences were detected between the two groups (Fig. 7C), nor was there a statistically significant correlation between NCTC1 methylation and H19 expression (Fig. 7F). Together, these results suggest a mechanism of let-7–mediated downregulation of H19 in the early-stage diabetic muscle.
When muscles were analyzed after 13 weeks of RC or HFD exposure, there was no longer a difference in let-7 levels between the two groups (Fig. 7G, left column), although the levels of both H19 (middle column) and DUSP27 (right column) remained significantly lower in the HFD group than in the RC group. The lack of let-7 difference between the two groups was consistent with impaired insulin signaling in the HFD group, as intact insulin signaling is required for let-7 upregulation (6). Moreover, whereas a negative correlation between let-7 and H19 was lost (Fig. 7I), a positive correlation between H19 and DUSP27 persisted (Fig. 7J), again consistent with an in vivo regulation of DUSP27 by H19. Remarkably, an increase in NCTC1 methylation (Fig. 7H), as well as a negative correlation between NCTC1 methylation and H19 expression (Fig. 7K), became evident, supporting the hypothesis that chronic hyperinsulinemia (a hallmark of T2D) leads to a stable increase in muscle NCTC1 gene body methylation and transcription, which in turn leads to decreased H19 transcription via promoter competition.
Discussion
This work identifies H19 lncRNA as a novel modulator of AMPK activity, with DUSP27 being an important downstream effector of H19 in muscle insulin sensitivity regulation. We show that DUSP27 is regulated by H19 at the posttranscriptional level and contributes, at least in part, to the ability of H19 to enhance AMPK activity. The demonstration that both H19 and DUSP27 expression are responsive to dietary cues and can influence signal transduction pathways related to T2D highlights their potential physiological importance in the regulation of systemic glucose metabolism. Further, our exploration of underlying mechanisms leading to H19 and DUSP27 reduction in diabetic muscles holds the promise of discovering new therapeutic targets for T2D and other metabolic diseases. Although we cannot exclude the possibility that tissues other than muscle may contribute to the observed in vivo effects (Fig. 1) due to the use of whole-body KO mice, our results from both cell and animal models together strongly point to the H19/DUSP27/AMPK pathway as a novel contributing mechanism of muscle insulin sensitivity regulation.
Notably, we observed a modest increase in Igf2 expression in the H19 KO muscle (data not shown), consistent with previous reports (10,11). However, it is very unlikely that this Igf2 increase contributes to the muscle insulin resistance seen in the H19 KO mice because it should exert effects opposite to those observed. Indeed, it has been well documented that Igf2 binds to INSR and activates the insulin-PI3K-mTOR signaling pathway in muscle to increase glucose uptake both in mice (37) and humans (38–40).
Based on our genome-wide transcriptome studies revealing extensive gene expression changes in KO versus WT muscle (GEO accession no. GSE103202) and on H19 being a multifunctional lncRNA, we are certain that DUSP27 is not the only downstream effector of H19. Nonetheless, DUSP27 likely plays an important part in the H19-mediated regulation of AMPK activity for the following reasons. First, both H19 and DUSP27 are evolutionarily conserved and both are highly (and almost exclusively) expressed in human and mouse skeletal muscle. Second, there is a strong in vivo positive correlation of expression between H19 and DUSP27 in the context of KO muscle (Fig. 2G and H) and prediabetic and chronic diabetic muscle (Fig. 7E and J). Third, H19 siRNA knockdown in vitro leads to decreased DUSP27 expression via the H19/let-7 axis (Fig. 3B–G). Fourth, siRNA knockdown of either H19 or DUSP27 alone is sufficient to decrease AMPK signaling (Fig. 5A and B). Finally, exogenous expression of DUSP27 under the condition of H19 knockdown led to partial restoration of AMPK phosphorylation and insulin-stimulated glucose uptake to control levels (Fig. 5G and H). Collectively, our study supports a role of DUSP27 in linking H19 to AMPK activation and glucose uptake. Future studies using KO and transgenic DUSP27 mouse models will be necessary to firmly establish the physiological role of DUSP27 in H19-mediated muscle insulin sensitivity regulation.
DUSPs have been implicated as major modulators of critical signaling pathways that are dysregulated in various diseases (25). Unlike many DUSPs that have been extensively studied, little is known about the regulation of expression, substrate specificity, mode of action, and especially the biological and physiological functions of DUSP27 (24), except for one study showing that DUSP27 can dephosphorylate and deactivate ERK and p38 MAPKs in ovarian cells (26). In the current study, we identify DUSP27 as a target of the H19/let-7–mediated posttranscriptional regulation and AMPK as a potentially physiologically relevant target of DUSP27. Although we show evidence that DUSP27 and AMPK interact, the exact mode of action of DUSP27 in AMPK regulation is unclear, which warrants future investigation.
Despite the identification of numerous lncRNAs in the human genome, the physiological role of the vast majority has remained elusive. In the mouse, a number of lncRNAs have been shown to play important roles in metabolism (41–47). For example, the liver-specific triglyceride regulator (lncLSTR) regulates systemic lipid homeostasis by inhibiting triglyceride uptake into adipose tissues and skeletal muscle (42). The fasting-induced liver glucokinase (GCK) repressor (LncLGR) inhibits hepatic glycogen synthesis and lipogenesis by downregulating transcription of GCK (43). The brown fat lncRNA1 (Blnc1) forms ribonucleoprotein complexes with transcription factor EBF2 to stimulate thermogenic gene expression, thereby promoting thermogenic adipocyte differentiation (44). Finally, the dietary-inducible liver LeXis regulates cholesterol biosynthetic gene expression through interaction with the heterogeneous ribonucleoprotein Raly (47). Notably, all these lncRNAs (lncLSTR, LncLGR, Blnc1, and LeXis) are nuclear and function by forming ribonucleoprotein complexes to affect gene expression at the transcriptional level, albeit with distinct modes of action. Although no human counterparts have yet been found, the identification and characterization of these mouse lncRNAs have shed critical light on our understanding of mechanisms of how lncRNAs impact metabolism. Our findings are of particular importance to the field for the following reasons. First, they represent the first example of an evolutionarily conserved, abundantly expressed lncRNA in muscle metabolic regulation. Second, the regulation of AMPK, an evolutionarily conserved key metabolic fuel gauge, by an lncRNA has not been previously documented. Finally, the way by which H19 regulates AMPK is unique in that it involves a little-studied dual-specificity protein phosphatase DUSP27, which is highly expressed in skeletal muscle with previously unknown function.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Luisa Dandolo for providing the H19 KO mice and the Yale Mouse Metabolic Phenotyping Center In Vivo Metabolism Core for performing hyperinsulinemic-euglycemic clamp studies and measurements of muscle TAG contents.
Funding. This work was supported by the American Diabetes Association (1-15-BS-084), the National Institutes of Health (R01-HD-072418 to G.G.C.), the National Natural Science Foundation of China (81402130 to D.L.), and the Albert Mckern Foundation (GE001347 to Y.H.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. T.G. designed and performed the experiments, collected and analyzed the data, and wrote the manuscript. Y.L., Y.X., Y.J., and N.Z. performed the experiments and collected and analyzed the data. Z.W. performed the experiments and collected the data. G.G.C. and H.S.T. provided intellectual insights and critical discussion of the project. D.L. performed the experiments, collected and analyzed the data, and provided intellectual insights and critical discussion of the project. Y.H. designed the experiments and analyzed the data. All authors revised the manuscript critically for important intellectual content and read and approved the final version. Y.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-0370/-/DC1.
References
- 1.Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab 2009;9:407–416 [DOI] [PubMed] [Google Scholar]
- 2.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol 2015;33:1–7 [DOI] [PubMed] [Google Scholar]
- 3.Salminen A, Kaarniranta K, Kauppinen A. Age-related changes in AMPK activation: role for AMPK phosphatases and inhibitory phosphorylation by upstream signaling pathways. Ageing Res Rev 2016;28:15–26 [DOI] [PubMed] [Google Scholar]
- 4.Cantó C, Gerhart-Hines Z, Feige JN, et al. . AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009;458:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem 2010;47:69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y, Wu F, Zhou J, et al. . The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res 2014;42:13799–13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toda C, Kim JD, Impellizzeri D, Cuzzocrea S, Liu ZW, Diano S. UCP2 regulates mitochondrial fission and ventromedial nucleus control of glucose responsiveness. Cell 2016;164:872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurczak MJ, Lee AH, Jornayvaz FR, et al. . Dissociation of inositol-requiring enzyme (IRE1α)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem 2012;287:2558–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays 2010;32:473–480 [DOI] [PubMed] [Google Scholar]
- 10.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 1997;11:1596–1604 [DOI] [PubMed] [Google Scholar]
- 11.Martinet C, Monnier P, Louault Y, Benard M, Gabory A, Dandolo L. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development 2016;143:962–971 [DOI] [PubMed] [Google Scholar]
- 12.Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol 2014;6:a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu HY, Hong T, Wen GB, et al. . Increased basal level of Akt-dependent insulin signaling may be responsible for the development of insulin resistance. Am J Physiol Endocrinol Metab 2009;297:E898–E906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii N, Ho RC, Manabe Y, et al. . Ablation of AMP-activated protein kinase alpha2 activity exacerbates insulin resistance induced by high-fat feeding of mice. Diabetes 2008;57:2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schönke M, Myers MG Jr., Zierath JR, Björnholm M. Skeletal muscle AMP-activated protein kinase γ1(H151R) overexpression enhances whole body energy homeostasis and insulin sensitivity. Am J Physiol Endocrinol Metab 2015;309:E679–E690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullerton MD, Galic S, Marcinko K, et al. . Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med 2013;19:1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill HM, Lally JS, Galic S, et al. . AMPK phosphorylation of ACC2 is required for skeletal muscle fatty acid oxidation and insulin sensitivity in mice. Diabetologia 2014;57:1693–1702 [DOI] [PubMed] [Google Scholar]
- 18.Choi CS, Savage DB, Abu-Elheiga L, et al. . Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci U S A 2007;104:16480–16485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson DP, Pulinilkunnil T, Cline GW, Shulman GI, Lowell BB. Gene knockout of Acc2 has little effect on body weight, fat mass, or food intake. Proc Natl Acad Sci U S A 2010;107:7598–7603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodpaster BH, He J, Watkins S, Kelley DE. Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. J Clin Endocrinol Metab 2001;86:5755–5761 [DOI] [PubMed] [Google Scholar]
- 21.Kitessa SM, Abeywardena MY. Lipid-induced insulin resistance in skeletal muscle: the chase for the culprit goes from total intramuscular fat to lipid intermediates, and finally to species of lipid intermediates. Nutrients 2016;8:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brøns C, Grunnet LG. Mechanisms in endocrinology: skeletal muscle lipotoxicity in insulin resistance and type 2 diabetes: a causal mechanism or an innocent bystander? Eur J Endocrinol 2017;176:R67–R78 [DOI] [PubMed] [Google Scholar]
- 23.Tao H, Cao W, Yang JJ, et al. . Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc Pathol 2016;25:381–389 [DOI] [PubMed] [Google Scholar]
- 24.Friedberg I, Nika K, Tautz L, et al. . Identification and characterization of DUSP27, a novel dual-specific protein phosphatase. FEBS Lett 2007;581:2527–2533 [DOI] [PubMed] [Google Scholar]
- 25.Patterson KI, Brummer T, O’Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J 2009;418:475–489 [DOI] [PubMed] [Google Scholar]
- 26.Devi YS, Seibold AM, Shehu A, et al. . Inhibition of MAPK by prolactin signaling through the short form of its receptor in the ovary and decidua: involvement of a novel phosphatase. J Biol Chem 2011;286:7609–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallen AN, Zhou XB, Xu J, et al. . The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013;52:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan L, Zhou J, Gao Y, et al. . Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 2015;34:3076–3084 [DOI] [PubMed] [Google Scholar]
- 29.Ghazal S, McKinnon B, Zhou J, et al. . H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med 2015;7:996–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuckerwise L, Li J, Lu L, et al. . H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TβR3 in placentae with fetal growth restriction. Oncotarget 2016;7:38398–38407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou MH, Hou XY, Shi CM, et al. . Activation of 5′-AMP-activated kinase is mediated through c-Src and phosphoinositide 3-kinase activity during hypoxia-reoxygenation of bovine aortic endothelial cells. Role of peroxynitrite. J Biol Chem 2003;278:34003–34010 [DOI] [PubMed] [Google Scholar]
- 32.Ganesan R, Hos NJ, Gutierrez S, et al. . Salmonella typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy. PLoS Pathog 2017;13:e1006227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Yang L, Zhong T, et al. . H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun 2015;6:10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong T, Men Y, Lu L, et al. . Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene 2017;36:2345–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eun B, Sampley ML, Good AL, Gebert CM, Pfeifer K. Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus. Nucleic Acids Res 2013;41:817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner N, Kowalski GM, Leslie SJ, et al. . Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia 2013;56:1638–1648 [DOI] [PubMed] [Google Scholar]
- 37.Zhu H, Shyh-Chang N, Segrè AV, et al.; DIAGRAM Consortium; MAGIC Investigators . The Lin28/let-7 axis regulates glucose metabolism. Cell 2011;147:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chode S, Albert SG, Shoemaker JD, Green AL. Estimation of glucose utilization in a type 2 diabetes mellitus patient on insulin analogs with tumor hypoglycemia induced by IGF-II. Growth Horm IGF Res 2016;26:8–10 [DOI] [PubMed] [Google Scholar]
- 39.Bodnar TW, Acevedo MJ, Pietropaolo M. Management of non-islet-cell tumor hypoglycemia: a clinical review. J Clin Endocrinol Metab 2014;99:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dynkevich Y, Rother KI, Whitford I, et al. . Tumors, IGF-2, and hypoglycemia: insights from the clinic, the laboratory, and the historical archive. Endocr Rev 2013;34:798–826 [DOI] [PubMed] [Google Scholar]
- 41.Sun L, Goff LA, Trapnell C, et al. . Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A 2013;110:3387–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li P, Ruan X, Yang L, et al. . A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 2015;21:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan X, Li P, Cangelosi A, Yang L, Cao H. A long non-coding RNA, lncLGR, regulates hepatic glucokinase expression and glycogen storage during fasting. Cell Rep 2016;14:1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 2014;55:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Li P, Yang W, et al. . Integrative transcriptome analyses of metabolic responses in mice define pivotal LncRNA metabolic regulators. Cell Metab 2016;24:627–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato M, Wang M, Chen Z, et al. . An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 2016;7:12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallam T, Jones MC, Gilliland T, et al. . Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016;534:124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.