Abstract
Background
Identification of selection signatures can provide a direct insight into the mechanism of artificial selection and allow further disclosure of the candidate genes related to the animals’ phenotypic variation. Domestication and subsequent long-time selection have resulted in extensive phenotypic changes in domestic pigs, involving a number of traits, like behavior, body composition, disease resistance, reproduction and coat color. In this study, based on genotypes obtained from PorcineSNP60 Illumina assay we attempt to detect both diversifying and within-breed selection signatures in 530 pigs belonging to four breeds: Polish Landrace, Puławska, Złotnicka White and Złotnicka Spotted, of which the last three are a subject of conservative breeding and substantially represent the native populations.
Results
A two largely complementary statistical methods were used for signatures detection, including: pairwise FST and relative extended haplotype homozygosity (REHH) test. Breed-specific diversifying selection signals included several genes involved in processes connected with fertility, growth and metabolism which are potentially responsible for different phenotypes of the studied breeds. The diversifying selection signals also comprised PPARD gene that was previously found to have a large effect on the shape of the external ear in pigs or two genes encoding neuropeptide Y receptors (Y2 and Y5) involved in fat deposition and stress response which are important features differentiating the studied breeds. REHH statistics allowed detecting several within-breed selection signatures overlapping with genes connected with a range of functions including, among others: metabolic pathways, immune system response or implantation and development of the embryo.
Conclusions
The study provides many potential candidate genes with implication for traits selected in the individual breeds and gives strong basis for further studies aiming at identification of sources of variation among the studied pig breeds.
Electronic supplementary material
The online version of this article (10.1186/s12863-018-0681-0) contains supplementary material, which is available to authorized users.
Keywords: FST, Pig, Polish landrace, Puławska, REHH, Selection signatures, Złotnicka
Background
Numerous studies have shown applicability of genomics in the field of quantitative genetics and identification of sources of variation of important phenotypic features such as production traits [1]. Unfortunately, research in this field in most cases requires numerous study populations, well characterized in terms of phenotype, relatedness structure and genome features. Due to the high cost of conducting such research, generated by the need to establish genotypes and phenotypes of large animal populations, attempts have been made to identify traces left in animal genomes by selection pressure directed at the consolidation or improvement of particular phenotypic traits [2]. The identification of selection signatures assumes detection of genomic regions in which gene variants subjected to a rapid increase in allele frequencies under the influence of selective pressure (ongoing selection) are located or detection of genome regions that are fixed in a population with well-established phenotypical features. Detection of selection signals in conjunction with subsequent candidate gene identification approach may indicate the location of major genes responsible for selected traits [3]. The advantage of such approach is that it is independent of the availability of detailed information on the phenotype of individual animals and is applicable to relatively small study populations [2].
Similarly, as other livestock species, pigs have been under long term selection during domestication, breeds formation and further improvement of production and functional traits. To analyze the mechanism underlying phenotypic differentiation caused by selection in pigs, the evidence of selection has been searched in genomes of various pig breeds using whole genome genotype data or high-throughput sequencing [4–9]. The studies allowed detecting several selection signals associated with growth traits, reproduction traits, coat color or ear phenotype and to indicate several genes with major effects on these traits [5, 7]. Nevertheless, selection patterns in pig breeds differ depending on their evolution and breeding histories, so exploration of selection signatures in possibly the largest number of different breeds will help to better understand the genetic variation underlying the traits of interest.
In the present study, we detected selection signatures at the whole genome level in three conserved pig breeds derived from the native pig populations (Puławska, Złotnicka White and Złotnicka Spotted) and a commercial Polish Landrace breed, differing in terms of production, reproduction characteristics and exterior features. Among the native breeds we included Puławska breed which is valued for good resistance to harsh environmental conditions and diseases, taste of meat (its aroma and juiciness) and usefulness for extensive breeding in ecological farms. Two other studied Złotnicka pig breeds (White and Spotted) are characterized by similar functional features but have worse productivity than Puławska breed and have more primitive character. Especially Złotnicka Spotted is being considered as a meat-lard type breed and is characterized by high subcutaneous fat content and relatively low fertility [10–13]. The detailed characteristic of the studied breeds is presented in Additional file 1.
To identify selection signatures in the analyzed breeds, we applied two different methods: first, based on FST (classical measure of population differentiation) [14] and aimed at identification of among-breeds diversifying selection and second, based on relative extended haplotype homozygosity (REHH) [15] statistics allowing for detection of mainly within-breed ongoing selection. Both FST and REHH statistics were shown to be useful to detect selection signatures [16] and they are largely complementary, because REHH test has good power to detect selection signatures within breeds and is more accurate in case of ongoing selection, while FST is useful to detect selection signatures across breeds, represented mainly by loci that were differentially fixed in different breeds [17].
Despite the lack of typical selection in terms of production in the case of conservative breeding, the animals’ qualification into the conservation program (based on breed standard and aimed at consolidation of breed-specific traits and stabilization of exterior) may also be considered as an extensive selection and lead to similar but less pronounced changes in the frequency of alleles. The applied combination of breeds and statistical methods allowed us to search for selection signals associated with both fixed traits of individual breeds and features that are still under improvement, which may help to better understand adaptation of breeds to local environmental conditions and help to evidence processes behind good health, longevity and low environmental requirements of native pigs.
Methods
Animals and genotyping
The material of the study was genomic DNA obtained from blood or hair bulbs of 530 animals sampled from each of the four pig breeds: Polish Landrace (PL, n = 135), Puławska (PUL; n = 155), Złotnicka White (ZW; n = 141), Złotnicka Spotted (ZS; n = 99) differing in terms of production, reproduction and exterior features. The animals were selected to be unrelated for at least two generations and originating from different herds. Each population sample included at least 7% of males. This was because we analyzed breeding population (reproductive studs) in which share of males is matched to the number of boars which are designed to natural matting. All animal procedures were approved by the Local Animal Care Ethics Committee No. II in Kraków - permission number 1293/2016 in accordance with EU regulations. The genomic DNA was purified using a Sherlock AX kit (A&A Biotechnology) and after quality control was genotyped with the use of the PorcineSNP60 BeadChip assay (Illumina) according to the standard Infinium Ultra protocol. The obtained genotypes were controlled for quality by evaluation of call rates and only samples with more than 97% of called genotypes were used for further analysis. Of the 61,565 assayed SNPs, a panel of 50,485 markers was further obtained by removing SNPs mapped to contigs, located on the sex chromosomes (Sscrofa10.2 genome assembly) or classified as intensity-only probes.
Data analysis
The initially filtered SNP set was further reduced by applying population-wide polymorphism filters. The filtering included removal of SNPs with MAF lower than 5% and SNPs with more than 20% of missing genotypes across all breeds. MAF cutoff used for SNPs filtering was applied to the whole population (all breeds). This allowed to retain small proportion of SNPs that are monomorphic only in some breeds (presumably fixed for some reason, including selection and inbreeding). MAF value of 0.01 was used to characterize remaining SNP polymorphisms. SNPs deviating from HWE with critical P-value of 1.0E-06 in each breed separately were also removed resulting in a final panel of 43,923 common SNPs with average inter-marker distance of 55.7 kb (±78.0). The signals of diversifying selection were detected using pairwise Wright’s FST [18], the classical measure of population genetic differentiation. The FST values obtained for pairwise comparisons at each SNP were treated according to a methodology proposed by Akey et al. [19] and further applied by other studies [7]. In brief, standardized FST values were calculated (di) as:
where and denote the expected value and standard deviation of FST between breeds i and j calculated from all analyzed 43,923 SNPs. This allowed to make comparison of each breed against all other breeds under study. To account for stochasticity in locus-by-locus variation, a 10-SNP sliding window was further implemented on the obtained values. Candidate selected regions were then defined as the 99.9th percentile of the empirical distributions of window-averaged di values. The adjacent regions under selection were merged and (while searching for gene content) regions were expanded on both ends by 25 kb to detect neighboring, potentially linked genes.
The signals of positive selection within single breeds were detected using REHH statistics implemented in the Sweep v.1.1 software [6]. First, the obtained genotypes were phased and imputed using the fastPhase software [20]. The phased genotypes were then used to detect core haplotypes with minimum of three and no more than twenty SNPs. The detected longest non-overlapping core haplotypes were then subjected to EHH test, which is based on comparing a core haplotype with both higher frequency and higher EHH with other core haplotypes at the same locus. Subsequently, a probability that two randomly selected haplotypes within a core region are identical-by-descent for the entire interval spanning the core region to a given locus was computed [15, 21]. Finally, considering variation in recombination rates across the genome, the relative extended haplotype homozygosity (REHH) statistics was used [15] and calculated at about 1 cM (approximated to 1 Mb) distance [22] on both upstream and downstream directions (with exception of chromosome ends) from each core against all other cores within the region. To determine REHH significance, haplotypes were allocated to twenty frequency bins and the REHH values were compared between equally frequent core haplotypes found within the region. REHH P-values were ultimately obtained by a logarithmic transformation of the REHH values within these bins (to reach normality) and calculation of mean and standard deviation. The core haplotypes with the most extreme P-values (extended by 0.5 Mb in each direction) were filtered for frequency (> 0.25) and screened for overlapping pig ENSEMBL genes with the use of UCSC Genome Browser.
The functional annotation of detected genes was performed using the KOBAS 3.0 web server [23] and WebGestalt (WEB-based GEneSeTAnaLysis Toolkit) [24]. A gene list enrichment analysis was done according to all known pig genes applying a correction for multiple testing.
The population differentiation was additionally visualized using the principal component analysis (PCA) based on SNP genotypes and a cladogram of mean pairwise FST distances created using the neighbor joining (NJ) method [25].
Results
SNPs polymorphism parameters and breeds genetic differentiation
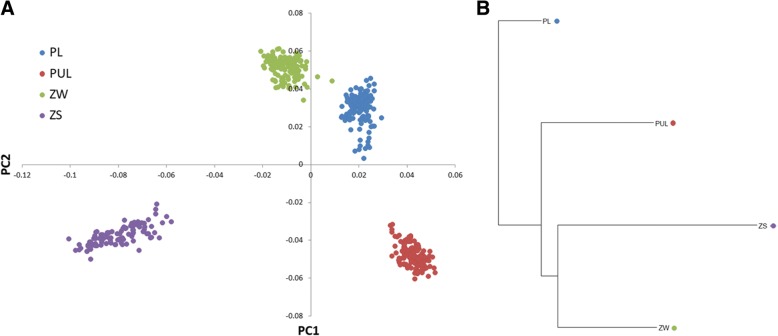
The applied SNPs filtering criteria allowed obtaining a common set of 43,923 SNPs polymorphic across the whole population with mean inter-marker distance of 55.7 kb (±78.0). The number of polymorphic SNPs (MAF > 0.01) per breed ranged from 37,423 to 43,567 in ZS and PL breeds, respectively. The average MAF across all SNPs was the lowest in ZS (0.212) and the highest in PL (0.279). The averaged observed heterozygosity per breed ranged from 0.3 to 0.367 for the same breeds (Table 1). Mean and weighted overall pairwise FST distances were the highest between PUL and ZS (0.143 and 0.173) and the lowest level of genetic differentiation was found between PL and ZW (0.085 and 0.097) (Table 2). The visualization of pig breeds genetic diversity using PCA and NJ methods is presented in Fig. 1.
Table 1.
SNPs panel polymorphism parameters
| Breed | SNPs No | Monomorphic SNPs (No) | Low frequency variants (0 < MAF < 0.01) | Polymorphic SNPs (MAF > 0.01) | Mean MAF | Observed heterozygosity |
|---|---|---|---|---|---|---|
| PL | 43,923 | 157 | 199 | 43,567 | 0.279 | 0.367 |
| PUL | 288 | 71 | 43,564 | 0.274 | 0.361 | |
| ZW | 1008 | 1254 | 41,661 | 0.251 | 0.344 | |
| ZS | 4638 | 1862 | 37,423 | 0.212 | 0.300 |
Table 2.
Mean (above diagonal) and weighted (below diagonal) pairwise FST distances between the studied pig breeds
| Breed | PL | PUL | ZW | ZS |
|---|---|---|---|---|
| PL | – | 0.0897 | 0.0853 | 0.1423 |
| PUL | 0.1014 | – | 0.1097 | 0.1432 |
| ZW | 0.0973 | 0.1276 | – | 0.1353 |
| ZS | 0.1731 | 0.1733 | 0.1685 | – |
Fig. 1.
Genetic differentiation of the analyzed pig breeds based on (a) principal component analysis and (b) the neighbor joining method on mean pairwise FST distances
Signals of diversifying selection
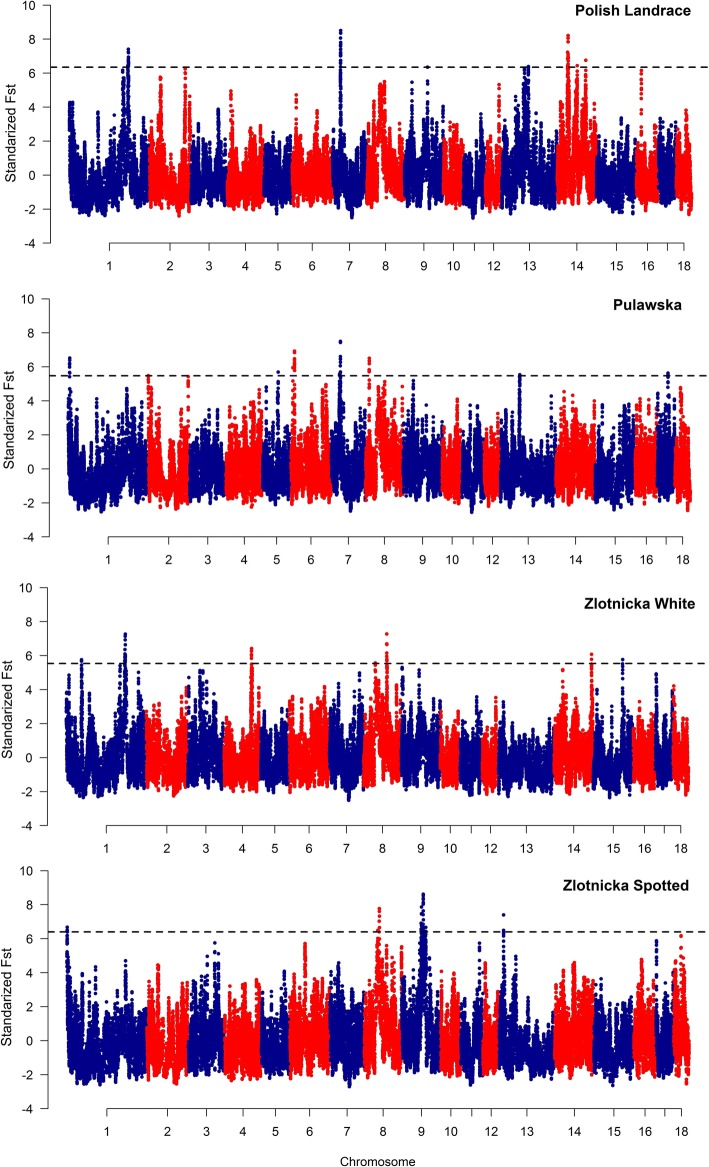
Signals of diversifying selection among the studied pig breeds were detected based on the breed-normalized pairwise FST distances (Additional file 2). After smoothing of the data by moving average, top 0.1% of the observations were considered as pinpointing breed-specific selection signals. After merging of overlapping signals, from 7 to 11 genome regions with strong selection signals were detected per breed with a size ranging from 266.8 kb to 2.9 Mb. The highest number of selection signals across all breeds was detected on SSC1, SSC7 and SSC14 (SSC9 only in ZS breed) and only single regions were detected on SSC2, 4, 5, 15 and 17. No signals (except closely positioned regions on SSC7, between 32 and 34 Mb of the genomic sequence in PL and PUL) were common for different breeds (Table 3, Fig. 2).
Table 3.
Genome regions spanning the strongest detected diversifying (FST-based) selection signatures
| Breed | CHR | START | END | SIZE | START-25 kb | END+ 25 kb |
|---|---|---|---|---|---|---|
| PL | 7 | 32,166,462 | 33,358,569 | 1,192,107 | 32,141,462 | 33,383,569 |
| 13 | 104,557,045 | 105,298,541 | 741,496 | 104,532,045 | 105,323,541 | |
| 14 | 40,707,564 | 41,060,913 | 353,349 | 40,682,564 | 41,085,913 | |
| 14 | 41,333,642 | 41,977,281 | 643,639 | 41,308,642 | 42,002,281 | |
| 14 | 42,237,338 | 43,688,399 | 1,451,061 | 42,212,338 | 43,713,399 | |
| 14 | 78,434,877 | 78,742,346 | 307,469 | 78,409,877 | 78,767,346 | |
| 14 | 111,740,597 | 112,008,662 | 268,065 | 111,715,597 | 112,033,662 | |
| PUL | 1 | 6,272,450 | 6,735,120 | 462,670 | 6,247,450 | 6,760,120 |
| 2 | 365,309 | 1,264,220 | 898,911 | 340,309 | 1,289,220 | |
| 5 | 60,978,291 | 61,354,749 | 376,458 | 60,953,291 | 61,379,749 | |
| 6 | 7,872,494 | 8,303,476 | 430,982 | 7,847,494 | 8,328,476 | |
| 6 | 14,097,672 | 14,905,615 | 807,943 | 14,072,672 | 14,930,615 | |
| 7 | 33,416,020 | 33,720,562 | 304,542 | 33,391,020 | 33,745,562 | |
| 7 | 34,856,640 | 35,251,345 | 394,705 | 34,831,640 | 35,276,345 | |
| 7 | 35,935,629 | 37,260,234 | 1,324,605 | 35,910,629 | 37,285,234 | |
| 8 | 15,073,006 | 15,600,021 | 527,015 | 15,048,006 | 15,625,021 | |
| 13 | 75,723,927 | 76,244,239 | 520,312 | 75,698,927 | 76,269,239 | |
| 17 | 41,777,060 | 42,101,006 | 323,946 | 41,752,060 | 42,126,006 | |
| ZW | 1 | 60,225,139 | 60,769,684 | 544,545 | 60,200,139 | 60,794,684 |
| 1 | 229,502,611 | 229,792,874 | 290,263 | 229,477,611 | 229,817,874 | |
| 1 | 230,021,584 | 231,098,497 | 1,076,913 | 229,996,584 | 231,123,497 | |
| 1 | 231,365,442 | 232,552,969 | 1,187,527 | 231,340,442 | 232,577,969 | |
| 4 | 107,135,195 | 107,586,557 | 451,362 | 107,110,195 | 107,611,557 | |
| 8 | 45,191,096 | 47,560,329 | 2,369,233 | 45,166,096 | 47,585,329 | |
| 8 | 90,972,549 | 91,828,187 | 855,638 | 90,947,549 | 91,853,187 | |
| 14 | 146,952,943 | 147,444,441 | 491,498 | 146,927,943 | 147,469,441 | |
| 15 | 115,092,422 | 115,767,070 | 674,648 | 115,067,422 | 115,792,070 | |
| ZS | 1 | 1,300,464 | 2,153,932 | 853,468 | 1,275,464 | 2,178,932 |
| 8 | 52,565,396 | 55,529,571 | 2,964,175 | 52,540,396 | 55,554,571 | |
| 8 | 58,625,849 | 61,180,840 | 2,554,991 | 58,600,849 | 61,205,840 | |
| 9 | 75,593,700 | 75,973,705 | 380,005 | 75,568,700 | 75,998,705 | |
| 9 | 76,589,514 | 77,680,603 | 1,091,089 | 76,564,514 | 77,705,603 | |
| 9 | 78,073,285 | 78,534,222 | 460,937 | 78,048,285 | 78,559,222 | |
| 9 | 81,370,401 | 82,465,204 | 1,094,803 | 81,345,401 | 82,490,204 | |
| 9 | 82,868,790 | 84,766,063 | 1,897,273 | 82,843,790 | 84,791,063 | |
| 9 | 93,709,999 | 94,139,393 | 429,394 | 93,684,999 | 94,164,393 | |
| 13 | 17,239,351 | 17,506,216 | 266,865 | 17,214,351 | 17,531,216 |
Fig. 2.
The genomic distribution of diversifying selection signals for all studied breeds. Dashed line indicates the top 0.1% of the highest standardized FST values
To analyze gene content of the genome regions spanning the detected selection signals, each region was expanded by 25 kb on each end to account for potentially linked genes. This allowed for detection from 61 (ZS) to 116 (PUL) ENSEMBL genes per breed (Additional file 3). To manage variety of genes found within the selection signals we performed a functional analysis aimed at the identification of enriched processes. Keeping in mind that the detected selection signatures are associated with a number of phenotypic features differing the studied breeds (which are conditioned by complex and very different molecular mechanisms), we expected only very few genes connected with separate biological processes in the enrichment analysis and low statistical significance of the obtained results. This analysis was rather treated as a supporting method which helped us to manage the obtained extensive gene content. Nevertheless, this analysis allowed to reduce the complexity of the obtained data and allowed us to search for processes and underlying genes potentially being the targets of diversifying selection. The functional classification of well-annotated genes showed that the genes were mainly involved in GO biological processes connected with: metabolic processes, cellular processes, biological regulation, response to stimulus and developmental processes. Among the top ten enriched (pointwise P < 0.05) GO biological processes there were inter alia those connected with: lipid binding, fatty acid metabolic process, cellular senescence or response to muscle stretch. When functional classification was performed for individual breeds, visible differences in the enriched GO categories were detected (Table 4). In PL pigs, a large share of genes was engaged in metabolic pathways and lipid binding, and included e.g.: COQ5, GATC, COX6A1, PLA2G1B, HK1, SDSL, PRIM2, ALDH2, SDS, GLTP and RPH3A genes. In PUL breed, the genes involved in several GO categories were detected, including those connected with: striated muscle cell proliferation, skeletal muscle tissue regeneration (PPARD and MAPK14) and embryo implantation (ARHGDIB, PPARD). The diversifying selection signals in ZW breed encompassed genes enriching very general categories of biological processes, like e.g. transcriptional repressor activity, intracellular processes or processes in nuclear lumen, however, some of the genes were connected with more specific pathways like: tryptophan metabolism (TDO2, AOX1) or salivary secretion (GUCY1A3, GUCY1B3). In ZS breeds, selection signals were associated with genes responsible for e.g.: neuropeptide Y receptor activity and feeding behavior (NPY1R, NPY5R), metabolic processes (PON1, PON2, PON3) and cortisol metabolic process (REST).
Table 4.
Top ten biological processes enriched in genes detected in the genome regions under diversifying selection in separate pig breeds
| #Term | ID | Input number | Background number | P-Value | Corrected P-Value |
|---|---|---|---|---|---|
| PL | |||||
| Lipid binding | GO:0008289 | 5 | 150 | 0.0002 | 0.1501 |
| Single-organism transport | GO:0044765 | 9 | 658 | 0.0004 | 0.1501 |
| Cilium morphogenesis | GO:0060271 | 3 | 45 | 0.0006 | 0.1501 |
| Single-organism localization | GO:1902578 | 9 | 708 | 0.0007 | 0.1501 |
| Organic substance transport | GO:0071702 | 8 | 571 | 0.0007 | 0.1501 |
| Macromolecule localization | GO:0033036 | 8 | 590 | 0.0009 | 0.1567 |
| Regulation of DNA biosynthetic process | GO:2000278 | 2 | 14 | 0.0013 | 0.1574 |
| Cell projection organization | GO:0030030 | 5 | 239 | 0.0014 | 0.1574 |
| p53 binding | GO:0002039 | 2 | 15 | 0.0015 | 0.1574 |
| Transport | GO:0006810 | 11 | 1158 | 0.0017 | 0.1574 |
| PUL | |||||
| Skeletal muscle tissue regeneration | GO:0043403 | 2 | 9 | 0.0015 | 0.4425 |
| Embryo implantation | GO:0007566 | 2 | 11 | 0.0021 | 0.4425 |
| Tissue regeneration | GO:0042246 | 2 | 13 | 0.0027 | 0.4425 |
| Cellular senescence | GO:0090398 | 2 | 14 | 0.0031 | 0.4425 |
| Striated muscle cell proliferation | GO:0014855 | 2 | 16 | 0.0040 | 0.4425 |
| Connective tissue development | GO:0061448 | 3 | 63 | 0.0051 | 0.4425 |
| Regulation of myoblast differentiation | GO:0045661 | 2 | 19 | 0.0054 | 0.4425 |
| Plasma membrane region | GO:0098590 | 4 | 133 | 0.0059 | 0.4425 |
| Regeneration | GO:0031099 | 2 | 21 | 0.0064 | 0.4425 |
| Cell aging | GO:0007569 | 2 | 22 | 0.0070 | 0.4425 |
| ZW | |||||
| Transcriptional repressor activity, RNA polymerase II transcription factor binding | GO:0001191 | 2 | 24 | 0.0033 | 0.4411 |
| Intracellular | GO:0005622 | 22 | 3854 | 0.0034 | 0.4411 |
| Nuclear part | GO:0044428 | 9 | 989 | 0.0048 | 0.4411 |
| RNA polymerase II transcription cofactor activity | GO:0001104 | 2 | 36 | 0.0069 | 0.4411 |
| Nuclear lumen | GO:0031981 | 8 | 863 | 0.0071 | 0.4411 |
| Transcription factor activity, RNA polymerase II transcription factor binding | GO:0001076 | 2 | 44 | 0.0100 | 0.4411 |
| intracellular part | GO:0044424 | 20 | 3671 | 0.0101 | 0.4411 |
| Nucleus | GO:0005634 | 12 | 1785 | 0.0121 | 0.4411 |
| membrane-enclosed lumen | GO:0031974 | 8 | 971 | 0.0139 | 0.4411 |
| intracellular organelle lumen | GO:0070013 | 8 | 971 | 0.0139 | 0.4411 |
| ZS | |||||
| Neuropeptide Y receptor activity | GO:0004983 | 2 | 8 | 0.0001 | 0.0677 |
| Neuropeptide receptor activity | GO:0008188 | 2 | 20 | 0.0007 | 0.1715 |
| Feeding behavior | GO:0007631 | 2 | 31 | 0.0016 | 0.2289 |
| Neuropeptide signaling pathway | GO:0007218 | 2 | 36 | 0.0021 | 0.2289 |
| Peptide receptor activity | GO:0001653 | 2 | 64 | 0.0061 | 0.2289 |
| G-protein coupled peptide receptor activity | GO:0008528 | 2 | 64 | 0.0061 | 0.2289 |
| Circulatory system development | GO:0072359 | 3 | 218 | 0.0069 | 0.2289 |
| Cardiovascular system development | GO:0072358 | 3 | 218 | 0.0069 | 0.2289 |
| Cortisol metabolic process | GO:0034650 | 1 | 5 | 0.0106 | 0.2289 |
| Response to electrical stimulus | GO:0051602 | 1 | 5 | 0.0106 | 0.2289 |
Selection signals between breeds with different phenotypical features
To detect genome regions bearing variants potentially responsible for the most pronounced phenotypical differences between breeds, the pig breeds with white (PL, ZW) and spotted (PUL, ZS) coat color patterns were compared using FST statistics. The comparison between single color and spotted pig breeds revealed the strongest (99.9th percentile of the observations) selection signals on SSC3, SSSC7, SSC8 and SSC9 (Table 5). The most pronounced selection signal, involving five separate regions (between 50.5 and 58.6 Mb of sequence) was located on SSC8 in close vicinity to the KIT gene locus (SSC8, 41.4–41.5 Mb). Altogether, the regions spanned 72 different ENSEMBL genes which together did not enrich any biological processes of pathways, however, again included PPARD gene.
Table 5.
Genomic regions spanning the strongest detected diversifying selection signals between single-color and spotted pig breeds
| CHR | START | END | SIZE | START-25 kb | END+ 25 kb |
|---|---|---|---|---|---|
| 3 | 110,615,540 | 111,359,515 | 743,975 | 110,590,540 | 111,384,515 |
| 7 | 32,456,532 | 33,142,614 | 686,082 | 32,431,532 | 33,167,614 |
| 8 | 50,537,893 | 52,513,888 | 1,975,995 | 50,512,893 | 52,538,888 |
| 8 | 51,235,665 | 53,929,233 | 2,693,568 | 51,210,665 | 53,954,233 |
| 8 | 52,301,582 | 55,181,102 | 2,879,520 | 52,276,582 | 55,206,102 |
| 8 | 52,565,396 | 55,647,917 | 3,082,521 | 52,540,396 | 55,672,917 |
| 8 | 58,598,344 | 61,863,927 | 3,265,583 | 58,573,344 | 61,888,927 |
| 8 | 69,065,977 | 71,405,594 | 2,339,617 | 69,040,977 | 71,430,594 |
| 9 | 90,863,475 | 91,514,134 | 650,659 | 90,838,475 | 91,539,134 |
Within-breed selection signatures
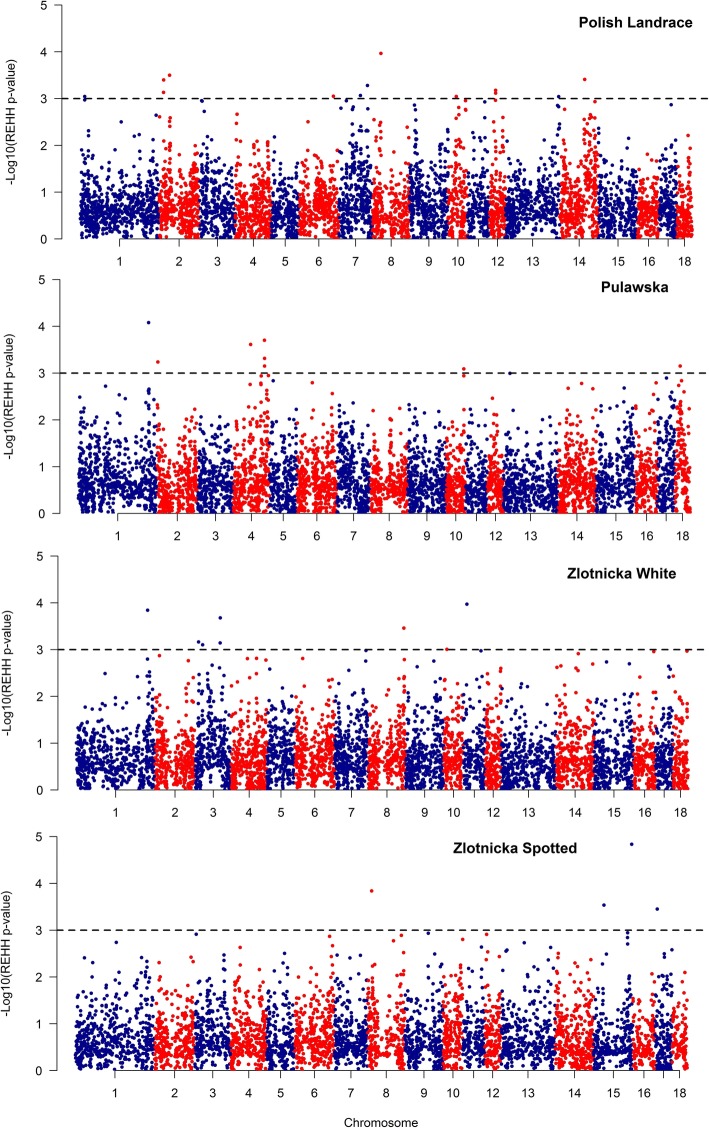
The analysis performed with the Sweep v.1.1 software allowed detection from 4546 (ZS) to 5638 (PUL) longest non-overlapping core regions (CR – a unique genome region spanning locus specific core haplotypes) consisting of minimum three and no more than 20 SNPs with average lengths ranging from 193.7 kb (PL) to 394.3 kb (ZS). For all these core haplotypes, from 37,809 (ZS) to 46,162 (PUL) EHH tests were performed – from 8.2 to 8.3 per core region, on average (Additional file 4). Considering that alleles being under positive selection are fixed or going to be fixed and hence core haplotypes harboring these alleles should be frequent [21], core haplotypes with the frequency < 0.25 were removed. This allowed selection from 14,079 (ZS) to 17,580 (PUL) REHH tests per breed for which from 118 (ZS) to 176 (PL) core haplotypes were outliers at the significance level of 0.01. The obtained REHH P-values were –log10 transformed and plotted against the chromosomal positions to visualize outlying core haplotypes and selection patterns across the breeds genome (Fig. 3, Additional file 5). This analysis showed clear non-random distribution of selection signals across the breeds genome with visible overrepresentation of long and common haplotypes on e.g.: SSC2 and 14 in PL or SSC4 and 18 in PUL breed. The selection patterns also clearly differed between the analyzed breeds. To minimize the number of genes potentially associated with significant core haplotypes, only cores with P-value lower than 0.001 (representing the strongest selection signals) were further analyzed. The positions, haplotype frequency and other statistics for the most significant haplotypes belonging to separate core regions are presented in Table 6.
Fig. 3.
The genomic map of REHH P-values for all core haplotypes showing a frequency > 0.25. Dashed line represents the significance threshold of 0.001
Table 6.
Statistics for the most significant core haplotypes being under selection and belonging to separate core regions in the analyzed breeds
| Breed | Chr | Start Base | End Base | Size (kb) | Hap frequency | EHH | REHH | REHH P-Value | Genes directly overlapping with core haplotypesa |
|---|---|---|---|---|---|---|---|---|---|
| PL | 1 | 17,474,143 | 17,532,523 | 58.38 | 0.33 | 0.77 | 10.45 | 0.000902 | LOC100152956, LOC100621885, LOC100512136, HOXB2, ZNF22, LOC100739279, C10orf10, RASSF4, LOC100157415, HOXB1, ALG6, HOXB3, ALOX5, LOC100523276, HOXB4, MARCH8 |
| 2 | 18,722,196 | 18,775,121 | 52.925 | 0.31 | 0.53 | 10.91 | 0.00074 | ||
| 2 | 18,850,395 | 18,872,659 | 22.264 | 0.31 | 0.45 | 12.46 | 0.0004 | ||
| 2 | 42,642,528 | 42,708,543 | 66.015 | 0.64 | 0.60 | 9.68 | 0.000318 | ||
| 6 | 137,335,517 | 137,534,028 | 198.511 | 0.27 | 0.79 | 10.35 | 0.000892 | ||
| 7 | 87,029,852 | 87,066,927 | 37.075 | 0.59 | 0.88 | 7.79 | 0.000858 | ||
| 7 | 115,164,638 | 115,233,642 | 69.004 | 0.39 | 0.89 | 10.21 | 0.000526 | ||
| 8 | 33,740,426 | 33,760,858 | 20.432 | 0.34 | 0.88 | 16.21 | 0.000108 | ||
| 10 | 32,485,201 | 32,589,926 | 104.725 | 0.60 | 0.53 | 7.74 | 0.000902 | ||
| 12 | 24,427,829 | 24,643,031 | 215.202 | 0.27 | 0.64 | 11.03 | 0.000668 | ||
| 12 | 24,696,740 | 24,812,751 | 116.011 | 0.27 | 0.64 | 10.70 | 0.000768 | ||
| 13 | 211,907,795 | 211,989,807 | 82.012 | 0.48 | 0.48 | 9.93 | 0.00091 | ||
| 14 | 98,887,138 | 99,642,277 | 755.139 | 0.42 | 0.93 | 10.94 | 0.00039 | ||
| PUL | 1 | 279,545,606 | 279,701,487 | 155.881 | 0.52 | 0.83 | 5.87 | 0.000374 | PRKCQ, TERF1, FAM71F2, SBSPON, IMPDH1, LRP5 |
| 1 | 279,545,606 | 279,701,487 | 155.881 | 0.52 | 0.74 | 7.52 | 8.35E-05 | ||
| 2 | 3,166,230 | 3,236,832 | 70.602 | 0.58 | 0.58 | 5.09 | 0.000578 | ||
| 4 | 68,321,268 | 68,385,012 | 63.744 | 0.58 | 0.63 | 5.93 | 0.000245 | ||
| 4 | 123,472,417 | 123,553,728 | 81.311 | 0.48 | 0.99 | 7.18 | 0.000199 | ||
| 4 | 123,712,634 | 123,819,209 | 106.575 | 0.48 | 0.92 | 5.73 | 0.000706 | ||
| 4 | 123,838,086 | 123,907,205 | 69.119 | 0.48 | 0.90 | 6.14 | 0.000485 | ||
| 10 | 70,638,252 | 70,669,410 | 31.158 | 0.35 | 0.52 | 8.00 | 0.000812 | ||
| 18 | 20,991,435 | 21,077,884 | 86.449 | 0.77 | 0.55 | 4.22 | 0.00071 | ||
| ZW | 1 | 283,594,245 | 283,667,732 | 73.487 | 0.93 | 0.65 | 2.91 | 0.000144 | C1orf115, SUSD1, PRKCE, MARC2 |
| 3 | 14,090,668 | 14,221,604 | 130.936 | 0.32 | 0.98 | 6.04 | 0.000686 | ||
| 3 | 30,457,204 | 30,733,237 | 276.033 | 0.45 | 1.00 | 5.01 | 0.000792 | ||
| 3 | 100,417,119 | 100,448,894 | 31.775 | 0.81 | 0.55 | 2.95 | 0.000719 | ||
| 3 | 100,521,767 | 100,536,654 | 14.887 | 0.71 | 0.69 | 4.08 | 0.000211 | ||
| 3 | 100,521,767 | 100,536,654 | 14.887 | 0.71 | 0.70 | 3.88 | 0.000295 | ||
| 8 | 141,594,565 | 141,644,208 | 49.643 | 0.46 | 0.80 | 5.73 | 0.000349 | ||
| 8 | 141,594,565 | 141,644,208 | 49.643 | 0.46 | 0.98 | 5.49 | 0.000457 | ||
| 10 | 12,274,858 | 12,388,946 | 114.088 | 0.27 | 1.00 | 5.98 | 0.000988 | ||
| 11 | 14,256,201 | 14,501,882 | 245.681 | 0.32 | 1.00 | 8.03 | 0.000107 | ||
| ZS | 8 | 16,617,477 | 16,681,729 | 64.252 | 0.32 | 1.00 | 6.07 | 0.000145 | UBE2F, RAMP1 |
| 15 | 41,476,357 | 41,710,320 | 233.963 | 0.36 | 1.00 | 4.98 | 0.000291 | ||
| 15 | 151,930,390 | 152,024,615 | 94.225 | 0.39 | 0.95 | 7.12 | 1.46E-05 | ||
| 17 | 9,863,762 | 9,926,558 | 62.796 | 0.27 | 0.64 | 5.67 | 0.000355 |
agenes with unassigned name were removed from the table
After extension of the detected significant (P < 0.001) core haplotypes to 1 cM on both ends, 171 unique genes potentially being under selection were found in PL, 116 in PUL, 84 in ZW and 35 in ZS (Additional file 6). The functional annotation of the genes allowed credible GO categories enrichment analysis only for PL and PUL pigs, with only single genes connected with separate GO categories in ZW and ZS pigs (Table 7). Nevertheless, among the top 10 enriched biological processes, the genes associated inter alia with skeletal system development and morphogenesis (HOXB1, − 2, − 5, − 6, − 7, − 9) were found in PL pigs and regulation of protein transport and localization (AIP, TBC1D10C, GCC1, LEP) or muscle fiber development (FLNC, SMO) in PUL pigs. In ZW breed the suggestive biological processes were involved in e.g.: metal ion homeostasis and regulation (TTC7A, SLC30A10), negative regulation of innate immune response (DUSP10) and negative regulation of T cell apoptotic process (SLC46A2). In ZS breed, the most commonly represented processes included: galactosidase/galactosidase activity and lipid catabolic process (LOC102167689, LOC102167689).
Table 7.
Top ten biological processes enriched in genes detected in the core haplotypes under positive selection in individual pig breeds
| #Term | ID | Input number | Background number | P-Value | Corrected P-Value |
|---|---|---|---|---|---|
| PL | |||||
| Embryonic skeletal system development | GO:0048706 | 6 | 37 | 8.68E-07 | 0.001 |
| Embryonic skeletal system morphogenesis | GO:0048704 | 5 | 29 | 5.69E-06 | 0.0037 |
| Skeletal system morphogenesis | GO:0048705 | 6 | 65 | 1.71E-05 | 0.0074 |
| Anterior/posterior pattern specification | GO:0009952 | 6 | 70 | 2.53E-05 | 0.0081 |
| Regionalization | GO:0003002 | 7 | 118 | 4.95E-05 | 0.0127 |
| Skeletal system development | GO:0001501 | 7 | 123 | 6.35E-05 | 0.0136 |
| Pattern specification process | GO:0007389 | 7 | 131 | 9.26E-05 | 0.0170 |
| Sequence-specific DNA binding | GO:0043565 | 10 | 305 | 0.00015 | 0.0251 |
| Chordate embryonic development | GO:0043009 | 7 | 171 | 0.0004 | 0.0603 |
| Embryo development ending in birth or egg hatching | GO:0009792 | 7 | 173 | 0.0005 | 0.0603 |
| PUL | |||||
| Regulation of hair cycle | GO:0042634 | 2 | 5 | 0.0005 | 0.2658 |
| Establishment of protein localization to organelle | GO:0072594 | 5 | 151 | 0.0010 | 0.2658 |
| Regulation of protein import | GO:1904589 | 3 | 41 | 0.0013 | 0.2658 |
| Regulation of protein import into nucleus | GO:0042306 | 3 | 41 | 0.0013 | 0.2658 |
| Protein targeting | GO:0006605 | 5 | 162 | 0.0014 | 0.2658 |
| Regulation of nucleocytoplasmic transport | GO:0046822 | 3 | 46 | 0.0018 | 0.2658 |
| Regulation of protein localization to nucleus | GO:1900180 | 3 | 49 | 0.0021 | 0.2658 |
| Negative regulation of peptide hormone secretion | GO:0090278 | 2 | 13 | 0.0024 | 0.2658 |
| Negative regulation of peptide secretion | GO:0002792 | 2 | 13 | 0.0024 | 0.2658 |
| Muscle fiber development | GO:0048747 | 2 | 14 | 0.0028 | 0.2658 |
| ZW | |||||
| Cellular transition metal ion homeostasis | GO:0046916 | 2 | 26 | 0.0032 | 0.4776 |
| Transition metal ion homeostasis | GO:0055076 | 2 | 35 | 0.0056 | 0.4776 |
| Nucleotidase activity | GO:0008252 | 1 | 5 | 0.0180 | 0.4776 |
| Zinc ion homeostasis | GO:0055069 | 1 | 6 | 0.0210 | 0.4776 |
| Negative regulation of innate immune response | GO:0045824 | 1 | 6 | 0.0210 | 0.4776 |
| Glycosphingolipid biosynthetic process | GO:0006688 | 1 | 6 | 0.0210 | 0.4776 |
| Glucosyltransferase activity | GO:0046527 | 1 | 6 | 0.0210 | 0.4776 |
| Negative regulation of T cell apoptotic process | GO:0070233 | 1 | 6 | 0.0210 | 0.4776 |
| Cellular zinc ion homeostasis | GO:0006882 | 1 | 6 | 0.0210 | 0.4776 |
| Response to zinc ion | GO:0010043 | 1 | 6 | 0.0210 | 0.4776 |
| ZS | |||||
| Galactosidase activity | GO:0015925 | 1 | 5 | 0.0087 | 0.3709 |
| Sphingolipid catabolic process | GO:0030149 | 1 | 5 | 0.0087 | 0.3709 |
| Membrane lipid catabolic process | GO:0046466 | 1 | 5 | 0.0087 | 0.3709 |
| Glucosidase activity | GO:0015926 | 1 | 5 | 0.0087 | 0.3709 |
| Activation of NF-kappaB-inducing kinase activity | GO:0007250 | 1 | 6 | 0.0101 | 0.3709 |
| snRNA processing | GO:0016180 | 1 | 7 | 0.012 | 0.3709 |
| Postreplication repair | GO:0006301 | 1 | 8 | 0.0130 | 0.3709 |
| snRNA metabolic process | GO:0016073 | 1 | 9 | 0.0144 | 0.3709 |
| Positive regulation of NIK/NF-kappaB signaling | GO:1901224 | 1 | 9 | 0.0144 | 0.3709 |
| Protein K63-linked ubiquitination | GO:0070534 | 1 | 10 | 0.0159 | 0.3709 |
Discussion
Natural or artificial selection are the major mechanisms driving differentiation of the populations. Pig domestication resulted in considerable changes in the phenotypes and behavior of the animals. In the early stages of domestication, unconscious selection for behavioral traits was applied and this early stage was followed by methodical selection in which specific traits were selected based on breeding goals [26, 27]. This resulted in development of specialized breeds, improved to produce desired animal products or to represent a desired morphological standard. The artificial selection increased the variation between domesticated animals and their wild ancestors and generated a variety of different populations, differing in phenotypical features related to their specialization [28]. Identifying recent positive selection signatures in domesticated animals can provide valuable information on genomic regions that are under the influence of both artificial and natural selection, and thus, can help to identify beneficial variants and underlying biological pathways influencing economically important traits.
In this study, by using two largely complementary methods we detected selection signatures in four pig breeds, including Landrace and three conserved native breeds representing a maternal breeding component. The native pig populations can be considered as unselected, because the conservation programs are more focused on preserving genetic diversity and breed standard than improvement of production traits. The analysis of breeds’ genetic differentiation based on a cladogram obtained with the neighbor-joining method on the averaged FST values (accounting for all pairwise distances) showed the highest genetic similarity between both Złotnicka pig breeds. Such result was expected, due to the fact that both breeds originated from the same geographic background and were created in a similar time period based on the same genetic group. The clear separation of Landrace from the Polish conserved breeds was also observed, which probably reflects its origin from German and Swedish pig breeds and its high speciation in terms of production. The applied PCA method showed the highest genetic similarity between Polish Landrace and Złotnicka White pigs which both represent white, meat-type pigs with dropping ears.
In the studied pig breeds, we detected several selection signals, representing genome regions that were differentially fixed in different breeds or representing within-breed selection signatures. The diversifying selection signals were detected based on the comparison of a specific breed with all other breeds under study, presuming that these signals will be characteristic for unique breed features or strongly fixed breed traits being poorly developed in the other compared breeds. This can be an explanation for the lack of selection signals being common for different breeds. With the FST-based method, we detected large (over 1 Mb in size) selection sweeps on SSC7 (32.1–33.3 Mb) and 14 (42.2–43.7 Mb) among the top 0.1% of strongest selection signals in Polish Landrace. Several selection signals associated with these chromosomes were previously reported in other pig breeds [7, 29], however, their direct comparison is difficult since diversifying section signals are strongly dependent on structure of breeds used for comparisons. Several GWAS showed the association of closely positioned SNPs on SSC7 with backfat thickness [30], fatty acid metabolic indices [31], number of teats or body weight and height [32]. A similar to ours region on SSC14 was shown to be associated with a number of teats [33, 34]. The animals of conserved breeds included in this study considerably differed in terms of this trait, ranging from on average 13.71 (±0.97) teats in ZS to 14.22 (±0.88) in PUL breed. Unfortunately, no reproductive performance records were available for the PL pigs analyzed in this study. Nevertheless, according to the national evaluation, the average number of teats in PL breed is visibly higher than this observed in the conserved breeds - 15.02 (Additional file 7). This suggests that the detected strong selection signals on these autosomes may be a result of long-term selection for conformation and reproductive performance traits in PL breed.
While screening detected selection signals for gene content we extended the detected regions by additional bases. Extension of the detected regions was motivated by a need to account for linkage disequilibrium and the applied sliding-window approach. While moving average smooths the data, some surrounding genes may not fall directly into the selection signals. Because of some accidental differences in allele frequencies (e.g. genetic drift), selection signal peaks can be slightly shifted in relation to real functional variant locations. To not to lose this information we extended FST - based regions by additional 25 kb, which is an approximated half of mean distance between SNPs in this study. In the REHH method we extended the detected signals by 0.5 Mb in each direction. In our data, we found that core haplotype domains (their extended homozygosity) on average spans over 0.7 Mb in upstream and downstream directions from cores. We have narrowed these regions by adding 0.5 Mb to each core haplotype to account only for genes within the region of relatively high linkage with the core SNPs. Similar approach was previously proposed by Qanbari et al. [21]. The size of selection signals detected using the two approaches was comparable, suggesting that thanks to the linkage disequilibrium, selection signals are extensive and should not be narrowed down only to the detected signal peaks.
Within the strongest differential (FST-based) selection signals detected in Polish Landrace we found several candidate genes which may be responsible for the well-developed breed characteristics like good fertility and high growth rate. One of the interesting detected genes is RASAL1 gene (RAS Protein Activator Like 1), acting as a suppressor of RAS function. The protein enhances the weak intrinsic GTPase activity of RAS proteins resulting in the inactive GDP-bound form of RAS, thereby allowing control of cellular proliferation and differentiation [35]. A large share of detected genes in PL breed was engaged in metabolic pathways and included e.g. HK1 (Hexokinase 1) gene which phosphorylates glucose to produce glucose-6-phosphate which is the first step in glucose metabolism pathways. Glucose metabolism is important for energy homeostasis but has also an effect on growth and meat quality traits [36]. Two other genes were also detected within the identified selection signals in PL pigs, such as: LC8 (LC8 dynein light chain – DYNLL1) and KHDRBS2 (KH RNA Binding Domain Containing, Signal Transduction Associated 2) – related with reproduction traits. In mice, it was established that dynein regulates meiotic checkpoint during oocyte maturation [37] and KHDRBS2 was found to be associated with the number of teats in the GWA study in Large White pigs [33]. The other gene – PRIM2 (which encodes a DNA primase – large subunit) – was previously found in Bayesian GWAS to be in a gene network associated both with the number of stillborn piglets (SB) and the number of teats (NT) [33].
Among the strongest diversifying selection signals in PUL pig breed, we detected one with length over 1 Mb on SSC7 (35.9–37.3 Mb). This region was previously described to bear several QTLs for health, meat and carcass traits and exterior (Pig QTLdb, [38]). Within all diversifying selection signals in PUL pigs we detected candidate genes associated with inter alia striated muscle cell proliferation, skeletal muscle tissue regeneration (PPARD and MAPK14 - GO:0043403) and embryo implantation (ARHGDIB, PPARD - GO:0007566) important for fertility traits. The most interesting diversifying selection candidate in this breed seems to be PPARD gene (Peroxisome proliferator-activated receptor beta/delta) located on SSC7, which was shown to be associated both with ear morphology and backfat thickness [39, 40]. A previous study identified a missense mutation in the PPARD gene that significantly reduces its transcription activity, and consequently causes enlarged external ears in pigs [40, 41]. This is concordant with the phenotype of PUL breed which is the only breed with prick ear morphology in this study and the detected signal potentially results from diversifying selection for ear morphology among the studied breeds.
In the ZW breed, large size sweeps were detected on SSC1 (230.0–232.5 Mb) and 8 (45.2–47.5 Mb). These regions were previously shown to carry mainly QTLs for meat and carcass traits (SSC1) or health (SSC8) (Pig QTLdb, [38]). Within the strongest diversifying section signals in this breed, we detected genes associated inter alia with tryptophan metabolism (TDO2, AOX1) or salivary secretion (GUCY1A3, GUCY1B3). Tryptophan (TRP), as a precursor of neurotransmitters, was shown to be involved in pale, soft, exudative (PSE) pork syndrome [42] and dietary TRP deficiency was shown to correlate with decline of the appetite leading to reduced growth performance [43]. The TRP metabolism-associated genes may be then related with high stress resistance reported in the ZW breed and suggest its potential association with good meat quality and growth in this breed.
The largest genomic regions associated with differential selection in ZS breed were detected on SSC9. The regions were located between 75.6 Mb and 84.8 Mb of the chromosome sequence and were previously shown to carry mainly QTLs for exterior and reproduction features (Pig QTLdb, [38]). In ZS breed, the strongest detected selection signals were associated e.g. with genes responsible for: neuropeptide Y receptor activity and feeding behavior (NPY1R, NPY5R), metabolic processes (PON1, PON2, PON3) and cortisol metabolic process (REST). Our interest focused especially on genes associated with neuropeptide Y (NPY) activity. NPY has been implicated in several human diseases involving fat deposition aberrations and obesity [44]. This has special importance taking into account that Złotnicka Spotted breed is characterized by relatively high fat content in the carcass and, in comparison to other breeds, the meat of the ZS pigs is characterized by specific marbling resulting from higher intra muscular fat content. It was found that NPY reduces energy expenditure by decreasing adipose tissue thermogenesis [45, 46] and the obesity of mice was attenuated when NPY was knocked out [47, 48]. NPY deficiency also impaired responses to a palatable high fat diet in mice [49] and animals in which proopiomelanocortin neurons were knocked out were obese and hyperphagic [50–52]. These studies show the strong association of NPY with body fat generation by acting through several NYP receptors and suggest that two genes detected in this study that code for NPY receptors (2 and 5) may be connected with higher fat content and general higher fatness in the ZS breed when compared to the other studied breeds. The analysis of average back-fat thickens in the studied ZS animals confirmed general observations on this breed and showed that the average back-fat thickens was statistically significantly higher (P < 0.001) in ZS pigs (23.13 ± 3.53) than in three other analyzed breeds (in a range of 9.27 in PL to 18.03 in ZW breed) (Additional file 7).
Our FST-based comparison of white and spotted pig breeds revealed an ambiguous selection signal in SSC8 positioned in close vicinity to KIT gene – a known coat color modulator [53]. However, a few other strong signals were detected on SSC3, SSSC7 and SSC9, which did not overlap with any of genes commonly implicated in coat pattern in mammals, like e.g.: EDNRB, KITLG or MC1R [53]. Nevertheless, in a previous work similar to ours, QTL regions on chromosomes 7 and 9 were detected [54] which were shown to have effects on coat color extension in crossbreed pigs. Significant QTL loci affecting black spotting were also detected on SSC3 and SSC9 in exotic pig crosses [55]. This suggests that at least a few other coat color-related variants can be present in the studied pigs’ genomes. Another explanation of the observed signatures pattern can be that the phenotypes of both studied spotted breeds (PUL and ZS) are not yet fully stabilized or the combination of phenotypes used for comparisons was not fully correct, because PUL pigs can have black or reddish spots on slightly pigmented skin, whereas no skin pigmentation is characteristic for the other studied breeds, which may bias the obtained results.
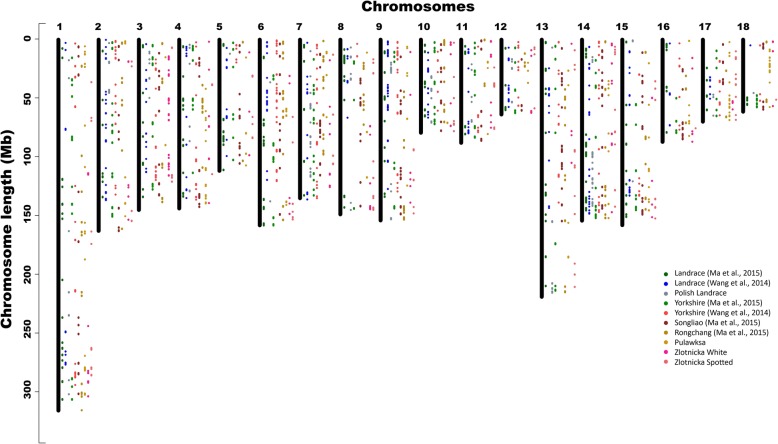
At least some of the detected divergent selection signals could be easily associated with genes potentially responsible for traits especially pronounced or fixed in the studied breeds and suggest the detection of genome regions connected with mechanisms underlying differentiating traits. To supplement this data with information on within-breed selection signatures, which are independent of breeds differentiation, we additionally used REHH statistics [15], which was shown to be the most powerful for detecting ongoing selection for which the target allele has a moderate to high frequency [56]. The use of such data allowed us to search for selection signals connected with traits that are selected across all studied breeds, representing variants that are still segregating and are under ongoing selection. With this approach we detected from 118 (ZS) to 176 (PL) core haplotypes per breed being the outliers at the significance level of 0.01. The general genomic pattern of these signals had visible similarity among the studied breeds and was comparable with previously analyzed pig breeds (Fig. 4). The visible clusters of strong selection signals were found on several chromosomes, however, the particularly visible were those located in distal parts of SSC1, SSC14 and SSC15 (Fig. 4). These chromosomal regions were previously associated with meat color density (SSC14) [3], growth, reproduction and immune responses (SSC1, SSC15) [29], which seems reasonable as most of pig breeds are selected towards the improvement of these traits.
Fig. 4.
The comparison of the detected, significant (P < 0.01) within-breed selection signatures with previous results obtained using similar methods in different breeds
To analyze in details the genomic regions with the strongest signs of selection, we took a closer look at regions bearing haplotypes with P-value lower than 0.001. After extending them to 1 cM on both sides, we detected from 35 (ZS) to 171 (PL) unique genes per breed potentially being under positive selection. Several of those genes are connected with important production features like growth and fertility. Exemplarily, in PL pigs we identified 10 genes belonging to the Hox family (HOX1 to HOX9 and HOX13), which are key factors in the implantation and development of the embryo and regulate the function of the endometrium [57–62]. In PL pigs, we also detected a gene belonging to the ESR receptor signaling pathway – STAT5 – responsible for the signal transduction from the estrogen receptor. The STAT5 gene itself is broadly tested for association with fertility and milk production traits [63–65]. The STAT5 protein plays a key role in signaling in the estrogen receptor pathway and therefore can play an important role in modulating the role of estrogen [66]. A gene encoding the estrogen receptor itself – ER – recognized as a key component responsible for female reproduction traits [67] and reproductive-related behavior in female [68] was also detected within the selection signatures found in the studied PL pigs. Furthermore, our analysis allowed identification of CXCL12 gene (C-X-C Motif Chemokine Ligand 12) which as a ligand for the G-protein coupled receptor plays an important role in many biological processes including embryogenesis [69]. In pigs, CXCL12 gene was identified as one of differentially expressed genes up-regulated in endometrium at 18th day of pregnancy, which can indicate its potential role in embryogenesis [70].
In the regions overlapping with the strongest within-breed selection signals in PUL breed as many as seven genes involved in lipid metabolism were detected (DHCR7, CLPS, PNPLA1, PPARD, BPIFB5, CLPSL2, BPIFB1) which may be associated with former selection of this breed toward reduced fat content in carcass. We also detected genes connected with immune system processes related to resistance to bacterial (FLNC, TCIRG1) and virus (POLD4, SND1) diseases, which fits to the characteristics of this native breed which is known to have a good disease resistance and adaptation to harsh environmental conditions. In Złotnicka White pigs, the strongest within-breed selection signals encompassed genes associated with inter alia the immune system processes and adaptation to environmental conditions or stress response. The first group of genes included those connected with inflammation mediated by chemokine and cytokine: GNG10, PRKCE, SOCS5 or inflammatory mediator regulation of TRP channels – MAPK10 (mitogen-activated protein kinase 10) and PRKCE (protein kinase C epsilon), which may contribute to the good health and disease resistance observed in this breed. The second group included MCFD2 gene engaged in response to a stimulus and other genes responsible for response to stress (GO:00069 50)/stress-activated MAPK cascade, like: MARC1, ERCC4, DUSP10, MAPK10.
In Złotnicka Spotted breed, some predicted genes involved in lipid and carbohydrates metabolism were detected and were engaged in processes connected with: sphingolipid catabolism (LOC102167689), membrane lipid catabolism or glucosidase/galactosidase activity (LOC102167689) which as metabolic processes are often associated with selection signals in pigs [3]. The detection of the SCLY gene encoding isoform X1 of selenocysteine lyase associated with selenium metabolism and viral mRNA translation seems to be interesting because of the fact that it was established that SCLY selenocysteine plays a key role in lipid metabolism and can be related to selenium concentration in meat [71].
Conclusions
Summarizing, in this study we used two different but largely complementary statistical approaches, REHH and pairwise FST, to detect selection signatures in four Polish pig breeds, including three native unselected breeds. FST-derived signatures were useful to detect diversifying selection signals across breeds (represented by loci for which alleles were differentially fixed in different breeds) and allowed us to indicate genes connected e.g. with fertility, growth and metabolism which are potentially responsible for phenotypes of the studied breeds. These selection signals also comprised PPARD gene that contributes to the shape of external ears in pigs and genes encoding neuropeptide Y receptors involved in fat deposition and stress response which are important features differentiating the studied breeds. REHH statistics pointed several within-breed selection signals overlapping with genes connected with a broad range of functions, including e.g.: metabolic pathways, immune system response or implantation and development of the embryo. The study provides many potential candidate genes responsible for traits selected in the individual breeds and gives a strong basis for further studies aimed at closing the gap between the genotype and phenotype of the studied pig breeds.
Additional files
Breed characteristics. (DOCX 12 kb)
Standardized FST values (di) for all studied breeds averaged within ten-SNP sliding windows. (XLSX 4722 kb)
Genes detected within the strongest diversifying selection signals detected in the studied pig breeds. (XLSX 71 kb)
Summary characteristics of core regions (CR) or core haplotypes (CH) in the studied pig breeds. (XLSX 13 kb)
REHH P-values for the detected core haplotypes with the highest frequency. (XLSX 730 kb)
Genes detected within the strongest within-breed selection signals detected in the studied pig breeds. (XLSX 99 kb)
Comparison of selected phenotypic traits for the studied pig breeds. (DOCX 19 kb)
Acknowledgments
Funding
The study was financed from funds of the project: “Directions for use and conservation of livestock genetic resources in sustainable development” co-financed by the National Research and Development Center (Poland) under the Strategic Research and Development Program: “Environment, Agriculture and Forestry” – BIOSTRATEG, the decision number BIOSTRATEG2/297267/14/NCBR/2016.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- cM
Centimorgan
- GWAS
Genome-wide association study
- HWE
Hardy-Weinberg equilibrium
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- PCA
Principal component analysis
- PL
Polish Landrace
- PUL
Puławska pig
- QTL
Quantitative trait locus
- REHH
Relative extended haplotype homozygosity
- SNP
Single nucleotide polymorphism
- SSC
Sus scrofa (chromosome)
- ZS
Złotnicka Spotted pig
- ZW
Złotnicka White pig
Authors’ contributions
AG, IJ, KPT, ESG, TB, KS, ES and MSN provided material and performed the experiments. AG, TS, KRM and MBP interpreted the results and wrote the manuscript. JK supervised study and acquired funding. All authors read and approved the final manuscript.
Ethics approval
All animal procedures were approved by the Local Animal Care Ethics Committee No. II in Kraków - permission number 1293/2016 in accordance with EU regulations.
The animals in this study have been sourced from private studs supervised by organs responsible for stud books (Polish Pig Breeders and Producers Association – POLSUS and Poznań University of Life Sciences). The material was collected based on cooperation agreements between these Organizations and the National Research Institute of Animal Production, dated September 9, 2004 and July 18, 2016, respectively.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Artur Gurgul, Email: artur.gurgul@izoo.krakow.pl.
Igor Jasielczuk, Email: igor.jasielczuk@izoo.krakow.pl.
Katarzyna Ropka-Molik, Email: katarzyna.ropka@izoo.krakow.pl.
Ewelina Semik-Gurgul, Email: ewelina.semik@izoo.krakow.pl.
Klaudia Pawlina-Tyszko, Email: klaudia.pawlina@izoo.krakow.pl.
Tomasz Szmatoła, Email: tomasz.szmatola@izoo.krakow.pl.
Magdalena Szyndler-Nędza, Email: magdalena.szyndler@izoo.krakow.pl.
Monika Bugno-Poniewierska, Email: monika.bugno@izoo.krakow.pl.
Tadeusz Blicharski, Email: t.blicharski@ighz.pl.
Karolina Szulc, Email: karolasz@up.poznan.pl.
Ewa Skrzypczak, Email: ewa.skrzypczak@up.poznan.pl.
Jędrzej Krupiński, Email: jedrzej.krupinski@izoo.krakow.pl.
References
- 1.Minozzi G, Nicolazzi EL, Stella A, Biffani S, Negrini R, Lazzari B, et al. Genome wide analysis of fertility and production traits in Italian Holstein cattle. PLoS One. 2013;8:e80219. doi: 10.1371/journal.pone.0080219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Simoni Gouveia JJ, da Silva MVGB, Paiva SR, de Oliveira SMP. Identification of selection signatures in livestock species. Genet Mol Biol. 2014;37:330–342. doi: 10.1590/S1415-47572014000300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, Ding X, Qanbari S, Weigend S, Zhang Q, Simianer H. Properties of different selection signature statistics and a new strategy for combining them. Heredity. 2015;115:426–436. doi: 10.1038/hdy.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaral AJ, Ferretti L, Megens H-J, Crooijmans RP, Nie H, Ramos-Onsins SE, et al. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One. 2011;6:e14782. doi: 10.1371/journal.pone.0014782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin C-J, Megens H-J, Barrio AM, Maqbool K, Sayyab S, Schwochow D, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai H, Huang L, Ren J. Genetic diversity, linkage disequilibrium and selection signatures in Chinese and Western pigs revealed by genome-wide SNP markers. PLoS One. 2013;8:e56001. doi: 10.1371/journal.pone.0056001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson S, Lu ZH, Megens H-J, Archibald AL, Haley C, Jackson IJ, et al. Signatures of diversifying selection in European pig breeds. PLoS Genet. 2013;9:e1003453. doi: 10.1371/journal.pgen.1003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Zhang H, Zhang Q, Ding X. Identification of selection footprints on the X chromosome in pig. PLoS One. 2014;9:e94911. doi: 10.1371/journal.pone.0094911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S, Li X, Li K, Fan B, Tang Z. A genome-wide scan for signatures of selection in Chinese indigenous and commercial pig breeds. BMC Genetics. 2014;15:7. doi: 10.1186/1471-2156-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrowicz S, Czubak J, Ratajszczak M. Utilisation of crosses of native pig breeds to create breed groups of meat and meat-lard performance. Roczniki Nauk Rolniczych. 1954;68(B-4):369–395. [Google Scholar]
- 11.Florowski T, Pisula A, Adamczak L, Buczyński JT, Orzechowska B. Technological parametres of meat in pigs of two polish local breeds - Zlotnicka spotted and Pulawska. Anim Sci Pap Rep. 2006;24:217–224. [Google Scholar]
- 12.Szulc K, Skrzypczak E, Buczyński JT, Stanisławski D, Jankowska-Mąkosa A, Knecht D. Evaluation of fattening and slaughter performance and determination of meat quality in Złotnicka spotted pigs and their crosses with the Duroc breed. Czech J Anim Sci. 2012;57:95–107. doi: 10.17221/5561-CJAS. [DOI] [Google Scholar]
- 13.Bogucka J, Kapelański W. Microstructure of longissimus Lumborum muscle and meat quality of native polish pig breeds: Złotnicka spotted and Puławska. Ann Ani Sci. 2016;16:1199–1210. doi: 10.1515/aoas-2016-0029. [DOI] [Google Scholar]
- 14.Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 16.Zhao F, McParland S, Kearney F, Du L, Berry DP. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet Sel Evol. 2015;47:49. doi: 10.1186/s12711-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qanbari S, Pimentel EC, Tetens J, Thaller G, Lichtner P, Sharifi AR, et al. A genome-wide scan for signatures of recent selection in Holstein cattle. Anim Genet. 2010;41:377–389. doi: 10.1111/j.1365-2052.2009.02016.x. [DOI] [PubMed] [Google Scholar]
- 22.Tortereau F, Servin B, Frantz L, Megens H, Milan D, Rohrer G, et al. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics. 2012;13:586. doi: 10.1186/1471-164-13-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Jing, Vasaikar Suhas, Shi Zhiao, Greer Michael, Zhang Bing. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Research. 2017;45(W1):W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 27.Gregory TR. Artificial selection and domestication: modern lessons from Darwin’s enduring analogy. Evo Edu Out-reach. 2009;2:5–27. doi: 10.1007/s12052-008-0114-z. [DOI] [Google Scholar]
- 28.Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Yang S, Tang Z, Li K, Rothschild MF, Liu B, et al. Genome-wide scans to detect positive selection in large white and Tongcheng pigs. Anim Genet. 2014;45:329–339. doi: 10.1111/age.12128. [DOI] [PubMed] [Google Scholar]
- 30.Qiao R, Gao J, Zhang Z, Li L, Xie X, et al. Genome-wide association analyses reveal significant loci and strong candidate genes for growth and fatness traits in two pig populations. Genet Sel Evol. 2015;47:17. doi: 10.1186/s12711-015-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Yang B, Zhang J, Cui L, Ma J, et al. Genome-wide association studies for fatty acid metabolic traits in five divergent pig populations. Sci Rep. 2016;6:24718. doi: 10.1038/srep24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji J, Zhou L, Guo Y, Huang L, Ma J. Genome-wide association study identifies 22 new loci for body dimension and body weight traits in a white Duroc×Erhualian F2 intercross population. Asian Austral J Anim Sci. 2017;30:1066–1073. doi: 10.5713/ajas.16.0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verardo LL, Silva FF, Lopes MS, Madsen O, Bastiaansen JW, Knol EF, et al. Revealing new candidate genes for reproductive traits in pigs: combining Bayesian GWAS and functional pathways. Genet Sel Evol. 2016;48:9. doi: 10.1186/s12711-016-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le TH, Christensen OF. Nielsen B, Sahana G. Genome-wide association study for conformation traits in three Danish pig breeds. Genet Sel Evol. 2017;49:12. doi: 10.1186/s12711-017-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seto M, Ohta M, Ikenoue T, Sugimoto T, Asaoka Y, Tada M, et al. Reduced expression of RAS protein activator like-1 in gastric cancer. Int J Cancer. 2011;128:1293–1302. doi: 10.1002/ijc.25459. [DOI] [PubMed] [Google Scholar]
- 36.Shen LY, Luo J, Lei HG, Jiang YZ, Bai L, Li MZ, et al. Effects of muscle fiber type on glycolytic potential and meat quality traits in different Tibetan pig muscles and their association with glycolysis-related gene expression. Genet Mol Res. 2015;14:14366–14378. doi: 10.4238/2015.November.13.22. [DOI] [PubMed] [Google Scholar]
- 37.Zhang D, Yin S, Jiang MX, Ma W, Hou Y, Lian CG, et al. Cytoplasmic dynein participates in meiotic checkpoint inactivation in mouse oocytes by transporting cytoplasmic mitotic arrest-deficient (mad) proteins from kinetochores to spindle poles. Reproduction. 2007;133:685–695. doi: 10.1530/rep.1.01167. [DOI] [PubMed] [Google Scholar]
- 38.Hu Z-L, Park CA, Reecy JM. Developmental progress and current status of the animal QTLdb. Nucleic Acids Res. 2016;44(Database issue):D827–D833. doi: 10.1093/nar/gkv1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meidtner K, Schwarzenbacher H, Scharfe M, Severitt S, Blöcker H, Fries R. Haplotypes of the porcine peroxisome proliferator-activated receptor delta gene are associated with backfat thickness. BMC Gen. 2009;10:76. doi: 10.1186/1471-2156-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren J, Duan Y, Qiao R, Yao F, Zhang Z, Yang B, et al. A missense mutation in PPARD causes a major QTL effect on ear size in pigs. PLoS Genet. 2011;7:e1002043. doi: 10.1371/journal.pgen.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Duan Y, Wu Z, Zhang H, Ren J, Huang L. PPARD is an inhibitor of cartilage growth in external ears. Int J Biol Sci. 2017;13:669–681. doi: 10.7150/ijbs.19714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adeola O, Ball RO. Hypothalamic neurotransmitter concentrations and meat quality in stressed pigs offered excess dietary tryptophan and tyrosine. J Anim Sci. 1992;70:1888–1894. doi: 10.2527/1992.7061888x. [DOI] [PubMed] [Google Scholar]
- 43.Sève B. Physiological roles of tryptophan in pig nutrition. Adv Exp Med Biol. 1999;467:729–741. doi: 10.1007/978-1-4615-4709-9_95. [DOI] [PubMed] [Google Scholar]
- 44.Brothers SP, Wahlestedt C. Therapeutic potential of neuropeptide Y (NPY) receptor ligands. EMBO Mol Med. 2010;2:429–439. doi: 10.1002/emmm.201000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao PT, Yang L, Aja S, Moran TH, Bi S. Knockdown of NPY expression in the dorsomedial hypothalamus promotes development of brown adipocytes and prevents diet-induced obesity. Cell Metab. 2011;13:573–583. doi: 10.1016/j.cmet.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Cline MA, Gilbert ER. Hypothalamus-adipose tissue crosstalk: neuropeptide Y and the regulation of energy metabolism. Nutr Metab. 2014;11:27. doi: 10.1186/1743-7075-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erickson JC, Hollopeter G, Palmiter RD. Attenuation of the obesity syndrome of Ob/Ob mice by the loss of neuropeptide Y. Science. 1996;274:1704–1707. doi: 10.1126/science.274.5293.1704. [DOI] [PubMed] [Google Scholar]
- 48.Segal-Lieberman G, Trombly DJ, Juthani V, Wang X, Maratos-Flier E. NPY ablation in C57BL/6 mice leads to mild obesity and to an impaired refeeding response to fasting. Am J Physiol Endocrinol Metab. 2003;284:E1131–E1139. doi: 10.1152/ajpendo.00491.2002. [DOI] [PubMed] [Google Scholar]
- 49.Patel HR, Qi Y, Hawkins EJ, Hileman SM, Elmquist JK, Imai Y, Ahima RS. Neuropeptide Y deficiency attenuates responses to fasting and high-fat diet in obesity-prone mice. Diabetes. 2006;55:3091–3098. doi: 10.2337/db05-0624. [DOI] [PubMed] [Google Scholar]
- 50.Diané Abdoulaye, Pierce W. David, Russell James C., Heth C. Donald, Vine Donna F., Richard Denis, Proctor Spencer D. Down-regulation of hypothalamic pro-opiomelanocortin (POMC) expression after weaning is associated with hyperphagia-induced obesity in JCR rats overexpressing neuropeptide Y. British Journal of Nutrition. 2013;111(05):924–932. doi: 10.1017/S0007114513003061. [DOI] [PubMed] [Google Scholar]
- 51.Raffan E, Dennis RJ, O'Donovan CJ, Becker JM, Scott RA, Smith SP, et al. A deletion in the canine POMC gene is associated with weight and appetite in obesity-prone labrador retriever dogs. Cell Metab. 2016;23:893–900. doi: 10.1016/j.cmet.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang XJ, Xu SH, Liu L, Song ZG, Jiao HC, Lin H. Dietary fat alters the response of hypothalamic neuropeptide Y to subsequent energy intake in broiler chickens. J Exp Biol. 2017;220:607–614. doi: 10.1242/jeb.143792. [DOI] [PubMed] [Google Scholar]
- 53.Fontanesi L, Russo V. Molecular genetics of coat colour in pigs. Acta Argic Slovenica. 2013;Suppl 4:15–20. [Google Scholar]
- 54.Fernández AI. A classical QTL scan supports the complexity of the genetic basis of coat colour in pig. Acta Agric Slovenica. 2013;Suppl 4:49–52. [Google Scholar]
- 55.Hirooka H, de Koning DJ, van Arendonk JA, Harlizius B, de Groot PN, Bovenhuis H. Genome scan reveals new coat color loci in exotic pig cross. J Hered. 2002;93:1–8. doi: 10.1093/jhered/93.1.1. [DOI] [PubMed] [Google Scholar]
- 56.Simianer H. Statistical problems in livestock population genomics. In: Proceedings of the 10th World Congress on Genetics Applied to Livestock Production: 17–22 August 2014; Vancouver; 2014. http://www.wcgalp.org/system/files/proceedings/2014/statistical-problems-livestock-population-genomics.pdf.
- 57.Taylor HS. The role of HOX genes in human implantation. Hum Reprod Update. 2000;6:75–79. doi: 10.1093/humupd/6.1.75. [DOI] [PubMed] [Google Scholar]
- 58.Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006;12:731–746. doi: 10.1093/humupd/dml004. [DOI] [PubMed] [Google Scholar]
- 59.Vitiello D, Kodaman PH, Taylor HS. HOX genes in implantation. Semin Reprod Med. 2007;25:431–436. doi: 10.1055/s-2007-991040. [DOI] [PubMed] [Google Scholar]
- 60.Xu B, Geerts D, Bu Z, Ai J, Jin L, Li Y, et al. Regulation of endometrial receptivity by the highly expressed HOXA9, HOXA11 and HOXD10 HOX-class homeobox genes. Hum Reprod. 2014;29:781–790. doi: 10.1093/humrep/deu004. [DOI] [PubMed] [Google Scholar]
- 61.Du H, Taylor HS. The role of Hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb Perspect Med. 2015;6:a023002. doi: 10.1101/cshperspect.a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin R, Jia H, Chen S, Zhao S. Expression analysis of the porcine Homeobox A11 gene and its association with litter size in large white pigs. Czech J Anim Sci. 2016;61:243–249. doi: 10.17221/127/2014-CJAS. [DOI] [Google Scholar]
- 63.Khatib H, Monson RL, Schutzkus V, Kohl DM, Rosa GJ, Rutledge JJ. Mutations in the STAT5A gene are associated with embryonic survival and milk composition in cattle. J Dairy Sci. 2008;91:784–793. doi: 10.3168/jds.2007-0669. [DOI] [PubMed] [Google Scholar]
- 64.Oikonomou G, Michailidis G, Kougioumtzis A, Avdi M, Banos G. Effect of polymorphisms at the STAT5A and FGF2 gene loci on reproduction, milk yield and lameness of Holstein cows. Res Vet Sci. 2011;91:235–239. doi: 10.1016/j.rvsc.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Coizet Beatrice, Frattini Stefano, Nicoloso Letizia, Iannuzzi Leopoldo, Coletta Angelo, Talenti Andrea, Minozzi Giulietta, Pagnacco Giulio, Crepaldi Paola. Polymorphism of the STAT5A, MTNR1A and TNFα genes and their effect on dairy production in Bubalus bubalis. Italian Journal of Animal Science. 2017;17(1):31–37. doi: 10.1080/1828051X.2017.1335181. [DOI] [Google Scholar]
- 66.Yamashita H, Iwase H. The role of Stat5 in estrogen receptor-positive breast cancer. Breast Cancer. 2002;9:312–318. doi: 10.1007/BF02967610. [DOI] [PubMed] [Google Scholar]
- 67.Wang H, Eriksson H, Sahlin L. Estrogen receptors alpha and beta in the female reproductive tract of the rat during the estrous cycle. Biol Reprod. 2000;63:1331–1340. doi: 10.1095/biolreprod63.5.1331. [DOI] [PubMed] [Google Scholar]
- 68.Ogawa S, Eng V, Taylor J, Lubahn DB, Korach KS, Pfaff DW. Roles of estrogen receptor-alpha gene expression in reproduction-related behaviors in female mice. Endocrinology. 1998;139:5070–5081. doi: 10.1210/endo.139.12.6357. [DOI] [PubMed] [Google Scholar]
- 69.Zhao HB, Tang CL, Hou YL, Xue LR, Li MQ, Du MR, et al. CXCL12/CXCR4 Axis Triggers the Activation of EGF Receptor and ERK Signaling Pathway in CsA-Induced Proliferation of Human Trophoblast. PLoS One. 2012;7:e38375. doi: 10.1371/journal.pone.0038375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Li A, Chen W, Wei J, Fu J, Wang A. Differential gene expression in uterine endometrium during implantation in pigs. Biol Reprod. 2015;92:52. doi: 10.1095/biolreprod.114.123075. [DOI] [PubMed] [Google Scholar]
- 71.Seale LA, Gilman CL, Hashimoto AC, Ogawa-Wong AN, Berry MJ. Diet-induced obesity in the Selenocysteine Lyase knockout mouse. Antioxid Redox Signal. 2015;23:761–774. doi: 10.1089/ars.2015.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Breed characteristics. (DOCX 12 kb)
Standardized FST values (di) for all studied breeds averaged within ten-SNP sliding windows. (XLSX 4722 kb)
Genes detected within the strongest diversifying selection signals detected in the studied pig breeds. (XLSX 71 kb)
Summary characteristics of core regions (CR) or core haplotypes (CH) in the studied pig breeds. (XLSX 13 kb)
REHH P-values for the detected core haplotypes with the highest frequency. (XLSX 730 kb)
Genes detected within the strongest within-breed selection signals detected in the studied pig breeds. (XLSX 99 kb)
Comparison of selected phenotypic traits for the studied pig breeds. (DOCX 19 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.