ABSTRACT
The flagellum is an evolutionarily conserved appendage used for sensing and locomotion. Its backbone is the axoneme and a component of the axoneme is the radial spoke (RS), a protein complex implicated in flagellar motility regulation. Numerous diseases occur if the axoneme is improperly formed, such as primary ciliary dyskinesia (PCD) and infertility. Radial spoke head 6 homolog A (RSPH6A) is an ortholog of Chlamydomonas RSP6 in the RS head and is evolutionarily conserved. While some RS head proteins have been linked to PCD, little is known about RSPH6A. Here, we show that mouse RSPH6A is testis-enriched and localized in the flagellum. Rsph6a knockout (KO) male mice are infertile as a result of their short immotile spermatozoa. Observation of the KO testis indicates that the axoneme can elongate but is disrupted before accessory structures are formed. Manchette removal is also impaired in the KO testis. Further, RSPH9, another radial spoke protein, disappeared in the Rsph6a KO flagella. These data indicate that RSPH6A is essential for sperm flagellar assembly and male fertility in mice.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Radial spoke protein, Axoneme, Flagella, Spermatozoa, CRISPR/Cas9
Summary: Removal of RSPH6A via CRISPR/Cas9 results in short-tailed immotile spermatozoa and loss of fertilizing ability in male mice.
INTRODUCTION
While it was found that 15% of couples in the United States are infertile (Thoma et al., 2013), there are few studies that elucidate the causes of male infertility (Agarwal et al., 2015). Roughly half of those 15% are attributed to male infertility and these men are unable to have children without assisted reproductive technology. The main categories of infertility include spermatozoa with abnormal morphology (teratospermia), reduced sperm motility (asthenospermia) and no or low sperm count (azoospermia or oligospermia, respectively). Defects in the sperm flagellum can cause these different types of infertility.
The flagellum is a thin, thread-like appendage that is evolutionarily conserved from unicellular organisms to mammals (Carvalho-Santos et al., 2011) and is used for sensing and locomotion. Mammalian spermatozoan flagella are highly specialized to carry male genetic material into the female reproductive tract and fertilize the oocyte. Internal cross-sections show that the flagellum comprises a ‘9+2’ microtubule structure: a bundle of nine microtubule doublets that surround a central pair of single microtubules (Satir and Christensen, 2007). Called the axoneme, this structure consists of macromolecular complexes such as the outer and inner dynein arms and radial spokes (RSs) (Fig. 1A).
Fig. 1.
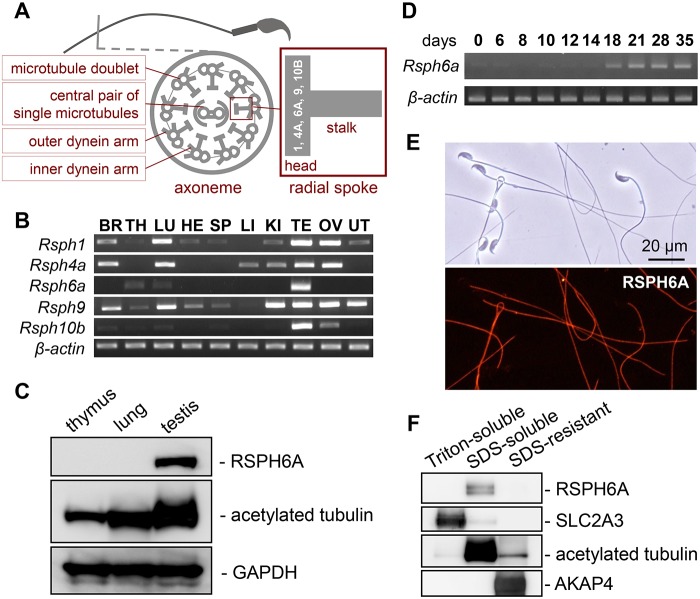
Characterization of mouse Rsph6a. (A) Schematic drawing of the flagella axoneme (end piece) and radial spoke structure. (B) The expression of mouse Rsph genes in various organs examined by RT-PCR. Rsph6a is testis-enriched but weak expression is also detected in the thymus and lung. β-actin was used as an expression control. BR, brain; TH, thymus; LU, lung; HE, heart; SP, spleen; LI, liver; KI, kidney; TE, testis; OV, ovary; UT, uterus. (C) Western blot analysis of RSPH6A. RSPH6A was detected in the testis, but not in the thymus or lung. GAPDH was detected as a loading control, and acetylated tubulin as a marker for stable microtubules including cilia and flagella. (D) The expression of mouse Rsph6a on indicated postnatal days in the testis was examined by RT-PCR. Rsph6a begins expression at postnatal day 18. β-actin was used as an expression control. (E) Immunofluorescence analysis of spermatozoa from WT mice labeled with antibodies against RSPH6A (red). Fluorescence is seen along the entire sperm flagella. (F) Fractionation of mouse spermatozoa. RSPH6A was found in the SDS-soluble fraction. SLC2A3, acetylated tubulin and AKAP4 were detected as makers for Triton-soluble, SDS-soluble and SDS-resistant fractions, respectively.
First characterized in sea urchins (Afzelius, 1959), the RS is a T-shaped protein complex that extends from the doublet microtubules towards the central pair of single microtubules, possessing an elongated ‘stalk’ that is bound to the doublet microtubules and terminates at a bulbous ‘head’. Through experiments on Chlamydomonas reinhardtii, much about the RS proteins has been uncovered. Purification of the RS complex resulted in the identification of 23 Chlamydomonas flagellar RS proteins (Piperno et al., 1981; Huang et al., 1981; Yang et al., 2006). RSs regulate flagellar motility, as elucidated by Chlamydomonas mutant strains. Chlamydomonas mutants that lack the entire RS become paralyzed and no longer possess the ability to propagate (Witman et al., 1978). Additional mutant lines were found to further understand the role of specific protein components within the RSs. Chlamydomonas mutants pf-1 and pf-17, missing RSP4 and RSP9, respectively, were deficient of all RS head proteins but not stalk proteins, causing paralysis (Huang et al., 1981). The temperature-sensitive mutant pf-26ts lacks RSP6, which is also localized in the spoke head, and mutant Chlamydomonas was found to be paralyzed when introduced into a restrictive temperature. Using these mutation analyses, research related to RS head proteins and its impact on mammalian systems have been explored. There are five RS head proteins found in Chlamydomonas (RSP1, -4, -6, -9 and -10) and all of these proteins are conserved in mice and humans (RSPH1, -4A, -6A, -9 and -10B, respectively).
In mammals, mutations in RS head proteins have been linked to primary ciliary dyskinesia (PCD) (Frommer et al., 2015). PCD is a condition that shows abnormal cilia movement, often leading to chronic respiratory tract infections and abnormally positioned internal organs. Human patients with mutations in RSPH4A and RSPH9 had PCD due to abnormalities in the RS and central microtubular pair (Castleman et al., 2009). Mouse knockout (KO) models confirm that RSPH4A is essential for normal ciliary motility (Shinohara et al., 2015). Loss-of-function mutations in RSPH1 showed a similar phenotype, with abnormal axoneme structures such as defects in the RS and central pair of microtubules (Kott et al., 2013; Knowles et al., 2014). In addition to ciliary motility defects, PCD patients can be infertile because of abnormal flagella. However, there is currently only one RS head KO mouse model – Rsph1 KO – that has been reported infertile. In this KO mouse line, spermatid formation was abnormal, as shown by deformed heads and stunted flagellum formation (Tokuhiro et al., 2008).
Another RSPH candidate gene related to male fertility is Rsph6a, the ortholog of Chlamydomonas RSP6 (Curry et al., 1992; Eriksson et al., 2001). Previous studies found that RSPH6A was specifically expressed in the testes in humans but little is known about the role it plays in the mammalian flagellar axoneme (Kott et al., 2013). Thus, this study utilized the CRISPR/Cas9 system to generate an Rsph6a KO mouse to uncover the function of RSPH6A.
RESULTS
RSPH6A is an evolutionarily conserved testis-enriched protein localized in sperm flagellum
RSPH6A is an evolutionarily conserved protein present in many eukaryotes, especially those that possess a flagellum. Sequence alignment amongst species shows that RSPH6A is highly conserved in eukaryotes such as Chlamydomonas, mice, rats and humans (Fig. S1). BLAST protein analysis showed 30% identity and 40% positive matches between Chlamydomonas RSP6 and mouse RSPH6A (89% query coverage with RSP6). Mouse RSPH6A contains two coiled-coil domains that may be involved in protein-protein interactions. In comparison, human and rat RSPH6A contain one coiled-coil domain, but Chlamydomonas RSP6 does not (SMART) (Letunic et al., 2015).
Quantitative RT-PCR reveals that Rsph6a is testis-specific in humans (Kott et al., 2013). To determine the expression pattern in mice, RT-PCR of Rsph6a as well as the other RS head genes in multi-tissues from adult mice was conducted (Fig. 1B). This analysis revealed that while Rsph6a is abundant in the testis, it also has minimal expression in the lungs and thymus, in contrast with the human expression pattern. Compared with other RS head genes, Rsph6a is one that is strongly enriched in the testis. As mentioned previously, Rsph1 KO mice are male infertile (Tokuhiro et al., 2008) but our RT-PCR results show strong expression of Rsph1 in the lung, ovary, brain and testis (Fig. 1B). To examine RSPH6A protein expression levels, we generated an antibody that recognized amino acid residues 145-163. Western blot analysis confirmed RSPH6A expression in the testis (Fig. 1C). In contrast, RSPH6A was not detected in the lung or thymus. To verify temporal expression of Rsph6a in relation to spermatogenesis, RT-PCR was performed using postnatal testis (Fig. 1D). Rsph6a is expressed starting from postnatal day 18, which corresponds to the production of secondary spermatocytes and round spermatids (Bellve et al., 1977). To analyze RSPH6A localization in the spermatozoa, we conducted immunofluorescence staining. We found that it was localized to the entire flagellum (Fig. 1E). Further, RSPH6A dissolved in an SDS buffer but not in a mild lysis buffer containing 1% Triton X-100, suggesting that RSPH6A is anchored to the internal structure, but is not strongly associated with the fibrous sheath like AKAP4 (Fig. 1F). These results are consistent with predicted localization of RSPH6A in the RS.
RSPH6A interacts with other RSPH proteins in HEK293T cells
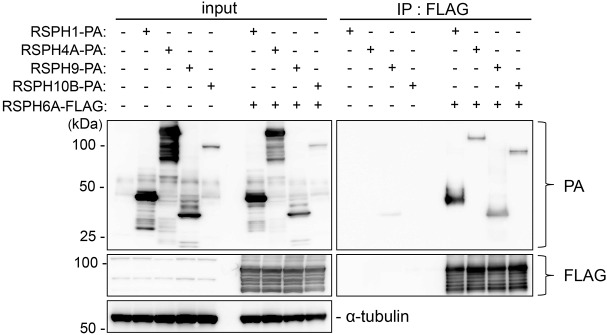
According to previous studies performed with Chlamydomonas RS proteins, RSP6 interacts with RSP9 and RSP10 (Kohno et al., 2011). To analyze the interaction between RS head proteins in mammals, we conducted co-immunoprecipitation (co-IP) studies using mouse recombinant RSPH proteins expressed in HEK293T cells because RSPH6A could not be dissolved from the mature spermatozoa using a mild lysis buffer (Fig. 1F) that is necessary for co-IP. RSPH6A (variant 1 with 708 aa) was tagged with FLAG while RSPH1 (301 aa), RSPH4A (716 aa), RSPH9 (276 aa) and RSPH10B (876 aa) were tagged with PA. We found that immunoprecipitation with RSPH6A could pull down RSPH1, RSPH4A, RSPH9 and RSPH10B, suggesting that RSPH6A can bind directly to other RS head proteins (Fig. 2).
Fig. 2.
Interaction of RS head proteins. RSPH6A interaction with other RS head proteins was examined by co-immunoprecipitation of the RSPH6A-FLAG and RSPH-PA complex using anti-FLAG antibody-conjugated beads. Input: whole cell lysate from transfected cells. IP: immunoprecipitation with FLAG-conjugated beads. The left lower panel shows α-tubulin as a loading control. RSPH6A can bind to all other RS head proteins in HEK293T cells.
Generation of Rsph6a mutant mice using CRISPR/Cas9
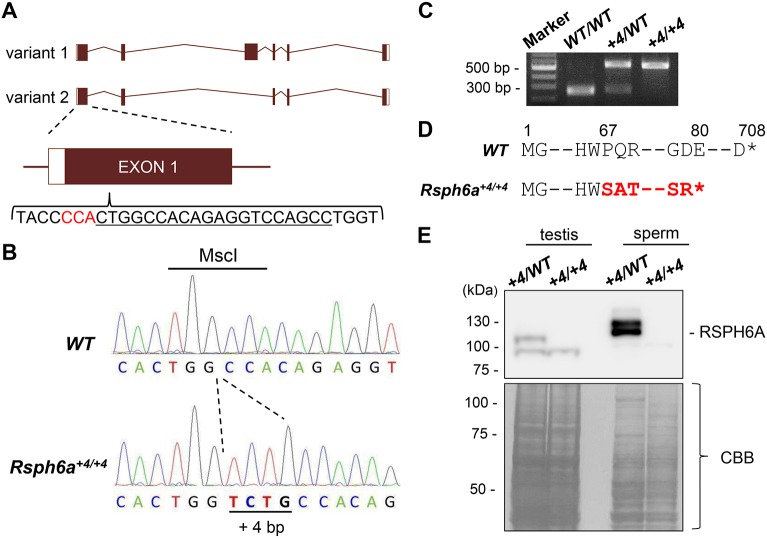
To uncover the role of Rsph6a in vivo, we produced an Rsph6a mutant mouse via zygotic injection of a CRISPR/Cas9-expressing plasmid. There are two transcriptional variants of Rsph6a as found via in silico analysis (UCSC Genome Browser). To ensure that both variants are mutated, we designed a guide RNA (gRNA) in the shared exon 1 (Fig. 3A). We inserted the target gRNA sequence into the pX330 plasmid, which also contains the humanized Cas9 sequence, and microinjected 5 ng/µl of the plasmid into the pronuclei of fertilized oocytes (Mashiko et al., 2013). Of the 127 fertilized oocytes that were injected, 75 two-cell embryos were transplanted into the oviducts of four pseudopregnant female mice and 11 pups were born. Two of the eleven pups had mosaic mutations, and one was caged with two wild-type (WT) females to obtain the next generation. Subsequent mating resulted in a KO mouse with a 4 bp insertion (Rsph6a+4/+4). This insertion was confirmed by PCR and sequencing analysis (Fig. 3B) as well as MscI digestion (Fig. 3C). The 4 bp insertion resulted in a frameshift mutation of P67S with a premature stop codon introduced 13 amino acids later (Fig. 3D).
Fig. 3.
Generation of Rsph6a mutant mice using the CRISPR/Cas9 system. (A) Structure of the Rsph6a and CRISPR/Cas9 targeting scheme. Rsph6a has two variants and both were targeted in exon 1. Black underline indicates gRNA sequence. Red characters indicate PAM (protospacer adjacent motif) sequence. (B) Wave pattern sequence of Rsph6a in WT and Rsph6a+4/+4 mice. (C) Genotyping Rsph6a+4/+4 mice by MscI digestion. The MscI recognition site (5′-TGGCCA-3′) was disrupted due to the 4 bp insertion. In WT mice, the PCR product was cut into two similar-sized sequences (resulting in one band of ∼250 bp) after MscI digestion. (D) The 4 bp insertion in the first exon in Rsph6a+4/+4 mice caused a P67S mutation resulting in a premature stop codon introduced 13 amino acids later. (E) Protein expression of RSPH6A in Rsph6aWT/+4 and Rsph6a+4/+4 testis and cauda epididymal spermatozoa. Coomassie Brilliant Blue (CBB) staining shows equal loading.
To confirm that the RSPH6A protein was deleted in Rsph6a+4/+4 mice, we performed western blot analysis using the RSPH6A antibody. A strong band at ∼110 kDa was present in the heterozygous testis but disappeared in the Rsph6a+4/+4 mutants (Fig. 3E). There was another band at ∼90 kDa in both Rsph6a+4/WT and Rsph6a+4/+4 testes. This band may be a shorter form of an Rsph6a variant (variant 2, Fig. 3A). However, the predicted molecular weight of variant 2 is 51 kDa and the band around 90 kDa may be non-specific. In spermatozoa, there were two bands strongly expressed at ∼120 kDa in the Rsph6a+4/WT sample, but those were absent in the Rsph6a+4/+4 mutant sample (Fig. 3E). Because only one RSPH6A signal was detected in the testis and the band sizes were different between the testis and spermatozoa, there may be RSPH6A modifications that occur in the epididymal spermatozoa.
Fertilizing ability of Rsph6a+4/+4 mutant male mice
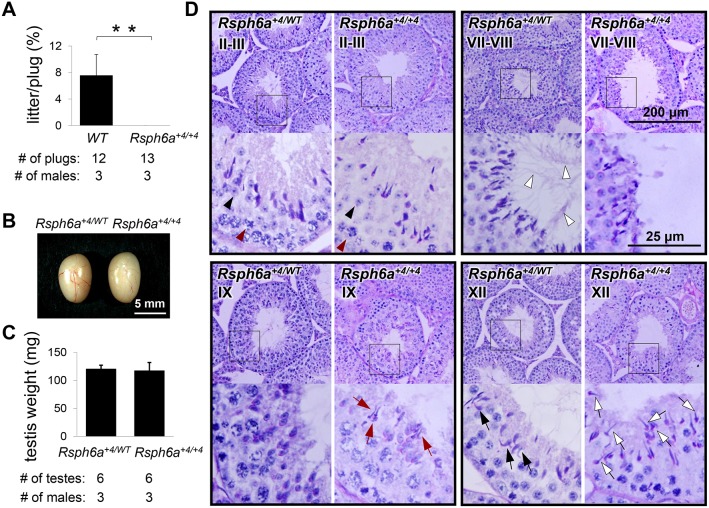
Heterozygous mutant pairs produced 26% homozygous offspring (10 out of 34 pups in 4 litters), suggesting that homozygous mice are not embryonically lethal. Rsph6a+4/+4 mice were viable and had no particular problems at first glance. To test the fertility of male mice, individual Rsph6a+4/+4 males as well as WT controls were cohabitated with WT females for 2 months. While the controls sired pups continuously, the three Rsph6a+4/+4 males were sterile despite the formation of 13 copulatory plugs in total (Fig. 4A). Because there were no differences in litter size or sperm motility between WT and heterozygous males (Fig. S2A,B), either WT or heterozygous males were used as the control for further phenotypic analysis.
Fig. 4.
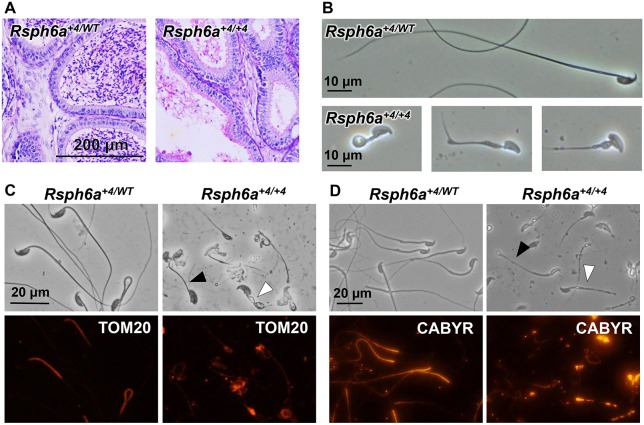
Fertilizing ability and testicular structure of Rsph6a+4/+4 mutant male mice. (A) Number of litters born per plug detected. N=3 males each for WT and Rsph6a+4/+4, mated with two WT females per male. **P<0.01, Student's t-test. (B) Testes of Rsph6a+4/WT and Rsph6a+4/+4 mice. (C) Testicular weights of Rsph6a+4/WT and Rsph6a+4/+4 mice were not significantly different (P=0.64, Student's t-test). N=3 males each for Rsph6a+4/WT and Rsph6a+4/+4. (D) PAS staining of testicular sections. Lower panels are magnified images of the boxes indicated in the figures above. Development of normal spermatocytes (red arrowheads) and spermatids (black arrowheads) can be seen in stage II-III tubules in both control and mutant mice. In stage VII-VIII seminiferous tubules of Rsph6a+4/WT mice, properly elongated spermatid tails are present in the lumen (white arrowheads), but similar elongated flagella are missing in Rsph6a+4/+4 tubules. Sections of Rsph6a+4/WT show no mature elongated spermatids remaining in the stage IX tubules but they are retained in Rsph6a+4/+4 mice (red arrows). Stage XII tubules of Rsph6a+4/WT mice have normal, hook-shaped heads (black arrows) whereas Rsph6a+4/+4 XII tubules have abnormal, club-shaped heads (white arrows).
Rsph6a+4/+4 mice exhibit abnormal sperm tail formation
There are many causes of male infertility, such as spermatogenic defects or impaired fertilization ability once the spermatozoa come into contact with the egg. To narrow down the possibilities, we examined spermatogenesis in Rsph6a+4/+4 males. Gross examination of the testis revealed little difference in morphology (Fig. 4B) or weight (Fig. 4C). However, periodic acid-Schiff (PAS) staining on testis sections showed that the lumen of the seminiferous tubules in Rsph6a+4/+4 testes lacked elongated sperm tails (stage VII-VIII) while spermatogonia and spermatocyte development were comparable to those of the control (stage II-III) (Fig. 4D), indicating that there is a defect in tail formation of elongating spermatids. Further, stage IX seminiferous tubules should not contain any elongated spermatids as they should have been released in the stage prior. However, retained elongated spermatids were frequently observed in stage IX tubules in Rsph6a+4/+4 males (Fig. 4D). In addition, nuclear condensation was impaired as abnormal head shapes were observed in stage XII tubules in Rsph6a+4/+4 mice (Fig. 4D).
Histological evaluation of the epididymis showed that while the cauda epididymis of heterozygous mice was filled with mature spermatozoa, the homozygous mutants lacked the same level of sperm content (Fig. 5A). When mature spermatozoa were obtained from the cauda epididymis and observed (Fig. 5B), they exhibited shorter tails at an average of length of 24.6±22.9 µm (Fig. S3) and misshapen heads, and were completely immotile (Movies 1 and 2). In addition, cell debris that resembled spermatogenic cells was seen in the epididymal fluid. Taken in conjunction with testis results, infertility can be attributed to abnormal sperm tail and head formation, which may lead to spermiation failure and the production of immotile spermatozoa.
Fig. 5.
Histological evaluation of the epididymis and spermatozoa. (A) PAS staining of cauda epididymis in Rsph6a+4/WT and Rsph6a+4/+4 mice. (B) Observation of spermatozoa obtained from cauda epididymis. Rsph6a+4/WT spermatozoa possessed full-length flagella and normal head shapes, while Rsph6a+4/+4 spermatozoa had truncated flagella and abnormal head shapes. (C) Immunofluorescence analysis of spermatozoa from control and mutant mice labeled with antibodies against TOM20 (red). TOM20 (mitochondria) localizes to the midpiece of control spermatozoa. Fluorescence in Rsph6a+4/+4 mouse spermatozoa was undetectable (black arrowhead) or spotty (white arrowhead). (D) Immunofluorescence analysis of spermatozoa from control and mutant mice labeled with antibodies against CABYR (red). The fibrous sheath protein was localized along the principal piece of control spermatozoa. Signal in Rsph6a+4/+4 mouse spermatozoa was undetectable (black arrowhead) or spotty (white arrowhead).
Generation and phenotypic analysis of Rsph6a ‘large deletion’ mice
To reaffirm that this phenotype was solely a result of the Rsph6a mutation via the 4 bp insertion and not due to any off-target mutations, we engineered mutant mice that had a large deletion (LD) using different gRNAs to that used for the 4 bp insertion. This deleted region also included the antigen region in exon 1 to confirm if the second set of bands in the testis western blot were truly non-specific (Fig. 3E). We designed two gRNAs, one 50 bp upstream of the antigen region and one in exon 4 of variant 1 (exon 3 of variant 2), and purchased them as CRISPR RNA (crRNA) (Fig. S4A). The two crRNAs were electroporated along with the CAS9 protein and trans-activating RNA (tracrRNA) into 168 fertilized oocytes. Of the surviving oocytes, 151 were transplanted into the oviducts of six pseudopregnant female mice and 47 pups were born. Of the 47 pups, four (two male, two female) received the heterozygous LD mutation. Subsequent mating resulted in a mutant mouse with homozygous LD alleles (Rsph6aLD/LD). This deletion was verified by PCR (Fig. S4B) and sequencing analysis. In the LD mutant mice, 12,068 bp were deleted, including amino acids 122-542 of variant 1 and 122-289 of variant 2.
Similar to the Rsph6a+4/WT western blot results, a strong band appeared at ∼110 kDa in the Rsph6aLD/WT testis but disappeared in the Rsph6aLD/LD (Fig. S4C). The extra set of bands at ∼90 kDa were still present in both the Rsph6aLD/WT and Rsph6aLD/LD testis, leading us to conclude that they were in fact non-specific. To corroborate the histological phenotype of spermatozoa, mature spermatozoa from the cauda epididymis were observed (Fig. S4D). Indistinguishable from Rsph6a+4/+4 mutant spermatozoa, the large deletion mutants exhibited shorter tails, misshapen heads and immotility. Therefore, we concluded that the phenotype is attributed to the mutation of Rsph6a.
Mitochondrial and fibrous sheath formation were abrogated
To analyze the structure of the flagella, we performed immunostaining using various flagella protein antibodies. To confirm that the RSPH6A protein was deleted in the mutant mice, immunofluorescence analysis using the RSPH6A antibody was conducted and, as expected, the signal was lost in the Rsph6a KO spermatozoa (Fig. S5A). To check if the mitochondrial sheath localized in the midpiece and the fibrous sheath localized in the principal piece were formed in the mutant spermatozoa, we stained the spermatozoa collected from the cauda epididymis using antibodies against TOM20 (outer mitochondrial membrane protein) and CABYR (fibrous sheath protein) (Young et al., 2016). While TOM20 clearly stained the mitochondrial sheath in the control spermatozoa, the TOM20 signal disappeared or was mislocalized in stunted flagella (Fig. 5C), indicating that the abnormality in tail formation occurs before the completion of mitochondrial sheath formation. To further understand the extent of the damage to the flagella, we also conducted immunostaining of CABYR, and found that the signal was not observed or was uneven (Fig. 5D). These results indicate that the formation of mitochondrial and fibrous sheaths is disrupted due to the deletion of RSPH6A.
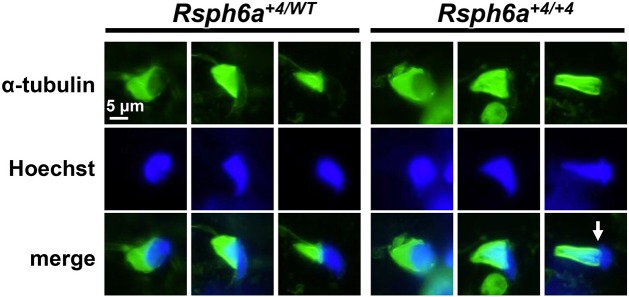
Manchette removal is disrupted in the Rsph6a+4/+4 mice
It is well understood that manchette formation and removal is important for the correct formation of sperm heads (Zhang et al., 2009; Lehti et al., 2013; Lehti and Sironen, 2016; Liu et al., 2015; Dunleavy et al., 2017). Because spermatozoa from Rsph6a+4/+4 mice had abnormal heads, we checked manchette formation with immunofluorescence using the anti-α-tubulin antibody to detect any defects. We found that while manchette formation was normal, removal (or caudal movement of the manchette) was atypical (Fig. 6), resulting in the club-shaped heads that were also observed in the testis sections (Fig. 4D, stage XII). Abnormal head morphology may be due to the perinuclear rings tightening in unusual places during manchette removal.
Fig. 6.
Immunofluorescence of α-tubulin in Rsph6a+4/WT and Rsph6a+4/+4 mice. Immunofluorescence analysis of spermatids from control and mutant mice labeled with antibodies against α-tubulin (green). α-tubulin localizes to the manchette of both control and Rsph6a+4/+4 mouse spermatids. Head elongation is shown progressively from left to right based on nuclear and manchette shapes. White arrow indicates abnormal caudal movement of the manchette. Hoechst staining (blue) indicates the nucleus.
Additionally, we examined the localization of RSPH6A in the spermatids using immunofluorescence (Fig. S5B). In the spermatid before manchette formation, RSPH6A was already localized in the flagellum, before accessory tubules are formed. We also found strong RSPH6A signals in the cytosol, but it was still found in the KO, suggesting that the signal in the cytosol was non-specific. We also saw RSPH6A signals in the manchette in both the heterozygous and KO, suggesting that fluorescence in the manchette was also non-specific. Because specific RSPH6A signal was not observed in the manchette, RSPH6A may not be directly involved in manchette removal. Rather, manchette removal may be impaired because the tail formation was disrupted.
Ultrastructural analysis of the spermatozoa from Rsph6a+4/+4 mice
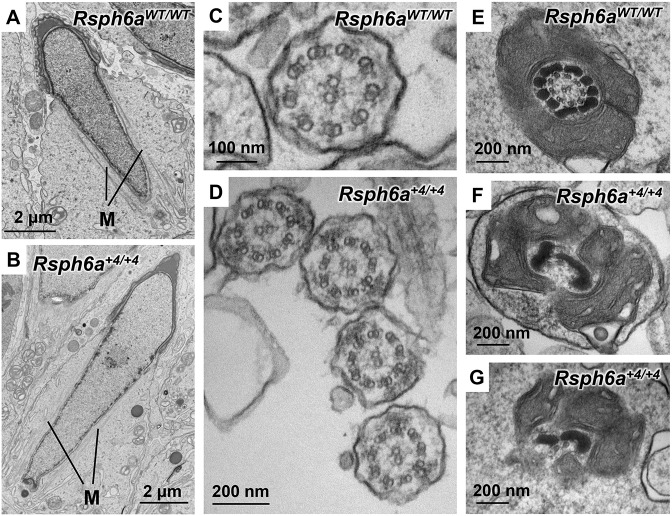
To further analyze defects in spermiogenesis, we observed the testis using transmission electron microscopy. We confirmed that manchette formation was normal even in the ultrastructure of Rsph6a+4/+4 KO mouse testis (Fig. 7A,B). Further, intact axonemes that were not surrounded by mitochondrial or fibrous sheaths were observed in Rsph6a+4/+4 KO testis (out of 18 axonemes examined, 18 axonemes exhibited intact 9+2 patterns; N=2 KO mice), suggesting that the axoneme can start forming normally (Fig. 7C,D). Yet, when we looked at the axoneme surrounded by the mitochondrial sheath, we found disrupted 9+2 patterns (out of 28 axonemes examined, 28 axonemes exhibited defects in both doublet microtubules and central pair; N=2 KO mice) in addition to disrupted mitochondrial localization (Fig. 7E-G, Fig. S6), which was consistent with immunostaining observation of TOM20 (Fig. 5C). These results suggest that the axoneme can elongate properly but is disrupted before the mitochondrial sheath is formed.
Fig. 7.
Ultrastructural analysis of the Rsph6a KO testicular spermatozoa. There is no difference in manchette formation (A,B: stage X) or axoneme structures that are not surrounded by mitochondrial or fibrous sheath (C: stage VI; D: stage VI) between control and Rsph6a+4/+4 mutant spermatozoa. However, compared with control mice (E: stage VIII), axoneme structures surrounded by a mitochondrial sheath and mitochondrial sheath formation were disrupted in Rsph6a+4/+4 mutant mice (F,G: stage VIII). M, manchette.
RSPH9 was not incorporated into the flagellum in the Rsph6a KO spermatozoa
To understand the localization patterns of other RSPHs in the Rsph6a+4/+4 KO, we analyzed RSPH9 via immunofluorescence and western blot. We chose RSPH9 because we have an RSPH9 antibody that recognizes mouse recombinant RSPH9 (Fig. S7A). In spermatids, we found that RSPH9 is localized in the tail similarly to RSPH6A, but the signal decreased in the KO (Fig. 8A), suggesting that RSPH9 cannot be incorporated properly into the tail. An RSPH9 signal was also detected in the manchette of the heterozygous spermatid and did not decrease in the KO. However, it should be noted that there is a possibility that the signal in the manchette was merely non-specific. Western blot analysis showed that RSPH9 decreases in the KO testis and disappears in mature KO spermatozoa (Fig. 8B), confirming our initial immunofluorescence results that RSPH9 cannot be incorporated into the flagella. Disappearance of RSPH9 in the flagellum of the Rsph6a+4/+4 KO mature spermatozoa was then corroborated with immunofluorescence (Fig. S7B). In contrast to the testis, RSPH9 signal did not decrease in the lung (Fig. S7C), suggesting that RSPH9 can be incorporated into the cilia. Because Rsph4a, a paralog of Rsph6a, is strongly expressed in the lung (Fig. 1B), RSPH4A may play similar roles in the cilia.
Fig. 8.
RSPH9 localization in control and Rsph6a KO mice. (A) Immunofluorescence analysis of spermatids from control and Rsph6a+4/+4 mutant mice labeled with antibodies against RSPH9 (red). RSPH9 signal was seen along the flagellum in Rsph6a+4/WT mice. In contrast, the signal decreased in the flagella of Rsph6a+4/+4 mutant mice. α-tubulin (green) stains both manchette and flagellum. Hoechst staining (blue) indicates the nucleus. W/o 1st Ab, Rsph6a+4/WT mouse spermatids processed without RSPH9 antibody staining. (B) Protein expression of RSPH9 in Rsph6aWT/+4 and Rsph6a+4/+4 testis and cauda epididymal spermatozoa. Acetylated tubulin signal decreased in the KO spermatozoa because the tail was short. In contrast, RSPH9 signal disappeared in the Rsph6a+4/+4 spermatozoa. CBB staining confirms equal loading.
Spermatozoa from Rsph6a+4/+4 mice can produce viable offspring with ICSI
While Rsph6a+4/+4 KO male mice are infertile due to short, immotile tails, we investigated if the nuclei can still produce viable embryos and attempted to rescue the phenotype using intracytoplasmic sperm injection (ICSI). Ninety-four WT oocytes were injected with the spermatozoa obtained from Rsph6a+4/+4 cauda epididymis and we found that 72 formed two-cell embryos, implying that KO spermatozoa can activate eggs. The embryos were transplanted into pseudopregnant female mice and four heterozygous mice were born. These results indicate that the infertility phenotype shown by Rsph6a+4/+4 males can be rescued via ICSI and the nuclei of KO spermatozoa have the ability to produce viable pups.
DISCUSSION
Rsph6a is an evolutionarily conserved gene, present in many eukaryotes including Chlamydomonas, mice, rats and humans. In this study, we showed that Rsph6a is expressed mainly in the mouse testis with little expression in the thymus and lungs. In the testis, Rsph6a is first detected during the haploid phase of spermatogenesis when secondary spermatocytes begin to appear at about postnatal day 18. Using the CRISPR/Cas9 system, we obtained a mouse that contained a 4 bp insertion in the coding region of Rsph6a and this led to the absence of RSPH6A in mature spermatozoa. Rsph6a+4/+4 male mice did not sire any pups due to flagellar deformation, and the spermatozoa were fully immotile. To confirm the phenotype, we also engineered Rsph6a large-deletion mice and found that these mice exhibited the same characteristics. Although Rsph6a KO spermatozoa exhibit abnormal flagellum formation, we cannot exclude the possibility that RSPH6A may also function in mature spermatozoa, as it was shown that RSPH6A is more phosphorylated during sperm capacitation (Paudel et al., 2018).
Misshapen sperm heads were also observed in Rsph6a+4/+4 mice. Using immunofluorescence and ultrastructural analysis, we found that manchette formation was normal but removal was not. Abnormal manchette removal can lead to the perinuclear ring tightening the nucleus incorrectly, resulting in club-shaped heads. This atypical manchette removal may be a secondary effect due to impaired tail formation because we could not detect a specific RSPH6A signal in the manchette. Further research is required to see if there is an interaction between manchette removal and tail formation.
Electron microscope (EM) data suggest that the 9+2 axoneme structures that are not surrounded by mitochondrial or fibrous sheath are normal but become disorganized as the flagellar formation process continues. Mitochondrial sheaths are misshapen and do not form properly. Immunofluorescence data confirm this, as mitochondrial protein TOM20 and fibrous sheath protein CABYR signals are undetectable or spotty. Without RSPH6A, sperm flagellar elongation may become unstable. As the axoneme is the important backbone for flagella, if the radial spoke is damaged, the axoneme may not be able to stabilize and elongation stops before proper mitochondrial and fibrous sheath formation. It is noteworthy that absence of Rsph4a, a paralog of Rsph6a, in mouse node cilia confers ultrastructural instability of the axoneme (Shinohara et al., 2015).
Co-IP analysis shows that RSPH6A interacts with RSPH1, RSPH4A, RSPH9 and RSPH10B in HEK293T cells. Further, we revealed that RSPH9 was depleted in the Rsph6a+4/+4 flagella, suggesting that the interaction of RSPH6A and RSPH9 is important to incorporate RSPH9 into the flagella. Further research about RSPH protein interactions may give clues as to how the radial spoke is assembled and incorporated into the sperm flagella. RSPH1 is a strong candidate that is associated with RSPH6A and RSPH9 during flagellar formation because Rsph1 KO mice have a similar phenotype to that of Rsph6a+4/+4 mice. It was found that Rsph1 mutant mice could develop normally until spermiogenesis step 9, but the rearrangement of mitochondria as well as flagellum and head formation was disturbed (Tokuhiro et al., 2008).
In addition to Rsph1, there are other short-tailed sperm phenotypes elucidated by KO mouse models. Another RS protein, DNAJB13, is the homolog of Chlamydomonas RSP16 located in the RS stalk. Dnajb13 KO mice exhibit hydrocephalus and KO spermatozoa are immotile as a result of flagellar defects (Oji et al., 2016). The Dnajb13 KO phenotype was similar to that of Rsph6a+4/+4 mice, as spermatozoa in both mutants had short tails. Mutations in DNAJB13 in humans have also been found and, as in the KO mouse model, spermatozoa had truncated flagellum and abnormal head morphology, suggesting that the disruption of the RSs in humans can also cause abnormal tail formation (El Khouri et al., 2016). In addition to RS proteins, there are axonemal proteins that when mutated exhibit abnormal sperm tail formation, such as IQCG (nexin-dynein regulatory complex) (Li et al., 2014), proteins in the central pair of microtubules [SPEF2 (Lehti et al., 2017), SPAG6 (Sapiro et al., 2002) and HYDIN (Oura et al., 2018)], and also proteins in the dynein arms [MNS1 (Zhou et al., 2012) and CCDC63 (Young et al., 2015)]. Similar phenotypes observed in the mutant lines of different axonemal components suggest that there may be a quality-control mechanism that triggers abortion of sperm tail formation when the axoneme structure is defective. Alternatively, it is possible that each component may play a distinct role in tail formation. Further experimentation is necessary to fully understand the molecular mechanism behind the role of RSPH6A in forming the flagellum.
In summary, our results show that Rsph6a is essential for male fertility due to its role in sperm flagellum formation in mice. Clinical research reports that human spermatozoa with short tails or stumps (also called multiple morphological abnormalities of the flagella or MMAF) were found almost 25 years ago (Baccetti et al., 1993) but the genes that were thus far revealed associated with MMAF account for only one-third of cases (Dong et al., 2018). Elucidating the interactions between the RS head proteins may give us a better understanding of the mechanism involved in sperm flagella formation and may lead us to better treatment for affected individuals.
MATERIALS AND METHODS
Animals
WT B6D2F1 or ICR mice were purchased from CLEA Japan (Tokyo, Japan) or Japan SLC (Shizuoka, Japan). All animal experiments were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University.
Antibodies
Antibodies against α-tubulin (#T5168), acetylated tubulin (#T7451) and FLAG (M2, #F1804) were purchased from Sigma-Aldrich (St Louis, MO, USA). AKAP4 antibody (#611564) from BD Biosciences (San Jose, CA, USA), CABYR antibody (#12351-1-AP) from Proteintech (Rosemont, IL, USA), FLAG antibody (#PM020) from MBL (Nagoya, Aichi, Japan), PA antibody (#016-25861) from FUJIFILM Wako Pure Chemical (Osaka, Japan), RSPH9 antibody (#HPA031703) from Atlas Antibodies (Bromma, Sweden), and TOM20 antibody (#sc-11415) and GAPDH antibody (#sc-25778) from Santa Cruz Biotechnology (Santa Cruz, CA, USA) were used. Rabbit polyclonal antibody was produced by immunization with mouse RSPH6A polypeptide (C plus REDEIFSQDTQHGPYLRDD). Produced RSPH6A antibody was purified using the RSPH6A polypeptide and SulfoLink coupling resin (Thermo Fisher Scientific, San Jose, CA, USA). Antibody against SLC2A3 (Fujihara et al., 2013) was described previously.
RT-PCR
RNA was prepared from multiple adult tissues and from the testes of ICR mice at different ages using TRIzol (#15596018, Thermo Fisher Scientific) according to the manufacturer's protocol. The obtained RNA was immediately reverse transcribed to cDNA with SuperScript III first-strand synthesis system (#18080051, Thermo Fisher Scientific) using an oligo (dT) primer. The amplification conditions for the subsequent PCR were 1 min at 94°C, followed by 35 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 30 s, with a final seven-minute extension at 72°C. The primers used are listed in Table S1 (primer #1-#12).
Immunofluorescence
Spermatozoa collected from the cauda epididymis were diluted in PBS, spread onto microscope slides and incubated at 37°C until dry. The samples were fixed with 4% paraformaldehyde in PBS for 20 min. The slides were subsequently fixed with 100% methanol at −30°C for 3 min. After two 10 min washes with PBS, the slides were blocked with 5% BSA and 10% goat serum diluted in PBS for 1 h at room temperature. Then, the slides were incubated with anti-RSPH6A (1:100), anti-TOM20 (1:25), anti-CABYR (1:200), anti-RSPH9 (1:50), anti-acetylated tubulin (1:500) or anti-α-tubulin (1:1000) diluted in blocking solution for 3 h at room temperature and then washed with PBS three times for 10 min each. After incubation with Alexa Fluor 488 or Alexa Fluor 546-conjugated secondary antibody (1:200) (Thermo Fisher Scientific) at room temperature for 90 min, the slides were washed with PBS three times for 10 min each. To stain the nucleus, the slides were incubated with Hoechst 33342 (1:5000) (#H3570, Thermo Fisher Scientific) for 15 min, washed with PBS three times for 10 min each and then mounted. Slides were viewed with an Olympus BX-50 or BX-53 microscope (Tokyo, Japan).
Mouse sperm protein fractionation
Sperm protein fractionation was performed as described previously with modifications (Cao et al., 2006). Spermatozoa were suspended in 1% Triton X-100 lysis buffer (50 mM NaCl, 20 mM Tris-HCl, pH 7.5, protease inhibitor mixture) and incubated for 2 h at 4°C. The sample was centrifuged at 15,000 g for 10 min to separate the Triton-soluble fraction (supernatant) and the Triton-resistant fraction (pellet). The pellet was resuspended in 1% SDS lysis buffer (75 mM NaCl, 24 mM EDTA, pH 6.0) and incubated for 1 h at room temperature. The sample was centrifuged at 15,000 g for 10 min to separate SDS-soluble fraction (supernatant) and SDS-resistant fraction (pellet). The pellet was dissolved in sample buffer (66 mM Tris-HCl, 2% SDS, 10% glycerol and 0.005% Bromophenol Blue) and boiled for 5 min.
Immunoblotting
Immunoblot analysis was performed as described previously (Yamaguchi et al., 2006). Protein lysates were resolved by SDS/PAGE under reducing condition (with 5% 2-mercaptoethanol) and transferred to PVDF membranes. After blocking with 10% skimmed milk, blots were incubated with primary antibodies overnight at 4°C and then incubated with secondary antibodies conjugated to horseradish peroxidase (1:10,000) (Jackson ImmunoResearch, West Grove, PA, USA). Antibodies used: anti-RSPH6A 1:1000; anti-acetylated tubulin 1:1000; ant-GAPDH 1:500; anti-SLC2A3 1:500; anti-AKAP4 1:5000; anti-PA 1:1000; anti-FLAG (#PM020) 1:500; anti-α-tubulin 1:10,000; and anti-RSPH9 1:200. The detection was performed using an ECL plus western blotting detection kit (GE Healthcare, Little Chalfont, UK). For Coomassie Brilliant Blue (CBB) staining, a Rapid Stain CBB Kit (#30035-14, Nacalai Tesque, Kyoto, Japan) was used.
Generation of RSPH recombinant proteins
To understand the interaction of RSPH proteins in HEK293T cells, expression plasmids were designed. Rsph1, Rsph4a, Rsph9 and Rsph10b were amplified from mouse testis cDNA, digested with EcoRI, HindIII or XbaI and ligated into PA-tagged (C-terminus) pCAG vectors that contain the CAG promoter and a rabbit globin poly(A) signal (Niwa et al., 1991). Rsph6a was ligated into a FLAG-tagged (C-terminus) pCAG vector. The primers used to amplify each gene are listed in Table S1 (primer #13-#22). Rsph6a and other Rsph vectors were cotransfected into HEK293T cells (Tiscornia et al., 2006), cultured for 24 h and then collected into 1.5 ml tubes. After centrifugation and removal of supernatant, 100 μl lysis buffer (1% Triton X-100, 50 mM NaCl, 20 mM Tris-HCl, pH 7.4, protease inhibitor mixture) was added to each tube and rotated at 4°C for 30 min. The tubes were subsequently centrifuged for 15 min at 15,000 g and the supernatant was used for immunoprecipitation.
Co-immunoprecipitation
Protein lysates (supernatants) were incubated with FLAG (M2) antibody-conjugated Dynabeads Protein G (#10003D, Thermo Fisher Scientific) for 60 min at 4°C. The samples were then washed three times with PBS containing 40 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Triton X-100 and 10% glycerol. The protein complexes were eluted with sample buffer containing 5% 2-mercaptoethanol before SDS-PAGE and immunoblot analysis.
gRNA design
Potential gRNA and PAM sites were found using the online source CRISPRdirect (Naito et al., 2014). gRNAs with fewer off-target sites were designed.
Plasmid and oligonucleotide preparation
Plasmids expressing hCas9 and gRNA were prepared by ligating oligonucleotides into the BbsI site of pX330 (Cong et al., 2013) (http://www.addgene.org/42230/). The pCAG-EGxxFP target plasmid was prepared as previously described (Mashiko et al., 2013) (http://www.addgene.org/50716/). The primers used for the pCAG-EGxxFP construction are in Table S1 (primer #23-#30). The gRNA target sequence for the 4 bp insertion was 5′-GGCTGGACCTCTGTGGCCAG-3′. The gRNA target sequences for large deletion were 5′-TAGACTCCAGTTGTTCTAAG-3′ (exon 1) and 5′-GAAGGCAGACGAGGCAATGG-3′ (exon 4 of variant 1).
gRNA reporter assay
Confirmation of gRNA/Cas9 cleavage activity was performed by transfecting HEK293T cells with pCAG-EGxxFP and pX330 plasmids, as previously reported (Mashiko et al., 2013).
Pronuclear injection for the 4 bp insertion
Superovulation-induced B6D2F1 female mice were mated with B6D2F1 males, and fertilized eggs were collected from the oviduct. The pX330 plasmid containing the target gRNA sequence was injected into one of the pronuclei at 5 ng/μl. The injected eggs were cultivated in potassium simplex optimization medium (KSOM) (Ho et al., 1995) overnight, and the two-cell-stage embryos were transferred into the oviducts of pseudopregnant ICR females. The pups obtained were genotyped by PCR and then subsequently confirmed by Sanger sequencing.
Electroporation for the large deletion
Superovulation-induced B6D2F1 female mice were mated with B6D2F1 males, and fertilized eggs were collected from the oviduct. Ordered crRNAs (Sigma-Aldrich), tracrRNA (#TRACRRNA05N-5NMOL, Sigma-Aldrich) and CAS9 protein (#B25640, Thermo Fisher Scientific) were incubated at 37°C for 5 min to make the CRISPR/Cas9 complex (16 ng/μl crRNA plus tracrRNA, 40 ng/μl CAS9). The obtained complex was electroporated into fertilized oocytes using a super electroporator NEPA21 (NEPA GENE, Chiba, Japan) (poring pulse, voltage: 225 V, pulse width: 2 ms, pulse interval: 50 ms, and number of pulses: +4; transfer pulse, voltage: 20 V, pulse width: 50 ms, pulse interval: 50 ms, and number of pulses: ±5). The eggs were cultivated in KSOM (Ho et al., 1995) overnight, and the two-cell-stage embryos were transferred into the oviducts of pseudopregnant ICR females. The pups obtained were genotyped by PCR and then subsequently confirmed by Sanger sequencing.
Genotyping of subsequent generations
PCR analysis of genomic DNA was performed for Rsph6a+4/+4 mutant mice using the same primer sets as those used for the construction of pCAG-EGxxFP. Direct Sanger sequencing of PCR products was then performed. Rsph6a+4/+4 mutant mice had a deletion of an MscI site, enabling faster genotypic analysis via PCR and subsequent enzyme digestion. Two sets of primers were used to genotype Rsph6aLD/LD (Fig. S4A, Table S1): one that amplified about 450 bp in exon 3 of variant 1 to check the WT allele (primers c and d) and one that only amplified 320 bp if the LD allele is present (primers a and b).
In vivo male fertility tests
Three sexually mature Rsph6a+4/+4 male mice were caged with two 8-week-old B6D2F1 female mice each for 2 months, and plugs were checked every morning. For controls, 3 WT and heterozygous mice were used. The number of pups was counted on the day of birth.
Protein extraction, testis histology, and sperm morphology and motility
To extract proteins from testis, thymus, lung or spermatozoa for western blotting, tissues or spermatozoa were homogenized in lysis buffer containing 6 M urea, 2 M thiourea and 2% sodium deoxycholate and then centrifuged with the supernatants before being collected. For testis histology, testes were fixed in Bouin's solution and were processed for paraffin embedding. Five micrometer paraffin sections were stained with periodic acid-Schiff (PAS) and then counterstained with Mayer hematoxylin solution (#193-08445, #131-09665, FUJIFILM Wako Pure Chemical). Cauda epididymal spermatozoa were dispersed in TYH (Toyoda et al., 1971) and observed under a phase contrast microscope (Olympus BX-50 or BX-53 microscope) to assess morphology and motility.
Sperm velocity analysis
Sperm velocity was analysed as described previously (Miyata et al., 2015). Cauda epididymal spermatozoa were suspended in TYH medium (Toyoda et al., 1971). Sperm velocity was measured using the CEROS sperm analysis system (software version 12.3; Hamilton Thorne Biosciences, Beverly, MA, USA) at 10 min and 2 h after incubation. More than 200 spermatozoa were analyzed for each male.
Manchette staining
Germ cells including spermatids were squeezed out from the seminiferous tubules onto slide glasses and air-dried at 37°C. The samples were fixed with 4% paraformaldehyde in PBS for 15 min and washed with PBS three times for 5 min each. The samples were then permeabilized with 0.1% Triton X-100 for 15 min, washed with PBS three times for 5 min each, and blocked with 5% BSA and 10% goat serum diluted in PBS for 1 h at room temperature. Reactions with antibodies were performed as described above.
Electron microscopy analysis of the sperm flagellum
To understand the ultrastructure of sperm flagella, testis samples were prepared for transmission electron microscopy analysis as previously described (Inoue et al., 2011). Sections were examined using a JEM-1400 plus electron microscope (JEOL, Tokyo, Japan) at 80 kV.
Intracytoplasmic sperm injection (ICSI)
ICSI was performed as previously reported (Kimura and Yanagimachi, 1995). Mature oocytes were collected from superovulated B6D2F1 mice 13–15 h after injection of human chorionic gonadotropin (hCG). After treatment with hyaluronidase to remove the cumulus oocyte complex, oocytes were placed in fresh KSOM medium at 37°C under 5% CO2 in air until subjected to ICSI. Mutant sperm heads were separated from the tail by applying a few piezo pulses, then injected into a mature oocyte using a piezo manipulator (PrimeTech, Ibaraki, Japan) (Kimura and Yanagimachi, 1995). The following day, two-cell embryos were counted and transferred to pseudopregnant females. Pups were genotyped at birth.
Statistics
Statistical analyses were performed using Student's t-test (two-tailed). Differences were considered significant at P<0.05 (*) or highly significant at P<0.01 (**). Error bars shown as standard deviation (s.d.) if not indicated otherwise.
Supplementary Material
Acknowledgements
We thank Dr Nobuyuki Sakurai, Seiya Oura and Eri Hosoyamada for technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: F.A., H.M., M.I.; Formal analysis: F.A., H.M., M.I.; Investigation: F.A., H.M., K.S., A.M., K.N., T.M., Z.X., P.P., M.I.; Writing - original draft: F.A., H.M.; Writing - review & editing: M.I.; Supervision: M.I.; Project administration: M.I.; Funding acquisition: M.I.
Funding
This work was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP17H04987 to H.M., JP17K17852 to K.S., JP17J09669 to T.M., JP17H01394 and JP 25112007 to M.I.); Japan Agency for Medical Research and Development (AMED) (JP18gm5010001 to M.I.); Takeda Science Foundation grants to H.M. and M.I.; National Institutes of Health (P01HD087157 and R01HD088412 to M.I.); and the Bill & Melinda Gates Foundation (Grand Challenges Explorations grant OPP1160866 to M.I.). Deposited in PMC for release after 12 months.
Data availability
Rsph6a+4/WT mice (B6D2-Rsph6aem1Osb) used in this study are available through the Riken BioResource Center, Japan (http://en.brc.riken.jp/) (RBRC number: RBRC09840) or Center for Animal Resources and Development (CARD), Kumamoto University, Japan (http://cardb.cc.kumamoto-u.ac.jp/transgenic/) (CARD ID: 2460). Rsph6aLD/WT mice (B6D2-Rsph6aem2Osb) are being processed for deposition.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.221648.supplemental
References
- Afzelius B. (1959). Electron microscopy of the sperm tail results obtained with a new fixative. J. Biophys. Biochem. Cytol. 5, 269-278. 10.1083/jcb.5.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Mulgund A., Hamada A. and Chyatte M. R. (2015). A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 13, 37 10.1186/s12958-015-0032-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccetti B., Burrini A. G., Capitani S., Collodel G., Moretti E., Piomboni P. and Renieri T. (1993). Notulae seminologicae. 2. The ‘short tail’ and ‘stump’ defect in human spermatozoa. Andrologia 25, 331-335. 10.1111/j.1439-0272.1993.tb02736.x [DOI] [PubMed] [Google Scholar]
- Bellve A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M. and Dym M. (1977). Spermatogenic cells of the prepuberal mouse: Isolation and morphological characterization. J. Cell Biol. 74, 68-85. 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Gerton G. L. and Moss S. B. (2006). Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol. Cell. Proteomics. 5, 801-810. 10.1074/mcp.M500322-MCP200 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B. and Bettencourt-Dias M. (2011). Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 195, 341-341 10.1083/jcb.2010111521952c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleman V. H., Romio L., Chodhari R., Hirst R. A., de Castro S. C. P., Parker K. A., Ybot-Gonzalez P., Emes R. D., Wilson S. W., Wallis C. et al. (2009). Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 84, 197-209. 10.1016/j.ajhg.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A. et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A. M., Williams B. D. and Rosenbaum J. L. (1992). Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol. Cell. Biol. 12, 3967-3977. 10.1128/MCB.12.9.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F. N., Amiri-Yekta A., Martinez G., Saut A., Tek J., Stouvenel L., Lorès P., Karaouzène T., Thierry-Mieg N., Satre V. et al. (2018). Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am. J. Hum. Genet. 102, 636-648. 10.1016/j.ajhg.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy J. E. M., Okuda H., O'Connor A. E., Merriner D. J., O'Donnell L., Jamsai D., Bergmann M. and O'Bryan M. K. (2017). Katanin-like 2 (KATNAL2) functions in multiple aspects of haploid male germ cell development in the mouse. PLoS Genet. 13, e1007078 10.1371/journal.pgen.1007078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khouri E., Thomas L., Jeanson L., Bequignon E., Vallette B., Duquesnoy P., Montantin G., Copin B., Dastot-Le Moal F., Blanchonet S. et al. (2016). Mutations in DNAJB13, encoding an HSP40 family member, cause primary ciliary dyskinesia and male infertility. Am. J. Hum. Genet. 99, 489-500. 10.1016/j.ajhg.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Ansved T., Anvret M. and Carey N. A. (2001). Mammalian radial spokehead-like gene, RSHL1, at the myotonic dystrophy-1 locus. Biochem. Biophys. Res. Commun. 281, 835-841. 10.1006/bbrc.2001.4465 [DOI] [PubMed] [Google Scholar]
- Frommer A., Hjeij R., Loges N. T., Edelbusch C., Jahnke C., Raidt J., Werner C., Wallmeier J., Große-Onnebrink J., Olbrich H. et al. (2015). Immunofluorescence analysis and diagnosis of primary ciliary dyskinesia with radial spoke defects. Am. J. Respir. Cell Mol. Biol. 53, 563-573. 10.1165/rcmb.2014-0483OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara Y., Tokuhiro K., Muro Y., Kondoh G., Araki Y., Ikawa M. and Okabe M. (2013). Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc. Natl. Acad. Sci. USA 110, 8111-8116. 10.1073/pnas.1222166110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Wigglesworth K., Eppig J. J. and Schultz R. M. (1995). Preimplantation development of mouse embryos in KSOM: Augmentation by amino acids and analysis of gene expression. Mol. Reprod. Dev. 41, 232-238. 10.1002/mrd.1080410214 [DOI] [PubMed] [Google Scholar]
- Huang B., Piperno G., Ramanis Z. and Luck D. J. (1981). Radial spokes of Chlamydomonas flagella: Genetic analysis of assembly and function. J. Cell Biol. 88, 80-88. 10.1083/jcb.88.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N., Satouh Y., Ikawa M., Okabe M. and Yanagimachi R. (2011). Acrosome-reacted mouse spermatozoa recovered from the perivitelline space can fertilize other eggs. Proc. Natl. Acad. Sci. USA 108, 20008-20011. 10.1073/pnas.1116965108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y. and Yanagimachi R. (1995). Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 121, 2397-2405. [DOI] [PubMed] [Google Scholar]
- Knowles M. R., Ostrowski L. E., Leigh M. W., Sears P. R., Davis S. D., Wolf W. E., Hazucha M. J., Carson J. L., Olivier K. N., Sagel S. D. et al. (2014). Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care Med. 189, 707-717. 10.1164/rccm.201311-2047OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T., Wakabayashi K., Diener D. R., Rosenbaum J. L. and Kamiya R. (2011). Subunit interactions within the Chlamydomonas flagellar spokehead. Cytoskeleton 68, 237-246. 10.1002/cm.20507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott E., Legendre M., Copin B., Papon J.-F., Dastot-Le Moal F., Montantin G., Duquesnoy P., Piterboth W., Amram D., Bassinet L. et al. (2013). Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 93, 561-570. 10.1016/j.ajhg.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti M. S. and Sironen A. (2016). Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 151, R43-R54. 10.1530/REP-15-0310 [DOI] [PubMed] [Google Scholar]
- Lehti M. S., Kotaja N. and Sironen A. (2013). KIF3A is essential for sperm tail formation and manchette function. Mol. Cell. Endocrinol. 377, 44-55. 10.1016/j.mce.2013.06.030 [DOI] [PubMed] [Google Scholar]
- Lehti M. S., Zhang F.-P., Kotaja N. and Sironen A. (2017). SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. Development 144, 2683-2693. 10.1242/dev.152108 [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T. and Bork P. (2015). SMART: recent updates, new development and status. Nucleic Acids Res. 43, D257-D260. 10.1093/nar/gku949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.-K., Tan J.-L., Chen L.-T., Feng J.-S., Liang W.-X., Guo X.-J., Liu P., Chen Z., Sha J.-H., Wang Y.-F. et al. (2014). Iqcg is essential for sperm flagellum formation in mice. PLoS ONE 9, e98053 10.1371/journal.pone.0098053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Deboer K., de Kretser D. M., O'Donnell L., O'Connor A. E., Merriner D. J., Okuda H., Whittle B., Jans D. A., Efthymiadis A. et al. (2015). LRGUK-1 is required for basal body and manchette function during spermatogenesis and male fertility. PLoS Genet. 11, e1005090 10.1371/journal.pgen.1005090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A. and Ikawa M. (2013). Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3, 3355 10.1038/srep03355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H., Satouh Y., Mashiko D., Muto M., Nozawa K., Shiba K., Fujihara Y., Isotani A. and Ikawa M. (2015). Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science 350, 442-445. 10.1126/science.aad0836 [DOI] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H. and Ui-Tei K. (2014). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120-1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K. and Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193-199. 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- Oji A., Noda T., Fujihara Y., Miyata H., Kim Y. J., Muto M., Nozawa K., Matsumura T., Isotani A. and Ikawa M. (2016). CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci. Rep. 6, 31666 10.1038/srep31666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura S., Miyata H., Noda T., Shimada K., Matsumura T., Morohoshi A., Isotani A. and Ikawa M. (2018). Chimeric analysis with newly established EGFP/DsRed2-tagged ES cells identify HYDIN as essential for spermiogenesis in mice. Exp. Anim. 10.1538/expanim.18-0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel B., Gervasi M. G., Porambo J., Caraballo D., Tourzani D., Mager J., Platt M. D., Salicioni A. M. and Visconti P. E. (2018). Sperm capacitation is associated with phosphorylation of the testis-specific radial spoke protein Rsph6a. Biol. Reprod. ioy202 10.1093/biolre/ioy202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Ramanis Z. and Luck D. J. L. (1981). Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88, 73-79. 10.1083/jcb.88.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapiro R., Kostetskii I., Olds-Clarke P., Gerton G. L., Radice G. L. and Strauss J. F. III (2002). Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 22, 6298-6305. 10.1128/MCB.22.17.6298-6305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P. and Christensen S. T. (2007). Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69, 377-400. 10.1146/annurev.physiol.69.040705.141236 [DOI] [PubMed] [Google Scholar]
- Shinohara K., Chen D., Nishida T., Misaki K., Yonemura S. and Hamada H. (2015). Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev. Cell 35, 236-246. 10.1016/j.devcel.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Thoma M. E., McLain A. C., Louis J. F., King R. B., Trumble A. C., Sundaram R. and Louis G. M. (2013). Prevalence of infertility in the United States as estimated by the current duration approach and a traditional constructed approach. Fertil. Steril. 99, 1324-1331. 10.1016/j.fertnstert.2012.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G., Singer O. and Verma I. M. (2006). Production and purification of lentiviral vectors. Nat. Protoc. 1, 241-245. 10.1038/nprot.2006.37 [DOI] [PubMed] [Google Scholar]
- Tokuhiro K., Hirose M., Miyagawa Y., Tsujimura A., Irie S., Isotani A., Okabe M., Toyama Y., Ito C., Toshimori K. et al. (2008). Meichroacidin containing the membrane occupation and recognition nexus motif is essential for spermatozoa morphogenesis. J. Biol. Chem. 283, 19039-19048. 10.1074/jbc.M708590200 [DOI] [PubMed] [Google Scholar]
- Toyoda Y., Yokoyama M. and Hosi T. (1971). Studies on the fertilization of mouse eggs in vitro. Jpn. J. Anim. Reprod. 16, 152-157. 10.1262/jrd1955.16.152 [DOI] [Google Scholar]
- Witman G. B., Plummer J. and Sander G. (1978). Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76, 729-747. 10.1083/jcb.76.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R., Yamagata K., Ikawa M., Moss S. B. and Okabe M. (2006). Aberrant distribution of ADAM3 in sperm from both angiotensin-converting enzyme (Ace)- and calmegin (Clgn)-deficient mice1. Biol. Reprod. 75, 760-766. 10.1095/biolreprod.106.052977 [DOI] [PubMed] [Google Scholar]
- Yang P., Diener D. R., Yang C., Kohno T., Pazour G. J., Dienes J. M., Agrin N. S., King S. M., Sale W. S., Kamiya R. et al. (2006). Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119, 1165-1174. 10.1242/jcs.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S., Miyata H., Satouh Y., Kato H., Nozawa K., Isotani A., Aitken R. J., Baker M. A. and Ikawa M. (2015). CRISPR/Cas9-mediated rapid generation of multiple mouse lines identified ccdc63 as essential for spermiogenesis. Int. J. Mol. Sci. 16, 24732-24750. 10.3390/ijms161024732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S. A. M., Miyata H., Satouh Y., Aitken R. J., Baker M. A. and Ikawa M. (2016). CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J. Cell Sci. 129, 4379-4387. 10.1242/jcs.193151 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shen X., Gude D. R., Wilkinson B. M., Justice M. J., Flickinger C. J., Herr J., Eddy E. M. and Strauss J. F. (2009). MEIG1 is essential for spermiogenesis in mice. Proc. Natl. Acad. Sci. USA 106, 17055-17060. 10.1073/pnas.0906414106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Yang F., Leu N. A. and Wang P. J. (2012). MNS1 is essential for spermiogenesis and motile ciliary functions in mice. PLoS Genet. 8, e1002516 10.1371/journal.pgen.1002516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.