Abstract
Background
While most cancer cells preferentially express the M2 isoform of the glycolytic enzyme pyruvate kinase (PKM2), PKM2 is dispensable for tumor development in several mouse cancer models. PKM2 is expressed in human pancreatic cancer, and there have been conflicting reports on the association of PKM2 expression and pancreatic cancer patient survival, but whether PKM2 is required for pancreatic cancer progression is unknown. To investigate the role of PKM2 in pancreatic cancer, we used a conditional allele to delete PKM2 in a mouse model of pancreatic ductal adenocarcinoma (PDAC).
Results
PDAC tumors were initiated in LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-Cre (KP−/−C) mice harboring a conditional Pkm2 allele. Immunohistochemical analysis showed PKM2 expression in wild-type tumors and loss of PKM2 expression in tumors from Pkm2 conditional mice. PKM2 deletion had no effect on overall survival or tumor size. Loss of PKM2 resulted in pyruvate kinase M1 (PKM1) expression, but did not affect the number of proliferating cells. These findings are consistent with results in other cancer models.
Conclusions
PKM2 is not required for initiation or growth of PDAC tumors arising in the KP−/−C pancreatic cancer model. These findings suggest that, in this mouse PDAC model, PKM2 expression is not required for pancreatic tumor formation or progression.
Electronic supplementary material
The online version of this article (10.1186/s40170-018-0188-1) contains supplementary material, which is available to authorized users.
Keywords: PKM2, PDAC, Pyruvate kinase, Pancreatic cancer
Introduction
Pyruvate kinase is an enzyme that catalyzes the final step of glycolysis to convert phosphoenolpyruvate and ADP to pyruvate and ATP. Pyruvate kinase has four isoforms encoded by two genes. The PKLR gene encodes PKL, which is expressed primarily in the liver, and PKR, which is expressed in erythrocytes [1]. The PKM gene encodes both the PKM1 and PKM2 isoforms, with isoform selected determined by alternate mRNA splicing to include either exon 9 or 10 [2, 3]. Both PKM isoforms catalyze the same reaction in glycolysis, but PKM1 has constitutively high catalytic activity, whereas the catalytic activity of PKM2 is allosterically regulated [2, 4]. As a glycolytic enzyme, PKM2 can exist in either a low activity state that promotes biosynthesis or in a high activity state that is similar to that of PKM1 and promotes oxidative glucose metabolism [2].
PKM2 is expressed in a variety of cancer types, and both metabolic and non-metabolic functions for PKM2 in cancer have been proposed [5–10]. However, accumulating evidence suggests that PKM2 is not required for the growth or progression of most tumors, and non-metabolic roles for PKM2 remain controversial [11–18]. For instance, deletion of PKM2 has been shown to promote tumor progression in breast cancer and medulloblastoma models [14, 15] and a recent study found that systemic PKM2 depletion (while maintaining PKM1 expression) promoted tumorigenesis [18]. However, other studies have found that PKM2 is dispensable for development of leukemia, liver cancer, colon cancer, lymphoma, lung cancer, and squamous cell carcinoma [16–18]. Thus, the role of PKM2 in cancer may depend on the genetic and/or environmental context.
Reports regarding the role of PKM2 in pancreatic cancer have also been conflicting. PKM2 has been reported to promote proliferation, migration, invasion, and angiogenesis and to decrease apoptosis in pancreatic cancer cell lines [19–21]. Other cell line studies have reported that PKM2 expression is important for gemcitabine resistance [22–26]. High PKM2 expression in human pancreatic tumors has been associated with larger tumor size, worse overall survival, and shorter recurrence-free survival of pancreatic cancer patients [20, 24, 27, 28]. However, one study reported that higher PKM2 expression was associated with longer overall survival of pancreatic cancer patients [29], and other analyses found that expression of PKM2 had no effect on overall survival in pancreatic cancer [30, 31].
Given the controversy surrounding the role of PKM2 in pancreatic tumors, we evaluated the importance of PKM2 in PDAC by crossing mice harboring a conditional Pkm2 allele [14] to the LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-Cre (KP−/−C) mouse PDAC model [32]. By comparing mice with and without PKM2 conditional alleles, we aimed to determine the requirement for PKM2 in pancreatic cancer.
Results
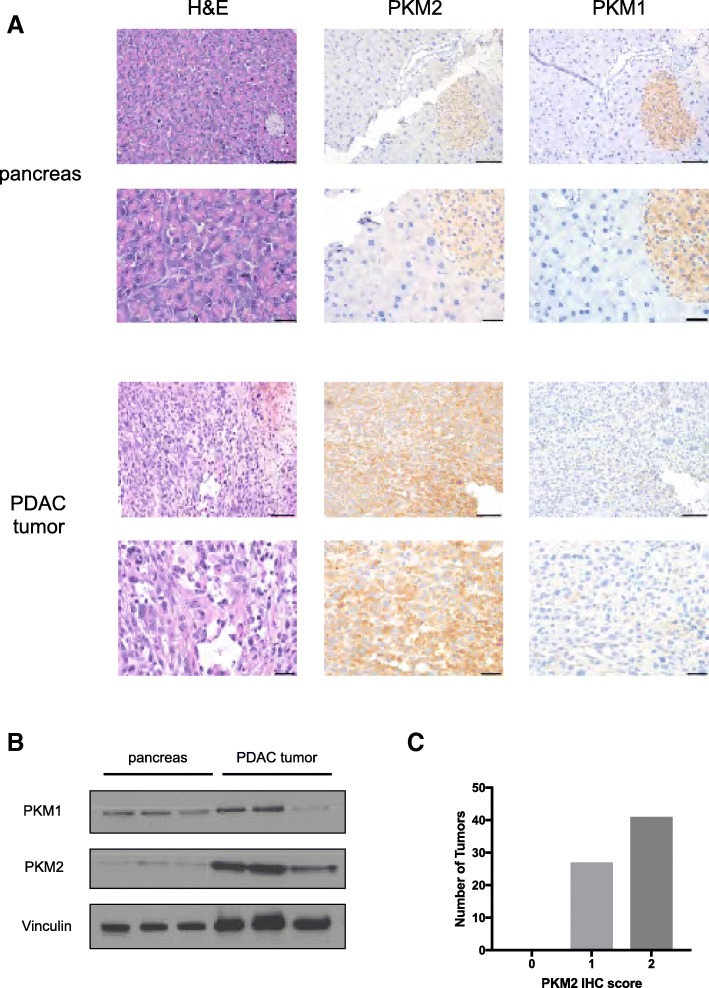
To examine PKM2 isoform expression in the pancreas and in pancreatic tumors, we performed immunohistochemistry on normal mouse pancreatic tissue and mouse pancreatic tumor tissue sections using PKM2 or PKM1 isoform-specific antibodies [14]. PKM1 and PKM2 staining was prominent in the islets of normal mouse pancreas, whereas the rest of the pancreas exhibited minimal PKM1 or PKM2 expression (Fig. 1a), consistent with previously published reports [1]. Compared to the normal pancreas, end-stage PDAC tumors from LSL-KrasG12D/+; Trp53flox/flox;Pdx-1-Cre (KP−/−C) [32] (Fig. 1a) or LSL-KrasG12D/+;LSL–Trp53R172H/+;Pdx-1-Cre (KPC) [33] (Additional file 1: Figure S1A) mice showed increased staining for PKM2 and minimal PKM1 staining. This expression pattern was further verified by Western blot analysis of tissue lysates (Fig. 1b). To examine PKM2 expression in human pancreatic tumors, we performed PKM2 immunohistochemistry on 68 pancreatic tumor tissue samples using a tissue array. All of the samples analyzed stained positive for PKM2 expression (Fig. 1c).
Fig. 1.
PKM2 is expressed in normal pancreas and in PDAC tumors. a Normal mouse pancreas and mouse PDAC tumor sections from end-stage LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-Cre (KP−/−C) mice were stained with hematoxylin and eosin (H&E) and isoform-specific antibodies against PKM2 or PKM1 as shown. Scale bars represent 50 μm for all images at × 20 magnification (top) and 20 μm for all images at × 40 magnification (bottom). b Western blot of normal mouse pancreas and mouse PDAC tumor lysates (from KP−/−C mice) performed using isoform-specific antibodies against PKM1 and PKM2 as well as an antibody against vinculin as a control. c Distribution of PKM2 staining intensities from a tissue microarray containing sections from 68 human pancreatic adenocarcinoma tumors
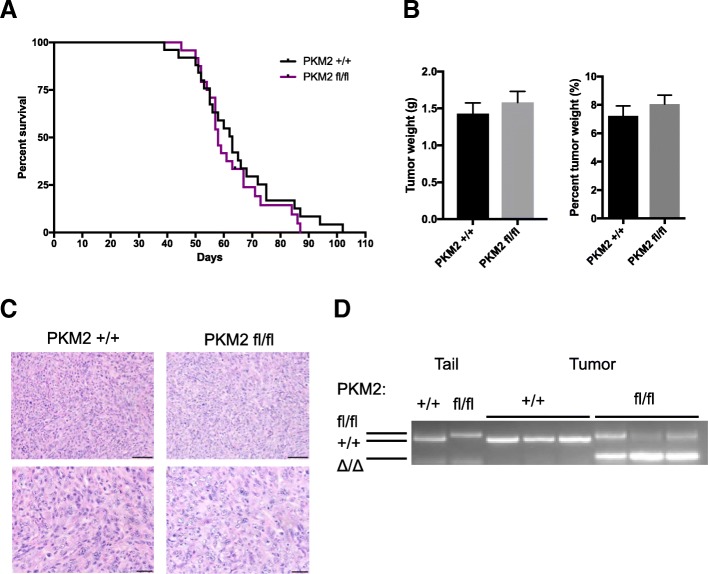
To delete Pkm2 in PDAC tumors, mice harboring a conditional Pkm2 allele (Pkm2flox/flox) [14] were crossed to KP−/−C mice [32]. Overall survival was not significantly different between the KP−/−C;Pkm2+/+ mice and the KP−/−C;Pkm2flox/flox mice (Fig. 2a), and there were no significant differences in weight of end-stage tumors in the KP−/−C;Pkm2+/+ mice compared to the KP−/−C;Pkm2flox/flox mice (Fig. 2b). Histological analysis revealed no major differences between the KP−/−C;Pkm2+/+ tumors and KP−/−C;Pkm2flox/flox tumors (Fig. 2c). To verify that Pkm2 was deleted in tumors from KP−/−C;Pkm2flox/flox mice, we analyzed genomic DNA from KP−/−C;Pkm2+/+ and KP−/-C;Pkm2flox/flox pancreatic tumors and confirmed that PKM2 deletion had occurred in tumors arising in KP−/−C;Pkm2flox/flox mice (Fig. 2d).
Fig. 2.
PKM2 deletion in PDAC tumors does not affect mouse survival, tumor weight, or tumor histology. a Kaplan-Meier curve showing survival of the KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mouse cohorts. No statistically significant difference in survival was observed between the two cohorts (n = 24 Pkm2flox/flox mice, 25 Pkm2+/+ mice per cohort, p = 0.3862, log-rank (Mantel-Cox) test). b The weight of tumors isolated from end-stage KP−/−C Pkm2+/+ and KPC Pkm2flox/flox mice, and the percentage of the tumor weight relative to whole body weight is shown. c Hematoxylin and eosin (H&E) staining of PDAC tumors from KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice. d PCR genotyping of the PKM2 allele in tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice is shown. Analysis of tail DNA from Pkm2+/+ and PKM2flox/flox mice is also shown as a control
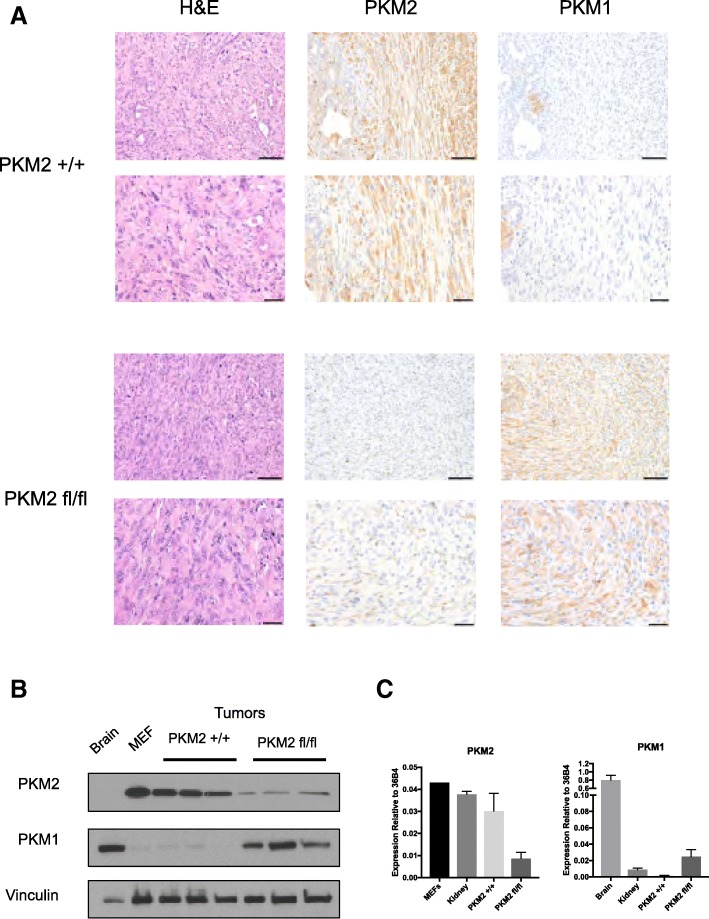
We also analyzed pyruvate kinase expression in tumors isolated from KP−/−C;Pkm2+/+ and KP−/−C;Pkm2flox/flox mice using immunohistochemistry with pyruvate kinase isoform-specific antibodies. As expected, the analysis of pancreatic tumors showed that KP−/−C;Pkm2+/+ tumors expressed almost exclusively PKM2 (Fig. 3a), whereas consistent with our genomic DNA analysis, tumors from KP−/−C;Pkm2flox/flox mice showed no evidence of PKM2 expression in most cells (Fig. 3a). Tumors from KP−/−C;Pkm2flox/flox animals showed increased PKM1 expression compared to KP−/−C;Pkm2+/+ wild-type tumors (Fig. 3a). To quantify pyruvate kinase expression in these tumors, we performed Western blot and qPCR analysis. Western blotting of KP−/−C;Pkm2+/+ tumor lysates showed that these tumors almost exclusively expressed PKM2 (Fig. 3b); qPCR analysis of KP−/−C;Pkm2+/+ tumor RNA also showed PKM2 expression and very little PKM1 expression (Fig. 3c). Western blotting of KP−/−C;Pkm2flox/flox tumor lysates showed low PKM2 expression and high PKM1 expression (Fig. 3b). Similarly, qPCR analysis of KP−/−C;Pkm2flox/flox tumor RNA showed increased PKM1 expression and decreased PKM2 expression, although PKM1 mRNA expression was much lower than that observed in the brain, a tissue that normally expresses PKM1 [1] (Fig. 3c). To investigate whether PKLR expression was induced by PKM2 loss, we also performed Western blot and qPCR analysis for PKLR. Neither KP−/−C;Pkm2+/+ nor KP−/−C;Pkm2flox/flox tumor lysates exhibited evidence for PKLR expression at the RNA or protein level (Additional file 1: Figure S2A, B). qPCR analysis was also performed for PKM2 skip, a PKM isoform that excludes exons 9 and 10, which has been reported to be expressed in PKM2flox/flox mice in other models [14]. KP−/−C;Pkm2+/+ tumors did not express PKM2 skip, while KP−/−C;Pkm2flox/flox tumors expressed PKM2 skip message at detectable levels (Additional file 1: Figure S2C).
Fig. 3.
PKM2 deletion in PDAC tumors results in increased PKM1 expression relative to wild-type tumors. a Tissue sections from tumors arising in end-stage KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice were stained with hematoxylin and eosin (H&E) and isoform-specific antibodies against PKM2 or PKM1 as shown. Scale bars represent 50 μm for all images at × 20 magnification (top) and 20 μm for all images at × 40 magnification (bottom). b Western blot of tumor lysates from KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice was performed using isoform-specific antibodies against PKM2 or PKM1 as well as an antibody against vinculin as a control. c PKM2 and PKM1 expression was assessed by qPCR in tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice
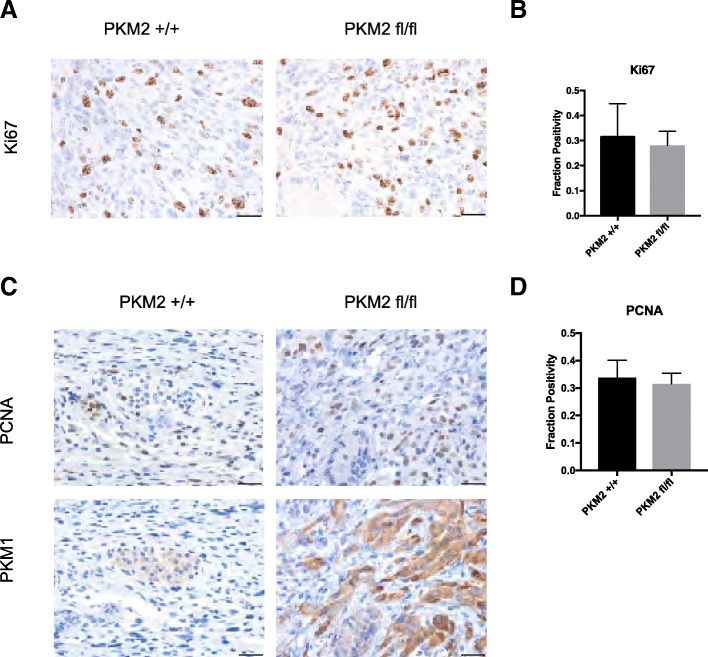
To compare cell proliferation in KP−/−C;Pkm2+/+ and KP−/−C;Pkm2flox/flox tumors, we performed immunohistochemical staining with antibodies against the proliferative markers Ki67 or PCNA (Fig. 4a–d). There was no statistically significant difference in Ki67 or PCNA staining between KP−/−C;Pkm2+/+ and KP−/−C;Pkm2flox/flox tumors (Fig. 4b, d). To determine if the proliferating cells expressed pyruvate kinase, we stained serial sections for PCNA or PKM1 (Fig. 4c). KP−/−C;Pkm2+/+ tumors had low PKM1 expression, and areas with high PKM1 expression did not correlate with areas of PCNA expression. KP−/−C;Pkm2flox/flox tumors had higher PKM1 expression, and these tumors had many regions with only either PCNA or PKM1 expression and some regions with both PKM1 and PCNA expression. Consistent with previous reports, proliferating cells in Pkm2flox/flox tumors may downregulate expression of pyruvate kinase to enable tumor growth.
Fig. 4.
PKM2 deletion does not alter PDAC tumor cell proliferation. a Tumor tissue sections from end-stage KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice were stained with an antibody against Ki67 as shown. Scale bars represent 50 μm for all images at × 20 magnification (top) and 20 μm for all images at × 40 magnification (bottom). b Fraction of nuclei positive for Ki67 was assessed as shown. No significant difference in Ki67 positivity was observed in tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice. c Tumor tissue sections from KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice were stained with isoform-specific antibodies against PKM1 and against PCNA as shown. Scale bars represent 50 μm for all images at × 20 magnification (top) and 20 μm for all images at × 40 magnification (bottom). d Fraction of nuclei positive for PCNA was assessed as shown. No significant difference in PCNA positivity was observed in tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice
Discussion
These results argue that PKM2 expression has no significant effect on tumor initiation or progression in a mouse pancreatic cancer model driven by mutant Kras and deletion of both copies of Trp53. Although PKM2 is highly expressed in both mouse and human PDAC tumors, deletion of PKM2 had no effect on tumor size or survival of PDAC tumor-bearing mice. Our findings are consistent with meta-analyses of published datasets examining PKM2 expression and survival of human pancreatic cancer patients, which found that expression of PKM2 had no effect on overall survival [30, 31].
Our findings that PKM2 expression is not required for pancreatic tumor growth are consistent with findings from other cancer models showing that PKM2 is similarly not required for tumor development. This includes mouse models of colon cancer [12], breast cancer [14], medulloblastoma [15], leukemia [16], hepatocellular carcinoma [17, 18], lymphoma [18], lung cancer [18], and squamous cell carcinoma [18]. While deletion of PKM2 accelerated breast cancer and medulloblastoma progression and promoted hepatocellular carcinoma [14, 15, 17], loss of PKM2 slowed leukemia progression [16] and had no effect on progression of colon cancer [12]. A recent report using PKM1 and PKM2 knock-in mouse models showed that mice that exclusively expressed PKM1 had higher tumor burden than PKM2-expressing mice in a lung adenocarcinoma model [18]. Additionally, these whole-body PKM1-expressing mice had higher tumor incidence than PKM2-expressing mice in a carcinogen-induced mouse model [18]. We find here that deletion of PKM2 does not impair PDAC tumorigenesis or affect mouse survival or tumor size, suggesting that whether loss of PKM2 accelerates or slows cancer depends on the tissue and environmental context.
PKM2 expression was relatively high in wild-type PDAC tumors, whereas PKM1 expression was low in these tumors. PKM2 deletion in tumors resulted in an isoform switch from PKM2 to PKM1 expression. This result is consistent with findings observed in colon cancer [12], breast cancer [14], and leukemia [16] and suggests that PKM2 is dispensable in a variety of tumor types. PKM1 expression has been associated with decreased proliferative capacity in some systems [5, 6, 14] although a recent study reported that PKM1 expression can also be associated with increased proliferation in tumors [18]. Although we observed higher PKM1 expression in PKM2 deleted tumors compared to wild-type tumors, this did not affect the percentage of proliferative cells in the tumor or overall animal survival.
Differences between our results and previously published reports may be attributed to the possibility that PKM2 was not efficiently deleted from all of the PDAC tumor cells, despite the fact that we observed a robust decrease in PKM2 expression in these tumors. Additionally, it is likely that tumor stromal cells, which make up a significant percentage of PDAC tumors [34], retained PKM2 expression. Furthermore, it will be interesting to investigate PKM2 loss in other mouse models of pancreatic cancer driven by Kras with wildtype, heterozygous, or point mutant Trp53 [32, 33]. Different metabolic requirements may exist in these tumors that arise more gradually, such as a differential dependence on autophagy [35]. Nonetheless, the loss of PKM2 expression in most tumor cells in a mouse PDAC model driven by mutant Kras and early homozygous loss of Trp53 supports the notion that PKM2 is not generally required for PDAC initiation and progression. Although PKM2 is highly expressed in mouse and human PDAC, our findings support a model where PKM2 is dispensable for the growth of these cancers.
Conclusions
Our study found that PKM2 is not required for KP−/−C pancreatic tumor growth or progression. Although PKM2 was found to be highly expressed in mouse and human PDAC tumors, loss of PKM2 in KP−/−C mice did not significantly affect overall survival, tumor size, or proliferative index. Loss of PKM2 in PDAC tumors resulted in higher PKM1 expression, but did not change cell proliferation, further arguing that PKM2 is not required for pancreatic tumor growth.
Methods
Mouse model and survival curve
Pkm2flox/flox mice [14] were bred to the LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-Cre (KP−/−C) mice [32] to generate animals of relevant genotypes. Male Pkm2flox/flox and Pkm2+/+ mice were used for Kaplan-Meier survival curve and tumor weight analysis.
Western blot
Western blots were performed using primary antibodies against PKM2 (Cell Signaling Technologies #4053, 1:1000 dilution), PKM1 (Cell Signaling Technologies #7067, 1:1000 dilution), PKLR (Santa Cruz #sc-133222, 1:1000 dilution), or vinculin (Abcam ab18058, 1:1000 dilution).
Immunohistochemistry
Sections from formalin-fixed paraffin-embedded mouse tissue were stained with hematoxylin and eosin or with antibodies against PKM2 (Cell Signaling Technologies #4053, 1:800 dilution), PKM1 (Cell Signaling Technologies #7067, 1:500 dilution), Ki67 (BD Biosciences #550609, 1:40 dilution), or PCNA (Cell Signaling Technologies #2586, 1:4000 dilution). Percent positivity of Ki67 or PCNA staining was calculated using a positive pixel count algorithm in ImageScope software (Leica Biosystems). A human pancreatic cancer tissue microarray (Biomax PA961a) was stained for PKM2 positivity and scored independently by a pathologist (D.D.V.).
qPCR
qPCR reactions were performed using SYBR Green Master Mix (Sigma) and primers for PKM1 (Forward: 5′- GTC TGG AGA AAC AGC CAA GG -3′, Reverse: 5′- TCT TCA AAC AGC AGA CGG TG -3′), PKM2 (Forward: 5′-GTC TGG AGA AAC AGC CAA GG -3′, Reverse: 5′- CGG AGT TCC TCG AAT AGC TG -3′), PKLR (Forward: 5′- AAG GGT CCC GAG ATA CGC A -3′, Reverse: 5′- CTG CAA CGA CCT GGG TGA TA -3′), and PKM-skip (Forward: 5′- ATG CTG GAG AGC ATG ATC AAG AAG CCA -3′ Reverse: 5′- CAA CAT CCA TGG CCA AGT T -3′) using 36B4 (Forward: 5′- TCC AGG CTT TGG GCA TCA -3′, Reverse: 5′- CTT TAT CAG CTG CAC ATC ACT CAG A -3′) as a control.
Additional file
Figure S1. PKM2 is expressed in KPC tumors. A) Sections from tumors arising in LSL-KrasG12D/+;LSL–Trp53R172H/+;Pdx-1-Cre (KPC) mice were stained with Hematoxylin & Eosin (H&E) and isoform-specific antibodies against PKM2 or PKM1 as shown. Scale bars represent 50 μm for all images at × 20 magnification (top) and 20 μm for all images at × 40 magnification (bottom). Figure S2. PKM2 deletion leads to expression of PKM skip, but does not induce PKLR expression. A) Western blot analysis of lysates from tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice performed using an isoform-specific antibody against PKLR and an antibody against vinculin as a control. B) PKLR expression was measured by qPCR of mRNA isolated from tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice. C) PKM-skip expression was measured by qPCR of mRNA isolated from tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice. (PDF 165 kb)
Acknowledgements
The authors thank the Swanson Biotechnology Facility for assistance with histology and immunohistochemistry.
Funding
A.N.L. is a Robert Black Fellow of the Damon Runyon Cancer Research Foundation, DRG-2241-15. This research was supported in part by the MIT Center for Precision Cancer Medicine, SU2C, the Ludwig Center at MIT, and the National Cancer Institute including NIH 5P30CA1405141 and R01CA168653. M.G.V.H. acknowledges additional support as a Howard Hughes Medical Institute Faculty Scholar.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- PDAC
Pancreatic ductal adenocarcinoma
- PKLR
Pyruvate kinase, L/R isoform
- PKM1
Pyruvate kinase, M1 isoform
- PKM2
Pyruvate kinase, M2 isoform
Authors’ contributions
ALH, ANL, CXD, TLD, LVD, and DDV performed and/or supervised experiments. MGVH, ALH, and ANL conceived and designed the study. ALH, ANL, and MGVH wrote the manuscript with input from all authors who read and approved the final manuscript.
Ethics approval
All mouse procedures were approved by the animal care and use committee at MIT.
Consent for publication
Not applicable.
Competing interests
M.G.V.H. is a consultant and scientific advisory board member for Agios Pharmaceuticals and Aeglea Biotherapeutics. The other authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alissandra L Hillis, Email: ahillis@mit.edu.
Allison N Lau, Email: anlau@mit.edu.
Camille X Devoe, Email: cxdevoe@mit.edu.
Talya L Dayton, Email: t.dayton@hubrecht.eu.
Laura V Danai, Email: ldanai@umass.edu.
Dolores Di Vizio, Email: Dolores.Divizio@cshs.org.
Matthew G Vander Heiden, Email: mvh@mit.edu.
References
- 1.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17(12):1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gui DY, Lewis CA, Vander Heiden MG. Allosteric regulation of PKM2 allows cellular adaptation to different physiological states. Sci Signal. 2013;6(263):pe7. doi: 10.1126/scisignal.2003925. [DOI] [PubMed] [Google Scholar]
- 3.Mazurek S, Hugo F, Zwerschke W. PKM2 (pyruvate kinase isoenzyme type M2). Atlas Genet Cytogenet Oncol Haematol. 2009;13(4):276–81.
- 4.Ikeda Y, Tanaka T, Noguchi T. Conversion of non-allosteric pyruvate kinase isozyme into an allosteric enzyme by a single amino acid substitution. J Biol Chem. 1997;272(33):20495–501. doi: 10.1074/jbc.272.33.20495. [DOI] [PubMed] [Google Scholar]
- 5.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 6.Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57(1):95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53(1):75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150(4):685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao X, Wang H, Yang JJ, Liu X, Liu Z-R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45(5):598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;478(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of evidence for PKM2 protein kinase activity. Mol Cell. 2015;59(5):850–857. doi: 10.1016/j.molcel.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau AN, Israelsen WJ, Roper J, Sinnamon MJ, Georgeon L, Dayton TL, et al. PKM2 is not required for colon cancer initiated by APC loss. Cancer Metab. 2017;5(1):e7. doi: 10.1186/s40170-017-0172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. 2012;1–6. [DOI] [PMC free article] [PubMed]
- 14.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155(2):397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tech K, Tikunov AP, Farooq H, Morrissy AS, Meidinger J, Fish T, et al. Pyruvate kinase inhibits proliferation during postnatal cerebellar neurogenesis and suppresses medulloblastoma formation. Cancer Res. 2017;77(12):3217–3230. doi: 10.1158/0008-5472.CAN-16-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y-H, Israelsen WJ, Lee D, Yu VWC, Jeanson NT, Clish CB, et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158(6):1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016;30(9):1020–1033. doi: 10.1101/gad.278549.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita M, Sato T, Nomura M, Sakamoto Y, Inoue Y, Tanaka R, et al. PKM1 confers metabolic advantages and promotes cell-autonomous tumor cell growth. Cancer Cell. 2018;33(3):355–357. doi: 10.1016/j.ccell.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Feng J, Ma T, Ge Z, Lin J, Ding W, Chen H, et al. PKM2 gene regulates the behavior of pancreatic cancer cells via mitogen-activated protein kinase pathways. Mol Med Rep. 2015;11(3):2111–2117. doi: 10.3892/mmr.2014.2990. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Zhao Z, Zhou Z, Liu R. PKM2 promotes cell survival and invasion under metabolic stress by enhancing Warburg effect in pancreatic ductal adenocarcinoma. Dig Dis Sci. 2016;61(3):767–773. doi: 10.1007/s10620-015-3931-2. [DOI] [PubMed] [Google Scholar]
- 21.Azoitei N, Becher A, Steinestel K, Rouhi A, Diepold K, Genze F, et al. PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Mol Cancer. 2016;15(3):1–15. doi: 10.1186/s12943-015-0490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Zhao Z, Zhou Z, Liu R. Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother Pharmacol. 2016;78(6):1199–1207. doi: 10.1007/s00280-016-3178-4. [DOI] [PubMed] [Google Scholar]
- 23.Pandita A, Kumar B, Manvati S, Vaishnavi S, Singh SK, Bamezai RNK. Synergistic combination of gemcitabine and dietary molecule induces apoptosis in pancreatic cancer cells and down regulates PKM2 expression. PLoS One. 2014;9(9):e107154–e107111. doi: 10.1371/journal.pone.0107154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35(16):2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DJ, Park YS, Kang MG, You Y-M, Jung Y, Koo H, et al. Pyruvate kinase isoenzyme M2 is a therapeutic target of gemcitabine-resistant pancreatic cancer cells. Exp Cell Res. 2015;336(1):119–129. doi: 10.1016/j.yexcr.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Tian S, Li P, Sheng S, Jin X. Upregulation of pyruvate kinase M2 expression by fatty acid synthase contributes to gemcitabine resistance in pancreatic cancer. Oncol Lett. 2018:1–7. [DOI] [PMC free article] [PubMed]
- 27.Mohammad GH, Olde Damink SWM, Malago M, Dhar DK, Pereira SP. Pyruvate kinase M2 and lactate dehydrogenase a are overexpressed in pancreatic cancer and correlate with poor outcome. PLoS One. 2016;11(3):e0151635–e0151617. doi: 10.1371/journal.pone.0151635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa H, Nagano H, Konno M, Eguchi H, Koseki J, Kawamoto K, et al. The combination of the expression of hexokinase 2 and pyruvate kinase M2 is a prognostic marker in patients with pancreatic cancer. Mol Clin Oncol. 2015;3(3):563–571. doi: 10.3892/mco.2015.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockney NA, Zhang M, Lu Y, Sopha SC, Washington MK, Merchant N, et al. Pyruvate kinase muscle isoenzyme 2 (PKM2) expression is associated with overall survival in pancreatic ductal adenocarcinoma. J Gastrointest Canc. 2015;46(4):390–398. doi: 10.1007/s12029-015-9764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Luo H, Zhu X, Hu X, Zheng L, Zhu X. Pyruvate kinase M2 (PKM2) expression correlates with prognosis in solid cancers: a meta-analysis. Oncotarget. 2016:1–13. [DOI] [PMC free article] [PubMed]
- 31.Wu J, Hu L, Chen M, Cao W, Chen H, He T. Pyruvate kinase M2 overexpression and poor prognosis in solid tumors of digestive system: evidence from 16 cohort studies. OncoTargets. 2016;9:4277–4288. doi: 10.2147/OTT.S106508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bardeesy N, Aguirre AJ, Chu GC, Cheng K-H, Lopez LV, Hezel AF, et al. Both p16Ink4a and the p19Arf-p53 pathway constrain progression of the pancreatic adenocarcinoma. 2006; 1–6. [DOI] [PMC free article] [PubMed]
- 33.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 34.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18(16):4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PKM2 is expressed in KPC tumors. A) Sections from tumors arising in LSL-KrasG12D/+;LSL–Trp53R172H/+;Pdx-1-Cre (KPC) mice were stained with Hematoxylin & Eosin (H&E) and isoform-specific antibodies against PKM2 or PKM1 as shown. Scale bars represent 50 μm for all images at × 20 magnification (top) and 20 μm for all images at × 40 magnification (bottom). Figure S2. PKM2 deletion leads to expression of PKM skip, but does not induce PKLR expression. A) Western blot analysis of lysates from tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice performed using an isoform-specific antibody against PKLR and an antibody against vinculin as a control. B) PKLR expression was measured by qPCR of mRNA isolated from tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice. C) PKM-skip expression was measured by qPCR of mRNA isolated from tumors arising in KP−/−C Pkm2+/+ and KP−/−C Pkm2flox/flox mice. (PDF 165 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article.