ABSTRACT
Midface dysgenesis is a feature of more than 200 genetic conditions in which upper airway anomalies frequently cause respiratory distress, but its etiology is poorly understood. Mouse models of Apert and Crouzon craniosynostosis syndromes exhibit midface dysgenesis similar to the human conditions. They carry activating mutations of Fgfr2, which is expressed in multiple craniofacial tissues during development. Magnetic resonance microscopy of three mouse models of Apert and Crouzon syndromes revealed decreased nasal passage volume in all models at birth. Histological analysis suggested overgrowth of the nasal cartilage in the two Apert syndrome mouse models. We used tissue-specific gene expression and transcriptome analysis to further dissect the structural, cellular and molecular alterations underlying midface and upper airway dysgenesis in Apert Fgfr2+/S252W mutants. Cartilage thickened progressively during embryogenesis because of increased chondrocyte proliferation in the presence of Fgf2. Oral epithelium expression of mutant Fgfr2, which resulted in a distinctive nasal septal fusion defect, and premature facial suture fusion contributed to the overall dysmorphology. Midface dysgenesis in Fgfr2-related craniosynostosis is a complex phenotype arising from the combined effects of aberrant signaling in multiple craniofacial tissues.
KEY WORDS: Apert syndrome, Crouzon syndrome, Midface dysgenesis, Fibroblast growth factor, Nasal cartilage, Suture, Mouse
Summary: We define the tissue-specific structural, cellular and molecular effects of activating Fgfr2 mutations in bone, cartilage and epithelium that combine to cause midfacial dysgenesis in mouse models of craniosynostosis.
INTRODUCTION
Midface developmental anomalies feature in more than 200 genetic conditions (Online Mendelian Inheritance in Man database, www.omim.org). These include the common conditions of nasal septum deviation and midface hypoplasia or retrusion. Physical consequences of upper airway dysmorphogenesis range from nasopharyngeal narrowing to choanal defects such as stenosis or atresia. Clinical consequences include obstructive sleep apnea and failure to thrive (Cielo et al., 2016; Tan et al., 2016). Although syndromic craniosynostosis is typically defined by calvarial suture fusion, midface dysgenesis and upper airway complications are common findings (Cunningham et al., 2007; Lesciotto et al., 2018). Obstructive sleep apnea, for example, occurs in the majority of patients with syndromic craniosynostosis (Driessen et al., 2013; Inverso et al., 2016). In severe cases, lower airway involvement can present as a cartilaginous tracheal sleeve, with high rates of morbidity and mortality (Cohen and Kreiborg, 1992; Stater et al., 2015; Wenger et al., 2017). Surgical correction of midface and upper airway disorders requires significant reconstructive procedures, including rhinoplasty and midface advancement that are performed by multidisciplinary teams of surgeons and are supported by a variety of medical specialists and social service providers (Bohm et al., 2016; Nash et al., 2015). To improve the precision of treatment and prevention modalities for midface and upper airway dysgenesis, the structural, cellular and molecular pathogenesis of these conditions requires investigation, as little is currently known.
Activating mutations of fibroblast growth factor receptors (FGFRs) are a major cause of midface abnormalities in association with syndromic craniosynostosis (Cunningham et al., 2007; Heuzé et al., 2014a; Wilkie et al., 2010). FGFR genes are expressed in a variety of craniofacial tissues during development, including bone, cartilage, brain, and skin, with distinct isoforms expressed between mesenchymal and epithelial tissues (Bansal et al., 2003; Delezoide et al., 1998; Orr-Urtreger et al., 1991, 1993; Rice et al., 2003). Various FGFR mutant mouse models of syndromic craniosynostosis, including Apert, Crouzon, Pfeiffer, Beare–Stevenson, and Muenke syndromes, have been created and reproduce key dysmorphologies of the analogous human syndromes, including coronal suture synostosis and midfacial dysgenesis (Chen et al., 2003; Eswarakumar et al., 2004; Mai et al., 2010; Twigg et al., 2009; Wang et al., 2005, 2010, 2012; Yin et al., 2008; Zhou et al., 2000). They provide model systems to examine the structural, cellular and molecular changes that cause these dysmorphologies (Holmes, 2012).
Previous studies from our group suggest that the distinct cranial dysmorphologies of syndromic craniosynostosis result from the effect of specific FGFR mutations in tissues beyond bone (Martínez-Abadías et al., 2013b; Motch Perrine et al., 2014). Here, we test this hypothesis specifically for midface and upper airway dysgenesis. To identify these changes we have used three mouse models of syndromic craniosynostosis that carry activating Fgfr2 mutations. They are two Apert syndrome mouse models, Fgfr2+/S252W (Wang et al., 2005) and Fgfr2+/P253R (Wang et al., 2010), and a mouse model that has a mutation common to both Crouzon and Pfeiffer syndromes, Fgfr2cC342Y/+ (Eswarakumar et al., 2004). The Apert syndrome Fgfr2S252W and Fgfr2P253R mutations affect both mesenchymal and epithelial receptor isoforms. The Crouzon and Pfeiffer syndrome Fgfr2cC342Y mutation (referred to herein as Crouzon) is restricted to the mesenchymal isoform. The midface dysgenesis of these models spans a range of craniofacial malformation severity (Martínez-Abadías et al., 2010, 2011, 2013a,b; Motch Perrine et al., 2014, 2017). We show that nasal cavity volume reduction and cartilage thickening are key features of the midface phenotype in Apert syndrome mouse models, with multifactorial etiologies.
RESULTS
Nasal passage volume is decreased in craniosynostosis mouse mutants
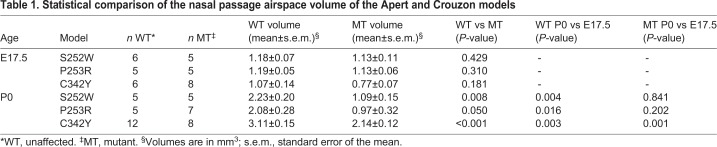
Nasal cavity airspace volumes were estimated using magnetic resonance microscopy (MRM) endocast volumes in three craniosynostosis mouse models (Fig. 1A, Fig. S1 and Table 1). In the two Apert syndrome models, the Fgfr2S252W and Fgfr2P253R alleles affect both the mesenchymal and epithelial FGFR2 isoforms, and their expression is conditional (Wang et al., 2005, 2010). Heterozygous expression of each was achieved using the EIIA-Cre driver line, which is expressed from the zygotic stage (Lakso et al., 1996). In the Crouzon syndrome model, the Fgfr2cC342Y allele affects only the mesenchymal Fgfr2 isoform and its expression is not conditional (Eswarakumar et al., 2004) (see Table S1 for a summary of features of these craniosynostosis mouse models). All three mutants had a significantly reduced nasal passage volume on the day of birth (P0), compared with unaffected littermates. There was a reduction >50% in both Apert mutants and >30% in the Crouzon Fgfr2cC342Y/+ mutants. At embryonic day (E) 17.5 there was no significant difference between mutants and unaffected littermates, indicating that the relative decrease in volume occurred between E17.5 and P0 (Table 1). Nasal passage volumes increased significantly between E17.5 and P0 for Crouzon Fgfr2cC342Y/+ mutants and unaffected mice in all three lines, but not in mutants of either Apert line (Table 1).
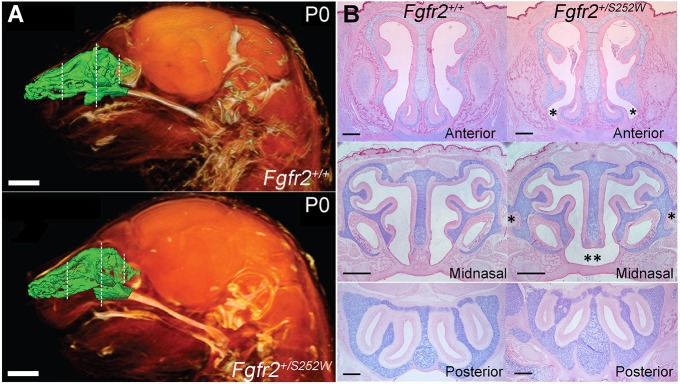
Fig. 1.
Nasal airspace and three regions of investigation in representative Fgfr2 mutant mice. (A) Lateral view of 3D reconstructions of MRM images of P0 Fgfr2+/+ and Apert Fgfr2+/S252W heads showing soft tissues (orange) and an endocast of the segmented nasal passages (green). Dashed lines indicate the three locations shown in (B) and in Movies 1 and 2. Scale bars: 1 mm. (B) Alcian Blue/Eosin staining of sections in the pars anterior (anterior), pars intermedia (midnasal) and pars posterior (posterior) regions of P0 Fgfr2+/+ and Apert Fgfr2+/S252W mice. These locations were used for quantitative analysis. Asterisks in the anterior mutant panel indicate flaring of the paraseptal region and pronounced narrowing of the adjacent nasal space. Single asterisks in the midnasal mutant panel indicate ectopic cartilage projecting from the paries nasi; double asterisks indicate lack of fusion between septum and anterior secondary palate. Scale bars: 500 µm.
Table 1.
Statistical comparison of the nasal passage airspace volume of the Apert and Crouzon models
The reduction in nasal passage volume seen in all three mouse mutants could result from occlusion or elimination of compartments within the passages, or localized or overall narrowing of the nasal passages, and may involve structural, cellular and molecular changes in surrounding tissues such as the cartilage and mucosa. To address these possibilities, we compared the nasal passage morphology of mutants with those of unaffected littermates histologically at P0 using paraffin-embedded and sectioned crania. Examination of sections throughout the anteroposterior length of the nasal passage in all mouse models revealed no gross disturbances of the turbinate morphology or occlusion of nasal spaces (Fig. 1B, Apert Fgfr2+/S252W mouse data is shown).
Of note, a structural defect of the septum was found in both Apert mutants. In mice, the nasal septum connects ventrally to the primary and secondary palates. This connection terminates at the ‘septal window’, anterior to the entrance of the nasopharyngeal passage (Kelemen, 1947, 1953). In both Apert mutants, the ventral epithelium of the septum failed to connect to the dorsal epithelium of the anterior secondary palate, so that the left and right nasal passages were not fully separated (Fig. 1B). In the Apert Fgfr2+/S252W line, 13/13 mutants lacked connection in this region, compared with 0/13 unaffected littermates. In the Apert Fgfr2+/P253R line, 4/5 mutants lacked connection in this region, compared with 0/8 unaffected littermates. Anteriorly, the septum did connect to the primary palate in both Apert mutants, although the cross-section of this region, which included the paraseptal cartilages, was broader and more flared in mutants compared with unaffected littermates (Fig. 1B). Crouzon Fgfr2cC342Y/+ mutants did not show these phenotypes, which may reflect the restriction of this mutation to the mesenchymal FGFR2 isoform (Eswarakumar et al., 2004). If connection between the cartilaginous septum and the bony secondary palate as they expand is required for complete facial outgrowth (Hall and Precious, 2013), loss of this connection in Apert mouse models could contribute to midface dysgenesis.
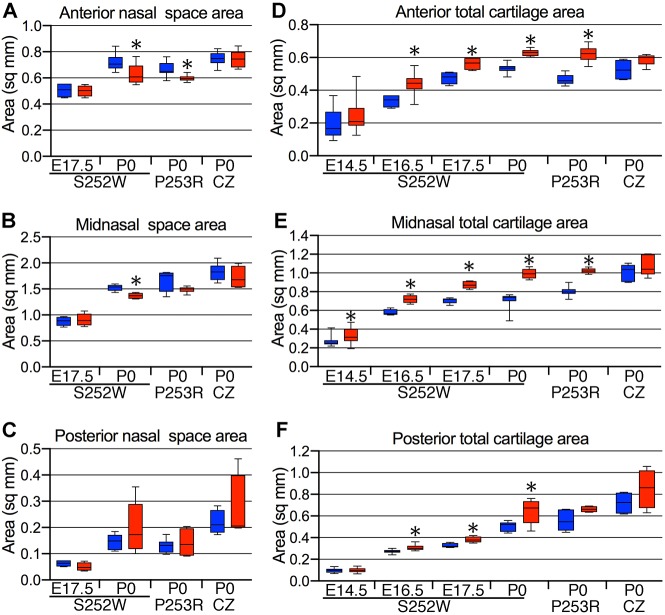
We then compared the cross-sectional areas of the nasal passages of mutants with those of unaffected littermates at P0. A representative location within each of the three embryonic divisions of the nasal capsule (pars anterior, pars intermedia and pars posterior) (De Beer, 1937; Smith and Rossie, 2008), each with a distinct cross-sectional profile of nasal passage space and capsular cartilage, was chosen for this comparison and designated as ‘anterior’, ‘midnasal’ and ‘posterior’, respectively (Fig. 1B). Significant decreases in the cross-sectional area of the anterior nasal passages were seen in both Apert mutants, but not in Crouzon Fgfr2cC342Y/+ mutants, compared with their unaffected littermates (Fig. 2A). The cross-sectional area was also decreased in the midnasal location of both Apert mutants compared with unaffected littermates, although this only reached significance for Apert Fgfr2+/S252W mutants (Fig. 2B). No decrease was seen in cross-sectional area of the posterior nasal passages of any mutant (Fig. 2C).
Fig. 2.
Nasal airspace and cartilage changes in Fgfr2 mutant mice. (A) Anterior, (B) midnasal and (C) posterior nasal space area measurements. (D) Anterior, (E) midnasal and (F) posterior total cartilage area measurements. S252W, Apert Fgfr2+/S252W; P253R, Apert Fgfr2+/P253R; CZ, Crouzon Fgfr2C342Y/+. Blue boxes, unaffected littermates (Fgfr2+/+; WT); red boxes, mutant. n=4-15. Data are presented as the median, 25th and 75th percentiles; whiskers indicate minimum and maximum values. *P<0.05 (Student's t-test).
As the Apert Fgfr2+/S252W mutants were the most consistently affected of the three mutants at P0, we further characterized the lack of nasal cavity expansion in this line. Histologically, no differences were seen in cross-sectional areas of nasal passages at E17.5 between Apert Fgfr2+/S252W mutants and unaffected littermates at the three representative locations (Fig. 2A-C). The reduction of anterior and midnasal cross-sectional areas in P0 Apert Fgfr2+/S252W mutants was confirmed on equivalent segments from the three-dimensional (3D) MRM endocasts (Movies 1, 2 and Fig. S2). We took linear measurements of nasal passage length, width and height from 3D MRM endocasts at E17.5 and P0 (Table S2 and Fig. S1). At E17.5, no differences were seen between Apert Fgfr2+/S252W mutants and unaffected littermates. At P0, the width and height of the nasal cavity was significantly reduced in Apert Fgfr2+/S252W mutants compared with unaffected littermates, and the width and height failed to increase significantly in Apert Fgfr2+/S252W mutants between E17.5 and P0 (Table S2). There were no significant differences in nasal passage length between Apert Fgfr2+/S252W mutants and unaffected littermates at E17.5 and P0 (Table S2). Our histological and MRM findings show that reduction of the anterior and midnasal passages reflects a defect in mediolateral and dorsoventral expansion of the nasal cavity and accounts significantly for the overall decrease in nasal passage volume in Apert Fgfr2+/S252W mutants.
Cartilage thickening significantly contributes to dysmorphogenesis of nasal passages
Changes in the tissues surrounding the nasal passages, particularly the cartilaginous nasal capsule or the mucosa, could contribute to local decreases in nasal passage cross-sectional area and overall volume. Previous studies of the Apert syndrome mouse models remarked on thickened cranial and nasal cartilage, and ectopic cartilage in the midline sagittal suture and interparietal foramen (Holmes et al., 2009; Holmes and Basilico, 2012; Wang et al., 2005, 2010; Yin et al., 2008). Aberrant cartilage formation that resulted in a cartilaginous tracheal sleeve was also noted in Apert Fgfr2+/S252W mutants and homozygous Crouzon Fgfr2cC342Y/C342Y mutants (Eswarakumar et al., 2004; Peskett et al., 2017; Wang et al., 2005). We therefore quantified the cross-sectional area of nasal cartilage in the three representative locations.
The total cartilage cross-sectional area was increased significantly in all three representative locations in Apert Fgfr2+/S252W mutants compared with unaffected littermates at P0 (Fig. 2D-F). The increases were 18%, 41% and 27% in the anterior, midnasal and posterior locations, respectively. Increases were also significant in the anterior and midnasal nasal locations in Apert Fgfr2+/P253R mutants (33% and 27%, respectively) (Fig. 2D-F). An additional cartilage phenotype, noted in both Apert mutants, was ectopic cartilage extending from the external surface of the nasal capsule in midnasal sections near the junctions of the lamina horizontalis and frontoturbinate 2 with the paries nasi (Fig. 1B). In Crouzon Fgfr2cC342Y/+ mutants there was a trend towards increased cartilage cross-sectional area compared with unaffected littermates, but this did not reach statistical significance (Fig. 2D-F).
To determine when the cartilage began to thicken we measured the cross-sectional cartilage area in all three representative locations of the nasal capsule during embryonic development. For all further studies, we focused on Apert Fgfr2+/S252W mutants because out of the three mutations, the S252W mutation had the greatest effect on total cartilage area (Fig. 2D-F), which correlates with the more severe craniofacial phenotypes in both mice and humans (Heuzé et al., 2014b; Martínez-Abadías et al., 2010; Motch Perrine et al., 2014; von Gernet et al., 2000), and it is the most common mutation associated with Apert syndrome (Lajeunie et al., 1999; Park et al., 1995; Wilkie et al., 1995). At E16.5 and E17.5 in each location, the cross-sectional area was increased significantly in Apert Fgfr2+/S252W mutants compared with unaffected littermates (Fig. 2D-F). At E14.5, the cross-sectional area was also increased significantly in the midnasal region in Apert Fgfr2+/S252W mutants compared with unaffected littermates (Fig. 2D-F).
As each embryonic division of the nasal capsule consists of distinct elements (septum, paries, tecta and paraseptals) with individual growth trajectories, we sought to determine whether the cartilage increase was a generalized effect or whether it was restricted to specific elements of the nasal capsule. Cross-sectional areas of the nasal septum and paries nasi were measured in each of the three representative locations. In addition, cross-sectional areas of the tectum nasi, which has a regular morphology throughout the anterior nasal capsule, and the paraseptal cartilages, which end around the midnasal location, were measured in the anterior location (Fig. S3A). Between E16.5 and P0, the cross-sectional areas of the nasal septum and paries nasi were increased significantly in Apert Fgfr2+/S252W mutants compared with unaffected littermates in most locations (Fig. S3B,C). Significant increases also were seen in the cross-sectional areas of the anterior nasal septum and paries nasi, the midnasal paries nasi, and the posterior nasal septum at P0 in Apert Fgfr2+/P253R mutants. The cross-sectional area of the tectum nasi was significantly larger in both Apert mutants compared with unaffected littermates at P0, but not in Crouzon Fgfr2cC342Y/+ mutants (Fig. S3D). Between E16.5 and P0, the cross-sectional area of the paraseptal cartilages was increased significantly in Apert Fgfr2+/S252W mutants compared with unaffected littermates. Also, a significant increase in paraseptal cross-sectional area was seen at P0 in both Apert Fgfr2+/P253R and Crouzon Fgfr2cC342Y/+ mutants (Fig. S3E). In summary, cross-sectional areas of the septum, paries and tecta were significantly increased in both Apert mutants. Paraseptal cartilages were increased significantly in all mouse models at P0.
Increased proliferation contributes to cartilage thickening
We next considered the potential causes of the observed increase in cartilage thickness throughout the nasal capsule in both Apert mutants, including increased mesenchymal cell recruitment into the early anlagen of the nasal capsule (Ornitz and Marie, 2015), decreased chondrocyte apoptosis, increased chondrocyte proliferation or increased extracellular matrix production. Again we focused our study of these processes on the more severely affected Apert Fgfr2+/S252W model.
The E12.5 nasal capsule anlagen at anterior and posterior locations were stained by immunohistochemistry for SOX9 and FGFR2, but no clear differences in the extent or intensity of their expression were evident between Apert Fgfr2+/S252W mutants and unaffected littermates (Fig. S4A). Apoptosis, determined by the TUNEL assay at E14.5 and P0, was not obvious within the nasal septum or paries nasi in either Apert Fgfr2+/S252W mutants or in unaffected littermates at these ages (Fig. S4B).
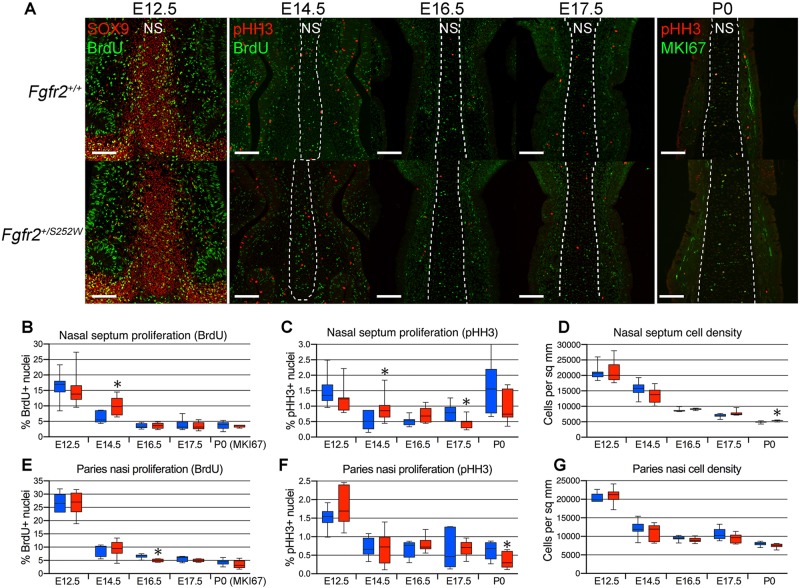
Chondrocyte proliferation was quantified at E12.5, E14.5, E16.5, E17.5 and P0 in the anterior nasal septum and paries nasi, representing the major components of the nasal capsule, by staining for phosphohistone H3 (pHH3) expression and either bromodeoxyuridine (BrdU) incorporation (embryonic stages) or MKI67 expression (P0) (Fig. 3A). Increased chondrocyte proliferation was detected at E14.5 in the nasal septum of Apert Fgfr2+/S252W mutants compared with unaffected littermates, as determined by both BrdU incorporation and pHH3 expression (Fig. 3B,C). No increase was detected at the other ages examined in the nasal septum, or at any age examined in the paries nasi (Fig. 3B,C,E,F).
Fig. 3.
Increased chondrocyte proliferation occurs early in the Apert Fgfr2+/S252W anterior nasal septum. (A) Fgfr2+/+ and Apert Fgfr2+/S252W anterior nasal septa co-stained for SOX9 (red, to visualize chondrocytes) and BrdU (green) at E12.5, pHH3 (red) and BrdU (green) at E14.5, E16.5, and E17.5, and pHH3 (red) and MKI67 (green) at P0. Dashed lines indicate boundaries of nasal septum (NS). Scale bars: 100 µm. (B-D) BrdU incorporation (B), pHH3 expression (C) and chondrocyte density (D) in the E14.5 Fgfr2+/+ and Apert Fgfr2+/S252W nasal septum. (E-G) BrdU incorporation (E), pHH3 expression (F) and chondrocyte density (G) in the E14.5 Fgfr2+/+ and Apert Fgfr2+/S252W paries nasi. Blue boxes, WT; red boxes, mutant. n=4-17. Data are presented as the median, 25th and 75th percentiles; whiskers indicate minimum and maximum values. *P<0.05 (Student's t-test).
An increase in cross-sectional cartilage area also could reflect an increase in extracellular matrix production by chondrocytes. An increase in extracellular matrix production by each chondrocyte should result in a decreased cell density. However, no decrease in cell density was seen in either the nasal septum or paries nasi between E12.5 and P0 (Fig. 3D,G). Thus, increased chondrocyte proliferation appears to be the main cause of cartilage thickening in Apert mutants.
Decrease in thickness of nasal mucosa is limited to Apert Fgfr2+/S252W mutants
The nasal mucosa consists of the epithelium that lines the nasal passages and the underlying lamina propria that lines the cartilage of the nasal capsule (Fig. S5A). At P0, the mucosal area and width was significantly decreased in the anterior (Fig. S5B,C) and midnasal (Fig. S5E,F) locations of Apert Fgfr2+/S252W mutants compared with unaffected littermates. In the anterior mucosa the decrease in area was apparent at E17.5 (Fig. S5B). No changes were noted in Apert Fgfr2+/P253R or Crouzon Fgfr2cC342Y/+ mutants at P0 (Fig. S5B,C,E). Width measurements within different subregions of the anterior mucosa at P0 showed that the decrease in width was present throughout the mucosa of Apert Fgfr2+/S252W mutants, but was most significant in the subregions along the maxilloturbinates, nasoturbinates, lateral tectum nasi and nasal septum (Figs S5D and S6). These findings suggest that the decrease in nasal passage volume in these craniosynostosis mouse mutants is not due to mucosal expansion, and that mucosal growth is actually decreased in Apert Fgfr2+/S252W mutants.
Fusion of the frontomaxillary suture may contribute to the decreased anteroposterior midface length
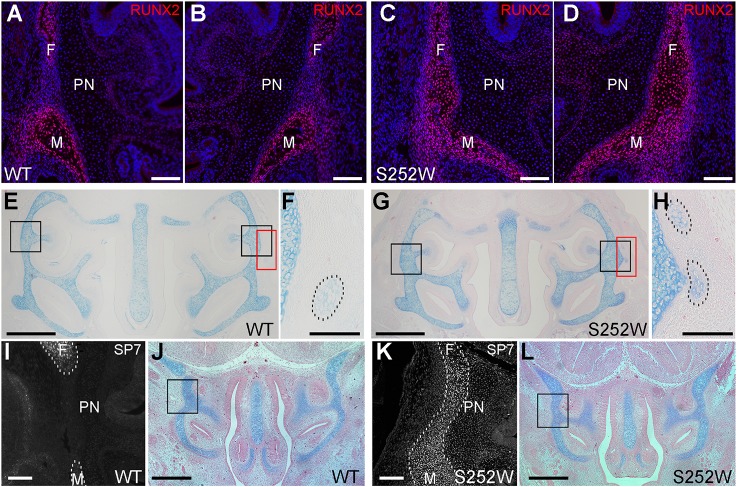
We also considered indirect causes that may lead to an increased cross-sectional area of cartilage and/or a reduction in nasal passage volume. Fusion of facial sutures and their elimination as growth sites could contribute to these phenotypes. We reported previously that in the Apert Fgfr2+/S252W and Fgfr2+/P253R mutants the initial stages of fusion are evident by micro-computed tomography imaging at E17.5 in the frontomaxillary and premaxillary-maxillary sutures, whereas partial or complete fusion of the frontomaxillary, premaxillary-maxillary, maxillary-palatine and zygomatic-maxillary sutures occurs by P0 (Table S1; see Fig. S7 for diagram of craniofacial sutures) (Martínez-Abadías et al., 2010, 2013a; Motch Perrine et al., 2014; Wang et al., 2005, 2010). In Crouzon Fgfr2cC342Y/+ mutants no facial sutures show complete fusion at P0 with a high frequency (Martínez-Abadías et al., 2013b) (Table S1). In the Apert mouse models, fusion of facial sutures around the nasal capsule could physically constrain its expansion, in particular between E17.5 and P0. If the cartilage receives no signal to restrain growth, expansion may be redirected mediolaterally and dorsoventrally, producing thicker cartilage. In this study we examined earlier embryonic stages and found that the frontomaxillary suture in Apert Fgfr2+/S252W mutants showed premature fusion as early as E16.5, with the formation of a continuous osteoid bridge secreted by RUNX2-expressing osteoblasts (Fig. 4A-E,G). Interestingly, small cartilage islands occur in the region in which ectopic cartilage outgrowths are seen at P0 in both Apert Fgfr2+/S252W mutants and unaffected littermates (Fig. 4E-H). At E14.5 we found ectopic expression of osterix (also known as SP7) protein within the frontomaxillary suture mesenchyme in Apert Fgfr2+/S252W mutants (Fig. 4I-L). The timing of frontomaxillary suture fusion is therefore consistent with a potential impact on mediolateral and dorsoventral expansion of the nasal capsule before E17.5. This impact would be increased between E17.5 and P0 by the previously documented fusion of other facial sutures described above.
Fig. 4.
Early fusion of the frontomaxillary suture in Apert Fgfr2+/S252W mice. (A-D) The expression of RUNX2 protein (red) in the left and right frontomaxillary sutures of WT (A,B; n=3) and Apert Fgfr2+/S252W (C,D; n=3) mice at E16.5 is shown. Sections are counterstained with Hoechst 33258. Scale bars: 100 µm. (E,G) Alcian Blue/Eosin-stained sections near-adjacent to the sections shown in A-D. Black boxes in E and G indicate the approximate locations of the frontomaxillary sutures of WT and Apert Fgfr2+/S252W mice, respectively, shown in A-D. Red boxes indicate the areas enlarged in F and H. Scale bars: 500 µm. (F,H) Small cartilages external to the paries nasi in WT (F) and Apert Fgfr2+/S252W (H) mice are indicated (dashed outlines). Scale bars: 100 µm. (I,K) The expression of SP7 protein in the frontomaxillary sutures of WT (I; n=7) and Apert Fgfr2+/S252W (K; n=4) mice at E14.5 is shown. Dashed lines outline the frontal and maxillary bones in I and the fused bones in K. Scale bars: 100 µm. (J,L) Alcian Blue/Eosin-stained sections near-adjacent to the sections shown in I and K. Black boxes indicate the approximate locations of the frontomaxillary sutures of WT and Apert Fgfr2+/S252W mice, respectively, shown in I and K. Scale bars: 500 µm. F, frontal bone; M, maxillary bone; PN, paries nasi.
Tissue-specific contributions to the dysmorphogenesis of the nasal passages
Fgfr2 is expressed in a variety of craniofacial tissues during embryonic development (Bansal et al., 2003; Delezoide et al., 1998; Orr-Urtreger et al., 1991, 1993; Rice et al., 2003). We sought to define the contributions of separate tissues to the midface phenotype by restricting expression of Apert Fgfr2S252W using tissue-specific Cre-expressing lines. We have previously shown that expression of Apert Fgfr2S252W in the neural crest-derived mesenchyme using a Wnt1-Cre driver is sufficient to reproduce the midface dysgenesis seen in heterozygous mutants (Heuzé et al., 2014c; Holmes and Basilico, 2012). Expression of Apert Fgfr2S252W in paraxial mesoderm using the Mesp1-Cre driver can also affect midface development secondarily to coronal suture synostosis in the calvaria (Heuzé et al., 2014c; Holmes and Basilico, 2012).
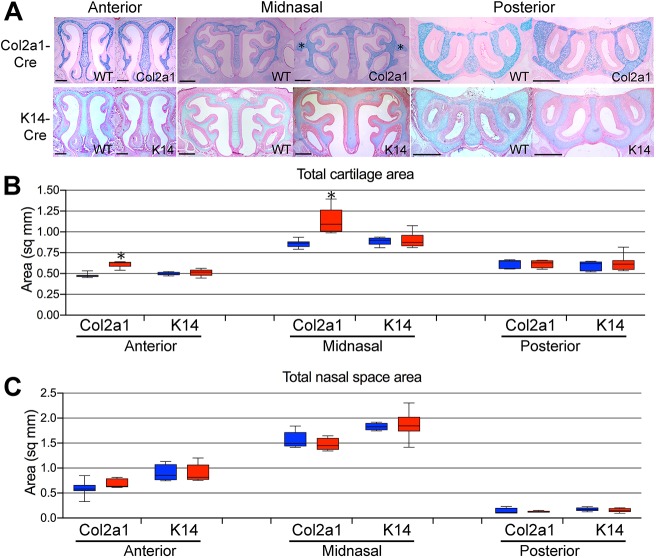
Cartilage-specific expression of Apert Fgfr2S252W using Col2a1-Cre was sufficient to cause an increase in cross-sectional cartilage area in the anterior and midnasal locations in Apert Col2a1-Cre+;Fgfr2+/S252W mutants compared with unaffected littermates at P0 (Fig. 5A,B). We also examined size changes of individual elements of the nasal capsule in each representative location. In the anterior location, the nasal septum, paries nasi, tectum nasi and paraseptal cartilage were all increased in size (Fig. S8A). In midnasal sections, the paries nasi were enlarged (Fig. S8A). As in fully heterozygous mutants, ectopic cartilage was seen in the midnasal location external to the nasal capsule at the level of the lamina horizontalis, but was typically more extensive in size (Fig. 5A). No difference was seen in the cross-sectional area of the nasal space (Fig. 5C) between Apert Col2a1-Cre+;Fgfr2+/S252W mutants and unaffected littermates. To determine the effect of cartilage-specific expression of Apert Fgfr2S252W on the total volume of the nasal cavity airspace at P0, MRM images were used to compare Apert Col2a1-Cre+;Fgfr2+/S252W with Col2a1-Cre+;Fgfr2+/+ mice. Despite a clear effect on the morphology of nasal capsule cartilage, nasal passage airspace volume was unaffected (Table S3). Col2a1-Cre expression has previously been reported in the nasal capsule at E16.5 (Sakai et al., 2001) and we confirmed expression throughout the nasal capsule at P0 (Fig. S9A).
Fig. 5.
Tissue-specific contributions to the P0 Apert Fgfr2+/S252W midfacial phenotype. (A) Alcian Blue/Eosin staining of sections in the anterior, midnasal and posterior regions of WT and indicated Cre+;Fgfr2+/S252W mice. Asterisks indicate ectopic cartilage projecting from the midnasal paries nasi in Col2a1-Cre+;Fgfr2+/S252W mice. Scale bars: 500 µm. (B) Total cross-sectional area of cartilage in representative anterior, midnasal and posterior locations in the indicated tissue-specific Cre driver lines. (C) Total cross-sectional area of nasal space in representative anterior, midnasal, and posterior locations in the indicated tissue-specific Cre driver lines. Blue boxes, WT; red boxes, mutant. n=4-11. Data are presented as the median, 25th and 75th percentiles; whiskers indicate minimum and maximum values. *P<0.05 (Student's t-test).
The decreased thickness of the mucosa in Apert Fgfr2+/S252W mutants suggested that this phenotype does not contribute to decreased nasal passage volume. However, the lack of connection between the septum and palate in mutants (Fig. 1B) may be a factor in facial shortening (Hall and Precious, 2013), and therefore may affect nasal passage volume. This defective connection does not occur when Apert Fgfr2S252W expression is restricted to neural crest-derived facial mesenchyme, suggesting that this is an epithelium-specific defect (Holmes and Basilico, 2012). Oral epithelium-specific expression of Apert Fgfr2S252W using K14-Cre did indeed prevent fusion of the septum to the anterior secondary palate, but had no effect on the total cross-sectional area of cartilage throughout the nasal capsule (Fig. 5A,B), although the paraseptal cartilages alone were larger in area (Fig. S8B). The cross-sectional area of nasal space was not affected in mutants (Fig. 5C).
To test whether thickened cartilage and decreased nasal passage volume could be produced by physical constraint after craniofacial suture fusion, we hoped to use the Col1a1(3.6 kb)-Cre-expressing line in which Cre is more broadly expressed in mesenchyme than with the Col1a1(2.3 kb)-Cre promoter (Liu et al., 2004). However, the skulls of Col1a1(3.6 kb)-Cre+;Fgfr2+/S252W mutants did not show fusion of craniofacial bones at P0. Col1a1(3.6 kb)-Cre expression in the E14.5 frontomaxillary suture mesenchyme was assessed by X-gal staining in Col1a1(3.6 kb)-Cre+;R26R+ mice. Recombination efficiency was approximately 50% (Fig. S9B-D), which may not be sufficient to generate a fusion phenotype. We also found that this Cre line is expressed in the nasal cartilage, making it unsuitable for creating bone-specific effects at this location (Fig. S9B-D).
RNA-seq analysis reinforces the significance of increased chondrocyte proliferation at E14.5
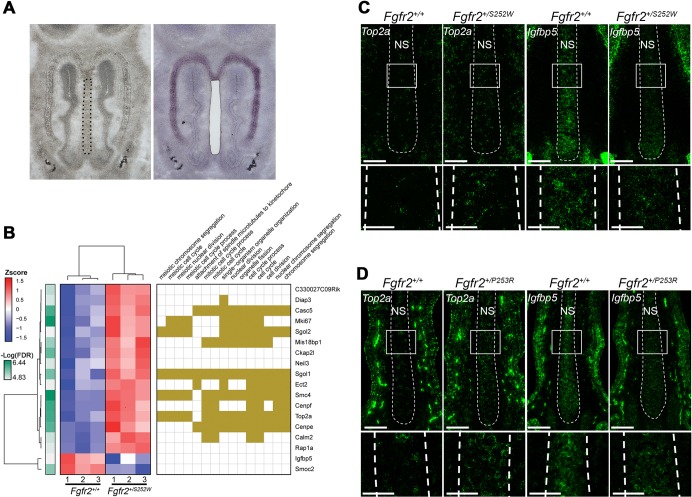
Given our finding of increased chondrocyte proliferation specifically at E14.5, we sought more detailed information on gene expression at this age. We performed laser capture microdissection of anterior nasal septum cartilage from Apert Fgfr2+/S252W mutants and unaffected littermates. This approach provides pure cartilage-specific RNA that is uncontaminated by surrounding tissue, such as facial mesenchyme or nasal mucosa, with which to construct RNA-seq libraries (Fig. 6A). Comparison of gene expression, using a stringent false discovery rate (FDR) adjusted P-value threshold of 0.01, revealed only 16 upregulated genes and two downregulated genes in Apert Fgfr2+/S252W septal cartilage compared with unaffected littermates (Fig. 6B). Using an FDR threshold of 0.05, 56 genes were upregulated and 13 genes were downregulated (Fig. S10 and Table S4). Gene ontology (GO) analysis of either the smaller or larger group of upregulated genes showed enrichment for biological process (BP) categories related to nuclear division, the cell cycle, and chromosome condensation and segregation. Of the larger group of 56 upregulated genes, 24 were assigned to these categories. However, inspection of the literature for the remaining 32 genes identified 17 with links to these categories, so that 41 of 56 genes (73%) were related to chromosome and cell cycle categories. These genes included cyclin B1 (Ccnb1), cyclin B2 (Ccnb2), and the gene encoding the antigen identified by monoclonal antibody Ki67 (Mki67) (Fig. 6B and Fig. S10). GO analysis of the 13 downregulated genes showed no enrichment for any BP categories (Fig. S10). Again, however, inspection of the literature identified links to the cell cycle for nine genes. Overall, 50 of 69 differentially expressed genes (72%) were related to chromosome and cell cycle categories. RNA-seq analysis thus validates our histological finding of increased proliferation at this location and age (Fig. 3), and reveals the broad impact of mutant FGFR2 activity on the chondrocyte cell cycle.
Fig. 6.
Differential gene expression at E14.5 in Apert Fgfr2+/S252W nasal cartilage and validation in both Apert mutants. (A) Anterior nasal septum cartilage was isolated from WT (n=3) and Apert Fgfr2+/S252W (n=3) embryos by LCM to obtain RNA for RNA-seq analysis. Images show a representative section with nasal septum outlined (left) and a nearby Cresyl Violet-stained section after LCM (right). (B) Heat map (left) and GO analysis for significantly (P<0.05) enriched BP categories (right) of differentially expressed genes in Apert Fgfr2+/S252W nasal cartilage compared with WT (FDR<0.01). (C,D) Fluorescent RNA ISH (green) for Top2a and Igfbp5 in E14.5 nasal septum cartilage (NS, within dashed lines) of Apert Fgfr2+/S252W (n=3) (C) and Apert Fgfr2+/P253R (n=3) (D) mice. White boxes in upper panels outline areas of enlargement shown in lower panels. Scale bars: 100 µm in upper panels; 50 µm in lower panels.
Various FGFs have been shown to affect chondrocyte differentiation and proliferation, in particular FGF2, FGF9 and FGF18 (Long and Ornitz, 2013; Ornitz and Marie, 2015; Su et al., 2014). Fgfr1, Fgfr2 and Fgfr3 were clearly expressed in septal cartilage (Table S5). The only FGF gene with appreciable expression was Fgf2 (Table S5). Fgf11, Fgf12 and Fgf13 were also expressed, but these encode FGF homologous factors that lack functional homology to FGFs (Mohammadi et al., 2005). We found that none of these receptors or ligands were differentially expressed between Apert Fgfr2+/S252W mutants and unaffected littermates (Table S5). Sox5, Sox6 and Sox9 encode transcription factors that are necessary for chondrocyte differentiation. Sox9 showed evidence of a decrease in expression in Apert Fgfr2+/S252W cartilage compared with unaffected littermates, and this decrease approached statistical significance (Table S5).
Significant differential expression of genes with high expression levels was validated by reverse transcription quantitative PCR (RT-qPCR) and RNA in situ hybridization (ISH). Using RT-qPCR, we found that expression of Neil3 and Mki67 was increased and expression of Igfbp5 was decreased in Apert Fgfr2+/S252W cartilage compared with unaffected littermates, in agreement with RNA-seq results, although statistical significance was not reached (Fig. S11). RNA ISH showed that Igfbp5 expression was again decreased and Top2a expression was increased in Apert Fgfr2+/S252W cartilage compared with unaffected littermates, in agreement with RNA-seq results (Fig. 6C). As both Apert mutants had similar cartilage phenotypes at P0, we determined whether similar transcriptional and phenotypic changes also occurred at E14.5 in Apert Fgfr2+/P253R mutants. Indeed, Igfbp5 expression was decreased and Top2a expression was increased in Apert Fgfr2+/P253R cartilage compared with unaffected littermates (Fig. 6D). Furthermore, as Top2a is a cell cycle gene we assayed chondrocyte proliferation in the E14.5 nasal septum of Apert Fgfr2+/P253R mutants by 5-ethynyl-2′-deoxyuridine (EdU) incorporation. Proliferation was significantly increased in Apert Fgfr2+/P253R nasal cartilage compared with unaffected littermates [22.0±1.4% (mean±s.d.) in n=3 Fgfr2+/P253R mutants; 15.3±0.1% in n=3 unaffected littermates; Student's t-test, P=0.001]. Increased chondrocyte proliferation in E14.5 nasal septum cartilage is therefore a phenotype common to both Apert mouse models.
DISCUSSION
In this study we demonstrate that whereas both Apert and Crouzon syndrome mutants show a marked reduction in nasal passage volume, they differ in associated changes to the mucosa and cartilage of the nasal capsule. Detailed analysis of the Apert Fgfr2+/S252W phenotype revealed that dysmorphic thickening of nasal cartilage occurs progressively throughout embryonic development, is associated with an early increase in cell proliferation that also occurs in Apert Fgfr2+/P253R mutants, and is cartilage-autonomous. Epithelium-specific changes and synostosis of facial sutures also contribute to the final Apert Fgfr2+/S252W midface phenotype.
Cartilage thickening was clearly evident in both Apert mutants at P0. In Apert Fgfr2+/S252W mutants this could be seen as early as E14.5 in the midnasal region and was consistent throughout subsequent development in all three regions of the nasal capsule, particularly in the septum and paries nasi. Increased chondrocyte proliferation was observed only at E14.5 in mutant septal cartilage. The initial size of the nasal capsule anlagen or subsequent cartilage extracellular matrix production did not differ between mutants and unaffected littermates. This suggests that a transient increase in mutant chondrocyte proliferation early in nasal capsule development may contribute to a persistent size and/or shape difference during later cartilage development. Although we did not observe this increase in the paries nasi, it is possible that significant transient increases in proliferation occurred at specific ages or varied with anteroposterior location at a given age.
The broader impact of FGFR2 S252W activity on cell proliferation at E14.5 was highlighted by our RNA-seq analysis of the nasal septum. Overall, 50 of 69 differentially expressed genes (72%) were related to chromosome and cell cycle regulation. Many of the genes, such as Ext2, Hipk2, Lbh, Nrxn1, Rap1a, Rpl10, Sema3a, Smoc2 and Wwp2, have also been linked to craniofacial or skeletal defects. For example, loss-of-function mutations in Wwp2 (WW domain-containing protein 2) (Zou et al., 2011) and Hipk2 (homeodomain interacting kinase 2) (Anzilotti et al., 2015) cause defects in craniofacial and/or cartilage development, including midfacial shortening. Wwp2 is an E3 ubiquitin ligase that regulates the cell cycle (Choi et al., 2015) and is a direct transcriptional target of SOX9 in mouse limb cartilage and chondrocytes (Nakamura et al., 2011; Zou et al., 2011). Hipk2 is also a cell cycle regulator (Iacovelli et al., 2009). Both Igfbp5 and H19 were downregulated in mutant septal cartilage. This suggests a link to the growth hormone/insulin-like growth factor 1 (GH/IGF1) axis that is a crucial regulator of skeletal development (Yakar and Isaksson, 2016). The insulin-like growth factor binding proteins are widely expressed in skeletal development and interact with IGF proteins in complex context-dependent ways to modulate their activity (Hoeflich et al., 2007). Deletion of H19 causes an increase in fetal and placental weight in mice through upregulation of Igf2 expression, and reduced H19 expression is associated with the Beckwith–Wiedemann overgrowth syndrome and macrosomia in gestational diabetes mellitus (Gabory et al., 2010; Jiang et al., 2014; Leighton et al., 1995; Su et al., 2016).
Tissue-specific expression of Apert Fgfr2S252W revealed novel observations of craniofacial cartilage development. Cartilage-specific expression using Col2a1-Cre was sufficient to cause an increase in cartilage thickening, but this was not as extensive as that seen in heterozygous Apert Fgfr2+/S252W mutants and these mutants did not show a difference in nasal passage volume compared with unaffected littermates. This may reflect differences in timing and efficiency of Cre recombinase activities, although EIIA-Cre and Col2a1-Cre are expressed in the relevant tissues, the zygote and the condensing mesenchyme of the cartilage anlagen, respectively (Lakso et al., 1996; Sakai et al., 2001). Oral epithelium-specific expression using K14-Cre did not produce a cartilage phenotype, despite reproducing the fusion defect between septum and secondary palate that is seen in heterozygous mutants.
We propose that the cartilage phenotypes in the Apert mice are primarily driven by mutant FGFR2 activity. The activity of Apert mutant FGFR2 remains ligand-dependent (Yu et al., 2000). FGF2 was the only functional FGF ligand for which we detected expression in E14.5 septal cartilage. FGF2 binds the mesenchymal FGFR2IIIC isoform, and Apert mutations are predicted to increase the binding affinity of FGFR2 for FGF2, especially for the S252W mutation (Ibrahimi et al., 2001, 2004). Excess FGF ligand signaling in vivo affects cartilage development. Overexpression of Fgf9 targeted to cartilage results in a shortened snout (Garofalo et al., 1999), and when targeted more generally to cranial mesenchyme can induce the cartilage cell fate (Govindarajan and Overbeek, 2006). FGF9/Fgf9 mutations in humans and mice that result in excess FGF9 activity (in multiple synostosis syndrome and elbow knee synostosis, respectively) cause ectopic cartilage formation and fusion of long bone joints (Harada et al., 2009; Wu et al., 2009). Constitutive overexpression of FGF2 in mice causes macrocephaly and a growth plate chondrocyte hyperplasia leading to dwarfism (Coffin et al., 1995; Sahni et al., 2001). Endogenous FGF2 may therefore be a key driver of differences between unaffected and mutant cartilage in Apert syndrome mouse models. FGFs acting on the nasal cartilage may also originate in the adjacent tissues. FGF20, for example, shows a higher binding affinity for both Apert mutant than wild-type (WT) receptors and recently has been shown to be expressed in immature regions of olfactory epithelium in a pattern associated with the growing turbinates of the nasal capsule, in which it can promote chondrocyte progenitor proliferation (Ibrahimi et al., 2004; Yang et al., 2018). Restricted expression of FGF20 therefore shapes the complex growth of the turbinates through a localized influence on proliferation. Although such focal and transient effects on proliferation were not observable in our broader survey of proliferation in the entire paries nasi, our results suggest FGF signaling as a potential regulator of chondrocyte proliferation and cartilage thickness. Mutant FGFR2-induced chondrocyte proliferation may promote the growth and persistence of ectopic cartilage seen in Apert syndrome mouse models. A recent study of nasal cartilage development suggests that cartilage thickness is determined by the number of cells in clonal chondrocyte columns that grow transversely within the cartilage sheet of the nasal capsule, perpendicular to the longitudinal axis (Kaucka et al., 2017).
Thickening of nasal capsule cartilage may also be accentuated as a secondary response to the overall reduction in midface size. In this scenario, shape changes in the facial bones and/or fusion of facial sutures in later embryonic development may impose a physical constraint to spatial expansion of the nasal capsule mediolaterally and dorsoventrally, as the cartilage itself continues to grow, restricting the nasal cavity space. We could not address the contribution of facial suture fusion to the cartilage phenotype because we could not identify an appropriate Cre driver that would be expressed in suture mesenchyme but not cartilage, and effectively cause premature fusion of the craniofacial bones. However, because thickened cartilage was seen in Col2a1-Cre+;Fgfr2+/S252W in the absence of FGFR2 S252W activity in osteoblasts, craniofacial suture fusion is not the sole mechanism for this cartilage phenotype.
However, suture fusion remains an important factor in midface dysgenesis in the Apert syndrome mouse mutants. The average degree of facial suture fusion in mutant mice correlates strongly with the intensity of facial dysmorphology in Apert syndrome mouse models (Motch Perrine et al., 2014). Those sutures that may adversely affect midface outgrowth show increasing fusion between E17.5 and P0, when nasal passage volume decreased in both Apert mutants (Martínez-Abadías et al., 2013a; Motch Perrine et al., 2014). In contrast, Crouzon Fgfr2cC342Y/+ mutants do not show complete fusion of these sutures at a high frequency, but still show a highly significant reduction in nasal passage volume compared with unaffected littermates between E17.5 and P0. Direct effects of mutant Fgfr2, specific to the particular syndromic mutation, are therefore likely to play independent roles in cartilage and bone. Changes in one tissue may, in turn, indirectly affect growth trajectories in the other. Together, direct and indirect effects will contribute to the final extent of nasal passage volume reduction and midface dysgenesis (Flaherty et al., 2016; Martínez-Abadías et al., 2013b; Motch Perrine et al., 2014). These intrinsic and secondary effects require further study and may require the use of computational models informed by experimental observation (Garzón-Alvarado et al., 2013; Lee et al., 2015, 2017).
The reasons for the differing phenotypes between Apert and Crouzon mutants is unclear. Although there are similarities between the effects of the different Fgfr2 mutations on facial morphology among these three craniosynostosis models, there are also fundamental differences. Most obvious is that the craniofacial complex of the Crouzon Fgfr2cC342Y/+ mouse is generally larger than their unaffected littermates at P0 (Martínez-Abadías et al., 2013b), whereas the facial skeletons of mice carrying the Apert syndrome mutations are generally smaller than their unaffected littermates (Martínez-Abadías et al., 2010; Motch Perrine et al., 2014; Wang et al., 2005, 2010). Despite this increase in size of the Crouzon Fgfr2cC342Y/+ mouse facial skeleton, their nasal passage volume is reduced by 30% relative to unaffected littermates at P0. Narrowing of the anterior and midnasal passages as measured by two-dimensional (2D) analyses occurs in both Apert mutants, but not in Crouzon Fgfr2cC342Y/+ mutants. There is a trend towards increased cartilage cross-sectional area compared with unaffected littermates in Crouzon Fgfr2cC342Y/+ mutants, though it did not achieve statistical significance. It is possible that the cartilage areas measured in 2D did not coincide with areas of significantly increased cartilage thickness in the Crouzon model. Finally, no facial sutures show complete fusion in the Crouzon Fgfr2cC342Y/+ mutants at P0 with high frequency (Martínez-Abadías et al., 2013b), reducing the potential impact of physical constraint as a potential cause of cartilage dysgenesis at this age. The potential for the FGFR2c C342Y mutation to behave differently and produce nasal airway volume reduction by mechanisms that differ from those responsible for the generally similar phenotypes in the two Apert syndrome mouse models may reflect the restriction of this mutation to the mesenchymal FGFR2 isoform (Eswarakumar et al., 2004). That it is necessary to maintain the Crouzon model on the outbred CD1 background to prevent embryonic lethality, whereas the two Apert models are maintained on the inbred C57BL/6J background, may also make a significant contribution to the phenotypic consequences of an FGFR2 mutation.
Our studies in Fgfr2-related craniosynostosis mouse models show that structural and cellular changes resulting in midfacial dysgenesis differ between specific Fgfr2 mutations. The Apert midface phenotype is a compound manifestation of defects within individual tissues. We identify decreased nasal airspace volume, thickened nasal capsule cartilage, decreased nasal mucosa, failure of fusion between the nasal septum and secondary palate, and facial suture synostosis as elements that contribute to this phenotype. Fgfr2 mutations create these defects, with the final upper airway phenotype resulting from the biophysical interactions between these tissues during embryonic craniofacial development.
MATERIALS AND METHODS
Study approval
All use of mice was in compliance with animal welfare guidelines approved by the Icahn School of Medicine at Mount Sinai and the Pennsylvania State University Institutional Animal Care and Use Committees. Mice were euthanized by isoflurane inhalation and cervical dislocation.
Mouse strains
The Fgfr2+/NeoS252W (Wang et al., 2005) and Fgfr2+/NeoP253R (Wang et al., 2010) Apert syndrome model mice were maintained as heterozygotes on the inbred C57BL/6J background. Fully heterozygous mutant mice expressing the activated mutant allele were generated by crossing male heterozygotes (Fgfr2+/NeoS252W or Fgfr2+/NeoP253R) with homozygous EIIA-Cre/EIIA-Cre female mice (also on the C57BL/6J background) (Lakso et al., 1996). For cartilage-specific expression of the S252W mutation, Col2a1-Cre+ mice (Sakai et al., 2001) were bred with Fgfr2+/NeoS252W mice to generate Col2a1-Cre+;Fgfr2+/S252W mice. For epithelium-specific expression of the S252W mutation, K14-Cre/K14-Cre mice (The Jackson Laboratory; 004782) (Dassule et al., 2000) were bred with Fgfr2+/NeoS252W mice to generate K14-Cre+;Fgfr2+/S252W mice. For osteoblast-specific expression of the S252W mutation, Col1a1(3.6 kb)-Cre+ mice (Liu et al., 2004) were bred with Fgfr2+/NeoS252W mice to generate Col1a1(3.6 kb)-Cre+;Fgfr2+/S252W mice. This mouse strain [Tg(Col1a1-Cre)1Bek/Mmucd, 015399-UCD] was obtained from the Mutant Mouse Regional Resource Center, a National Center for Research Resources-National Institutes of Health-funded strain repository, and was donated to the MMRRC by Dr Barbara Kream (University of Connecticut Health Center). To visualize Cre expression, Cre-expressing mice were crossed with R26R mice (Soriano, 1999). Crouzon Fgfr2cC342Y/+ mice (Eswarakumar et al., 2004) were maintained on the outbred CD1 background and generated by breeding Fgfr2cC342Y/+ males with CD1 females. Embryonic age was determined by the appearance of a vaginal plug in female mice indicating E0.5. For BrdU incorporation, pregnant mice were injected intraperitoneally with a 10 mg/ml solution of BrdU (100 µg/g body weight) and sacrificed 30 min later. For EdU incorporation, pregnant mice were injected intraperitoneally with a 5 mg/ml solution of EdU (25 µg/g body weight) and sacrificed 30 min later.
Magnetic resonance microscopy (MRM)
Specimens were fixed in 4% paraformaldehyde (PFA) for 48 h, washed in phosphate-buffered saline (PBS) and immersed in 2% Magnevist (gadopentetate dimeglumine, Bayer HealthCare) in PBS for 10 days to reduce the T1 and T2 relaxation time. All experiments were conducted on a vertical 14.1 Tesla (Varian) imaging system with direct drive technology. To prevent drying and to minimize magnetic susceptibility artifacts during scanning, specimens were immersed in a fluorinert liquid (FC-43, 3M). A standard imaging experiment with an isotropic resolution of 80 µm comprised a field of view of 15.4×14×11 mm3 and a matrix size of 192×132 (75% partial Fourier: 176×137). With eight averages and a repetition time of 75 ms (echo time 25 ms) the total scan time was 3 h. Matlab (The MathWorks) was used for image postprocessing. By zero-filling all directions by a factor of two, the pixel resolution of a standard imaging experiment was 40 µm3. The volume of the nasal passages was estimated for each specimen using a modification of previously described methods for segmenting nasopharyngeal volumes (Martínez-Abadías et al., 2013b). Briefly, 3D volume renderings of each specimen were visualized in Avizo 3D Analysis Software, v.8.1.1 (FEI, Thermo Fisher Scientific). For each specimen, the region of interest was segmented using a combination of standard imaging techniques and soft tissue landmarks to create planes that determined the most caudal end of the nasal passages. The nasal passages of each specimen were segmented and the volume estimated twice by the same observer using the material statistics module of Avizo as has been previously described (Martínez-Abadías et al., 2013b). Sample numbers (n=5-12) for each age and genotype are indicated in Table 1 and Table S3. To obtain the closest approximation to histological slices as possible for cross-sectional area measurements, MRM scans were reoriented into anatomical planes using the global axes and transform editor in Avizo 8.1.1. Data were then resampled to a new field using linear interpolation between the surrounding voxels while preserving voxel size. Individual slices were chosen based upon appropriate anatomical position (Fig. 1A,B) and anterior nasal space area, midnasal space area and posterior nasal space area were segmented and then quantified using the area per slice mode of material statistics (Movies 1 and 2). Analysis was performed on five Fgfr2+/+ and five Apert Fgfr2+/S252W P0 mice.
Micro-computed tomography
High-resolution micro-computed tomography images of mouse skulls were acquired in air at the Center for Quantitative Imaging at Pennsylvania State University (www.cqi.psu.edu) using an OMNI-X Universal HD600 industrial X-ray computed tomography system (Bio-Imaging Research). Solid hydroxyapatite phantoms (QRM) scanned with each set of skulls allowed us to linearly associate relative X-ray attenuation values with bone mineral density estimates. Image data were reconstructed on a 1024×1024 pixel grid as 16-bit TIFFs but reduced to 8 bit for image analysis.
Histology
For paraffin embedding, whole heads were dissected and fixed in 4% PFA overnight. Heads of P0 mice were demineralized in 10% EDTA/PBS for 4 days at 4°C. Heads were then paraffin-embedded and sectioned in the coronal plane at a thickness of 5 µm. For cryoembedding, whole heads were either embedded directly in Tissue-Tek optimal cutting temperature (OCT) Compound (4583, Sakura) in square plastic molds (Peel-A-Way embedding molds, S-22, Polysciences) or fixed in 4% PFA overnight, washed in PBS and placed in 30% sucrose/PBS for several hours before embedding. Heads were sectioned in the coronal plane at a thickness of 10 µm on an Avantik QS11 cryostat. Cryoembedding was performed for E12.5 Apert Fgfr2+/S252W embryos, E14.5 Apert Fgfr2+/S252W and Fgfr2+/P253R embryos used for fluorescent RNA ISH, and E14.5 Apert Fgfr2+/P253R embryos used for EdU incorporation.
Immunohistochemistry
Paraffin sections were dewaxed, rehydrated, permeabilized in 0.3% Triton X-100/Tris-buffered saline (TBS) for 15 min, and boiled for 10 min in 10 mM sodium citrate at pH 6.0 for antigen retrieval. Sections were blocked in 10% goat serum/TBS and treated with primary antibodies either overnight at 4°C or 1 h at room temperature. After washing in TBS, secondary antibodies were applied for 1 h at room temperature. Nuclei were stained with Hoechst 33258 (1:50,000; H1398, Invitrogen), and slides were mounted with fluorescent mounting medium (Agilent Dako). Primary antibodies used were rabbit anti-FGFR2 (1:100; sc-122, Santa Cruz), rabbit anti-RUNX2 (1:200; HPA022040, Sigma-Aldrich), rabbit anti-SOX9 (1:500; AB5535, Millipore), mouse anti-SOX9 (1:200; 76997, Abcam), rabbit anti-SP7/Osterix (1:5000; 22552, Abcam), mouse anti-BrdU (RPN202, GE Healthcare Life Sciences), rabbit anti-Ki67 (1:200; VPRM04, Vector Laboratories) and rabbit anti-Histone H3 (phospho S10) (1:1000; 5176, Abcam). Secondary antibodies used were fluorophore-conjugated donkey anti-rabbit IgG and anti-mouse IgG Alexa Fluor 488 and 594 (1:400; A21202, A21203, A21206, A21207, Invitrogen). Sample number ranges (n) are indicated in the respective figures. EdU staining was performed with the Click-iT Plus EdU Alexa Fluor 488 Imaging Kit (C10637, Thermo Fisher Scientific) as described by the manufacturer.
Cartilage staining
Paraffin sections were dewaxed, rehydrated and stained in 1% Alcian Blue/0.5% acetic acid (A5268, Sigma-Aldrich) for 2 min. Slides were rinsed with deionized water and counter-stained with 0.5% Eosin/0.5% acetic acid (E511, Fisher Scientific) for 2 min. Slides were quickly rinsed once in 95% ethanol, once in 100% ethanol, and were then passed through three changes of xylene. Slides were mounted with permanent mounting medium (H-5000, Vector Laboratories).
X-gal staining
To assess Col2a1-Cre and Col1a1(3.6 kb)-Cre expression, timed matings were performed with the R26R reporter strain to obtain E14.5 embryos or P0 pups as required. Whole E14.5 heads or P0 heads with skin removed were glutaraldehyde-fixed [2% glutaraldehyde (G5882, Sigma-Aldrich), 2% PFA, 2 mM MgCl2, 5 mM EGTA, 0.02% NP40 in PBS] for 4 h, washed with PBS, equilibrated overnight in 30% sucrose and cryoembedded for sectioning. Sections were cut at 10 µm and stored at −80°C. For X-gal staining, sections were dried at room temperature for 30 min, rehydrated in PBS, and stained in X-gal solution [1 mg/ml X-gal (B4254, Sigma-Aldrich), 5 mM K3Fe(CN)6 and K4Fe(CN)6, 2 µm MgCl2, 0.02% NP40, in PBS]. For quantification of Col1a1(3.6 kb)-Cre recombination, sections were further stained for alkaline phosphatase (ALP) activity to identify frontal and maxillary bones, and with DAPI. For ALP staining, X-gal-stained sections were washed 3× in PBS, preincubated for 5 min in 0.1 M Tris-Maleate buffer (pH 9.2) (T3128, Sigma-Aldrich), then stained with 0.4 mg/ml Fast Red TR (F8764, Sigma-Aldrich) and 0.02% Napthol AS-MX phosphate (855, Sigma-Aldrich) in 0.1 M Tris-Maleate buffer, followed by staining with DAPI for 5 min. Slides were mounted in fluorescent mounting medium. Alternatively, X-gal-stained sections were counterstained with Eosin Y, dehydrated and mounted in permanent mounting medium.
Slide selection
Three representative locations throughout the nasal capsule, referred to as anterior, midnasal and posterior, were selected for analysis in coronal plane sections between E16.5 and P0. The anterior location is situated within the primary palate to intersect the beginning of the palatal extensions of the premaxillary bones and the posterior roots of the maxillary incisors. The anterior nasal capsule features include the nasal septum, tectum nasi, paries nasi, nasoturbinates, maxilloturbinates, lamina infraconchalis and paraseptal cartilages. The midnasal location is situated within the secondary palate immediately anterior to the eyes, to intersect with the first maxillary molars and the anterior olfactory bulbs. The midnasal nasal capsule features include the nasal septum, paries nasi, lamina horizontalis with associated pars anterior and posterior of ethmoturbinate 1, frontoturbinate 2 and the posterior ends of the paraseptal cartilages. The nasal space features include the recessus frontoturbinalis that communicates with the main nasal passage through the hiatus semilunaris, and the recessus maxillaris, which is separate from the main nasal passage at this location. The posterior location is situated to intersect the second maxillary molars and anterior palatine shelves, and is within the murine olfactory recess. The posterior nasal capsule features include the nasal septum and paries nasi with attached ethmoturbinate 3. The nasal space features consist of four spaces, or ethmoidal recesses, with two on each side of the nasal septum, separated by ethmoturbinate 3. At E14.5 these locations are similarly situated. The anterior location is situated within the primary palate, and nasal capsule features include the nasal septum, paries nasi, tectum nasi and paraseptal cartilages. The midnasal location is situated immediately anterior to the eyes. The nasal capsule features include nasal septum, paries nasi and lamina horizontalis with associated pars anterior and posterior of ethmoturbinate 1. The posterior location is situated within the eyes to intersect with the upper and lower first molar primordia. The nasal capsule features include the septum and paries nasi. At E12.5 one anterior and one posterior representative location were selected for analysis. The anterior location was within the primary palate and intersected the anlagen of the nasal septum and paraseptal cartilages, and vomeronasal organs. The posterior location was situated immediately posterior to the vomeronasal organs. Care was taken to orient specimens similarly for sectioning.
Image processing and cell counting
Images of histological sections were taken using a Nikon Eclipse 80i microscope equipped with a Q Imaging Retiga 4000R digital camera and Q Capture software. Cell counts were performed using Adobe Photoshop CS5.
Histological measurements
All cartilage, nasal cavity space and mucosa area measurements were performed using Adobe Photoshop CS5 on images of histological sections oriented with the anatomically dorsal surface at the top of the image, using either the magnetic lasso tool or magic wand tool. Mucosa width measurements using ten landmarks were done in Adobe Photoshop CS5 with the ruler tool. Mucosa width measurements between landmarks were done in MetaMorph (Molecular Devices). All measurement analysis was carried out after calibrating from pixels to micrometers. Area and width measurements from the left and right sides of each section were averaged per individual animal. The mean area was then averaged across unaffected littermates and mutant mice. Sample number ranges (n) are indicated in the respective figures.
Mucosa width and area
The anterior nasal mucosa width was first determined by measuring the distance between cartilage and the outer surface of the epithelia lining the nasal cavity at ten landmark locations (Figs S5A and S6). For each landmark, the mucosa width was measured using Adobe Photoshop CS5 with the ruler tool. Pixels were calibrated to micrometers and the ten width measurements from each side of the section were averaged per mouse. These widths were then averaged for unaffected littermates and mutant mice. To further investigate the differences in mucosa width in specific subregions, the mucosa width between each consecutive landmark was measured in the representative anterior location. Using the multi-line tool in MetaMorph, a line was created along the cartilage between two consecutive landmarks. The line was then segmented into 20 µm regions (Fig. S6A). Mucosa width was then measured at the horizontal boundary of each colored segment, along a perpendicular line from the cartilage to the nasal space. The width measurements in specific subregions were averaged per mouse. These widths were then averaged for unaffected littermates and mutant mice. The anterior nasal mucosa area was measured by tracing around the nasal mucosa from landmark 1 to landmark 10 with the magnetic lasso tool in Adobe Photoshop CS5.
For mucosal widths in the midnasal location, a line was traced along the nasal septum cartilage beginning dorsally and ending ventrally at the point at which the nasal septum widens. The line was then segmented into 20 µm regions (Fig. S6B). The mucosal width was then measured and averaged as described above. The midnasal mucosa area was measured from the pars posterior of ethmoturbinate 1, around the pars anterior of ethmoturbinate 1 and frontoturbinate, and along the nasal septum until the point at which it begins to widen ventrally (Fig. S5A). Pixels were calibrated to micrometers and area measurements from each side were averaged per individual animal. This area was then averaged across unaffected littermates and mutant mice. For additional information see Fig. S6.
Laser capture microdissection
Embryos at E14.5 were dissected and heads rapidly embedded without fixation in OCT using a dry ice/methyl-2-butane bath. For laser capture microdissection (LCM), the entire nasal capsule from female unaffected littermates (n=3) and female Apert Fgfr2+/S252W (n=3) embryos was sectioned in the coronal plane onto Leica PEN membrane slides, with approximately 10 sections per slide, at a thickness of 12 µm, and stored at −80°C. Four consecutive slides from anterior regions equivalent to that used for proliferation analysis were chosen from each unaffected littermate and Fgfr2+/S252W embryo. To prepare slides for LCM, they were thawed briefly by placing them against a gloved hand, then washed twice for 30 s each in 45 ml of 80% ethanol (BP2818, Fisher Scientific) with agitation in 50 ml Falcon tubes to remove OCT. Slides were then stained with 0.1% Cresyl Violet in 50% ethanol for 30 s, washed in 45 ml of 80% ethanol, then dehydrated by passage for 30 s each through 45 ml of 95% ethanol, 100% ethanol and xylene (214736, Sigma-Aldrich) in 50 ml Falcon tubes. Slides were air dried before proceeding to LCM. The nasal septum cartilage across all four slides for each embryo was isolated using a Leica LMD6500, and RNA was prepared using the Arcturus Picopure RNA Isolation Kit (Thermo Fisher Scientific) as described by the manufacturer and eluted in a final volume of approximately 16 µl of elution buffer. RNA quality and concentration were assessed using the Agilent RNA Pico kit and Agilent Bioanalyzer 2100 as described by the manufacturer. RNA integrity number values of RNA samples used were between 7.7 and 8.6.
RNA-seq libraries
Library preparation was performed by the Gene Expression Core Facility at the Cincinnati Children's Hospital Medical Center. Briefly, an initial amplification step from 10 ng of RNA from each of six samples was performed with the NuGEN Ovation RNA-Seq System v2 (Edge Biosystems) to create double-stranded cDNA. Concentrations were measured using the Qubit dsDNA BR assay. The cDNA size was determined by using an Agilent DNA 1000 Chip. The Nextera XT DNA Sample Preparation Kit (Illumina) was used to create DNA library templates from the double-stranded cDNA as described by the manufacturer. The concentrations were measured using the Qubit dsDNA HS assay. The size of the libraries for each sample was measured using the Agilent HS DNA chip.
Next-generation sequencing
Library sequencing was performed by the Genetic Resources Core Facility at the Johns Hopkins School of Medicine. Briefly, DNA sequencing was performed on an Illumina HiSeq 2500 instrument using standard protocols for paired-end 100 bp sequencing. All six samples were multiplexed for running on a single lane. The average yield was ∼15 Gb of raw sequencing data per lane, or ∼300 million reads per lane. As per Illumina's recommendation, 5% PhiX was added to each lane as a control and to assist the analysis software with any library diversity issues.
Differential gene expression analysis
After adapter removal with cutadapt (Martin, 2011) and base quality trimming to remove 3′ read sequences if more than 20 bases with Q<20 were present, paired-end reads were mapped to the mouse (mm10) reference genome using STAR (Dobin et al., 2013) and gene count summaries were generated using featureCounts (Liao et al., 2014). Raw fragment (i.e. paired-end read) counts were then combined into a numerical matrix, with genes in rows and experiments in columns, and used as input for differential gene expression analysis with the Bioconductor Limma package (Ritchie et al., 2015) after multiple filtering steps to remove low-expressed genes. First, gene counts were converted to fragments per kb per million reads (FPKM) using the RSEM package (Li and Dewey, 2011) with default settings in strand specific mode, and only genes with expression levels above 1 FPKM in at least 50% of samples were retained for further analysis. Additional filtering removed genes with less than 50 total reads across all samples or less than 200 nucleotides in length. Finally, normalization factors were computed on the filtered data matrix using the weighted trimmed mean of M-values method, followed by voom (Law et al., 2014) mean-variance transformation in preparation for Limma linear modeling. Data were fitted to a design matrix that contained all sample groups and pairwise comparisons were performed between sample groups (mutants versus unaffected littermates). eBayes adjusted P-values were corrected for multiple testing using the Benjamin-Hochberg method and were used to select genes with significant expression differences (q≤0.01 or q≤0.05).
Gene ontology enrichment analyses
Gene ontology (GO) enrichment was computed using the R package ‘topGO’ and the ‘org.Mm.eg.db’ annotation database as reference for the GO and annotations. The ‘elim’ algorithm and ‘Fisher exact’ test were used to identify statistically over-represented GO BP categories at an FDR corrected P-value threshold of 0.05.
RT-qPCR
Independent E14.5 nasal septum RNA samples were isolated by laser capture microdissection, and RNA amplification and cDNA synthesis were performed with the NuGEN Ovation RNA-Seq System v2 as described above. RT-qPCR target genes were amplified from 50 ng of cDNA per reaction in triplicate using SYBR Green and Platinum Taq polymerase (Life Technologies) on a 7900HT Real-Time PCR instrument (Life Technologies). Primer sequences were 5′-GCTGCCTCAACACCTCAAC-3′ and 5′-AGGTGACAGCATTGCTTCTC-3′ (Actb); 5′-CAGATTCCGAGACGCCTACC-3′ and 5′-AGAGGACAGAGCTACGGTGT-3′ (Igfbp5); 5′-TGTGGAAGAGCAGGTTAGCAC-3′ and 5′-AACTTGGGCCTTGGCTGTTT-3′ (Mki67); and 5′-AGGGGAGGCAGTTTTATGCC-3′ and 5′-TTCATAATGGAGCGCTTGCC-3′ (Neil3). The relative expression of each gene was calculated using the ΔΔCt method, with Actb as the reference.
Fluorescent RNA ISH
Fluorescent RNA ISH was performed using the RNAscope Fluorescent Multiplex Reagent Kit (320850, Advanced Cell Diagnostics) as described by the manufacturer. Probes were for Top2a (Probe-Mm-Top2a, 491221) and Igfbp5 (Probe-Mm-Igfbp5, 425731). Apert Fgfr2+/S252W sections were from cryoembedded fresh frozen embryos and Apert Fgfr2+/P253R sections were from cryoembedded PFA-fixed embryos.
Statistics
For MRM analyses, mean volumes from two trials were used in statistical tests (Mann–Whitney U-tests of independent samples) conducted in SPSS v. 24 (IBM SPSS Statistics for Windows, Version 22.0, IBM). For histological analyses, statistical tests were performed in Windows Excel using the unpaired two-tailed Student's t-test. Results were plotted in GraphPad Prism 7 as box-and-whisker plots. Whiskers indicate maximum and minimum values. RT-qPCR results were plotted as scatter plots with mean and standard deviation indicated. P≤0.05 were considered significant.
Supplementary Material
Acknowledgements
Col2a1-Cre+ mice were a kind gift from Dr Attila Aszodi. LCM was performed within the Mount Sinai Biorepository and Pathology Core. RT-qPCR was performed by the Mount Sinai qPCR Core.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.T.R., E.W.J.; Methodology: G.H., S.M.M.P., J.T.R., E.W.J.; Validation: G.H., S.M.M.P., N.L.; Formal analysis: G.H., C.O., S.M.M.P., N.L., H.v.B.; Investigation: G.H., C.O., S.M.M.P., N.L.; Data curation: G.H., C.O., S.M.M.P., N.L., H.v.B.; Writing - original draft: G.H., S.M.M.P.; Writing - review & editing: G.H., C.O., S.M.M.P., N.L., H.v.B., J.T.R., E.W.J.; Visualization: G.H., C.O., S.M.M.P., H.v.B.; Supervision: G.H., J.T.R., E.W.J.; Project administration: G.H., J.T.R., E.W.J.; Funding acquisition: J.T.R., E.W.J.
Funding
This work was supported by a grant from the National Institute of Dental and Craniofacial Research [R01 DE022988 to J.T.R. and E.W.J.], and in part by grants from the National Institute of Dental and Craniofacial Research [U01 DE024448 to G.H., H.v.B. and E.W.J.; R01 DE027677 to J.T.R.], the National Institute of Child Health and Human Development [P01 HD078233 to J.T.R. and E.W.J.], and the National Institutes of Health Office of Research Infrastructure Programs [S10 OD018522]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Deposited in PMC for release after 12 months.
Data availability
RNA-seq data have been deposited in GEO under the accession number GSE97828.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.166488.supplemental
References
- Anzilotti S., Tornincasa M., Gerlini R., Conte A., Brancaccio P., Cuomo O., Bianco G., Fusco A., Annunziato L., Pignataro G. et al. (2015). Genetic ablation of homeodomain-interacting protein kinase 2 selectively induces apoptosis of cerebellar Purkinje cells during adulthood and generates an ataxic-like phenotype. Cell Death Dis. 6, e2004 10.1038/cddis.2015.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R., Lakhina V., Remedios R. and Tole S. (2003). Expression of FGF receptors 1, 2, 3 in the embryonic and postnatal mouse brain compared with Pdgfralpha, Olig2 and Plp/dm20: implications for oligodendrocyte development. Dev. Neurosci. 25, 83-95. 10.1159/000072258 [DOI] [PubMed] [Google Scholar]
- Bohm L. A., Sidman J. D. and Roby B. (2016). Early airway intervention for craniofacial anomalies. Facial Plast. Surg. Clin. North Am. 24, 427-436. 10.1016/j.fsc.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Chen L., Li D., Li C., Engel A. and Deng C.-X. (2003). A Ser252Trp [corrected] substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone 33, 169-178. 10.1016/S8756-3282(03)00222-9 [DOI] [PubMed] [Google Scholar]
- Choi B. H., Che X., Chen C., Lu L. and Dai W. (2015). WWP2 is required for normal cell cycle progression. Genes Cancer 6, 371-377. 10.18632/genesandcancer.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cielo C. M., Montalva F. M. and Taylor J. A. (2016). Craniofacial disorders associated with airway obstruction in the neonate. Semin. Fetal Neonatal Med. 21, 254-262. 10.1016/j.siny.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. D., Florkiewicz R. Z., Neumann J., Mort-Hopkins T., Dorn G. W. II, Lightfoot P., German R., Howles P. N., Kier A., O'toole B. A. et al. (1995). Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol. Biol. Cell 6, 1861-1873. 10.1091/mbc.6.12.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. M. Jr. and Kreiborg S. (1992). Upper and lower airway compromise in the Apert syndrome. Am. J. Med. Genet. 44, 90-93. 10.1002/ajmg.1320440121 [DOI] [PubMed] [Google Scholar]
- Cunningham M. L., Seto M. L., Ratisoontorn C., Heike C. L. and Hing A. V. (2007). Syndromic craniosynostosis: from history to hydrogen bonds. Orthod. Craniofac. Res. 10, 67-81. 10.1111/j.1601-6343.2007.00389.x [DOI] [PubMed] [Google Scholar]
- Dassule H. R., Lewis P., Bei M., Maas R. and Mcmahon A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775-4785. [DOI] [PubMed] [Google Scholar]
- De Beer G. (1937). The Development of the Vertebrate Skull. Oxford: Clarendon Press. [Google Scholar]
- Delezoide A.-L., Benoist-Lasselin C., Legeai-Mallet L., Le Merrer M., Munnich A., Vekemans M. and Bonaventure J. (1998). Spatio-temporal expression of FGFR 1, 2 and 3 genes during human embryo-fetal ossification. Mech. Dev. 77, 19-30. 10.1016/S0925-4773(98)00133-6 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen C., Joosten K. F. M., Bannink N., Bredero-Boelhouwer H. H., Hoeve H. L. J., Wolvius E. B., Rizopoulos D. and Mathijssen I. M. J. (2013). How does obstructive sleep apnoea evolve in syndromic craniosynostosis? A prospective cohort study. Arch. Dis. Child. 98, 538-543. 10.1136/archdischild-2012-302745 [DOI] [PubMed] [Google Scholar]
- Eswarakumar V. P., Horowitz M. C., Locklin R., Morriss-Kay G. M. and Lonai P. (2004). A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc. Natl. Acad. Sci. USA 101, 12555-12560. 10.1073/pnas.0405031101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K., Singh N. and Richtsmeier J. T. (2016). Understanding craniosynostosis as a growth disorder. Wiley Interdiscip. Rev. Dev. Biol. 5, 429-459. 10.1002/wdev.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A., Jammes H. and Dandolo L. (2010). The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays 32, 473-480. 10.1002/bies.200900170 [DOI] [PubMed] [Google Scholar]
- Garofalo S., Kliger-Spatz M., Cooke J. L., Wolstin O., Lunstrum G. P., Moshkovitz S. M., Horton W. A. and Yayon A. (1999). Skeletal dysplasia and defective chondrocyte differentiation by targeted overexpression of fibroblast growth factor 9 in transgenic mice. J. Bone Miner. Res. 14, 1909-1915. 10.1359/jbmr.1999.14.11.1909 [DOI] [PubMed] [Google Scholar]
- Garzón-Alvarado D. A., González A. and Gutiérrez M. L. (2013). Growth of the flat bones of the membranous neurocranium: a computational model. Comput. Methods Programs Biomed. 112, 655-664. 10.1016/j.cmpb.2013.07.027 [DOI] [PubMed] [Google Scholar]
- Govindarajan V. and Overbeek P. A. (2006). FGF9 can induce endochondral ossification in cranial mesenchyme. BMC Dev. Biol. 6, 7 10.1186/1471-213X-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. K. and Precious D. S. (2013). Cleft lip, nose, and palate: the nasal septum as the pacemaker for midfacial growth. Oral Surg Oral Med. Oral Pathol. Oral Radiol. 115, 442-447. 10.1016/j.oooo.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Harada M., Murakami H., Okawa A., Okimoto N., Hiraoka S., Nakahara T., Akasaka R., Shiraishi Y., Futatsugi N., Mizutani-Koseki Y. et al. (2009). FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat. Genet. 41, 289-298. 10.1038/ng.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuzé Y., Holmes G., Peter I., Richtsmeier J. T. and Jabs E. W. (2014a). Closing the gap: genetic and genomic continuum from syndromic to nonsyndromic craniosynostoses. Curr. Genet. Med. Rep. 2, 135-145. 10.1007/s40142-014-0042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuzé Y., Martínez-Abadías N., Stella J. M., Arnaud E., Collet C., García Fructuoso G., Alamar M., Lo L.-J., Boyadjiev S. A., Di Rocco F. et al. (2014b). Quantification of facial skeletal shape variation in fibroblast growth factor receptor-related craniosynostosis syndromes. Birth Defects Res. A Clin. Mol. Teratol. 100, 250-259. 10.1002/bdra.23228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuzé Y., Singh N., Basilico C., Jabs E. W., Holmes G. and Richtsmeier J. T. (2014c). Morphological comparison of the craniofacial phenotypes of mouse models expressing the Apert FGFR2 S252W mutation in neural crest- or mesoderm-derived tissues. Bone 63, 101-109. 10.1016/j.bone.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich A., Götz W., Lichanska A. M., Bielohuby M., Tönshoff B. and Kiepe D. (2007). Effects of insulin-like growth factor binding proteins in bone – a matter of cell and site. Arch. Physiol. Biochem. 113, 142-153. 10.1080/13813450701531193 [DOI] [PubMed] [Google Scholar]
- Holmes G. (2012). The role of vertebrate models in understanding craniosynostosis. Childs Nerv. Syst. 28, 1471-1481. 10.1007/s00381-012-1844-3 [DOI] [PubMed] [Google Scholar]
- Holmes G. and Basilico C. (2012). Mesodermal expression of Fgfr2S252W is necessary and sufficient to induce craniosynostosis in a mouse model of Apert syndrome. Dev. Biol. 368, 283-293. 10.1016/j.ydbio.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G., Rothschild G., Roy U. B., Deng C.-X., Mansukhani A. and Basilico C. (2009). Early onset of craniosynostosis in an Apert mouse model reveals critical features of this pathology. Dev. Biol. 328, 273-284. 10.1016/j.ydbio.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovelli S., Ciuffini L., Lazzari C., Bracaglia G., Rinaldo C., Prodosmo A., Bartolazzi A., Sacchi A. and Soddu S. (2009). HIPK2 is involved in cell proliferation and its suppression promotes growth arrest independently of DNA damage. Cell Prolif. 42, 373-384. 10.1111/j.1365-2184.2009.00601.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi O. A., Eliseenkova A. V., Plotnikov A. N., Yu K., Ornitz D. M. and Mohammadi M. (2001). Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc. Natl. Acad. Sci. USA 98, 7182-7187. 10.1073/pnas.121183798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi O. A., Zhang F., Eliseenkova A. V., Itoh N., Linhardt R. J. and Mohammadi M. (2004). Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum. Mol. Genet. 13, 2313-2324. 10.1093/hmg/ddh235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inverso G., Brustowicz K. A., Katz E. and Padwa B. L. (2016). The prevalence of obstructive sleep apnea in symptomatic patients with syndromic craniosynostosis. Int. J. Oral Maxillofac. Surg. 45, 167-169. 10.1016/j.ijom.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Jiang H., Yu Y., Xun P., Zhang J., Luo G. and Wang Q. (2014). Maternal mRNA expression levels of H19 are inversely associated with risk of macrosomia. Arch. Med. Sci. 10, 525-530. 10.5114/aoms.2014.43746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaucka M., Zikmund T., Tesarova M., Gyllborg D., Hellander A., Jaros J., Kaiser J., Petersen J., Szarowska B., Newton P. T. et al. (2017). Oriented clonal cell dynamics enables accurate growth and shaping of vertebrate cartilage. eLife 6, e25902 10.7554/eLife.25902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen G. (1947). The junction of the nasal cavity and the pharyngeal tube in the rat. Arch. Otolaryngol. 45, 159-168. 10.1001/archotol.1947.00690010168002 [DOI] [PubMed] [Google Scholar]
- Kelemen G. (1953). Nonexperimental nasal and paranasal pathology in hereditarily obese mice. AMA Arch. Otolaryngol. 57, 143-151. 10.1001/archotol.1953.00710030162003 [DOI] [PubMed] [Google Scholar]
- Lajeunie E., Cameron R., El Ghouzzi V., De Parseval N., Journeau P., Gonzales M., Delezoide A.-L., Bonaventure J., Le Merrer M. and Renier D. (1999). Clinical variability in patients with Apert's syndrome. J. Neurosurg. 90, 443-447. 10.3171/jns.1999.90.3.0443 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J. G., Gorman J. R., Sauer B., Okamoto Y., Lee E., Alt F. W. and Westphal H. (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93, 5860-5865. 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W. and Smyth G. K. (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Richtsmeier J. T. and Kraft R. H. (2015). A computational analysis of bone formation in the cranial vault in the mouse. Front. Bioeng. Biotechnol. 3, 24 10.3389/fbioe.2015.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Richtsmeier J. T. and Kraft R. H. (2017). A computational analysis of bone formation in the cranial vault using a coupled reaction-diffusion-strain model. J. Mech. Med. Biol. 17, 1750073 10.1142/S0219519417500737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton P. A., Ingram R. S., Eggenschwiler J., Efstratiadis A. and Tilghman S. M. (1995). Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375, 34-39. 10.1038/375034a0 [DOI] [PubMed] [Google Scholar]
- Lesciotto K. M., Heuzé Y., Jabs E. W., Bernstein J. M. and Richtsmeier J. T. (2018). Choanal atresia and craniosynostosis: development and disease. Plast. Reconstr. Surg. 141, 156-168. 10.1097/PRS.0000000000003928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. and Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu F., Woitge H. W., Braut A., Kronenberg M. S., Lichtler A. C., Mina M. and Kream B. E. (2004). Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int. J. Dev. Biol. 48, 645-653. 10.1387/ijdb.041816fl [DOI] [PubMed] [Google Scholar]
- Long F. and Ornitz D. M. (2013). Development of the endochondral skeleton. Cold Spring Harb. Perspect. Biol. 5, a008334 10.1101/cshperspect.a008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai S., Wei K., Flenniken A., Adamson S. L., Rossant J., Aubin J. E. and Gong S.-G. (2010). The missense mutation W290R in Fgfr2 causes developmental defects from aberrant IIIb and IIIc signaling. Dev. Dyn. 239, 1888-1900. 10.1002/dvdy.22314 [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10-12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martínez-Abadías N., Percival C., Aldridge K., Hill C. A., Ryan T., Sirivunnabood S., Wang Y., Jabs E. W. and Richtsmeier J. T. (2010). Beyond the closed suture in Apert syndrome mouse models: evidence of primary effects of FGFR2 signaling on facial shape at birth. Dev. Dyn. 239, 3058-3071. 10.1002/dvdy.22414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abadías N., Heuzé Y., Wang Y., Jabs E. W., Aldridge K. and Richtsmeier J. T. (2011). FGF/FGFR signaling coordinates skull development by modulating magnitude of morphological integration: evidence from Apert syndrome mouse models. PLoS ONE 6, e26425 10.1371/journal.pone.0026425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abadías N., Holmes G., Pankratz T., Wang Y., Zhou X., Jabs E. W. and Richtsmeier J. T. (2013a). From shape to cells: mouse models reveal mechanisms altering palate development in Apert syndrome. Dis. Model. Mech. 6, 768-779. 10.1242/dmm.010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abadías N., Motch S. M., Pankratz T. L., Wang Y., Aldridge K., Jabs E. W. and Richtsmeier J. T. (2013b). Tissue-specific responses to aberrant FGF signaling in complex head phenotypes. Dev. Dyn. 242, 80-94. 10.1002/dvdy.23903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M., Olsen S. K. and Ibrahimi O. A. (2005). Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor. Rev. 16, 107-137. 10.1016/j.cytogfr.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Motch Perrine S. M., Cole T. M. III, Martínez-Abadías N., Aldridge K., Jabs E. W. and Richtsmeier J. T. (2014). Craniofacial divergence by distinct prenatal growth patterns in Fgfr2 mutant mice. BMC Dev. Biol. 14, 8 10.1186/1471-213X-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motch Perrine S. M., Stecko T., Neuberger T., Jabs E. W., Ryan T. M. and Richtsmeier J. T. (2017). Integration of brain and skull in prenatal mouse models of Apert and Crouzon syndromes. Front. Hum. Neurosci. 11, 369 10.3389/fnhum.2017.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Yamamoto K., He X., Otsuki B., Kim Y., Murao H., Soeda T., Tsumaki N., Deng J. M., Zhang Z. et al. (2011). Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat. Commun. 2, 251 10.1038/ncomms1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash R., Possamai V., Manjaly J. and Wyatt M. (2015). The management of obstructive sleep apnea in syndromic craniosynostosis. J. Craniofac. Surg. 26, 1914-1916. 10.1097/SCS.0000000000002097 [DOI] [PubMed] [Google Scholar]
- Ornitz D. M. and Marie P. J. (2015). Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 29, 1463-1486. 10.1101/gad.266551.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Urtreger A., Givol D., Yayon A., Yarden Y. and Lonai P. (1991). Developmental expression of two murine fibroblast growth factor receptors, flg and bek. Development 113, 1419-1434. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A., Bedford M. T., Burakova T., Arman E., Zimmer Y., Yayon A., Givol D. and Lonai P. (1993). Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev. Biol. 158, 475-486. 10.1006/dbio.1993.1205 [DOI] [PubMed] [Google Scholar]
- Park W. J., Theda C., Maestri N. E., Meyers G. A., Fryburg J. S., Dufresne C., Cohen M. M. Jr. and Jabs E. W. (1995). Analysis of phenotypic features and FGFR2 mutations in Apert syndrome. Am. J. Hum. Genet. 57, 321-328. [PMC free article] [PubMed] [Google Scholar]
- Peskett E., Kumar S., Baird W., Jaiswal J., Li M., Patel P., Britto J. A. and Pauws E. (2017). Analysis of the Fgfr2C342Y mouse model shows condensation defects due to misregulation of Sox9 expression in prechondrocytic mesenchyme. Biol. Open 6, 223-231. 10.1242/bio.022178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D. P. C., Rice R. and Thesleff I. (2003). Fgfr mRNA isoforms in craniofacial bone development. Bone 33, 14-27. 10.1016/S8756-3282(03)00163-7 [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W. and Smyth G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni M., Raz R., Coffin J. D., Levy D. and Basilico C. (2001). STAT1 mediates the increased apoptosis and reduced chondrocyte proliferation in mice overexpressing FGF2. Development 128, 2119-2129. [DOI] [PubMed] [Google Scholar]
- Sakai K., Hiripi L., Glumoff V., Brandau O., Eerola R., Vuorio E., Bösze Z., Fässler R. and Aszódi A. (2001). Stage-and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol. 19, 761-767. 10.1016/S0945-053X(00)00122-0 [DOI] [PubMed] [Google Scholar]
- Smith T. D. and Rossie J. B. (2008). Nasal fossa of mouse and dwarf lemurs (primates, cheirogaleidae). Anat. Rec. 291, 895-915. 10.1002/ar.20724 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70-71. 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Stater B. J., Oomen K. P. Q. and Modi V. K. (2015). Tracheal cartilaginous sleeve association with syndromic midface hypoplasia. JAMA Otolaryngol. Head Neck Surg. 141, 73-77. 10.1001/jamaoto.2014.2790 [DOI] [PubMed] [Google Scholar]
- Su N., Jin M. and Chen L. (2014). Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res. 2, 14003 10.1038/boneres.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Wang C., Feng H., Lin L., Liu X., Wei Y. and Yang H. (2016). Alteration in expression and methylation of IGF2/H19 in placenta and umbilical cord blood are associated with macrosomia exposed to intrauterine hyperglycemia. PLoS ONE 11, e0148399 10.1371/journal.pone.0148399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.-L., Kheirandish-Gozal L., Abel F. and Gozal D. (2016). Craniofacial syndromes and sleep-related breathing disorders. Sleep Med. Rev. 27, 74-88. 10.1016/j.smrv.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg S. R. F., Healy C., Babbs C., Sharpe J. A., Wood W. G., Sharpe P. T., Morriss-Kay G. M. and Wilkie A. O. M. (2009). Skeletal analysis of the Fgfr3(P244R) mouse, a genetic model for the Muenke craniosynostosis syndrome. Dev. Dyn. 238, 331-342. 10.1002/dvdy.21790 [DOI] [PubMed] [Google Scholar]
- Von Gernet S., Golla A., Ehrenfels Y., Schuffenhauer S. and Fairley J. D. (2000). Genotype-phenotype analysis in Apert syndrome suggests opposite effects of the two recurrent mutations on syndactyly and outcome of craniofacial surgery. Clin. Genet. 57, 137-139. 10.1034/j.1399-0004.2000.570208.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiao R., Yang F., Karim B. O., Iacovelli A. J., Cai J., Lerner C. P., Richtsmeier J. T., Leszl J. M., Hill C. A. et al. (2005). Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development 132, 3537-3548. 10.1242/dev.01914 [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun M., Uhlhorn V. L., Zhou X., Peter I., Martinez-Abadias N., Hill C. A., Percival C. J., Richtsmeier J. T., Huso D. L. et al. (2010). Activation of p38 MAPK pathway in the skull abnormalities of Apert syndrome Fgfr2(+P253R) mice. BMC Dev. Biol. 10, 22 10.1186/1471-213X-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou X., Oberoi K., Phelps R., Couwenhoven R., Sun M., Rezza A., Holmes G., Percival C. J., Friedenthal J. et al. (2012). p38 Inhibition ameliorates skin and skull abnormalities in Fgfr2 Beare-Stevenson mice. J. Clin. Invest. 122, 2153-2164. 10.1172/JCI62644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger T. L., Dahl J., Bhoj E. J., Rosen A., Mcdonald-Mcginn D., Zackai E., Jacobs I., Heike C. L., Hing A., Santani A. et al. (2017). Tracheal cartilaginous sleeves in children with syndromic craniosynostosis. Genet. Med. 19, 62-68. 10.1038/gim.2016.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie A. O. M., Slaney S. F., Oldridge M., Poole M. D., Ashworth G. J., Hockley A. D., Hayward R. D., David D. J., Pulleyn L. J., Rutland P. et al. (1995). Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat. Genet. 9, 165-172. 10.1038/ng0295-165 [DOI] [PubMed] [Google Scholar]
- Wilkie A. O. M., Byren J. C., Hurst J. A., Jayamohan J., Johnson D., Knight S. J. L., Lester T., Richards P. G., Twigg S. R. F. and Wall S. A. (2010). Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics 126, e391-e400. 10.1542/peds.2009-3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.-L., Gu M.-M., Huang L., Liu X.-S., Zhang H.-X., Ding X.-Y., Xu J.-Q., Cui B., Wang L., Lu S.-Y. et al. (2009). Multiple synostoses syndrome is due to a missense mutation in exon 2 of FGF9 gene. Am. J. Hum. Genet. 85, 53-63. 10.1016/j.ajhg.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakar S. and Isaksson O. (2016). Regulation of skeletal growth and mineral acquisition by the GH/IGF-1 axis: lessons from mouse models. Growth Horm. IGF Res. 28, 26-42. 10.1016/j.ghir.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. M., Huh S.-H. and Ornitz D. M. (2018). FGF20-expressing, Wnt-responsive olfactory epithelial progenitors regulate underlying turbinate growth to optimize surface area. Dev. Cell 46, 1-17. 10.1016/j.devcel.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Du X., Li C., Xu X., Chen Z., Su N., Zhao L., Qi H., Li F., Xue J. et al. (2008). A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone 42, 631-643. 10.1016/j.bone.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Yu K., Herr A. B., Waksman G. and Ornitz D. M. (2000). Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc. Natl. Acad. Sci. USA 97, 14536-14541. 10.1073/pnas.97.26.14536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.-X., Xu X., Chen L., Li C., Brodie S. G. and Deng C.-X. (2000). A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 and premature fusion of calvarial sutures. Hum. Mol. Genet. 9, 2001-2008. 10.1093/hmg/9.13.2001 [DOI] [PubMed] [Google Scholar]
- Zou W., Chen X., Shim J.-H., Huang Z., Brady N., Hu D., Drapp R., Sigrist K., Glimcher L. H. and Jones D. (2011). The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat. Cell Biol. 13, 59-65. 10.1038/ncb2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.