ABSTRACT
Poly(U) polymerases (PUPs) catalyze 3′ uridylation of mRNAs and small RNAs, a modification often correlating with decreased RNA stability. We have investigated the importance of three proteins with in vitro PUP activity, PUP-1/CDE-1, PUP-2 and PUP-3, in C. elegans germline development. Genetic analysis indicates that PUP-1/CDE-1 and PUP-2 are developmentally redundant under conditions of temperature stress during which they ensure germline viability and development. Multiple lines of evidence indicate that pup-1/-2 double mutant germ cells fail to maintain their identity as distinct from soma. Consistent with phenotypic data, PUP-1 and PUP-2 are expressed in embryonic germ cell precursors and throughout germline development. The developmental importance of PUP activity is presumably in regulating gene expression as both a direct and indirect consequence of modifying target RNAs. PUP-3 is significantly overexpressed in the pup-1/-2 germline, and loss of pup-3 function partially suppresses pup-1/-2 germline defects. We conclude that one major function of PUP-1/-2 is to limit PUP-3 expression. Overall, the balance of PUP-1, PUP-2 and PUP-3 activities appears to ensure proper germline development.
KEY WORDS: Poly(U) polymerase, C. elegans germline development, PUP-1, PUP-2, PUP-3, RNA stability, Transgenerational inheritance
Summary: Poly(U) polymerase activity is crucial throughout C. elegans germline development, impacting viability, maintenance of germ cell identity and formation of fertilization-competent gametes capable of supporting embryonic development.
INTRODUCTION
The germline is a unique tissue responsible for gamete production and continuation of the species. Germ cell formation in the embryo and subsequent growth and survival of the germline require patterns of gene expression distinct from somatic cells. One conundrum is that the germline is a highly specialized tissue producing unique cell types – sperm and oocytes – that, upon fusion at fertilization, produce a totipotent zygote. Gene expression studies in animals ranging from nematodes to mammals have identified repressive mechanisms acting at the transcriptional and post-transcriptional levels as essential for primordial germ cell formation and, subsequently, for germline viability and development (e.g. Cinalli et al., 2008; Lai and King, 2013; Mu et al., 2014). In Caenorhabditis elegans, certain chromatin regulators and translational repressors limit somatic gene expression in the germline and promote fertility (Mello et al., 1992; Ciosk et al., 2006; Strome and Lehmann, 2007; Gaydos et al., 2012; Updike et al., 2014). Interestingly, defects in some of these processes result in immediate sterility, whereas defects in others cause a gradual reduction in fertility over successive generations leading to eventual sterility (a ‘mortal’ germline phenotype; Smelick and Ahmed, 2005). Presumably, the Mrt phenotype results from accumulated mis-regulation of gene expression essential for maintaining fertility.
RNA stability is regulated by numerous 5′ and 3′ modifications (Kwak and Wickens, 2007; Wickens and Kwak, 2008; Norbury, 2013; Scott and Norbury, 2013). The best-studied example is addition of a 3′ poly(A) ‘tail’, a modification well documented to increase mRNA stability. In addition, mRNAs can be 3′ mono- or poly-uridylated, and these modifications generally correlate with reduced mRNA abundance (Lim et al., 2014). Interestingly, uridyl transferase activity may have distinct roles in different cellular contexts (Kim et al., 2015).
Recent studies have uncovered roles for uridylation in multiple aspects of RNA metabolism in diverse species. In Arabidopsis and mammalian cells, uridylation was first noticed at the 3′ ends of microRNA (miRNA)-directed cleavage products (Shen and Goodman, 2004). A sequence of one to nine uridine nucleotides is added at the 3′ ends of cleaved mRNAs downstream of the corresponding miRNA cleavage site, suggesting one role of uridylation is to enhance decay of mRNA cleavage products. Uridylation has been detected on human replication-dependent histone mRNAs specifically at the end of S phase and when DNA synthesis is inhibited; uridylation presumably facilitates a rapid decrease in histone synthesis in response to the completion of DNA synthesis (Mullen and Marzluff, 2008). Terminal uridyl transferases, TUT1, TUT3 and TUT4 (Mullen and Marzluff, 2008; Schmidt et al., 2011; Su et al., 2013), have been identified as responsible for histone mRNA uridylation. The clearest insight into mRNA uridylation has been acquired from using a uridylation-optimized deep sequencing method to compare the mRNA sequence signatures in mammalian TUT mutants (Lim et al., 2014). The results were consistent with poly(U) tails serving as a general molecular signal for mRNA decay.
Uridylation is implicated in miRNA biogenesis and stability in a variety of organisms, including human cell culture, zebrafish and C. elegans (Thornton et al., 2014; Kim et al., 2015). In C. elegans, 3′ uridylation functions in regulating the stability of LIN-28-blockaded let-7 pre-miRNA (Lehrbach et al., 2009). The let-7 ortholog in human is a candidate tumor suppressor and a regulator of stem cell differentiation; hence, uridylation may be a factor in cancer biology and stem cell biology (Lehrbach et al., 2009; Heo et al., 2012; Kim et al., 2015; Balzeau et al., 2017). 3′ uridylation may promote degradation of siRNA (van Wolfswinkel et al., 2009; Ibrahim et al., 2010). In contrast, 3′ methylation prevents uridylation and correlates with increased steady state levels of small RNAs in many species (Kamminga et al., 2010; Ren et al., 2014). The widespread occurrence of 3′ RNA uridylation in diverse species indicates its general importance as a regulatory mechanism.
Enzymes that add terminal uridine monophosphate groups are members of the DNA polymerase beta-like nucleotidyl transferase family, the domain structure of which is conserved from yeast to mammals and includes an upstream nucleotidyl transferase domain and downstream PAP-associated domain (Norbury, 2013). In C. elegans, these enzymes include: conventional poly(A) polymerases, GLD-2 and GLD-4; enzymes with demonstrated in vitro 3′ uridylation activity, PUP-1 (aka CID-1, CDE-1), PUP-2 and PUP-3 (Kwak and Wickens, 2007), and USIP-1 (Rüegger et al., 2015); and several other proteins whose nucleotidyl transferase specificity is not known (Norbury, 2013). Initially, 3′ uridyl transferases that add short oligo(U) tails were called terminal uridyl transferases (TUTases) and those that add longer poly(U) tails were called poly(U) polymerases (PUPs) (Wickens and Kwak, 2008). The terms are used interchangeably in the current literature (e.g. Norbury, 2013); here, we use the C. elegans designation, PUP.
Among C. elegans PUPs, PUP-1/CDE-1 is reported to target endogenous siRNAs that bind to CSR-1 and/or WAGO-4 Argonaute (van Wolfswinkel et al., 2009), and PUP-2 is reported to target let-7 miRNA. Although PUP-3 has validated uridyl transferase activity, targets are not known. By analogy with other systems, one or more of these proteins may also target mRNAs. USIP-1 (U six snRNA-interacting protein 1) has a distinct role in U6 snRNA accumulation (Rüegger et al., 2015). Our current understanding of PUP function is based mainly on biochemical and structural data, and the developmental functions of these proteins remain underappreciated. Given their roles in RNA regulation and the global prevalence of uridylation in most eukaryotic species, a developmental role for PUP activity seems likely. Here, we have used genetic and molecular tools to investigate the developmental function of PUP-1, PUP-2 and PUP-3. We report that C. elegans PUP-1 and PUP-2 function redundantly to ensure that the germline maintains its identity as distinct from soma, produces functional gametes and is sustained through successive generations under conditions of temperature stress. PUP-1 and PUP-2 colocalize to perinuclear foci in embryonic germ cell precursors and throughout larval development. Later, PUP-2 shifts to cytoplasmic foci. PUP-3 also promotes germline development. Intriguingly, its abundance is limited by PUP-1/PUP-2 activity and overexpression of PUP-3 contributes to the pup-1/-2 loss-of-function phenotype. We propose that germline survival, identity and development require the correct balance of PUP activity.
RESULTS
PUP-1 and PUP-2 promote germline and embryonic development
To evaluate the impact of PUP activity on germline development and the possibility of redundant developmental functions among these three enzymes, we compared the germline phenotypes of three deletion alleles, pup-1(tm1021), pup-2(tm4344) and pup-3(tm5089), with each other as well as with double and triple pup mutant combinations. Each allele causes a shift in the open reading frame and is predicted to be null for function (www.wormbase.org, Fig. S1A). For simplicity, we refer to these alleles as pup-1(0), pup-2(0) and pup-3(0) for the remainder of the paper, except where specified.
pup-1 and pup-2 single mutants exhibit germline and embryonic lethal phenotypes that primarily result from an absence of maternal gene product and are more penetrant at a culture temperature of 25°C than at 20°C (Table 1, Fig. 1A,B, Table S1). pup-1(0) and pup-2(0) F1 hermaphrodites are viable and fertile, although these M+Z− animals (where M indicates maternal genotype and Z indicates embryonic genotype) have reduced brood sizes and exhibit low-penetrance gamete defects (Table 1, Table S2, Fig. 1B). In contrast, germline development of pup-1(0) and pup-2(0) F2 (M−Z−) hermaphrodites is significantly impaired: some individuals are sterile (Fig. 1), and fertile individuals produce fewer embryos than their M+Z− parents (Table 1). We observe two other phenotypes in the F2 population: some embryos are nonviable; and the frequency of XO male progeny is elevated (a Him phenotype) (Table 1). Embryonic viability depends strictly on maternal expression of pup-1 and primarily on maternal expression of pup-2, although zygotic pup-2 expression has a minor role (Table S2). The Him phenotype of each gene is strictly maternal effect. As a consequence of the maternal-effect nature of these phenotypes, we maintain the mutations in a heterozygous state using a balancer chromosome, even at ‘permissive’ temperatures.
Table 1.
pup mutations impair germline development and embryogenesis
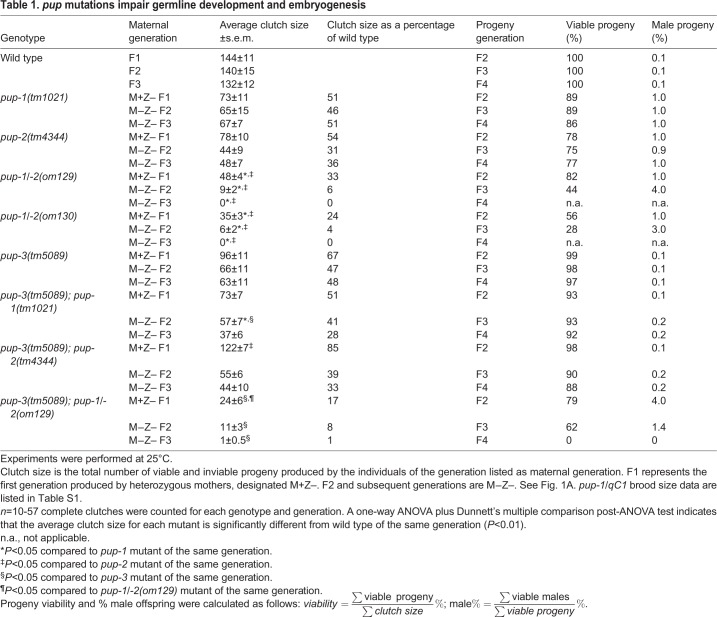
Fig. 1.
PUP activity is crucial for germline development. (A) Schematic representation of nomenclature used to designate presence/absence of maternally inherited pup gene product. (B) Histograms indicate the percentage of fertile and sterile adults produced in the F1 (M+Z–), F2 (M−Z–) and F3 (M−Z–) generations at 25°C. Adult sterile hermaphrodites were classified as containing: (i) germ cells, including sperm and/or oocytes; (ii) germ cells, but no gametes; or (iii) no germ cells. (C) Representative composite images of DAPI-stained F3 pup-1/-2(0) hermaphrodites raised at 25°C. Top panel, L3 (∼27 h post-L1) gonad containing germ cells is outlined with a dotted line. Middle panel, L4 larva (∼35 h post-L1) lacks germ cells. Bottom panel, adult without germ cells. n=36 L3 gonad arms, 50 L4 gonad arms, 114 adult gonad arms. Germ cell loss results from apoptosis during larval development; see Fig. S4. (D,E) Examples of defects observed in F2 pup-1/-2(0) hermaphrodites at 25°C. Wild type is shown for comparison. (D) Disorganized germ cells observed at the distal end of the pup-1/-2(0) gonad. Asterisk indicates distal tip; gonad arms are outlined with a dotted line. (E) Nuclei observed in the cytoplasmic core (rachis) of pup-1/-2(0) sterile adults. Scale bars: 16 µm.
After completing our analysis, another deletion allele, pup-1(gg519), was described (Spracklin et al., 2017), and its reported phenotype was more severe than that observed for pup-1(tm1021). We obtained pup-1(gg519), outcrossed and balanced the mutation, and evaluated the phenotype using our criteria described above (see supplementary Materials and Methods). In our hands, pup-1(tm1021) and pup-1(gg519) had similar phenotypes (compare Table 1 and Fig. S2). We suspect the pup-1(gg519) phenotype is milder when the mutation is maintained in a balanced state (discussed below).
We further evaluated germline development by examining germ cell morphology in pup-1(0) and pup-2(0) hermaphrodites (Fig. 1B). Consistent with reduced brood sizes, some F1 (M+Z–) animals contained oocytes that appeared to be polyploid (Fig. S3). Polyploid oocytes can arise in the oviduct as a consequence of impaired ovulation (also called endomitotic oocytes) (Iwasaki et al., 1996; McCarter et al., 1997) or in the uterus as a consequence of an impaired sperm-egg interaction, e.g. in the spe-38 mutant (Chatterjee et al., 2005). We observed polyploid oocytes in the pup-1 and pup-2 uterus but not in the oviduct; therefore, we assume the phenotype reflects a fertilization defect.
We observed three general phenotypes among sterile F2 (M−Z–) adults (Fig. 1B). (1) Some sterile adults contained germ cells and sperm and/or oocytes. When both gamete types are present, at least one type is presumably fertilization defective and, indeed, many of these animals had polyploid oocytes. (2) Other sterile adults contained germ cells, but no gametes. (3) A third class of sterile adults contained no germ cells. Together, these results indicate that PUP-1 and PUP-2 promote germline development, especially under conditions of temperature stress.
PUP-1 and PUP-2 have redundant developmental functions
To evaluate the pup-1 pup-2 double mutant phenotype, we used CRISPR-Cas9 genome editing to delete the adjacent pup-1 and pup-2 genes (Fig. S1B). We analyzed two independent deletions, pup-1/-2(om129) and pup-1/-2(om130), and observed a similar phenotype with respect to brood size, embryonic viability and production of male progeny (Table 1). We used pup-1/-2(om129) in subsequent studies reported here and for brevity refer to it as pup-1/-2(0).
The pup-1/-2(0) double mutant has the same general pattern of defects observed in pup-1 and pup-2 single mutants; however, the penetrance and severity of these defects are significantly worse in the pup-1/-2(0) double mutant, particularly at 25°C (Table 1, Fig. 1B). The data indicate a synergistic interaction between pup-1 and pup-2 that has a catastrophic impact on fertility. Strikingly, in the F3 generation at 25°C, 97% of adult pup-1/-2(0) hermaphrodites lacked germ cells altogether compared with ∼9% of F3 pup-1(tm1021) and ∼7% of F3 pup-2(0) adult hermaphrodites (Table 1, Fig. 1B,C).
An absence of germ cells might reflect failure of germ cell precursors to form in the embryo or to remain viable during larval development. To distinguish between these alternatives, we raised synchronized pup-1/-2(0) F2 animals at 25°C and DAPI stained their progeny at different developmental stages. All the late L3 larvae contained germ cells, whereas mid-late L4 larvae contained either no (60%) or very few (40%) germ cells (Fig. 1C). Consistent with these data, we observed elevated apoptosis in larval pup-1/-2(0) hermaphrodite gonads (Fig. S4). We assessed apoptosis by monitoring sheath cell engulfment of apoptotic germ cells using CED-1::GFP cell membrane protein as a marker (Zhou et al., 2001). We counted CED-1::GFP-positive cells in mid-L4 pup-1/-2(0) and wild-type control gonads. pup-1/-2(0) F1 gonads contained significantly more CED-1::GFP-positive cells than wild type, and the number increased significantly again in F2 gonads compared with F1 (Fig. S4). We conclude that germ cell precursors form in pup-1/-2(0) embryos and proliferate in early larval development, but then undergo apoptosis during later larval development.
pup-1/-2 sperm are fertilization defective
We sought to determine the importance of PUP-1/-2 activity for male germline development. We tested sperm function by mating single pup-1/-2(0) M+Z– males, raised at 25°C, to fog-1 (feminization of the germline) females and counting cross-progeny number. Typically, XX C. elegans develop as hermaphrodites and XO C. elegans develop as males. FOG-1 is required for sperm production in XX animals; XX fog-1 mutants produce only oocytes and are female (Barton and Kimble, 1990). Fifty-six percent of pup-1/-2(0) M+Z– males produced no cross-progeny and the remainder yielded on average fewer cross-progeny than wild-type males mated in parallel under the same conditions (Fig. 2A). We DAPI stained the adults in the non-productive crosses and evaluated their gametes. All of the pup-1/-2(0) males contained sperm. Ninety-three percent of the mated females contained polyploid oocytes in the uterus (Fig. 2B), a phenotype not observed in the unmated fog-1 female controls under our assay conditions. Importantly, we observed sperm in 54% of these females, indicating that male sperm had transferred during mating (Fig. 2B). In many males, sperm were not clustered in the seminal receptacle as is typical for wild type, but instead were observed more distally within the gonad (Fig. 2C,D). pup-1/-2(0) sperm nuclei were variably sized compared with wild type, especially those that did not move to the seminal receptacle (Fig. 2C,D, insets). Therefore, pup-1/-2(0) M+Z– males typically produce sperm, but many of those sperm may be fertilization defective.
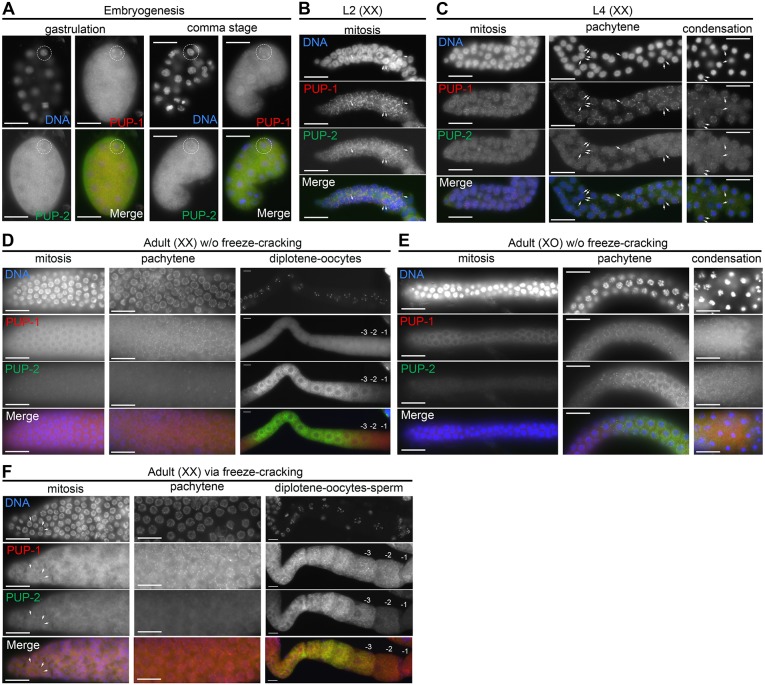
Fig. 2.
pup-1/-2(0) males exhibit fertility defects at 25°C. (A) Box-and-whisker plots of viable offspring produced by fog-1 females mated with wild-type or pup-1/-2(0) males, as indicated (n=18 experimental and 7 control crosses). Box represents the middle 50% of values; horizontal lines represent the 50th percentile (median) value; bars indicate the full range of values. (B) Representative image of gametes in a mated fog-1 female that did not produce cross-progeny. Arrowheads indicate polyploid oocyte nuclei in the uterus; arrows indicate sperm transferred in the mating. The permatheca is located to the left of the image. (C-D″) Representative DAPI-stained germlines of a (C,C′) wild-type control and (D-D″) pup-1/-2(0) male. Gonads are outlined. The pup-1/-2(0) male is one that did not sire progeny. Most sperm have not migrated to the seminal receptacle and their nuclei are variable in size (D′); those few sperm in the seminal receptacle are more uniform in size (D″). Scale bars: 16 µm.
We observed a similar germline phenotype in pup-1/-2(0) F2 (M−Z–) and F3 males raised at 25°C. The majority of F3 adult male germlines had an overall normal distal-to-proximal germ cell organization, although a minority (25%) lacked germ cells altogether. Hence, the pup-1/-2(0) germ cell loss phenotype was less penetrant in males than in hermaphrodites. Unfortunately, the 100% sterility of F3 hermaphrodites precludes our analysis of male germ cell viability in a later generation. We conclude that the XO germline can better tolerate loss of PUP activity than the XX germline.
Germ cells express somatic genes in the absence of PUP-1/-2 activity
Many pup-1/-2(0) sterile hermaphrodites have a disorganized germline where nuclei are not present in orderly rows and are sometimes found in the cytoplasmic core (rachis) (e.g. Fig. 1D,E). This phenotype is reminiscent of P granule-depleted germlines (Updike et al., 2014; Knutson et al., 2017). P granules are germline-specific ribonucleoprotein particles that assemble on the outer face of the nuclear envelope, typically spanning a nuclear pore, and associate with RNA molecules as they are exported from the nucleus (Wang and Seydoux, 2014). P granules are structurally similar to germline RNP particles described in other animal species and are thought to function in regulating RNA stability. A striking outcome of P granule depletion is inappropriate expression of some somatic genes in the germline (Updike et al., 2014; Knutson et al., 2017).
To test whether PUP-1/PUP-2 activity represses expression of somatic genes in the germline, we evaluated expression of unc-33p::gfp and unc-119::gfp in the pup-1/-2(0) germline. For these assays, we maintained the strains at 22°C and evaluated expression in the F2 generation to maximize the development of sterile animals that retained a germline (see Materials and Methods). UNC-33 is required for correct nervous system development in C. elegans and typically not expressed in the germline (Table 2, Fig. 3A) (Altun-Gultekin et al., 2001). In pup-1 and pup-2 F2 (M–Z–) single mutants, where a low percentage of animals fail to maintain a germline, we observed weak unc-33p::gfp expression in ∼9% of pup-1 (n=58) and ∼27% pup-2 (n=48) germlines (Table 2, Fig. 3B,C). In contrast, unc-33p::gfp was expressed in 53% of fertile and 100% of sterile pup-1/-2(0) germlines (Table 2, Fig. 3D-F). Quantification indicates that the intensity of GFP expression was variable among pup-1/-2(0) sterile germlines (Fig. 3G). Upregulation of unc-33p::gfp presumably occurs at the transcriptional level, as the construct does not contain the unc-33 3′UTR, and therefore we hypothesize that its expression is an indirect effect of losing PUP-1/-2 activity. In contrast to unc-33, we did not observe expression of either of two unc-119::gfp transgenes or another somatic reporter, rab-3p::rfp (see Materials and Methods). We conclude that the combined loss of PUP-1 and PUP-2 activity allows expression of some soma-specific genes in the germline, and we hypothesize that this inappropriate gene expression contributes to the developmental defects.
Table 2.
Summary of unc-33p::gfp expression in mutant germ cells
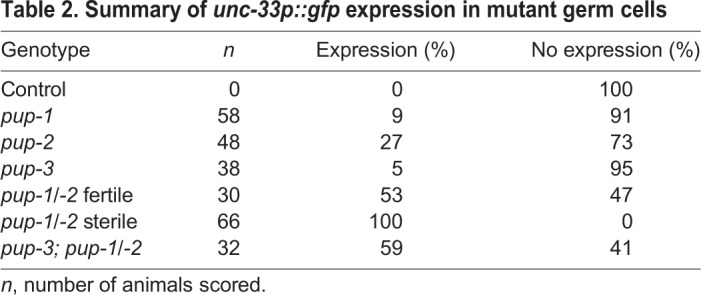
Fig. 3.
Expression of a pan-neuronal reporter in pup mutant germlines. Diagram shows the mid-late pachytene region of the germline represented in A-F,H-J. (A) unc33p::gfp expression was not detected in otherwise wild-type germ cells. Relatively low abundance unc33p::gfp expression was detected in (B) 9% of pup-1(0) F2 germlines and (C) 27% of pup-2(0) F2 germlines. For pup-1, GFP was limited to localized sets of a few germ cells; for pup-2, GFP extended more broadly. In all cases, GFP was observed in cortical cytoplasm and not in the rachis. unc-33p::gfp expression was detected in (D) 53% of fertile and (E,F) 100% of sterile pup-1/-2 F2 (M–Z–) germlines. (G) Quantification of GFP expression in control (n=5) and pup-1/-2(0) mutant (n=30) germlines. Each bar represents a single germline, and asterisk indicates the individuals pictured in E and F, respectively (see Materials and Methods). CTCF, corrected total cell fluorescence. (H) Very low cortical unc-33p::gfp expression was observed in 95% of pup-3(0) germlines; localized regions of stronger expression were observed in 5% of pup-3(0) germlines, as pictured. (I) Cortical unc-33p::gfp expression was observed in 59% of pup-3;pup-1/-2 F2 (M−Z–) germlines. (J) GFP puncta were also observed in the cytoplasmic core (rachis) of the germline in 65% of pup-3;pup-1/-2 F2 (M−Z–) gonad arms. Scale bars: 16 µm.
We further evaluated the impact of pup-1/-2(0) on germ cell fate identity by examining expression of two core P granule components: GLH-1 (germline helicase 1) and PGL-1 (P granule component). Expression of GLH-1 and PGL-1 in wild type is strictly germline specific and used as a diagnostic tool to evaluate germline versus somatic identity (Gruidl et al., 1996; Kawasaki et al., 1998). At 25°C, GLH-1::GFP and PGL-1::GFP foci were variable in size and relative intensity in the pup-1/-2(0) M+Z– germline compared with controls; regions of the M-Z– germline lacked foci altogether (Fig. 4A,B). DAPI-staining of glh-1::gfp; pup-1/-2(0) animals revealed atypical nuclear morphology in cells with reduced or unusual GLH-1::GFP signal (Fig. S5). We conclude that the combined absence of PUP-1 and PUP-2 leads to loss of P granule assembly, a phenotype consistent with inappropriate expression of the somatic marker, unc-33p::gfp.
Fig. 4.
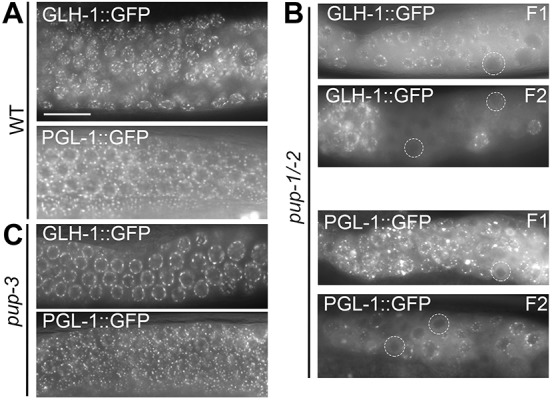
Expression of P granule proteins PGL-1::GFP and GLH-1::GFP is reduced in pup-1/-2(0) animals raised at 25°C. Expression of P granule components in (A) wild-type (n=10 PGL-1, 10 GLH-1) and (B) pup-1/-2(0) F1 (M+Z–) (n=28 PGL-1, 32 GLH-1) and F2 (M−Z–) (n=44 PGL-1, 32 GLH-1) germlines. Expression of both P granule proteins is reduced and less uniform in pup-1/-2(0) relative to controls. Circles indicate nuclei where PGL-1 or GLH-1 expression is very low/absent. F2 (M−Z–) germlines contain large patches with little/no detectable GLH-1 or PGL-1 expression. Nuclear morphology at P granule-defective regions is shown in Fig. S5. (C) PGL-1::GFP and GLH-1::GFP expression generally appear normal in pup-3(0), consistent with the >99% fertility of this strain (n=49 PGL-1, 15 GLH-1 gonad arms). Scale bars: 16 µm.
In addition to abnormal germline GLH-1::GFP and PGL-1::GFP expression in pup-1/-2(0) mutants, we observed GLH-1::GFP expression in 41% of pup-1/-2(0) F1 animals in the tail (Fig. S6), as well as very rare expression in the intestine. We did not observe somatic expression of PGL-1::GFP in pup-1/-2(0) somatic cells. Somatic expression of P granule components correlates with impaired activity of certain transcriptional regulators (Unhavaithaya et al., 2002; Wang et al., 2005) and in some cases only a subset of P granule proteins are expressed in somatic cells (Petrella et al., 2011). Our findings suggest that PUP-1 and PUP-2 activity limits GLH-1 expression in these somatic cells.
Loss of PUP-3 rescues developmental defects in pup-1/-2 double mutant
pup-3(0) mutants raised at 25°C have relatively subtle germline defects. Although >99% are fertile and P granule distribution appears normal, brood sizes are smaller than wild type and fertile individuals sometimes have developmental defects (Table 1, Fig. 1B, Fig. 4C). We observed ∼19% of fertile individuals where one gonad arm contained a disorganized germline and lacked gametes; in addition, ∼19% of fertile adults contained at least one polyploid oocyte in the uterus (n=32 animals). Very weak cortical expression of unc-33p::gfp was observed in 95% of pup-3(0);unc-33p::gfp gonad arms, and 5% gonad arms contained small domains (encompassing 2-4 nuclei) with moderate GFP signal (Fig. 3H). We conclude that PUP-3 activity promotes germline development.
To investigate the relationship of PUP-3 activity to PUP-1 and PUP-2, we evaluated pup-3(0); pup-2(0) and pup-3(0); pup-1(0) double mutants and pup-3(0); pup-1/-2(0) triple mutants. We were surprised to find that pup-3(0) suppressed the germline mortality phenotype associated with pup-1(0), pup-2(0) and pup-1/-2(0) mutations, including in pup-1/-2(0) F3 animals at 25°C (Fig. 1B). Moreover, although pup-3(0) did not completely suppress sterility, the range of germline defects was less severe in the triple mutant (Fig. 1B). Approximately 25% of pup-3(0); pup-1/-2(0) F3 hermaphrodites produced at least a few embryos (Table 1, Fig. 1B), although these embryos were not viable. Therefore, loss of PUP-3 activity compensated for the combined loss of PUP-1/PUP-2 with respect to germline viability and partially with respect to germline development.
We evaluated whether loss of PUP-3 function might decrease the inappropriate somatic expression in the pup-1/-2(0) germline. In pup-3(0);pup-1/-2(0);unc-33p::gfp hermaphrodites, GFP expression was indeed reduced compared with pup-1/-2(0);unc-33p::gfp controls. Approximately 59% of germlines contained low GFP in the cortex, similar to the GFP level observed in fertile pup-1/-2 double mutants (Fig. 3D,I). These arms also contained GFP puncta in the rachis, as did another ∼7% of germlines (65% total) (Fig. 3J). Hence, the loss of PUP-3 function reduced unc-33p::gfp expression in the pup-1/-2(0) germline.
We hypothesized that PUP-3 might play a role in limiting germline gene expression in the soma. In pup-3(0) mutants, we observed expression of PGL-1::GFP (but not GLH-1::GFP) in a subset of intestinal cells (Fig. S6B). A similar PGL-1 distribution has been observed in certain transcriptional regulatory mutants (Petrella et al., 2011). We did not observe PGL-1::GFP expression in other pup-3(0) somatic tissues besides the intestine. We conclude that PUP-3 activity plays a minor role in preventing germline gene expression in the intestine.
PUP-1 and PUP-2 are expressed throughout the developing germline
PUP-1 is reported to be expressed in germline cytoplasm and associated with embryonic P granules (van Wolfswinkel et al., 2009). To characterize the distribution of PUP-2 protein, including its localization relative to PUP-1, we epitope-tagged pup-1 and pup-2 via CRISPR-Cas9 (Fig. S1C), generating single- and double-tagged strains. Our pup-1::3xmyc 3xflag::pup-2 strain had an average clutch size of 252±10, essentially the same as wild type assayed in parallel (253±9). We concluded that the epitope tags did not appreciably interfere with gene function.
Because we observed developmental defects in pup-1/-2(0) beginning in mid-larval development and extending through adulthood, we evaluated PUP-1 and PUP-2 expression throughout this period. In embryonic germ cell precursors and larval germ cells, PUP-1 signal was primarily associated with perinuclear granules (Fig. 5A-C). At these stages, PUP-2 was diffusely distributed in the cytoplasm and weakly colocalized with PUP-1 perinuclear granules. Strikingly, the PUP-2 distribution shifted during development, becoming less prominently associated with perinuclear foci as larval development proceeded (Fig. 5B,C).
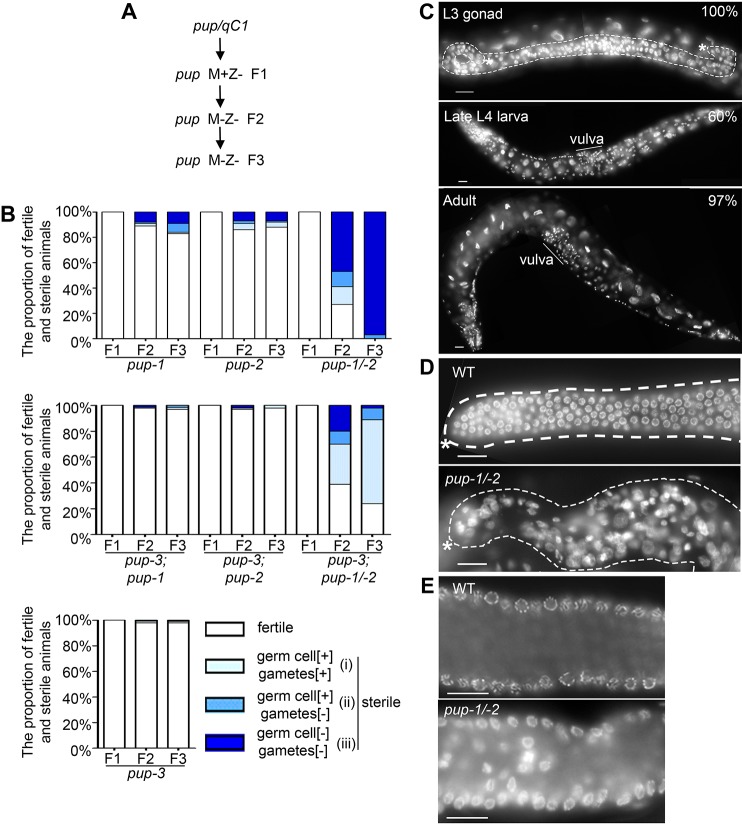
Fig. 5.
PUP-1 and PUP-2 expression in the germline. (A-C) Embryos and larvae were permeabilized by freeze-crack and co-labeled with anti-MYC and anti-FLAG to visualize the relative distribution of PUP-1::3xMYC (red) and 3xFLAG::PUP-2 (green) in germ cells. (A) Labeling of early embryos reveals PUP-1 and PUP-2 colocalized to the nuclear periphery in germ cell precursor (n=17). (B) In the L2-staged hermaphrodite (XX) gonad, PUP-1 and PUP-2 colocalize at perinuclear granules (arrow) (n=15). PUP-2 is also present in cytoplasm. (C) PUP-1 and PUP-2 expression in germ cells at mitosis, pachytene stage and condensation stage in an L4-staged hermaphrodite gonad (n=27). Perinuclear PUP-2 (arrows) is less prominent than in L2 larvae. (D,E) Adult gonads were dissected and labeled without freeze-crack (see Materials and Methods). (D,E) PUP-1 and PUP-2 expression in (D) mitotic, pachytene and diakinesis regions of the adult hermaphrodite (n=42), and in (E) mitotic, pachytene and condensation zones of the adult male germline (n=17). Perinuclear PUP-1 puncta are visible throughout the germline; perinuclear PUP-2 puncta are not visible. Instead, PUP-2 puncta are distributed throughout the cytoplasm, especially in oocytes and the male condensation zone. (F) Dissected hermaphrodite gonad subjected to freeze-crack and labeled with anti-MYC and anti-FLAG (n=39). Overall labeling is similar to D, except that perinuclear PUP-2 is visible throughout the proliferative region. -1, -2 and -3 refer to the oocyte position relative to the spermatheca. Scale bars: 16 µm.
In the adult germline, perinuclear PUP-1 foci were visible throughout the proliferative and meiotic regions (Fig. 5D-F). In maturing oocytes, these foci were distributed in the cytoplasm (Fig. 5D,F). Overall, this localization pattern resembles P granules and is similar to expression of GFP-tagged PUP-1 in a strain generated via bombardment by Zhong et al. (2010) (see Materials and Methods). In contrast, PUP-2 was barely detected in the distal germline and was relatively abundant in the proximal germline (Fig. 5D-F). In hermaphrodites, a region of elevated expression began in diplotene and peaked in maturing oocytes before decreasing sharply in late-stage oocytes at the 1, 2 and 3 positions (Fig. 5D,F). Elevated expression in males began in mid-pachytene and peaked in spermatocytes (Fig. 5E). When tissue was prepared by freeze-crack, weak perinuclear PUP-2 was observed in the adult proliferative region (Fig. 5F).
We investigated whether perinuclear PUP-1 and PUP-2 foci correspond to P granules by co-immunolabeling PUP-1::3xMYC and EKL-1 (enhancer of ksr-1 lethality), a component of the endogenous 22G RNA machinery (Gu et al., 2009; Claycomb et al., 2009) (Fig. S7A). In the course of other studies, we generated anti-EKL-1 antibody (see Materials and Methods) and determined that EKL-1 localizes to perinuclear foci and colocalizes with the core P granule component, GLH-2 (germline helicase 2) (Fig. S7B-D). Subsequent immunolabeling with anti-MYC and anti-EKL-1 detected PUP-1::3xMYC and EKL-1 colocalizing at the nuclear periphery (Fig. S7A), indicating that PUP-1 associates with P granules. By extension, we conclude that perinuclear PUP-2 is also associated with P granules. PUP-1 and PUP-2 localize to P granules during the developmental time period when activity of these proteins is essential for germline maintenance.
We compared the PUP-1 and PUP-2 distribution in developing oocytes and spermatocytes to determine whether they colocalize to cytoplasmic foci in these cells. The dramatic downregulation of PUP-2 cytoplasmic foci in 1, 2 and 3 oocytes is inconsistent with these being P granules, which are retained in oocytes and inherited by the embryo. Instead, they seem more likely to be a distinct structure. We observed only rare instances PUP-1::3xMYC and 3xFLAG::PUP-2 foci in close proximity in developing oocytes (Fig. 5D,F). Similarly, we observed only rare association of these foci in the proximal male germline (Fig. 5E). We conclude that PUP-1 and PUP-2 have largely non-overlapping distributions in maturing gametes.
We hypothesized that PUP-2 foci observed in developing oocytes might represent processing (P) body-like granules that have been described previously in the C. elegans hermaphrodite germline and implicated in mRNA stability and translation (Parker and Sheth, 2007; Voronina et al., 2011). However, immunolabeling indicated that PUP-2 did not colocalize with CGH-1 (conserved germline helicase 1), a core component of C. elegans processing body-like granules (Fig. S8A; Navarro and Blackwell, 2005; Jud et al., 2008; Noble et al., 2008). We conclude that PUP-2 foci are distinct from processing body-like granules. We also hypothesized that OMA-1/-2 activity might be required for PUP-2 repression in 1, 2 and 3 oocytes. The OMA-1/-2 translational regulators repress expression of numerous proteins in late-stage oocytes (Robertson and Lin, 2015). However, oma-1/-2(RNAi) did not prevent the downregulation of PUP-2 in late-stage oocytes (Fig. S8B), and we conclude that PUP-2 is downregulated via another mechanism.
PUP-3 expression is elevated in the absence of PUP-1/-2 activity
Although pup-3 mRNA has a low abundance in the wild-type gonad, its abundance increases substantially in P granule-depleted germlines (Updike et al., 2014; Knutson et al., 2017). Because of the observed similarities between pup-1/-2(0) and P granule-depleted germlines, we hypothesized that PUP-3 expression might be upregulated pup-1/-2(0) mutants. To test this idea, we epitope tagged the endogenous pup-3 gene (Materials and Methods, Fig. S1D). We readily detected epitope-tagged PUP-3 on protein blots, but not by immunolabeling. We therefore evaluated PUP-3 abundance in subsequent experiments by protein blot.
Comparison of 3xHA::PUP-3 abundance in pup-1/-2(+) versus pup-1/-2(0) animals raised at 25°C and 22°C revealed a significant increase in 3xHA::PUP-3 abundance in pup-1/-2(0) F2 (M–Z–) adults compared to controls (Fig. 6A,B lanes 4 and 5, Fig. 6C,D whole worms). We did not observe a significant increase in pup-1/-2(0) mutants raised at 20°C (Fig. S9A, lanes 4 and 5). This temperature sensitive increase in PUP-3 abundance is consistent with the general pattern of more severe pup-1/-2(0) defects at elevated culture temperature. In pup-1 and pup-2 single mutants, we observed an ∼15% and ∼30% increase in PUP-3 level, respectively (Fig. S9B). Although trending up, these values were not significantly different from controls. Our results are consistent with PUP-1 and PUP-2 acting redundantly to limit PUP-3 expression. We hypothesize that PUP-3 expression may gradually become more abundant in single mutant homozygotes when passaged for many generations, and this trend may explain the accumulation of severe germline defects reported for pup-1(gg519) homozygotes (Spracklin et al., 2017).
Fig. 6.
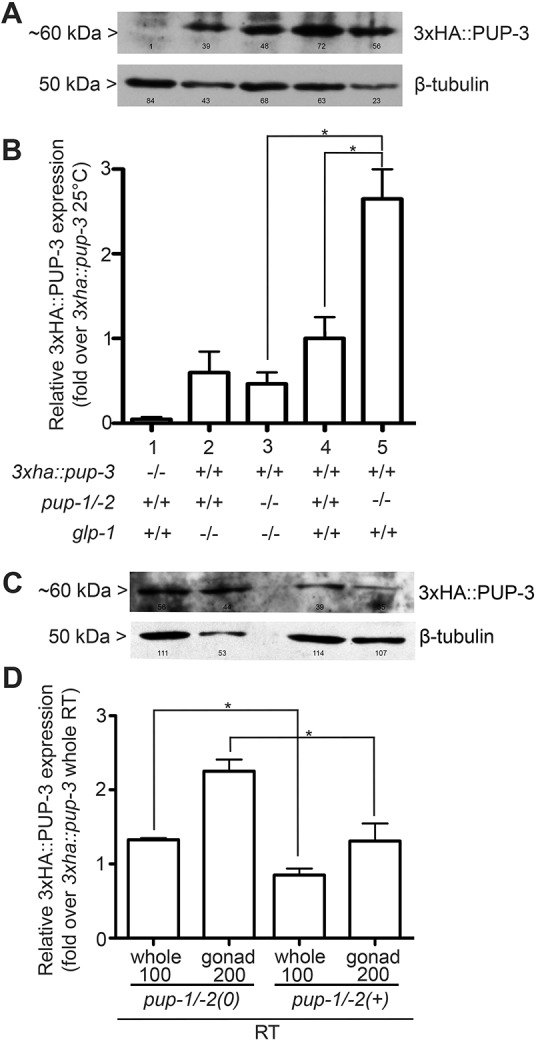
PUP-3 is upregulated in pup-1/-2(0) mutants. (A) Representative protein blot containing whole-protein extracts generated from adult hermaphrodites of the designated genotypes raised at 25°C. Blot was immunoprobed with anti-HA to visualize 3xHA::PUP-3 and re-probed with anti-β-tubulin as a loading control. Numbers indicate signal intensity minus background signal as measured by ImageJ software (see Materials and Methods). (B) Quantification of 3xHA::PUP-3 abundance in each strain. The 3xHA::PUP-3 signal in each genotype was normalized to the 3xha::pup-3; glp-1(+) pup-1/2(+) control. pup-1/-2(om129) was assayed in the F2 (M–Z–) generation. n=4 biological replicates. *P<0.05 by one-way ANOVA and Tukey's post-hoc test. (C) Representative protein blot generated with protein extracted from 200 isolated gonad arms and 100 whole animals of the designated genotypes raised at 22°C. pup-1/-2(om129) was assayed in the F2 (M–Z–) generation. (D) Quantification of 3xHA::PUP-3 abundance in gonads and intact animals. Gonad tissue is primarily germline. n=3 biological replicates. Error bars indicate ±s.e.m. *P<0.05 calculated by Student's t-test of whole-worm data and dissected gonad data, independently.
Given our phenotypic data, we hypothesized that 3xHA::PUP-3 abundance increases in the pup-1/-2(0) germline. We tested this idea in two ways. First, we assayed 3xHA::PUP-3 in a glp-1(ts) (germline proliferation defective) mutant background where the germline is very small. At 25°C, GLP-1/Notch signaling activity is severely reduced in glp-1(bn18ts) mutants; all germline stem cells enter meiosis prematurely during larval development and adults typically contain only mature sperm (Kodoyianni et al., 1992). In our experiments, L1 larvae were upshifted to 25°C and harvested as adults in order to facilitate a direct comparison of all genotypes, including the glp-1(bn18ts) strain that cannot be propagated at 25°C. PUP-3 abundance was not significantly reduced in glp-1(bn18) mutants compared with controls, suggesting PUP-3 expression is primarily somatic (Fig. 6A,B, lanes 2 and 4). Furthermore, PUP-3 abundance was not significantly elevated in glp-1(bn18) pup-1/-2(0) compared with pup-1/-2(0) mutants (Fig. 6A,B, lanes 3 and 5), indicating the increase in 3xHA::PUP-3 abundance in pup-1/-2(0) mutants is germline-dependent.
As a second test, we evaluated 3xHA::PUP-3 expression in dissected gonads compared with intact animals. It was difficult to recover sufficient material from M–Z– animals raised at 25°C because their gonads were often small and did not dissect well. Therefore, we evaluated M–Z– animals grown at 22°C where gonads are on average larger. We detected PUP-3 in dissected control and pup-1/-2(0) gonads, consistent with germline expression (Fig. 6C,D). Moreover, PUP-3 was significantly elevated in pup-1/-2(0) gonads compared with controls (Fig. 6C,D), consistent with increased PUP-3 in the germline. This increase is greater than in intact animals, and we think the explanation is that our dissected pup-1/-2(0) gonads were unavoidably biased toward larger gonad arms. We conclude that PUP-3 is elevated in the germline in the absence of PUP-1/-2 expression.
DISCUSSION
This study demonstrates the importance of PUP-1, PUP-2 and PUP-3 in C. elegans germline development. Under conditions of temperature stress, PUP-1 and PUP-2 activities together maintain germ cell identity and viability during larval development and ensure production of quality gametes and offspring viability. Presumably their direct function is to modify RNA targets, which ultimately impacts gene expression. Target RNAs may include both mRNAs and small RNAs. One outcome of PUP-1 and PUP-2 activity is to limit the expression of PUP-3 in the germline, and overexpression of PUP-3 in turn contributes to the loss of germline viability and identity (Fig. 7).
Fig. 7.
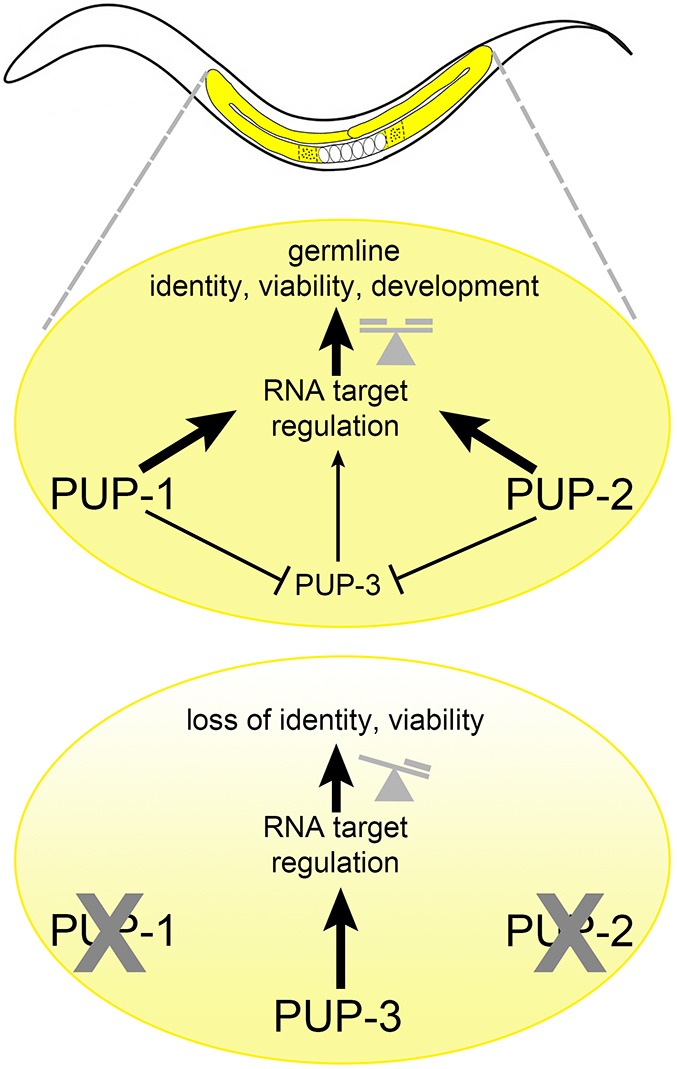
Model for regulation of germline gene expression by PUP proteins. (Top) Wild-type germline development requires a balance of PUP-1, PUP-2 and PUP-3 activities, and net PUP activity promotes the correct abundance of RNA targets to allow germline development. (Bottom) In pup-1/-2(0) mutants, PUP-3 abundance is elevated. Under conditions of temperature stress, the normal pattern of germline gene expression is compromised due to loss of PUP-1 and PUP-2, and overexpression of PUP-3. The net effect severely impairs germline development and leads to the loss of germline viability within three generations.
Germ cell identity and survival
A number of factors are known to ensure germ cell identity during larval development, as well as viability of the germline over successive generations, including certain histone modifying enzymes, e.g. the H3K4 demethylase, SPR-5 (Katz et al., 2009; Käser-Pébernard et al., 2014), the H3K4 methyltransferase, SET-2 (Xiao et al., 2011; Robert et al., 2014), and the heritable nuclear RNAi (NRDE) pathway (Buckley et al., 2012; Sakaguchi et al., 2014; Spracklin et al., 2017). PUP-1 and PUP-2 provide examples of RNA regulators that are important for germ cell viability and identity. Previous studies have linked RNA regulation to germ cell identity, although in these cases germline defects are severe and the animals are sterile. Notably, germ cell identity is severely compromised by the simultaneous loss of two broadly active translational regulators, MEX-3 and GLD-1 (Ciosk et al., 2006), and by depletion of P granules (Updike et al., 2014; Knutson et al., 2017). We hypothesize that the transgenerational germ cell loss we observe in pup-1/-2(0) mutants reflects a subtler misregulation of gene expression that compounds over successive generations until the germline senesces.
Germline development relies on the correct balance of PUP activity
Although redundant for developmental function, evidence suggests that PUP-1 and PUP-2 are not strictly redundant with respect to their target RNAs. PUP-1 and PUP-2 have previously been implicated as modifying certain siRNAs and miRNAs, respectively (Billi et al., 2014). We observed PUP-1 and PUP-2 colocalizing to P granules in the developing germline and (weakly) in the adult proliferative germline. SiRNAs – and components of the RNAi machinery generally – associate with P granules where they would be available as substrates for PUP-1/-2 (Billi et al., 2014). In adults, we observed distinct PUP-2 foci which may be alternate sites of RNA regulation. Alternative models consistent with our data are: (1) PUP-1 and PUP-2 share some targets that are essential for germline development; (2) PUP-1 and PUP-2 act in parallel to regulate distinct targets with complementary roles in germline development; or (3) a combination of the two. In any case, one consequence of PUP-1/PUP-2 activity is to limit accumulation of PUP-3 in the germline, and overexpression of PUP-3 contributes to the loss of germline viability.
In addition to small RNAs, PUP-1 and/or PUP-2 may also modify mRNAs – as do their orthologs in other species – and that regulation could occur either pre- or post-translation. Most mRNAs produced in germ cells are thought to associate with P granules upon nuclear export, where they would be available for modification. PUP-1 and PUP-2 may uridylate somatic mRNAs to limit/prevent their expression in the germline. In maturing oocytes, PUP-2 may modify target RNAs that have moved from the P granule to dispersed cytoplasmic foci. miRNAs – and some siRNAs – are found in RNA processing bodies, along with mRNAs whose translation is repressed (Parker and Sheth, 2007; Voronina et al., 2011). PUP-2 does not colocalize with the core component CGH-, and is presumably not located in RNA-processing bodies. PUP-2 may associate with and target RNAs as they are being shuttled to processing bodies and/or limit the expression of RNAs that escape PUP-1 control at the P granule.
Fertility is influenced by PUP-3 abundance. The wild-type level of PUP-3 promotes fertility, and the elevated level observed in pup-1/-2(0) mutants is detrimental to germline development. Our results are consistent with the hypothesis that PUP-3 targets are distinct from PUP-1/PUP-2, and germline development is highly sensitive to the balance of PUP-1/-2 versus PUP-3 activity. Comprehensive identification of PUP target RNAs in the future will resolve the relationship among these three enzymes with respect to germline gene expression.
MATERIALS AND METHODS
Nematode strains and culture
Strains were cultured using standard methods (Epstein and Shakes, 1995). The C. elegans wild-type Bristol variant (N2) and mutations used are as listed in Wormbase or described in the text. Mutations used were: LG (linkage group) I, fog-1(q253ts) and pup-3(tm5089); and LGIII, pup-1(tm1021), pup-1(gg519), pup-2(tm4344), pup-1/-2(om129) (this study) and pup-1/-2(om130) (this study). The following balancer was used: qC1[dpy-19(e1259ts) glp-1(q339) nIs189[myo-2::gfp]] (III). The following transgene insertions were used: wgIs428 [cid-1::TY1::EGFP::3xFLAG+unc-119(+)], otIs117 [unc-33p::gfp], edIs6 [unc-119::gfp] containing the unc-119 promoter and encoding amino acids 1-101 of UNC-119 fused to GFP, otIs45 [unc-119p::gfp], otIs355 [rab-3::NLS::tagRFP], DUP64 [glh-1::gfp] and DUP75 [pgl-1::gfp], ltIs37 [his-58::mCherry]. Endogenous genes were epitope-tagged via CRISPR-Cas9 genome editing to produce omIs7 [3xflag::pup-2], omIs8 [pup-1::3xmyc], omIs9 [3xHA::pup-3] and omIs10 [3xflag::pup-3] (see Fig. S1). Strain EL629 carries pup-1::3xmyc and 3xflag::pup-2. Strain EL655 carries all 3xha::pup-3; pup-1::3xmyc 3xflag::pup-2. Strains carrying multiple mutations were confirmed by PCR and/or sequencing, as appropriate.
CRISPR-Cas9 genome editing
Mutations were generated and epitope tags were added to endogenous genes via a CRISPR-Cas9 genome editing approach using a co-conversion strategy where a dominant co-inserted marker gene mutation, dpy-10(cn64), was used to enrich for F1 individuals containing genome-editing events (Arribere et al., 2014; Paix et al., 2014). In all injection mixtures, the Cas9 plasmid (pDD162; Dickinson et al., 2013) was present at 50 ng/μl, dpy-10(cn64) template DNA was present at 25 ng/μl, gene of interest template DNA was present at 50 ng/μl, and each sgRNA construct was present at 25 ng/μl. Progeny of injected animals were screened via DNA amplification using a method described previously (Arribere et al., 2014). Candidate CRISPR-Cas9 genome editing events were confirmed by sequencing. To generate pup-1 pup-2 double deletion alleles, we used a strategy designed to delete ∼14.7 kb of genomic DNA beginning in pup-1 exon 1 and ending in pup-2 exon 6 (Fig. S1A). We retained the 3′ region of pup-2 because we did not want to disturb the nearby downstream gene, K10D2.8. We characterized two deletions, pup-1/-2(om129) and pup-1/-2(om130), in detail and found that they have a similar phenotype (Table 1).
Brood size analysis
Brood size was assayed using standard methods as described previously (Safdar et al., 2016). Individual L4 larvae were placed onto single plates; once they became gravid adults, they were transferred to fresh plates daily until they no longer produced embryos. Embryos were counted immediately after the adult was moved. Viable progeny were counted as L3-L4 larvae. Animals were evaluated for fertility in the first day of adulthood.
RNAi
oma-1/-2 RNAi experiments were conducted using the feeding method as described previously (Timmons et al., 2001). L1 hermaphrodites were placed onto plates seeded with a mixture of oma-1 and oma-2 ‘feeding’ bacteria and allowed to develop at 25°C.
DAPI staining and characterization of germlines
For DAPI staining in intact animals, adults were fixed with −20°C methanol for 10 min, stained with 0.2 µg/ml DAPI for 10 min, mounted in VECTASHIELD medium (Vector Laboratories) and observed with a Zeiss Axioscope or Leica DM5500 microscope. Mitotic and meiotic nuclei were identified based on nuclear morphology as described previously (Francis et al., 1995; Qiao et al., 1995; Smardon et al., 2000; Shakes et al., 2009). For pan-neuronal reporters (i.e. unc-33p::gfp and unc-119::gfp), dissected gonads were fixed in 3% paraformaldehyde/1× PBS for 5 min prior to DAPI staining and visualization of the GFP signal. For rab-3p::gfp reporter, intact adults were mounted on 2% agar pads with 10% sodium azide and visualized using fluorescence microscope. Expression of unc-33p::gfp was quantified as described previously (McCloy et al., 2014; Ow et al., 2018).
Immunocytochemistry
Whole-mount immunolabeling experiments were performed as described previously (Maine et al., 2005; She et al., 2009; Guo et al., 2015), although with optimized fixation conditions. For single anti-FLAG, anti-MYC and double anti-FLAG/anti-MYC and anti-MYC/anti-EKL-1 labeling, the tissue was fixed in 3% PFA/PBS for 10 min and post-fixed in −20°C methanol for 10 min. For single anti-CGH-1 labeling and anti-FLAG/anti-CGH-1 co-labeling, the tissue was fixed in 3% PFA/PBS solution for 1 h and post-fixed in −20°C methanol for 5 min. Antibodies were used at the indicated dilution: mouse anti-FLAG (Sigma F1804), 1:200; rabbit anti-MYC (ThermoFisher Scientific PA1-981), 1:200; rabbit anti-CGH-1 (a gift from Dr David Greenstein, University of Minnesota, Minneapolis, USA), 1:5000; rabbit anti-EKL-1 (this study), 1:100; and chicken anti-GLH-2 (a gift from Dr Karen Bennett, University of Missouri School of Medicine, Columbia, USA), 1:100. Polyclonal antiserum against the C-terminal region of EKL-1 (amino acid residues 587-606, DKDEAVRAAFSQKEPIEWPN) was generated in rabbits and affinity purified (YenZym). Alexa Fluor 488-conjugated goat anti-mouse (1:200), Alexa Fluor 568-conjugated donkey anti-rabbit (1:200) and Alexa Fluor 568-conjugated donkey anti-chicken (1:1000) secondary antibodies were used.
Larvae and (when indicated) dissected gonads were permeabilized by freeze-crack, as follows. Staged larvae were washed twice with M9, transferred to a Superfrost slide (Fisher), covered with a microscope slide, inverted and gently tapped to rupture cells. Tissue was placed on a pre-chilled metal block on dry ice for ≥1 h. Slides were separated; tissue was fixed for 10 min in 3% PFA/PBS solution and post-fixed for 10 min in pre-chilled 100% methanol. Antibodies were used as indicated above. For dissected gonad freeze-crack, gonads were dissected in 30 μl egg buffer/0.1% Tween-2-/0.2 mM levamisole on a coverslip, and an equal volume of 6% PFA/PBS was added as fixative. After 10 min, most solution was removed, a Superfrost slide (Fisher) was placed on top of the tissue, and the tissue was frozen/cracked as described above. After freeze-crack, tissue was immediately placed in pre-chilled 100% methanol for 10 min and then washed three times with PBST.
Protein blot analysis
Protein extracts were generated as follows. For whole-worm extracts, synchronized adults were harvested in M9 buffer, pelleted, resuspended in 2× Laemmli buffer (Bio-Rad) with 5% (v/v) 2-mercaptoethanol, and boiled for 10 min. Tissue debris was removed by centrifugation and supernatants were transferred to new tubes. Dissected gonad extracts were prepared as described previously (Guo et al., 2015). Proteins were resolved by SDS-PAGE on a 10% separating gel and transferred to nitrocellulose membrane (Bio-Rad). Membrane was incubated overnight at 4°C with either rat anti-HA (Roche 11867423001) or mouse anti-β-tubulin (Developmental Studies Hybridoma Bank E7) antibody diluted 1:500 into 5% (w/v) powdered milk/PBS solution, incubated 2 h at room temperature with anti-rat or anti-mouse HRP-conjugated secondary antibody (Thermo Fisher) diluted 1:2000 in 5% milk solution, and visualized by Pierce SuperSignal West Femto (3xHA::PUP-3) or Pico (β-tubulin) detection substrate. Membranes were probed first for 3xHA::PUP-3 and then reprobed for β-tubulin. To reprobe, membranes were stripped in buffer containing 1.5% (w/v) glycine, 0.1% (w/v) SDS, 1% (v/v) Tween-20 (pH 2.2) at 37°C with shaking for 10 min, washed twice with PBS for 10 min, once with TBST for 5 min and blocked in 5% milk/PBS solution.
Quantification analysis was carried out using ImageJ software (NIH, version 1.51g). Background signal was subtracted from each 3xHA::PUP-3 signal, and the resulting value was normalized to the matching β-tubulin signal from that protein sample. To address the variation of overall signal intensity due to kinetics of the ECL enzymatic reaction across different biological replicates, we performed a second normalization relative to the 25°C 3xha::pup-3 strain signal on the same membrane. The normalized values for each genotype were averaged across replicates and the s.e.m. was calculated. To evaluate variation within the 3xHA::PUP-3 25°C control itself, we calculated the average and s.e.m. of the β-tubulin-normalized 3xHA::PUP-3 25°C signals.
Supplementary Material
Acknowledgements
We thank Geraldine Seydoux and Dave Pruyne for critical advice on methods; Sarah Hall and Bing Yang for comments on the manuscript; Sarah Hall, Bing Yang, Dave Pruyne, Tim Schedl, Matt Snyder, Yiqing Guo, Xia Xu, Maria Ow, Alexandra Nichitean and other members of the Maine and Hall labs for discussions during the course of this study; Dustin Updike and Scott Kennedy for providing strains; and Karen Bennett and David Greenstein for providing antibodies. Some strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs. Some strains were obtained from the National BioResource Project under the auspices of Shohei Mitani.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.M.M.; Methodology: Y.L., E.M.M.; Validation: Y.L., E.M.M.; Formal analysis: E.M.M.; Investigation: Y.L., E.M.M.; Resources: E.M.M.; Data curation: Y.L., E.M.M.; Writing - original draft: Y.L., E.M.M.; Writing - review & editing: Y.L., E.M.M.; Visualization: Y.L., E.M.M.; Supervision: E.M.M.; Project administration: E.M.M.; Funding acquisition: E.M.M.
Funding
This study was supported by a National Institutes of Health grant (R01 GM089818) and by funding from Syracuse University to E.M.M. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.165944.supplemental
References
- Altun-Gultekin Z., Andachi Y., Tsalik E. L., Pilgrim D., Kohara Y. and Hobert O. (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951-1969. [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S. and Fire A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837-846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzeau J., Menezes M. R., Cao S. and Hagan J. P. (2017). The LIN28/let-7 pathway in cancer. Front Genet 8, 31 10.3389/fgene.2017.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K. and Kimble J. (1990). fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125, 29-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi A. C., Fischer S. E. J. and Kim J. K. (2014). Endogenous RNAi pathways in C. elegans. In WormBook (ed. The C. elegans Research Community) doi/10.1895/wormbook.1.170.1, http://www.wormbook.org 10.1895/wormbook.1.170.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A. and Kennedy S. (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447-451. 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I., Richmond A., Putiri E., Shakes D. C. and Singson A. (2005). The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development 132, 2795-2808. 10.1242/dev.01868 [DOI] [PubMed] [Google Scholar]
- Cinalli R. M., Rangan P. and Lehmann R. (2008). Germ cells are forever. Cell 132, 559-562. 10.1016/j.cell.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., van Wolfswinkel J. C., Chaves D. A., Shirayama M., Mitani S., Ketting R. F. et al. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123-134. 10.1016/j.cell.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., DePalma M. and Priess J. R. (2006). Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311, 851-853. 10.1126/science.1122491 [DOI] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E. and Lin R. (2001). Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1, 187-199. 10.1016/S1534-5807(01)00026-0 [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J. and Goldstein B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein H. and Shakes D. (1995). Caenorhabditis Elegans: Modern Biological Analysis of an Organism, Vol. 48 Academic Press. [Google Scholar]
- Francis R., Maine E. and Schedl T. (1995). Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139, 607-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R. and Strome S. (2012). Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2, 1169-1177. 10.1016/j.celrep.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruidl M. E., Smith P. A., Kuznicki K. A., McCrone J. S., Kirchner J., Roussell D. L., Strome S. and Bennett K. L. (1996). Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 93, 13837-13842. 10.1073/pnas.93.24.13837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D. Jr, Vasale J., Batista P. J., Claycomb J. M., Moresco J. J., Yongman E. M., Keys J., Stoltz M. J. et al. (2009). Distinct Argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231-244. 10.1016/j.molcel.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Yang B., Li Y., Xu X. and Maine E. M. (2015). Enrichment of H3K9me2 on unsynapsed chromatin in Caenorhabditis elegans does not target de novo sites. G3 (Bethesda) 5, 1865-1878. 10.1534/g3.115.019828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I., Ha M., Lim J., Yoon M.-J., Park J.-E., Kwon S. C., Chang H. and Kim V. N. (2012). Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151, 521-532. 10.1016/j.cell.2012.09.022 [DOI] [PubMed] [Google Scholar]
- Ibrahim F., Rymarquis L. A., Kim E.-J., Becker J., Balassa E., Green P. J. and Cerutti H. (2010). Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl. Acad. Sci. USA 107, 3906-3911. 10.1073/pnas.0912632107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., McCarter J., Francis R. and Schedl T. (1996). emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J. Cell Biol. 134, 699-714. 10.1083/jcb.134.3.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jud M. C., Czerwinski M. J., Wood M. P., Young R. A., Gallo C. M., Bickel J. S., Petty E. L., Mason J. M., Little B. A., Padilla P. A. and Schisa J. A. (2008). Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev. Biol. 318, 38-51. 10.1016/j.ydbio.2008.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga L. M., Luteijn M. J., den Broeder M. J., Redl S., Kaaij L. J. T., Roovers E. F., Ladurner P., Berezikov E. and Ketting R. F. (2010). Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 29, 3688-3700. 10.1038/emboj.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käser-Pébernard S., Müller F. and Wicky C. (2014). LET-418/Mi2 and SPR-5/LSD1 cooperatively prevent somatic reprogramming of C. elegans germline stem cells. Stem Cell Rep. 2, 547-559. 10.1016/j.stemcr.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D. J., Edwards T. M., Reinke V. and Kelly W. G. (2009). A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308-320. 10.1016/j.cell.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.-H., Kirchner J., Kaminker J., Wood W. B. and Strome S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635-645. 10.1016/S0092-8674(00)81605-0 [DOI] [PubMed] [Google Scholar]
- Kim B., Ha M., Loeff L., Chang H., Simanshu D. K., Li S., Fareh M., Patel D. J., Joo C. and Kim V. N. (2015). TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 34, 1801-1815. 10.15252/embj.201590931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson A. K., Egelhofer T., Rechtsteiner A. and Strome S. (2017). Germ granules prevent accumulation of somatic transcripts in the adult Caenorhabditis elegans germline. Genetics 206, 163-178. 10.1534/genetics.116.198549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodoyianni V., Maine E. M. and Kimble J. (1992). Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Mol. Biol. Cell 3, 1199-1213. 10.1091/mbc.3.11.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J. E. and Wickens M. (2007). A family of poly(U) polymerases. RNA 13, 860-867. 10.1261/rna.514007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F. and King M. L. (2013). Repressive translational control in germ cells. Mol. Reprod. Dev. 80, 665-676. 10.1002/mrd.22161 [DOI] [PubMed] [Google Scholar]
- Lehrbach N. J., Armisen J., Lightfoot H. L., Murfitt K. J., Bugaut A., Balasubramanian S. and Miska E. A. (2009). LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16, 1016-1020. 10.1038/nsmb.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Ha M., Chang H., Kwon S. C., Simanshu D. K., Patel D. J. and Kim V. N. (2014). Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159, 1365-1376. 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., Hauth J., Ratliff T., Vought V. E., She X. and Kelly W. G. (2005). EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr. Biol. 15, 1972-1978. 10.1016/j.cub.2005.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T. and Schedl T. (1997). Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181, 121-143. 10.1006/dbio.1996.8429 [DOI] [PubMed] [Google Scholar]
- McCloy R. A., Rogers S., Caldon C. E., Lorca T., Castro A. and Burgess A. (2014). Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400-1412. 10.4161/cc.28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Draper B. W., Krause M., Weintraub H. and Priess J. R. (1992). The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell 70, 163-176. 10.1016/0092-8674(92)90542-K [DOI] [PubMed] [Google Scholar]
- Mu W., Starmer J., Fedoriw A. M., Yee D. and Magnuson T. (2014). Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 28, 2056-2069. 10.1101/gad.246124.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen T. E. and Marzluff W. F. (2008). Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22, 50-65. 10.1101/gad.1622708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro R. E. and Blackwell T. K. (2005). Requirement for P granules and meiosis for accumulation of the germline RNA helicase CGH-1. genesis 42, 172-180. 10.1002/gene.20136 [DOI] [PubMed] [Google Scholar]
- Noble S. L., Allen B. L., Goh L. K., Nordick K. and Evans T. C. (2008). Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 182, 559-572. 10.1083/jcb.200802128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury C. J. (2013). Cytoplasmic RNA: a case of the tail wagging the dog. Nat. Rev. Mol. Cell Biol. 14, 643-653. 10.1038/nrm3645 [DOI] [PubMed] [Google Scholar]
- Ow M. C., Borziak K., Nichitean A. M., Dorus S. and Hall S. E. (2018). Early experiences mediate distinct adult gene expression and reproductive programs in Caenorhabditis elegans. PLoS Genet. 14, e1007219 10.1371/journal.pgen.1007219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C.-Y. S., Calidas D., Lu T., Smith J., Schmidt H., Krause M. W. and Seydoux G. (2014). Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198, 1347-1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. and Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635-646. 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V. and Strome S. (2011). synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138, 1069-1079. 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Lissemore J. L., Shu P., Smardon A., Gelber M. B. and Maine E. M. (1995). Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141, 551-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G., Xie M., Zhang S., Vinovskis C., Chen X. and Yu B. (2014). Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. USA 111, 6365-6370. 10.1073/pnas.1405083111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., Mercier M. G., Bedet C., Janczarski S., Merlet J., Garvis S., Ciosk R. and Palladino F. (2014). The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans germline. Cell Rep. 9, 443-450. 10.1016/j.celrep.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Robertson S. and Lin R. (2015). The maternal-to-zygotic transition in C. elegans. Curr. Top. Dev. Biol. 113, 1-42. 10.1016/bs.ctdb.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Rüegger S., Miki T. S., Hess D. and Großhans H. (2015). The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling. Nucleic Acids Res. 43, 3344-3357. 10.1093/nar/gkv196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar K., Gu A., Xu X., Au V., Taylor J., Flibotte S., Moerman D. G. and Maine E. M. (2016). UBR-5, a conserved HECT-type E3 ubiquitin ligase, negatively regulates notch-type signaling in Caenorhabditis elegans. G3 (Bethesda) 6, 2125-2134. 10.1534/g3.116.027805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Sarkies P., Simon M., Doebley A.-L., Goldstein L. D., Hedges A., Ikegami K., Alvares S. M., Yang L., LaRocque J. R. et al. (2014). Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA 111, E4323-E4331. 10.1073/pnas.1406131111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.-J., West S. and Norbury C. J. (2011). The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17, 39-44. 10.1261/rna.2252511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. D. and Norbury C. J. (2013). RNA decay via 3′ uridylation. Biochim. Biophys. Acta 1829, 654-665. 10.1016/j.bbagrm.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Shakes D. C., Wu J.-C., Sadler P. L., LaPrade K., Moore L. L., Noritake A. and Chu D. S. (2009). Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5, e1000611 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Xu X., Fedotov A., Kelly W. G. and Maine E. M. (2009). Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 5, e1000624 10.1371/journal.pgen.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B. and Goodman H. M. (2004). Uridine addition after microRNA-directed cleavage. Science 306, 997 10.1126/science.1103521 [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke J. M., Stacey S. C., Klein M. E., Mackin N. and Maine E. M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169-178. 10.1016/S0960-9822(00)00323-7 [DOI] [PubMed] [Google Scholar]
- Smelick C. and Ahmed S. (2005). Achieving immortality in the C. elegans germline. Ageing Res. Rev. 4, 67-82. 10.1016/j.arr.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Spracklin G., Fields B., Wan G., Becker D., Wallig A., Shukla A. and Kennedy S. (2017). The RNAi inheritance machinery of Caenorhabditis elegans. Genetics 206, 1403-1416. 10.1534/genetics.116.198812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. and Lehmann R. (2007). Germ versus soma decisions: lessons from flies and worms. Science 316, 392-393. 10.1126/science.1140846 [DOI] [PubMed] [Google Scholar]
- Su W., Slepenkov S. V., Slevin M. K., Lyons S. M., Ziemniak M., Kowalska J., Darzynkiewicz E., Jemielity J., Marzluff W. F. and Rhoads R. E. (2013). mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA 19, 1-16. 10.1261/rna.034470.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J. E., Du P., Jing L., Sjekloca L., Lin S., Grossi E., Sliz P., Zon L. I. and Gregory R. I. (2014). Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 42, 11777-11791. 10.1093/nar/gku805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L. and Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y., Shin T. H., Miliaras N., Lee J., Oyama T. and Mello C. C. (2002). MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111, 991-1002. 10.1016/S0092-8674(02)01202-3 [DOI] [PubMed] [Google Scholar]
- Updike D. L., Knutson A. K., Egelhofer T. A., Campbell A. C. and Strome S. (2014). Germ-granule components prevent somatic development in the C. elegans germline. Curr. Biol. 24, 970-975. 10.1016/j.cub.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Claycomb J. M., Batista P. J., Mello C. C., Berezikov E. and Ketting R. F. (2009). CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139, 135-148. 10.1016/j.cell.2009.09.012 [DOI] [PubMed] [Google Scholar]
- Voronina E., Seydoux G., Sassone-Corsi P. and Nagamori I. (2011). RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 3, a002774 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. T. and Seydoux G. (2014). P granules. Curr. Biol. 24, R637-R638. 10.1016/j.cub.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D., Kim J. K., Gabel H. W., Kamath R. S., Mello C. C. and Ruvkun G. (2005). Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436, 593-597. 10.1038/nature04010 [DOI] [PubMed] [Google Scholar]
- Wickens M. and Kwak J. E. (2008). Molecular biology: a tail tale for U. Science 319, 1344-1345. 10.1126/science.1154946 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Bedet C., Robert V. J. P., Simonet T., Dunkelbarger S., Rakotomalala C., Soete G., Korswagen H. C., Strome S. and Palladino F. (2011). Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc. Natl. Acad. Sci. USA 108, 8305-8310. 10.1073/pnas.1019290108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Niu W., Lu Z. J., Sarov M., Murray J. I., Janette J., Raha D., Sheaffer K. L., Lam H. Y. K., Preston E. et al. (2010). Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6, e1000848 10.1371/journal.pgen.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E. and Horvitz H. R. (2001). CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43-56. 10.1016/S0092-8674(01)00190-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.