ABSTRACT
Stress granules (SGs) assemble under stress-induced conditions that inhibit protein synthesis, including phosphorylation of eIF2α, inhibition of the RNA helicase eIF4a proteins or inactivation of mTORC1. Classically defined SGs are composed of translation initiation factors, 40S ribosomes, RNA-binding proteins and poly(A)+ mRNAs. As such, they represent an important compartment for storage of mRNAs and regulation of their translation. Emerging work on SGs indicates that these structures might promote cellular survival in diverse disease states. Yet, much work on SG formation and function employs acute stress conditions, which might not accurately reflect the chronic stresses that manifest in human disease. Here, we used prolonged nutrient starvation to model and investigate SG formation and function during chronic stress in a human cell line and mouse embryonic fibroblasts. Surprisingly, we found that SGs that form under chronic nutrient starvation lack 40S ribosomes, do not actively exchange their constituent components with cytoplasmic pools and promote cell death. We named these SGs starvation-induced SGs (stSGs). Our results on stSGs imply that SG assembly and function in the context of prolonged nutrient starvation stress differ significantly from what has been described for acute stress conditions.
KEY WORDS: Stress granule, eIF2α, G3BP1, Starvation, Translation, Cell death
Summary: This work characterizes the mechanisms of formation of a novel type of stress granule that is induced in response to long-term starvation and, unlike previously described stress granules, functions in a pro-death capacity.
INTRODUCTION
Stress granules (SGs) are RNA granules that contain translation initiation factors, RNA-binding proteins, poly(A)+ RNA and 40S ribosomes (Reineke and Lloyd, 2013; Kedersha and Anderson, 2007). SGs assemble owing to accumulation of stalled translation initiation complexes, the latter of which can be induced by phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) or inhibition of the RNA helicase eIF4a proteins (Kedersha et al., 2013, 2002; Dang et al., 2006). eIF2α phosphorylation is a well-established precursor to SG assembly; the former process is regulated by the antagonistic functions of several eIF2α kinases and phosphatases. Four eIF2α kinases have been identified: HRI (also known as EIF2AK1), PERK (also known as EIF2AK3), PKR (also known as EIF2AK2) and GCN2 (also known as EIF2AK4). HRI is well characterized to be activated under conditions of low heme in erythrocytes, but can also act in response to oxidative stress, such as is experienced during arsenite exposure (McEwen et al., 2005). PERK primarily signals a state of endoplasmic reticulum (ER) stress (McQuiston and Diehl, 2017). PKR is activated in response to cytoplasmic double-stranded RNA during viral infection and might respond to other physiological stressors, including oxidative and ER stress, and cytokine signaling (Garcia et al., 2006). Finally, GCN2 is activated by amino acid deprivation and UV stress (Deng et al., 2002; Aulas et al., 2017). Thus, although each kinase is activated in response to a unique type of cellular stress, some commonality among their activation has been shown to exist, based on the type and duration of the stress. Two phosphatase complexes have been shown to ameliorate the function of the eIF2α kinases, one containing GADD34 (also known as PPP1R15A) and the other containing CrEP (also known as PPP1R15B) (Harding et al., 2009; Hetz et al., 2013). The regulation of these phosphatase complexes is not well characterized.
SG assembly has been studied extremely well in the context of acute stressors, including oxidative stress, ER stress, heat shock and UV irradiation. SGs induced by acute stress contain, by definition, translation initiation factors, 40S ribosomes and RNA-binding proteins, such as nucleating proteins G3BP1 and G3BP2, Tia1 and TiaR (also known as TIAL1). Some other SG-resident RNA-binding proteins include HuR (also known as ELAVL1), PABP (also known as PABPC3) and FMRP (also known as FMR1). Acute SGs are also extremely dynamic, and recent work characterizing core complexes within acute SGs has led to a model of a dynamic shell, from which various components are exchanged with the cytosol, surrounding a stable core complex (Jain et al., 2016). Acute SGs also contain many signaling molecules, and recent evidence suggests that they might modulate signaling through several pathways including mTORC1, NFκB and innate immune signaling through PKR and RIG-I (also known as DDX58) (Sfakianos et al., 2018; Reineke et al., 2015; Kim et al., 2005; Onomoto et al., 2012). SG assembly during acute stressors has also been shown to regulate MAPK signaling, resulting in the paradigm that SGs function in a pro-survival response (Arimoto et al., 2008). This paradigm has resulted in the assumption that all SGs function in this capacity, despite findings indicating that SGs forming in the broad range of acute stressors vary in composition (Aulas et al., 2017).
Given the above pathways that promote SG condensation and the paradigm that SGs function to promote pro-survival, we endeavored to investigate whether this paradigm also applies to chronic stress. We utilized a model of chronic nutrient starvation because of the commonality of this state within numerous pathologies, including tumor biology, cardiac ischemia and stroke. Nutrient starvation has been shown to inhibit protein synthesis through mTORC1 inhibition, which has been implicated in acute SG formation. Therefore, studying nutrient starvation also provides insight into crosstalk of mTORC1 signaling and eIF2α phosphorylation in promoting SG assembly. We characterized SG dynamics, composition and function in the regulation of cell fate during chronic nutrient starvation in a human cell line and mouse embryonic fibroblasts (MEFs). Our findings indicate that a novel type of granule forms under conditions of chronic nutrient starvation stress. In contrast to canonical acute SGs, these novel RNA granules are less dynamic, do not contain 18S small ribosomal RNA (rRNA) and act to promote cell death. Our findings indicate that different SGs function differently and the paradigm of SG as pro-survival might not apply to all stress conditions.
RESULTS
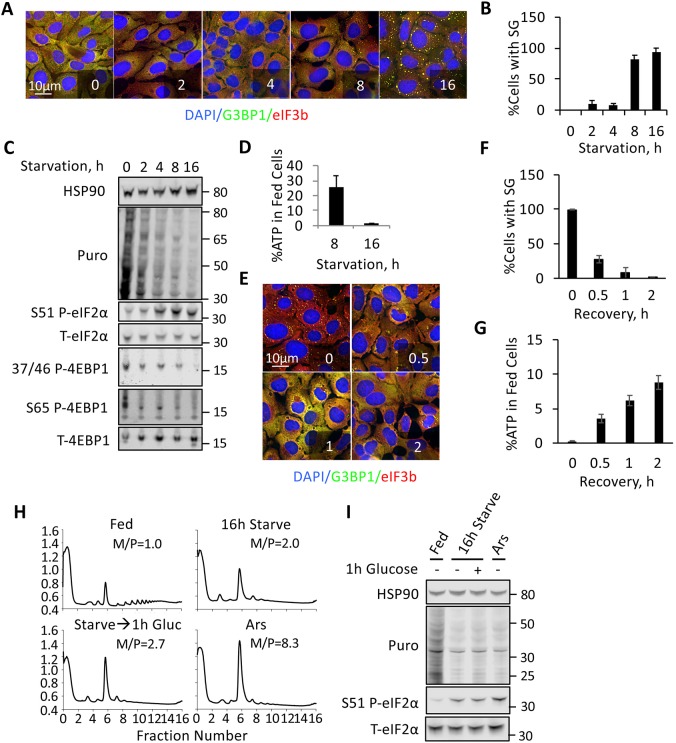
To model chronic stress, we employed an existing definition of this state within the literature as stress typically lasting a duration of greater than 4 h (Guan et al., 2017; Shelkovnikova et al., 2017). We utilized a chronic nutrient starvation stress, in which cells were starved of glucose, serum, glutamine and pyruvate. Under these conditions, amino acids are present, with the exception of glutamine. The conditions thus mimic starvation conditions experienced in pathological states characterized by restricted blood flow, during which amino acid catabolism is favored and the lack of oxygen impairs metabolism of glucose and glutamine (Drake et al., 2012; Vaupel et al., 1989). We first performed time-course experiments to investigate the formation of starvation-induced SGs (stSGs) in U2OS cells in response to these chronic nutrient starvation conditions. Strikingly, using the subcellular localization and aggregation of the SG nucleating RNA-binding protein G3BP1 and eIF3b as canonical markers of SGs, stSGs did not appear until 8 h of nutrient starvation (Fig. 1A,B). These kinetics are far slower than the kinetics previously described for acute stressors, where SG often appear within 1 h (Kedersha et al., 2002, 2000; Farny et al., 2009; Aulas et al., 2017). We thus asked whether translation was repressed concomitantly with stSG assembly, and which pathways are responsible for impaired protein synthesis during chronic nutrient starvation. We assessed levels of bulk translation using ribopuromycylation, as well as eIF2α phosphorylation and 4EBP1 (also known as EIF4EBP1) dephosphorylation, which repress translation under cellular stress conditions. 4EBP1 is a direct target of mTORC1, which senses nutrient availability and relays that information to the translation apparatus (Tee, 2018). Our experiments revealed that 4EBP1 phosphorylation at T37/46 and S65 is greatly reduced after 2 h of starvation, and that SG assembly within these conditions is itself closely correlated with the kinetics of eIF2α phosphorylation, beginning at 8 h (Fig. 1C). These results suggest that stSG assembly under chronic nutrient starvation is not directly dependent on decreased mTORC1 activity, but is likely to instead occur in response to eIF2α phosphorylation. Because the literature indicates that acute SGs depend on ATP for assembly and dynamics (Jain et al., 2016), and our starvation conditions were anticipated to deplete cells of ATP, we measured ATP levels using a luminescence-based assay. At 8 h of starvation, concomitant with stSG appearance, ATP levels had fallen to 25% of those measured in fed controls (Fig. 1D). After 16 h of starvation, the point at which all cells contain stSGs, ATP levels were ∼1% of those of fed controls. This indicates that the observed stSGs assemble concomitantly with declining ATP levels (Fig. 1D).
Fig. 1.
Chronic nutrient starvation induces SGs with slow assembly kinetics. (A) U2OS cells were starved of nutrients for the indicated times prior to fixation and staining with antibodies against G3BP1 (green) and eIF3b (red). (B) Quantification of the percentage of cells with stSGs from A. (C) Western blot analysis of starvation stress kinetics. Puromycin labeling was used to analyze ongoing translation, and pathways known to mediate translation repression were examined (mTORC1 and eIF2α), as indicated. Molecular weight standards are indicated. (D) ATP levels during starvation at 8 h and 16 h time points. ATP was detected using a luciferase-based luminescence assay. ATP levels are expressed as a percentage of control (fed) values at each time point. (E) U2OS cells were starved for 16 h prior to the addition of glucose. Cells were then incubated for the indicated times before harvesting and processing as in A. (F) Quantification of the percentage of cells with stSGs from E. (G) U2OS cells were treated as in E before ATP analysis. ATP levels are expressed as a percentage of control (fed) values at each time point. (H) U2OS cells were treated as indicated and harvested for polysome profiling. M/P represents the monosome/polysome ratio for the indicated graph. The y-axis is voltage units proportional to OD254 and the x-axis is fraction number. (I) Western blot analysis of the unfractionated extracts shown in H. Results from all panels are representative of a minimum of three experimental replicates, and data are presented as mean±s.d.
Reversibility of assembly is a notable feature of acute SGs. On the assumption that stSGs arise from ATP restriction, we aimed to assess whether stSGs are reversible by first stressing cells for 16 h, then added glucose to evaluate reversibility when a single ATP source is added back to the medium. We found that stSGs are reduced to ∼30% of cells with stSGs after just 30 min of glucose addition, and are only present in ∼10% of cells after 1 h of glucose addition (Fig. 1E,F). This decrease in stSGs coincides with a burst in ATP at 30 min and a steady rise in ATP thereafter (Fig. 1G). Because our data indicate that nutrient starvation induces translation repression through eIF2α phosphorylation, we sought to determine whether polysomes disappear (similar to acute stress conditions in which eIF2α phosphorylation is induced). During both acute arsenite and chronic nutrient starvation stress conditions, monosomes are greatly reduced, consistent with repression at the initiation stage of translation, and polysomes are absent (Fig. 1H). Addition of glucose to nutrient-starved cells for 1 h does not drastically rescue polysomes, despite the disappearance of stSGs (Fig. 1H), although the monosome peak is elevated, which might indicate loading of single ribosomes onto mRNAs. eIF2α phosphorylation was not significantly decreased after 1 h of glucose addition to starved cells, thus indicating that eIF2α dephosphorylation is not required for stSG disassembly (Fig. 1I). Taken together, these data indicate that stSGs induced by chronic starvation are coincident with a decrease in global translation; and that although the stSG induction can be reversed by increased availability of glucose, this increase in glucose is not sufficient to promote the full re-activation of the translation machinery.
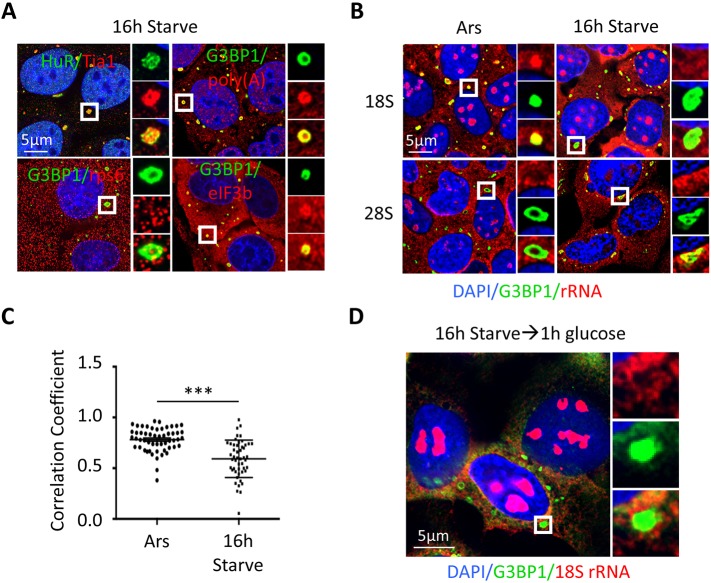
Although the kinetics of stSG assembly during extreme nutrient starvation are slower than with acute stressors, the temporal relationship between eIF2α phosphorylation and stSG assembly is reminiscent of a similar correlation during acute stress. This similarity suggests that parallels might exist in these two contexts in regard to the molecular control of SG formation. To more thoroughly understand the composition of stSGs, we analyzed the colocalization of other canonical SG proteins with G3BP1-containing stSGs under nutrient deprivation. Similar to acute SGs, we found colocalization of the RNA-binding proteins HuR and Tia1 within stSGs (Fig. 2A). Also similar to acute stress-induced SGs, chronic stSGs also stained positive for poly(A)+ RNA via fluorescence in situ hybridization (FISH) (Aulas et al., 2017). In contrast, stSGs lack rps6 (Fig. 2A) (Kedersha et al., 2002), suggesting that small ribosomal subunits are excluded from these structures. We therefore performed RNA FISH for 18S and 28S rRNA in cells subjected to both acute oxidative stress and chronic nutrient starvation (Fig. 2B,C) (Kedersha et al., 2002, 2016; Sfakianos et al., 2018; Reineke et al., 2017; Tsai et al., 2016). Surprisingly, 18S rRNA was strongly localized to arsenite-induced SG, whereas 18S was weak or absent in stSGs using two different 18S FISH probes (Fig. 2B,C and data not shown). Given that addition of glucose for 1 h following chronic nutrient starvation reduces the proportion of cells with stSGs to ∼10% and elevates ATP levels (Fig. 1), we asked whether glucose addition would be sufficient to allow recruitment of 18S rRNA to stSGs in the 10% of cells that still contain stSGs. Surprisingly, glucose addition did not result in recruitment of 18S to chronic stSGs (Fig. 2D). We next examined 18S colocalization following 8 h of chronic starvation (when ATP levels were higher), and under conditions in which arsenite was added to chronic nutrient-starved cells. We found that 18S did not accumulate in cells in either condition (Fig. S1), indicating that stSGs do not contain stalled 48S translation initiation complexes (Kedersha et al., 2002). This is consistent with a fundamental difference from canonical SGs. Consistent with previous work (Kedersha et al., 2002), 28S rRNA does not appear to concentrate in stSGs in the context of either arsenite or chronic nutrient starvation (Fig. 2B).
Fig. 2.
stSGs have similar composition to other types of stSG, but lack 40S ribosomes. (A) U2OS cells were starved for 16 h followed by fixation and staining for the indicated SG constituents. G3BP1 is the most well-studied RNA-binding protein within stSGs; HuR and Tia1 are other well-studied stSG constituents; poly(A)+ mRNAs are included in canonical, acute SGs; eIF3b is a translation initiation factor and its localization with stSGs defines stSGs; rps6 is a subunit of the 40S ribosome. (B) RNA FISH was performed with probes directed against 18S and 28S rRNA after U2OS cells were exposed to either 250 μM arsenite stress for 1 h or chronic nutrient starvation (as indicated). (C) Quantification of colocalization between 18S rRNA and G3BP1. Colocalization was measured using a Manders’ correlation test, where 0 indicates no colocalization and 1 indicates perfect colocalization. Each data point represents the Manders’ coefficient for each cell, spanning eight fields per condition. Microscopy was performed with a 100× objective. A two-tailed Student's t-test was used to measure statistical significance (***P<0.001), and data are presented as mean±s.e.m. (D) U2OS cells were starved for 16 h prior to the addition of glucose for 1 h. Cells were fixed and stained as in B with 18S probes. Results from all panels are representative of a minimum of three experimental replicates.
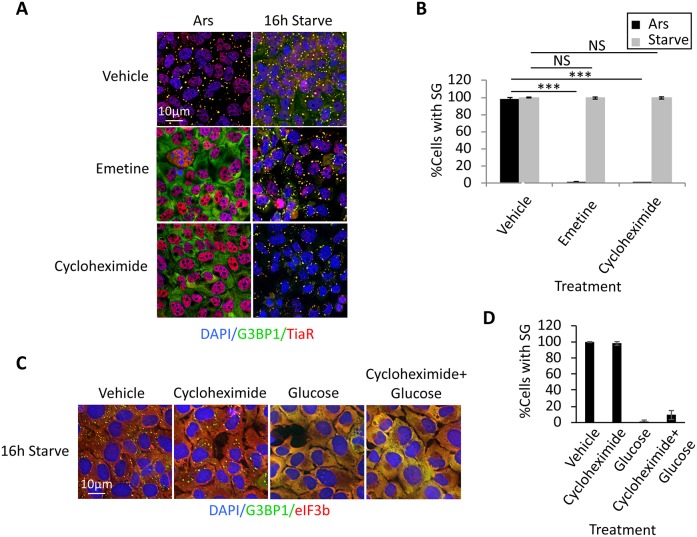
Acute stress-induced SGs are highly dynamic and reflect a shifted equilibrium away from polysomes and towards SG formation. Trapping translation complexes in polysomes using pharmacological drugs, such as cycloheximide and emetine, shifts the equilibrium and induces disassembly of dynamic SGs (Kedersha et al., 2000; Aulas et al., 2017). Because the lack of colocalization of 18S rRNA with stSGs suggested that the latter are fundamentally different than previously described SGs that assemble in response to acute stress, we were prompted to test whether stSGs would similarly disassemble in response to these drugs. As expected, arsenite-induced acute SGs could be disassembled when cells were co-treated with arsenite and either cycloheximide or emetine (Fig. 3A,B). Conversely, stSGs could not be disassembled with cycloheximide or emetine (Fig. 3A,B). To address the possibility that stSG disassembly is likely to be an ATP-dependent process, we evaluated stSG disassembly in cells co-treated with both cycloheximide and glucose. Within these conditions, stSG rapidly disassembled (Fig. 3C,D). However, this is likely to be due to increases in ATP and not trapping of complexes in polysomes, as stSGs are reduced with glucose alone and polysomes are not recovered (Figs 3D and 1G, respectively).
Fig. 3.
Chronic nutrient starvation induces static SGs that do not exchange material with translating mRNPs. (A) U2OS cells were either treated with 250 μM arsenite for 1 h or starved for 16 h prior to fixation and staining with antibodies against G3BP1 (green) and TiaR (red). During the last hour of stress, 20 μM emetine or 50 μg/ml cycloheximide were added to trap SG constituents within translating mRNPs by stalling elongating ribosomes on mRNAs to assess exchange of material with stSGs. (B) Quantification of results shown in A. The y-axis depicts the percentage of cells with stSGs for each condition. Statistical analysis was performed with a Chi-test (NS, not significant; ***P<0.001). (C) U2OS cells were starved for 16 h before addition of glucose, cycloheximide or glucose and cycloheximide, as indicated, for 1 h prior to fixation. Cells were stained as in A. (D) Quantification of the results shown in C. Results from all panels are representative of a minimum of three experimental replicates, and data are presented as mean±s.d.
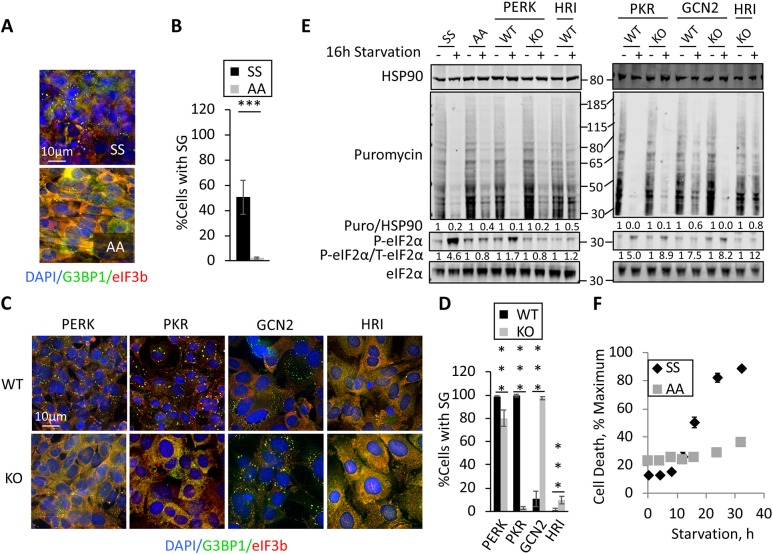
Because of the central role for eIF2α phosphorylation in SG induction for acute stressors, and the elevated eIF2α phosphorylation under these stress conditions (Fig. 1C), we sought to formally test the importance of eIF2α phosphorylation in induction of stSGs. To do this, we utilized MEFs homozygous for an allele encoding an S51A mutant of eIF2α (AA), which cannot be phosphorylated (Guan et al., 2014; Reineke et al., 2012). Similar to U2OS cells, control wild-type (SS) MEFs assembled stSGs under chronic nutrient starvation (Fig. 4A). stSGs were not abundant in the AA MEFs (Fig. 4A,B). These results support the importance of eIF2α phosphorylation in the pathway of stSG formation, leading us to ask which eIF2α kinase is responsible for eIF2α phosphorylation under chronic nutrient starvation conditions. We utilized knockout (KO) MEFs depleted of each of the previously described eIF2α kinases (HRI, PERK, PKR and GCN2). MEF genotypes were functionally validated using the distinct acute stressors that had been previously shown to selectively activate the various kinases (Fig. S2). Interestingly, PERK and PKR-null MEFs were characterized by reduced levels of stSG formation (Fig. 4C,D), although the impact of PERK was small (∼25% reduction). Depletion of GCN2 and HRI actually promoted stSG formation (Fig. 4C,D). However, given that HRI wild-type and KO MEFs did not robustly respond to the stress after 12 h, we evaluated stSG formation in these MEFs after longer periods of starvation. Interestingly, even at 24 h and 32 h starvation, we did not observe strong stSGs, although they were generally higher in the HRI KO MEFs. These results suggest that HRI is not a major contributor to eIF2α phosphorylation and stSG formation.
Fig. 4.
Role of eIF2α kinases in translation repression and stSG induction during chronic nutrient starvation. (A) Wild-type or S51A eIF2α mutant MEFs were stressed for 12 h prior to fixation and staining of stSGs with antibodies against G3BP1 (green) or eIF3b (red). (B) Quantification of the results shown in A. Statistical analysis was performed with a Chi-test (***P<0.001). (C) Wild-type or eIF2α kinase KO MEFs indicated were stressed as in A followed by fixation and staining. (D) Quantification of the results shown in C. Statistical analysis was performed with a Chi-test (***P<0.001). (E) MEFs were starved and lysates were prepared for western blotting for the puromycin-labeled polypeptides to assess ongoing translation, or the indicated proteins. Molecular weight standards are indicated. Quantification of western blots was performed using image studio for Li-Cor to avoid saturation effects, and ratios of total to phosphoproteins are indicated. (F) Wild-type and S51A eIF2α mutant MEFs were starved in the presence of ethidium homodimer-1, as described above. Ethidium fluorescence was normalized to 0 h and plotted against time. Results from all panels are representative of a minimum of three experimental replicates, and data are presented as mean±s.d.
Because stSG assembly is altered by eIF2α kinase depletion, we next attempted to correlate levels of translation via puromycin pulse experiments and eIF2α phosphorylation with stSGs in MEFs. Although translation is still repressed in the AA MEFs during nutrient starvation, we were surprised to find that it is slightly elevated during stress as compared with the SS control MEFs (∼35% for AA vs 25% for SS, Fig. 4E). This result indicates that phosphorylation of eIF2α is crucial for formation of SG precursor complexes, and that other translation inhibition activities, such as inhibition of eIF4f activity by mTORC1, are less important under these conditions. In the eIF2α kinase KO MEFs, we observed eIF2α phosphorylation under chronic nutrient starvation in all cases except PERK KO MEFs (Fig. 4E). We were surprised to see that eIF2α is phosphorylated in the PKR KO MEFs, yet stSGs are not formed (Fig. 4E). This suggests that PKR can act on SG components or assembly pathways independent of eIF2α phosphorylation.
We thus identified three factors important for SG condensation in MEFs under chronic nutrient starvation: eIF2α phosphorylation, PERK and PKR. Because multiple eIF2α kinases are activated under chronic nutrient starvation, we utilized the SS and AA MEFs to investigate the contribution of these factors to cellular survival during chronic nutrient starvation. We performed a kinetic experiment in which SS and AA MEFs were subjected to nutrient starvation and cell death was monitored with ethidium homodimer staining of nuclei. The AA MEFs were remarkably less sensitive to cell death from nutrient starvation as compared with the wild-type SS counterparts (Fig. 4F). We saw partial protection of both PERK- and PKR-null MEFs during chronic nutrient starvation, similar to our observations in the AA MEFs (Fig. S4), which mimicked the effect on stSGs (Fig. 4D).
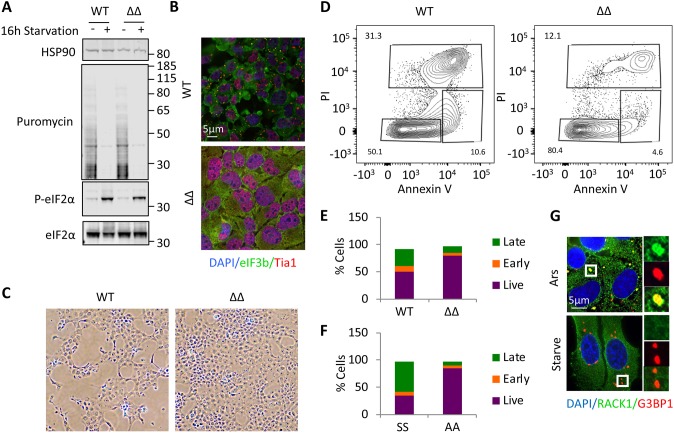
Previous work demonstrated that SGs are pro-survival through repression of MAPK signaling, or recruitment of ROCK1 (Arimoto et al., 2008; Tsai and Wei, 2010). Our findings suggested the intriguing possibility that, in contrast to pro-survival acute stress-induced SGs, some conditions induce pro-death SGs. However, our experiments to this point had not yet excluded the contribution of ongoing translation and eIF2α phosphorylation as critical factors underlying the process of cell death. Therefore, we attempted to test the importance of stSGs directly using U2OS cells, in which the SG nucleating family members G3BP1 and G3BP2 had been inactivated using CRISPR/Cas9 (Kedersha et al., 2016). We found that ΔΔ cells induce similar levels of eIF2α phosphorylation and translation repression during chronic nutrient starvation (Fig. 5A). ΔΔ cells are incapable of SG formation under chronic nutrient starvation, indicating that G3BP1 and G3BP2 are key proteins for both acute and chronic SG formation (Fig. 5B). However, we were surprised to find that the morphology of ΔΔ cell monolayers appeared considerably better under the chronic stress than in the wild-type U2OS control cells (Fig. 5C). As such, we evaluated cell death using annexin V/propidium iodide staining via flow cytometry, and found a significant reduction in dead and dying cells when the ΔΔ cells were compared with wild type (Fig. 5D,E). When the same analysis was performed on SS and AA MEFs, we saw that AA MEFs, which could not efficiently form SGs, had improved survival as well (Fig. 5F). Both U2OS and MEF cells appeared healthy and had low levels of dead cells under fed conditions (Fig. S5). Because RACK1 colocalization with SG has been linked with the pro-survival function of acute SG, we examined the localization of RACK1 in stSGs. Using arsenite-induced SGs as a control, we saw robust colocalization of RACK1 with acute stress SGs. In contrast, RACK1 did not colocalize with stSGs, suggesting that this pathway, at least in part, is altered under chronic nutrient starvation and might be responsible for the pro-death function of stSGs (Fig. 5G).
Fig. 5.
Chronic nutrient starvation induces pro-death stSG. (A) Wild-type and ΔΔ U2OS cells were starved for 16 h, followed by western blotting for puromycin or the indicated proteins. Molecular weight standards are indicated. (B) Cells treated as in A were stained for stSGs using eIF3b (green) and Tia1 (red) after 16 h starvation. (C) Phase-contrast images taken with a 10× objective, showing the appearance of the respective monolayers for wild-type and ΔΔ U2OS cells. (D) Flow cytometry was performed on fed and starved U2OS cells (Fig. S4) stained with annexin V and propidium iodide to monitor the proportion of cells undergoing cell death. (E) Graphical illustration of flow cytometry results in U2OS cells. The y-axis is the percentage of cells in each category, with the total equaling 100%. (F) Wild-type and S51A eIF2α mutant MEFs were subjected to the same analysis as in D and are represented as in E. (G) U2OS cells were stressed with arsenite or starved for 16 h, as indicated, before fixation and staining for RACK1 (green) and G3BP1 (red). Results from all panels are representative of a minimum of three experimental replicates.
DISCUSSION
We have characterized and delineated the importance of SGs during chronic nutrient starvation for the first time. Nutrient starvation is a stress encountered by many organisms and cells in various disease states, and upregulation of cell death under these conditions is crucial for limiting the deleterious effects of stress conditions. Cells that encounter chronic nutrient starvation are subjected to reactive oxygen species (Marambio et al., 2010; Filomeni et al., 2015), which can result in DNA mutations, and activation of epigenetic regulators, such as the histone deacetylase SIRT1, that can result in epigenetic changes promoting cancer progression (Woo et al., 2013). Although key pathways that regulate protein synthesis are well studied, their interplay under chronic stress conditions of multiple nutrients analogous to that experienced in disease is not well understood. Under the nutrient starvation conditions described here, mTORC1 activity is dramatically downregulated; however, stSG condensation is not visible until eIF2α is phosphorylated. Consistent with these kinetics, AA MEFs show strongly delayed stSG formation, despite robust translation repression. Protein synthesis is strongly repressed under all conditions examined here, with loss of polysomes and incorporation of puromycin into nascent polypeptides. However, it is notable that protein synthesis is slightly elevated in stressed AA MEFs compared with wild-type SS control MEFs, as well as in PERK and PKR-null MEFs. These results indicate that although mTORC1 signaling kinetics do not appear to be important in stSG assembly under these conditions, mTORC1 signaling is at least partly responsible for translation repression. Furthermore, the elevated level of protein synthesis in the AA MEFs during starvation suggests that a subset of mRNAs continue to be translated in the presence of inhibited cap-dependent translation.
SG induction is dependent on eIF2α phosphorylation. This observation leads to the critical question of which eIF2α kinase(s) are activated under chronic nutrient starvation conditions. PERK activation appears to be very important for eIF2α phosphorylation under these conditions, yet only a small (∼25%) effect on stSG condensation is observed. PKR-null MEFs still showed robust eIF2α phosphorylation during nutrient starvation, but had completely impaired stSG formation, similar to the effect in the AA MEFs. This suggests that PKR is important for SG condensation, independent of its activity on eIF2α. We previously showed that SGs activate PKR in a pathway involving Caprin1, which could be important in maintaining phase separation of SG components under these conditions (Reineke et al., 2012, 2015). These results indicate that PERK and PKR are important, through distinct pathways, for SG condensation during chronic starvation. PERK activation could suggest activation of an integrated stress response resulting from an unfolded protein response under these conditions, which will be the focus of future work. Interestingly, the integrated stress response downstream of the unfolded protein response is often associated with improved survival (Guan et al., 2017), but in this case it would suggest that the integrated stress response downstream of PERK activation is associated with elevated cell death, similar to systems in which sustained activation of the unfolded protein response is detrimental (Prola et al., 2017).
GCN2 is not activated under the nutrient starvation conditions used herein. This is not entirely surprising as the key amino acids that activate GCN2 are in sufficient abundance (Deng et al., 2002; Nikonorova et al., 2018). We cannot rule out HRI activation, as chronic nutrient starvation does induce oxidative stress, and oxidative stress was previously reported to induce SGs through HRI activation (McEwen, 2005). Although wild-type and HRI KO MEFs were intrinsically resistant to stSG, quantification did reveal enhanced stSG formation in the HRI KO MEFs, consistent with elevated eIF2α phosphorylation and previous work showing compensation between various eIF2α kinases (Koromilas, 2015). However, levels of stSGs never reached the levels observed in the other MEF cell lines, even at late 24 h and 32 h time points of nutrient starvation, possibly due to the background of the mice from which these MEFs were derived (Fig. S3, Table S1). We do not believe that HRI is a major regulator of eIF2α phosphorylation in the context of chronic nutrient starvation, because the HRI-null MEFs had robust eIF2α phosphorylation (∼12-fold, Fig. 4E) under these conditions. However, strong HRI expression patterns are largely restricted to erythrocytes and macrophages (Chen, 2014), suggesting that any large role for HRI in these pathways would be highly context dependent.
We show that stSGs form under starvation conditions depleted of glucose, glutamine, pyruvate and serum. stSGs are unique in that they do not contain 40S ribosomes, they are not disassembled with translation elongation inhibitors and they promote cell death. Previous work studying induction of SGs during selenite and hydrogen peroxide exposure has described the formation of noncanonical SGs that lack eIF3a or eIF3b and eIF4e, respectively (Fujimura et al., 2012; Emara et al., 2012). Although cell death under hydrogen peroxide treatment was not examined (Emara et al., 2012), selenite-induced SGs appear to function in a pro-death capacity (Fujimura et al., 2012). However, Fujimura and colleagues did not have the reagents to completely impair SG assembly to show that the SGs were indeed promoting cell death, and that cell death was not related to cap-dependent translation of a small pro-death subset of mRNAs. Therefore, our results under chronic nutrient starvation are the first example of stSGs that promote cell death. Given that chronic nutrient starvation induces stSGs containing eIF3b, it is not expected that eIF3 localization is a major contributor to translation of pro-death mRNAs under these conditions. However, it is important to note that several reports delineate specific eIF3 subcomplexes, and we did not investigate the colocalization of other eIF3 subunits in stSGs (Shah et al., 2016; Sha et al., 2009; Phan et al., 2001).
Although ATP levels are expected to influence SG assembly, previous reports show conflicting effects. Inhibition of mitochondrial respiration by treating cells with clotrimazole or carbonyl cyanide-4-phenylhydrazone (FCCP) has been shown to promote SG assembly, whereas other work showed that inhibition of ATP production by addition of the nonhydrolyzable 2-deoxyglucose and carbonyl cyanide m-chlorophenyl hydrazone (CCCP) inhibited the assembly of acute oxidative SGs (Jain et al., 2016; Kedersha et al., 2002). Our results support the notion that ATP depletion induces SG formation, suggesting that SG assembly can occur under low ATP concentrations. We show that SGs start appearing at 8 h of nutrient starvation, when ATP levels are ∼20% of those of fed control cells. By 16 h, when cells are beginning to die, ATP levels are a mere 1% of control cells. Therefore, the decreased ability of stSG-resident messenger ribonuclear proteins (mRNPs) to exchange with cytoplasmic pools could be a result of impairing the machinery responsible for all or part of the movement of stSG material. Addition of glucose rescues ATP levels and disassembles stSGs, but polysomes do not recover and eIF2α is not dephosphorylated (Fig. 1). In the future, we will investigate the importance of these aspects of translational control and stSG function during chronic nutrient starvation. We will also attempt to understand the mechanisms of pro-death functionality of stSGs.
Impaired cell death correlated with enhanced protein synthesis, although not at the levels of fed controls. We anticipated that enhanced translation during nutrient starvation would be detrimental to survival because cells are ATP depleted, and translation is a major consumer of cellular ATP (Stouthamer, 1973; Pontes et al., 2015). Strikingly, we found that ongoing translation and the ability to prevent SG formation improved cellular survival during chronic nutrient starvation stress using the AA mutant MEFs (Fig. 4A,E). We did not observe enhanced protein synthesis in ΔΔ U2OS cells during chronic nutrient starvation, similar to findings during acute stress (Kedersha et al., 2016). This suggests that the observed elevation of protein synthesis during chronic stress is not a function of the absence of SGs. However, this possibility cannot be excluded and will be the subject of continued work.
MATERIALS AND METHODS
Cell culture and treatments
All cells were cultured under standard conditions (5% CO2, 37°C) in Dulbecco's modified eagle medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) supplemented with 15 mM HEPES, pH 7.4. Cells were regularly tested for contamination using epifluorescence microscopy and PCR-based methods. Nutrient starvation was performed in DMEM minus glucose, FBS, glutamine and pyruvate for the indicated times. Nutrient starvation followed two washes in starvation medium. All cell lines used in this study were previously published (Kedersha et al., 2016; Reineke et al., 2012). Sodium arsenite was used at a concentration of 250 μM for 1 h. For glucose recovery experiments, glucose was added to a final concentration of 22 mM.
Immunoblotting
Immunoblots were performed on lysates using NuPAGE MOPS gels (Thermo Fisher Scientific) after protein quantification with BCA assays (Thermo Fisher Scientific). Blocking was performed in TBST with 5% bovine serum albumin (BSA). Immunoblots were conducted using the following antibodies: anti-HSP90 (1:4000, BD Biosciences), anti-puromycin (1:2000, Kerafast, 3RH11), anti-S51 P-eIF2α (1:2000, Cell Signaling Technology, 119A11), anti-eIF2α (1:2000, Cell Signaling Technology, D7D3), anti-T37/46 P-4EBP1 (1:2000, Cell Signaling Technology, 236B4), anti-S65 P-4EBP1 (1:2000, Cell Signaling Technology, 174A9) and anti-4EBP1 (1:2000, Cell Signaling Technology, 53H11). Secondary antibodies are conjugated with Dylight dyes (Cell Signaling Technology) and detected using the Li-Cor Odyssey imaging system.
Immunofluorescence microscopy
Cells were seeded onto poly-D-Lysine-coated glass coverslips the day before starvation. Starvation proceeded in accordance with the above description, and was followed by fixation in PBS with 4% formaldehyde for 30 min. Cells were permeabilized in PBS with 0.5% Triton X-100 followed by blocking with 5% BSA in PHEM buffer (Reineke et al., 2017). Cells were stained with antibodies against G3BP1 [Reineke et al. (2012) or Santa Cruz Biotechnology, 1:500, H10], eIF3b (1:500, Santa Cruz Biotechnology, N20), HuR (1:500, Santa Cruz Biotechnology, 3A2), Tia1 (1:200, Santa Cruz Biotechnology, C20), RACK1 (1:200, BD Biosciences, 20/RACK1) or rps6 (1:200, Cell Signaling Technology, 5G10), and mounted with medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Imaging was performed as previously described (Reineke et al., 2017). Secondary antibodies were all raised in donkey and labeled with Alexa Fluor 488 or Alexa Fluor dyes (1:1000, Thermo Fisher Scientific).
Polysome profiling
Polysome profiling was performed essentially as described previously (Pereboom et al., 9999). Briefly, cells were treated with cycloheximide at 100 µg/ml for 10 min at 37°C. Cells were then harvested by trypsinization, washed in ice-cold PBS and lysed by dounce homogenization in lysis buffer. Lysates were loaded onto stepwise sucrose gradients (17-50% sucrose) and fractionated as previously described (Nguyen et al., 2018). The areas of monosome and polysome regions were calculated using Prism software (GraphPad). For quantification, polysomes were defined as the region after the disome peak, under the rationale that this particular peak was concealed by the large size of the monosome during nutrient starvation or arsenite treatment.
In situ hybridization
Cells were seeded as above and starved for the indicated time points. Cells were fixed in PBS with 4% formaldehyde for 15 min, immediately followed by fixation in ice-cold methanol for 15 min. Cells were then processed as previously described (Aulas et al., 2017) using oligo(dT)40, 18S or 28S cy5-conjugated probes (Sigma-Aldrich). rRNA probe sequences used were as follows: 18S, 5′-TTGAGACAAGCATATGCTACTGGC-3′; 28S, 5′-TAGGTTGAGATCGTTTCGGCCCCAAGACCTCTAATCATTCGCTTTACCGGATAAAACTGCGTGG-3′. After RNA FISH, SGs were stained using standard immunofluorescence procedures prior to mounting coverslips.
ATP analysis
ATP levels were detected with the luciferase-based Celltiter 2.0 reagent (Promega) in accordance with the manufacturer’s protocol. Briefly, cells were seeded into 96-well plates and either fed or starved for the indicated time points. The Celltiter reagent was mixed with cell culture medium at a ratio of 1:1, incubated for 10 min and measured on a Tecan m200 plate reader. After starvation, the cell monolayer in a parallel set of wells was stained with 0.5% Crystal Violet as previously described (Feoktistova et al., 2016) to normalize luciferase values. Absorbance of Crystal Violet was monitored at 590 nm.
Cell death analysis
Cell death was monitored on a Tecan m200 plate reader equipped for fluorescence measurements. Cells were seeded into a 96-well black-walled tissue culture-treated plate and grown overnight. Cells were then washed and starvation medium with 4 μM ethidium homodimer-1 (Thermo Fisher Scientific) was added prior to measurement of fluorescence with 530/645 emission/excitation wavelengths. Measurements were then collected at the indicated time points. After 32 h, Triton X-100 was added to each well to a final concentration of 0.5%, and incubated for 15 min at 37°C prior to reading ethidium homodimer-1 fluorescence, to determine fluorescence when all cells are dead for normalization purposes.
Flow cytometry
Cells were starved as described above followed by harvesting the medium and trypsinization of the cells. Trypsinized cells were diluted in ice-cold DMEM with 10% FBS, and combined with starvation medium. Cells were then washed twice in PBS followed by resuspending cells at a concentration of 107/ml in annexin binding buffer containing 10 mM HEPES, pH 7.4, 140 mM NaCl and 25 mM CaCl2. Annexin V- fluorescein isothiocyanate (FITC) (BD Biosciences) and propidium iodide (Roche) were then added and incubated for 15 min on ice, prior to the dilution (five-fold) of cells in annexin binding buffer and filtration through mesh sieves. Single-color and unstained controls were conducted for each experiment, with the single-color annexin V-FITC control being treated for 15 min with 0.15% bleach on ice prior to staining. Flow cytometry was performed on an LSRII instrument (BD Biosciences) at the Cytometry and Cell Sorting Core Facility at Baylor College of Medicine.
Colocalization and SG quantification, and statistics
Colocalization analysis was performed using a custom-made MATLAB (MathWorks) script to measure the Manders’ colocalization coefficients (MCC) (Manders et al., 1993). Briefly, nuclei were segmented in the DAPI channel using a watershed algorithm. Cell boundaries were then approximated by dilating the nuclear mask. Within each cell, G3BP1 (SG) and 18S rRNA signal were segmented by intensity thresholding using the Otsu method. MCCs between each channel [G3BP1 (green, G), and 18S rRNA (red, R)] were then calculated as follows:
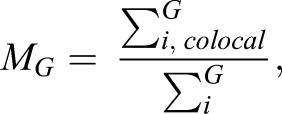 |
where Gi, colocal=Gi if Ri>0 and Gi, colocal=0 if Ri=0; and
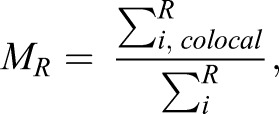 |
where Ri, colocal=Ri if Gi>0 and Ri, colocal=0 if Gi=0.
Only the MG is shown for simplicity, and because the 18S signal was diffuse, resulting in only a small fraction colocalizing with SG even under arsenite-treated conditions.
For quantification of the percentage of cells with stSG, ImageJ (https://imagej.nih.gov/ij/) was used. For this analysis, cell nuclei were counted after thresholding with the ‘analyze particles’ function. Subsequently, G3BP1 signal was detected with the ‘detect local maxima’ function as previously described (Reineke et al., 2017). This was performed in at least four fields per condition, representing over 200 cells. To determine the percentage of cells with stSGs, cells devoid of stSGs were manually counted in each field and tallied against the total number of cells. A Chi-test was performed for percentage of cells with stSGs, and a Student's t-test was performed on Manders’ correlation data. Each experiment was repeated at least three times.
Supplementary Material
Acknowledgements
We thank Drs Nancy Kedersha and Maria Hatzoglou for generous contributions of cell lines.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.C.R.; Methodology: J.D.; Formal analysis: L.C.R., J.D.; Investigation: S.A.C.; Writing - original draft: L.C.R.; Writing - review & editing: L.C.R., S.A.C., J.D., J.R.N.; Supervision: L.C.R., J.R.N.; Funding acquisition: J.R.N.
Funding
This work was supported by the National Institutes of Health [CA185769 and CA190467 to J.R.N.; CA125123, RR024574, NIDDK-56338-13/15], the American Cancer Society [RSG-15-088-01RMC to J.R.N.], the Cancer Prevention and Research Institute of Texas [RP150578] and the John S. Dunn Foundation. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.220244.supplemental
References
- Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H. and Takekawa M. (2008). Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324-1332. 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- Aulas A., Fay M. M., Lyons S. M., Achorn C. A., Kedersha N., Anderson P. and Ivanov P. (2017). Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130, 927-937. 10.1242/jcs.199240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-J. (2014). Translational control by heme-regulated eIF2α kinase during erythropoiesis. Curr. Opin Hematol. 21, 172-178. 10.1097/MOH.0000000000000030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P. and Liu J. O. (2006). Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281, 32870-32878. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- Deng J., Harding H. P., Raught B., Gingras A.-C., Berlanga J. J., Scheuner D., Kaufman R. J., Ron D. and Sonenberg N. (2002). Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 12, 1279-1286. 10.1016/S0960-9822(02)01037-0 [DOI] [PubMed] [Google Scholar]
- Drake K. J., Sidorov V. Y., Mcguinness O. P., Wasserman D. H. and Wikswo J. P. (2012). Amino acids as metabolic substrates during cardiac ischemia. Exp. Biol. Med. 237, 1369-1378. 10.1258/ebm.2012.012025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M. M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P. and Anderson P. (2012). Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763-769. 10.1016/j.bbrc.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny N. G., Kedersha N. L. and Silver P. A. (2009). Metazoan stress granule assembly is mediated by P-eIF2 -dependent and -independent mechanisms. RNA 15, 1814-1821. 10.1261/rna.1684009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M., Geserick P. and Leverkus M. (2016). Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, pdb.prot087379 10.1101/pdb.prot087379 [DOI] [PubMed] [Google Scholar]
- Filomeni G., De Zio D. and Cecconi F. (2015). Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 22, 377-388. 10.1038/cdd.2014.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K., Sasaki A. T. and Anderson P. (2012). Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40, 8099-8110. 10.1093/nar/gks566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C. and Esteban M. (2006). Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70, 1032-1060. 10.1128/MMBR.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B.-J., Krokowski D., Majumder M., Schmotzer C. L., Kimball S. R., Merrick W. C., Koromilas A. E. and Hatzoglou M. (2014). Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 289, 12593-12611. 10.1074/jbc.M113.543215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B.-J., Van Hoef V., Jobava R., Elroy-Stein O., Valasek L. S., Cargnello M., Gao X.-H., Krokowski D., Merrick W. C., Kimball S. R. et al. (2017). A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885-900.e6. 10.1016/j.molcel.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Scheuner D., Chen J.-J., Kaufman R. J. and Ron D. (2009). Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proc. Natl Acad. Sci. USA 106, 1832-1837. 10.1073/pnas.0809632106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Chevet E. and Harding H. P. (2013). Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 12, 703-719. 10.1038/nrd3976 [DOI] [PubMed] [Google Scholar]
- Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A. and Parker R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487-498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N. and Anderson P. (2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61-81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E. and Anderson P. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257-1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J. and Wickens M. P. (2002). Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195-210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P. and Anderson P. (2013). Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494-506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M. D., Achorn C. A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., Mcinerney G. M., Ivanov P. et al. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845-860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. J., Back S. H., Kim V., Ryu I. and Jang S. K. (2005). Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 25, 2450-2462. 10.1128/MCB.25.6.2450-2462.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas A. E. (2015). Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim. Biophys. Acta 1849, 871-880. 10.1016/j.bbagrm.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Manders E. M. M., Verbeek F. J. and Aten J. A. (1993). Measurement of co-localization of objects in dual colour confocal images. J. Microscopy. 169, 375-382. 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- Marambio P., Toro B., Sanhueza C., Troncoso R., Parra V., Verdejo H., García L., Quiroga C., Munafo D., Díaz-Elizondo J. et al. (2010). Glucose deprivation causes oxidative stress and stimulates aggresome formation and autophagy in cultured cardiac myocytes. Biochim. Biophys. Acta 1802, 509-518. 10.1016/j.bbadis.2010.02.002 [DOI] [PubMed] [Google Scholar]
- McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.-J., Anderson P. and Kaufman R. J. (2005). Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925-16933. 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- McQuiston A. and Diehl J. A. (2017). Recent insights into PERK-dependent signaling from the stressed endoplasmic reticulum. F1000Research 6, 1897 10.12688/f1000research.12138.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. M., Kabotyanski E. B., Dou Y., Reineke L. C., Zhang P., Zhang X. H., Malovannaya A., Jung S. Y., Mo Q., Roarty K. P. et al. (2018). FGFR1-activated translation of WNT pathway components with structured 5′ UTRs is vulnerable to inhibition of EIF4A-dependent translation initiation. Cancer Res. 78, 4229-4240. 10.1158/0008-5472.CAN-18-0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonorova I. A., Mirek E. T., Signore C. C., Goudie M. P., Wek R. C. and Anthony T. G. (2018). Time-resolved analysis of amino acid stress identifies eIF2 phosphorylation as necessary to inhibit mTORC1 activity in liver. J. Biol. Chem. 293, 5005-5015. 10.1074/jbc.RA117.001625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K., Jogi M., Yoo J. S., Narita R., Morimoto S., Takemura A., Sambhara S., Kawaguchi A., Osari S., Nagata K. et al. (2012). Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity A. Kanai, ed. PLoS ONE 7, e43031 10.1371/journal.pone.0043031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereboom T. C., Bondt A., Pallaki P., Klasson T. D., Goos Y. J., Essers P. B., Groot Koerkamp M. J. A., Gazda H. T., Holstege F. C. P., Costa L. D. et al. (9999). Translation of branched-chain aminotransferase-1 transcripts is impaired in cells haploinsufficient for ribosomal protein genes. Exp. Hematol. 42, 394-403.e4. 10.1016/j.exphem.2013.12.010 [DOI] [PubMed] [Google Scholar]
- Phan L., Schoenfeld L. W., Valásek L., Nielsen K. H. and Hinnebusch A. G. (2001). A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J. 20, 2954-2965. 10.1093/emboj/20.11.2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes M. H., Sevostyanova A. and Groisman E. A. (2015). When too much ATP is bad for protein synthesis. J. Mol. Biol. 427, 2586-2594. 10.1016/j.jmb.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prola A., Pires Da Silva J., Guilbert A., Lecru L., Piquereau J., Ribeiro M., Mateo P., Gressette M., Fortin D., Boursier C. et al. (2017). SIRT1 protects the heart from ER stress-induced cell death through eIF2α deacetylation. Cell Death Differ. 24, 343-356. 10.1038/cdd.2016.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C. and Lloyd R. E. (2013). Diversion of stress granules and P-bodies during viral infection. Virology 436, 255-267. 10.1016/j.virol.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C., Dougherty J. D., Pierre P. and Strome S. (2012). Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell 23, 3499-3510. 10.1091/mbc.e12-05-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C., Kedersha N., Langereis M. A., Van Kuppeveld F. J. M. and Lloyd R. E. (2015). Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. MBio 6, e02486 10.1128/mBio.02486-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L. C., Tsai W. C., Jain A., Kaelber J. T., Jung S. Y. and Lloyd R. E. (2017). Casein kinase 2 is linked to stress granule dynamics through phosphorylation of the stress granule nucleating protein G3BP1. Mol. Cell. Biol. 37, e00596-16 10.1128/MCB.00596-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos A. P., Mellor L. E., Pang Y. F., Kritsiligkou P., Needs H., Abou-Hamdan H., Désaubry L., Poulin G. B., Ashe M. P. and Whitmarsh A. J. (2018). The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 67, 225 10.1038/s41418-018-0076-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z., Brill L. M., Cabrera R., Kleifeld O., Scheliga J. S., Glickman M. H., Chang E. C. and Wolf D. A. (2009). The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell 36, 141-152. 10.1016/j.molcel.2009.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Su D., Scheliga J. S., Pluskal T., Boronat S., Motamedchaboki K., Campos A. R., Qi F., Hidalgo E., Yanagida M. et al. (2016). A transcript-specific eIF3 complex mediates global translational control of energy metabolism. Cell Reports 16, 1891-1902. 10.1016/j.celrep.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova T. A., Dimasi P., Kukharsky M. S., An H., Quintiero A., Schirmer C., Buã©E L., Galas M.-C. and Buchman V. L. (2017). Chronically stressed or stress-preconditioned neurons fail to maintain stress granule assembly. Cell Death Dis. 8, e2788 10.1038/cddis.2017.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer A. H. (1973). A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie van Leeuwenhoek 39, 545-565. 10.1007/BF02578899 [DOI] [PubMed] [Google Scholar]
- Tee A. R. (2018). The target of rapamycin and mechanisms of cell growth. Int. J. Mol. Sci. 19, 880 10.3390/ijms19030880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N.-P. and Wei L.-N. (2010). RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell. Signal. 22, 668-675. 10.1016/j.cellsig.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.-C., Gayatri S., Reineke L. C., Sbardella G., Bedford M. T. and Lloyd R. E. (2016). Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 10.1074/jbc.M116.739573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P., Kallinowski F. and Okunieff P. (1989). Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 49, 6449-6465. [PubMed] [Google Scholar]
- Woo S. R., Park J.-E., Kim Y. H., Ju Y.-J., Shin H.-J., Joo H.-Y., Park E.-R., Hong S. H., Park G. H. and Lee K.-H. (2013). SIRT1 suppresses activating transcription factor 4 (ATF4) expression in response to proteasome inhibition. J. Microbiol. Biotechnol. 23, 1785-1790. 10.4014/jmb.1309.09027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.