Abstract
Background:
The measurement of circulating miRNAs has proven to be a powerful bi-omarker tool for several disease processes. Current protocols for the detection of miRNAs usually in-volve an RNA extraction step, requiring a substantial volume of patient serum or plasma to obtain suffi-cient input material.
Objective:
Here, we describe a novel methodology that allows detection of a large number of miRNAs from a small volume of serum or plasma without the need for RNA extraction.
Methods:
Three μl of serum or plasma was subjected to three cycles of high and low temperatures (heat/freeze cycles) followed by miRNA arrays.
Results:
Our results indicate that miRNA detection following this process is highly reproducible when comparing multiple samples from the same subject. Moreover, this protocol increases the reproducibility of miRNA detection in samples that were previously subjected to multiple freeze-thaw cycles. Im-portantly, the detection of miRNAs from serum vs. plasma following heat/freeze cycling are highly comparable, indicating that this heat/freeze process effectively eliminates differences in detection be-tween serum and plasma samples that have been reported using other sample preparation methodologies.
Conclusion:
We propose that this method is a potent alternative to current RNA extraction protocols, substantially reducing the amount of sample necessary for miRNA detection while simultaneously im-proving miRNA detection and reproducibility
Keywords: Circulating miRNAs, liquid biopsy, miRNA array, plasma, serum, biomarker
1. INTRODUCTION
The detection and measurement of miRNAs from blood is rapidly becoming recognized for its potential to be a powerful biomedical tool for the early detection, diagnosis, and/or monitoring of a wide range of diseases [1, 2]. An explosion of published reports over the past decade have described the identification of circulating miRNA biomarkers for cancer [3-5], infectious disease [6], cardiovascular disease [7-9], and neurodegenerative disorders [10, 11]. Adding to the excitement surrounding circulating miRNAs as clinically relevant biomarkers, the ability to obtain patient samples with standard blood collection techniques provides a mechanism to gather critical patient data with common, non-invasive procedures. The field of circulating miRNA biomarkers is relatively young, however, and much variety exists in the way that patient samples are processed and evaluated for miRNA content [12, 13]. The efficient and reliable detection of miRNAs in plasma and serum is not trivial [14], and refining and/or standardizing methods to detect miRNAs in liquid biopsies is critical to ensuring repeatability and accuracy across platforms [14-16]. Furthermore, the low abundance of circulating miRNAs poses challenges for accurate quantification and detection [17]. In general, methods used for circulating miRNA detection require >100 μl of serum [16, 18, 19], which may pose an issue to pediatric patients.
Methods used by different groups to isolate miRNAs from samples obtained from blood vary greatly [12, 15, 16], which may contribute to reproducibility issues across and within miRNA detection platforms [14, 16, 20-22]. Traditional RNA extraction approaches involve phenol:chloro-form steps to separate RNAs from associated proteins, followed by precipitation and pelleting of RNA in alcohol. TRIzol and Tri-Reagent, commercially available products based on this approach, can be used for the isolation of miRNAs; however, the small size of miRNAs makes them difficult to efficiently pellet after precipitation in alcohol, introducing a potential source of sample-to-sample variability. Furthermore, miRNAs with low GC content have been observed to be selectively lost when preparing samples with TRIzol [23], indicating that inherent biases may be introduced in sample miRNA content using these methods.
Several commercial kits employing column-based purification techniques are also available, including the miRNeasy Mini Kit from Qiagen (Carlsbad, CA), mirVana PARIS miRNA Isolation Kit from ThermoFisher (Waltham, MA), and mirPremier miRNA Isolation Kit from Sigma (St. Louis, MO). These kits use multistep processes in which blood-derived samples are exposed to lysis buffers and denaturants, passed through RNA-binding filters, washed, and then eluted from the filters with specific buffers. In addition to requiring significant volumes of starting plasma or serum to obtain sufficient miRNA yields, high variability in array results can be attributed to the different isolation methods used [24]. As with TRIzol and Tri-Reagent methods, the incomplete removal of denaturants can influence downstream target amplification, affecting absolute and/or relative miRNA quantification [25]. Little appears to be reported about the efficiency of miRNA elution from columns, but incomplete and/or selective elution of miRNAs from these filter substrates would effectively alter sample composition and miRNA measurements.
Here, we describe a simple plasma and serum processing step that prepares samples for the reliable detection of miRNAs by qRT-PCR while avoiding pitfalls of commonly employed miRNA extraction protocols. Importantly, this process relies strictly on rapid temperature changes that are thought to release membrane-bound miRNAs and does not require the addition of chemical reagents that may affect downstream detection methods (qRT-PCR, next-generation sequencing, microarrays, etc.). Because samples processed by this method are not subjected to purification steps that can result in material loss (either selective or in general) or dilution/elution, downstream detection methods receive inputs that fairly represent the original sample without being compromised by purification processes that may or may not be completely efficient. While it would be generally thought that exposure of plasma and serum to cycles of elevated temperatures would reduce sample integrity, thereby decreasing miRNA stability and subsequent detection, our results demonstrate that miRNA detection and reproducibility is actually improved by this methodology.
2. MATERIALS AND METHODS
2.1. Blood Collection, Storage, and Freeze-thaw Cycling
Blood collection was performed using standard phlebotomy techniques in accordance with an approved IRB protocol at the University of Colorado Denver. Unless otherwise noted, BD vacutainers (Becton, Dickinson and Company, Franklin Lakes, NJ) were used for blood collection. Blood from a single control individual was used for experiments shown in Figs. (1-5). Serum was used in most experiments as it is less prone to hemolysis than plasma. Serum is prepared by the intentional activation of platelets and clotting cascades. In addition to releasing the contents of their granules, platelets become part of the fibrin network that is formed and are efficiently removed upon centrifugation. White blood cells likely get trapped in these networks as well and are pulled down during serum preparation.
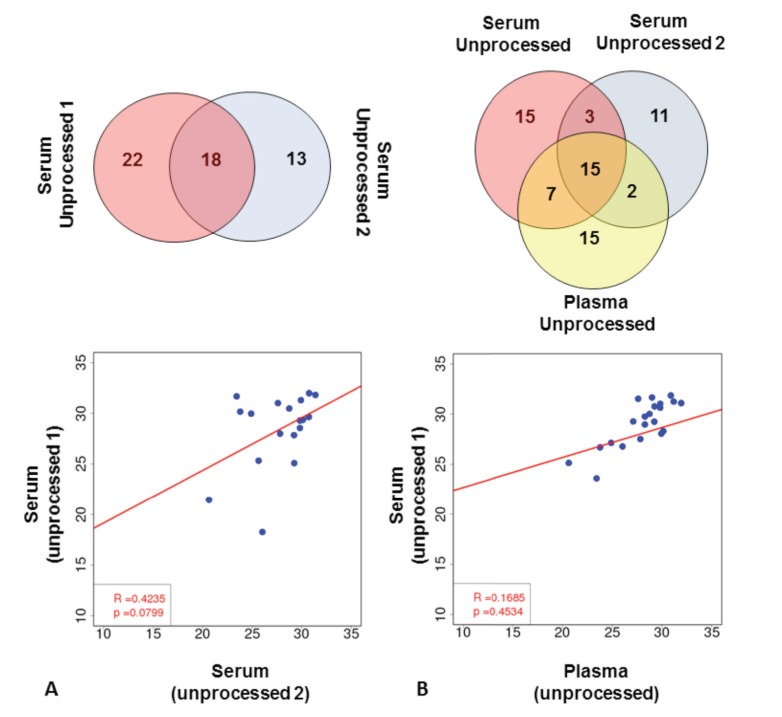
Fig. (1).
Extraction-free miRNA detection results in low number of detected miRNAs. (A) Comparison of array results in unprocessed serum samples. Top panel shows miRNAs detected in two independent serum samples from the same subject. Bottom panel shows regression analysis of miRNAs detected in both samples (based on Ct values). (B) Comparison of array results from two unprocessed serum and one unprocessed plasma sample. Top panel shows miRNAs detected in all three samples. Regression analysis of one unprocessed serum sample and unprocessed plasma sample is shown (based on Ct values).
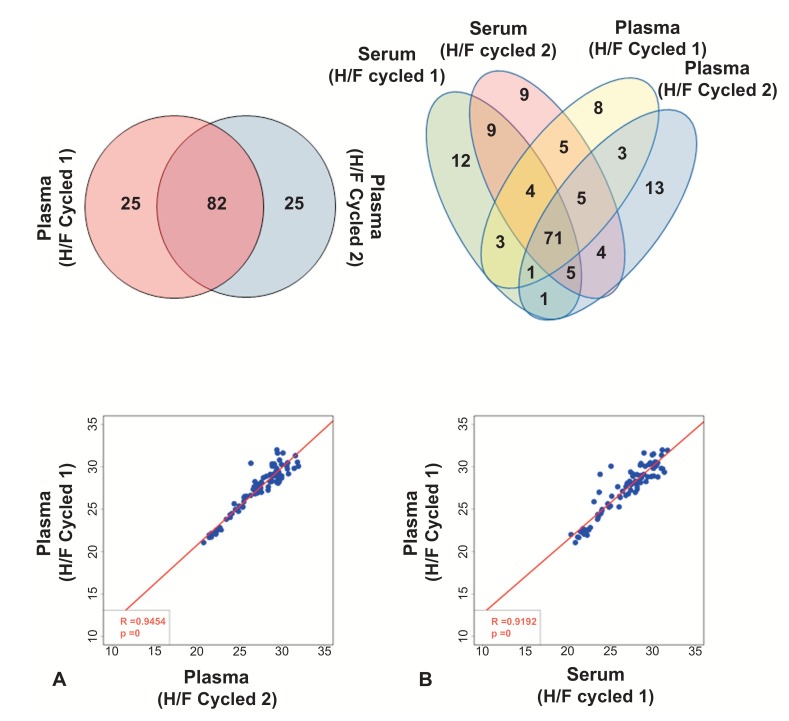
Fig. (5).
Heat/Freeze (H/F) cycle significantly improves miRNA detection in plasma samples. (A) Venn diagram is shown in the top panel, and regression analysis in the bottom panel (based on Ct values). (B) miRNA array from plasma and serum from the same subject after heat/freeze (H/F) cycle. Top panel depicts common miRNAs between 4 samples, and bottom panel show correlation between one plasma and one serum sample after H/F cycle (based on Ct values).
2.2. Sample Processing – Experimental Groups
Plasma (N=1) was prepared by collecting whole blood into vacutainers containing anticoagulant citrate dextrose (ACD). Samples were centrifuged at 850 x g for 20 minutes at 10°C. The supernatant, or plasma, was then aliquoted into 1.5ml microcentrifuge tubes and stored at -80°C.
Serum (N=2) was prepared by collecting whole blood into red-top clot-activating vacutainers. After allowing the blood to clot by incubation at room temperature for >10 minutes, samples were centrifuged at 850 x g for 20 minutes at 10°C. The supernatant, or serum, was then aliquoted into 1.5ml microcentrifuge tubes and stored at -80°C.
Serum miRNA (N=2) samples were prepared using Qiagen’s miRNeasy Plasma/Serum Mini Kit as per the manufacturer’s instructions. 250μl of unprocessed serum was used for each sample preparation, resulting in 50μl of purified serum miRNA. Eluted RNA was stored at -80oC and defrosted on ice before use.
5X Freeze/Thaw (F/T)-cycled Serum (N=1) samples were prepared by taking an aliquot of serum and subjecting it to a total of five F/T cycles. Serum was initially thawed on ice until no evidence of frozen material was observed, aliquoted into two tubes, and returned to a -80°C freezer for one hour. One of the two tubes was then removed from the freezer, thawed on ice, and returned to the -80°C freezer. The F/T-cycled sample was cycled through this freeze-thaw process a total of five times.
Heat/Freeze (H/F)-cycled Plasma and Serum (N=2 each) samples were prepared by taking an aliquot of plasma or serum and subjecting it to rapid H/F cycles. Briefly, plasma and serum samples were thawed on ice, and 10ul of each sample was aliquoted into a new microcentrifuge tube. The tube was placed into a 65°C heat block for 5 minutes and then placed immediately into a dry ice-ethanol bath to rapidly freeze the sample before being returned to the 65°C heat block. This heat-freeze cycle was repeated three times. Three μl of the H/F-cycled sample was then used for the Taqman array without further storage at -80oC.
5X F/T-cycled, then H/F-cycled (N=1) samples were prepared by subjecting 5X F/T-cycled serum to the H/F-cycling process as described above.
2.3. MegaPlex™ Pool and TaqMan® Low Density Array
A modified MegaPlex Pool and TaqMan Low Density Arrays protocol was performed for plasma and serum samples as recommended by the manufacturer (Applied Biosystems, Foster City, CA). Briefly, 7µl of reverse transcription reaction mix was prepared using a modified MegaPlex™ Pools protocol that consisted of 1µl MegaPlex RT primers, 3 mmol/liter deoxyribonucleoside triphosphates, 2µl MultiScribe Reverse Transcriptase, 0.2µl RNAse Inhibitor, 3 mmol/liter magnesium chloride, 1µl reverse transcription buffer and 1.3µl of nuclease free water. The reaction mix was completed with 3µl of serum, vortexed briefly, and then collected by brief centrifugation.
After completion of the reverse transcription cycle, complimentary DNA products were pre-amplified using MegaPlex™ PreAmp Primers (10x) to account for miRNAs expressed at low levels. 40µl of reaction mix was prepared consisting of 25µl of TaqMan PreAmp Master Mix (2x), 5µl MegaPlex PreAmp Primers, and 10µl of nuclease free water. To this 40µl reaction mix, all 10µl of the RT product was added.
Nine µl of pre-amplified product was diluted in 445µl TaqMan Universal PCR Master Mix, No UNG (2x) and 445µl nuclease free water for a total of 899µl of real-time PCR reaction mix. The reagents were mixed by brief vortexing. 100µl of real-time PCR reaction mix was pipetted into each of the 8 fill reservoirs of a TaqMan 384-Well Array pre-loaded with TaqMan Gene Expression Assays. The array plate was centrifuged twice consecutively for 1 minute at a time at 331xg. After centrifugation, the TaqMan Array card was placed in a sealer to isolate the wells of the array and then run in an ABI7900HT system for quantitative real-time PCR analysis.
The MegaPlex Pool and TaqMan Array system consists of two sets of MegaPlex RT and PreAmp primers, designated as Pool A and Pool B, as well as TaqMan arrays specific to each primer pool and designed together to test a total of 754 miRNAs. For this study, only Pool A was tested with the assumption that relative comparisons related to the number of targets detected and the correlation of these measurement would be a representative sampling of the miRNAs included in both arrays.
2.4. Data Analysis
Array analysis was performed using the Expression Suite Software version 1.0 (Life Technologies). Only miRNAs that displayed Ct values <32 were included in the analysis. Linear regression was determined using Pearson correlation and was based on Ct values of commonly detected miRNAs. miRNAs IDs and Ct values are presented in the Supplemental Table. Cohen Kappa statistics were used to determine reliability of miRNA detection between duplicate samples [26].
3. RESULTS
3.1. miRNA Measurement with Serum and Plasma
Given the potential for miRNA purification methods to alter target representation within blood-derived samples, the ability to use serum and plasma directly in downstream miRNA detection steps would be a significant advantage when considering both the volume of patient sample needed for analysis as well as the reduction in processing steps that could introduce sample artifacts and/or biases. To establish the validity of using plasma and serum directly for miRNA quantification, TaqMan miRNA array cards were used to measure miRNAs from the same original patient sample. Three μl of serum or plasma was loaded directly into reaction mixtures.
As shown in Fig. (1A), of the 373 targets included in the TaqMan miRNA array Pool A Card, a total of 53 miRNAs were detected in different aliquots of serum taken from the same patient sample. 18 miRNAs were detected in both aliquots, with 35 miRNAs measured in only one of the aliquots (22 unique miRNAs in one sample vs. 13 unique miRNAs in the other sample). These data indicate that, while it is possible to measure miRNA in serum without first performing extraction steps, the reproducibility of results is low (R2 value= 0.42; p-value=0.08). When comparing miRNA measurements from serum and plasma derived from the same patient, 24 miRNAs were detected in both plasma and serum samples, with 15 miRNAs detected only in plasma and 29 in serum samples (Fig. 1B). 39 miRNAs were detected in plasma using this methodology. No correlation between plasma and serum samples was observed (Serum-1 vs. Plasma: R2 value= 0.16; p-value=0.45 and Serum-2 vs. Plasma: R2 value= 0.07; p-value=0.78) (Fig. 1B and S1 (739.6KB, pdf) ).
3.2. RNA Extraction Improves miRNA Detection
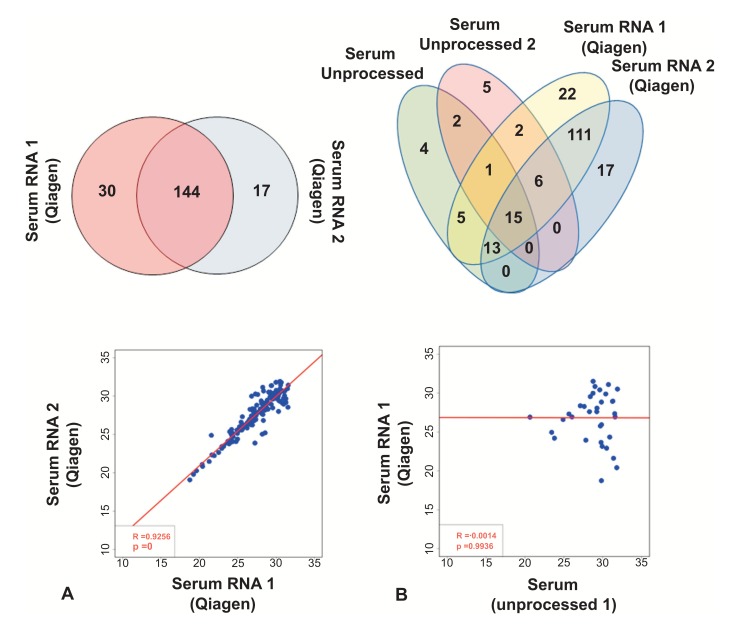
To investigate whether RNA extraction/purification from serum improves miRNA detection, RNA was purified using Qiagen’s miRNeasy kit from two aliquots of serum taken from the same patient sample. As shown in Fig. (2A), 145 miRNAs were detected in both samples, with 48 miRNAs measured in only one of the processed aliquots (31 unique miRNAs in one sample vs. 17 unique miRNAs in the other sample). The extraction of miRNA from serum using the miRNeasy kit substantially increased the number of miRNAs that were detected with the Taqman arrays (Fig. 2B), with only 42 miRNAs detected in both serum and miRNA extracted from the same serum sample. Of note, the reproducibility of miRNA quantification was high when comparing the two miRNA-extracted samples (R2 value= 0.93; p-value<0.0001, Fig. 2A) but not when comparing extracted RNA and unprocessed serum (RNA-1 vs. serum-1: R2 value= -0.001, p-value=0.99; RNA-2 vs. serum-1: R2 value= -0.11; p-value=0.57; RNA-1 vs. serum-2: R2 value= -0.17; p-value=0.46; RNA-2 vs. serum-2: R2 value= -0.1; p-value=0.64, Fig. 2B and S2).
Fig. (2).
miRNA extraction substantially increases the number of miRNAs. (A) Comparison of array results in RNA extracted from two independent samples from the same subject. Top panel shows number of detected miRNAs. Regression analysis between the two samples is shown in the bottom panel (based on Ct values). (B) Comparison of array results from Qiagen RNA prep and unprocessed serum. Venn diagram of two unprocessed samples and two RNA-extracted samples from the same subject is shown in the top panel. Regression analysis between one unprocessed serum sample and one RNA-extracted sample is shown in the bottom panel (based on Ct values).
3.3. miRNA Detection is Improved in Serum Samples Submitted to Rapid H/F Cycling when Compared to Unprocessed Samples
Given the limited reproducibility of miRNA detection when using plasma and serum directly in Taqman arrays, combined with the significant increase in detectable miRNAs following extraction with the miRNeasy kit, we hypothesized that miRNA detection in unprocessed samples was being affected by the encapsulation of miRNAs in circulating vesicles. In an attempt to release RNAs from circulating vesicles without damaging miRNAs and/or introducing foreign reagents that would alter sample composition (denaturants, diluents, etc), serum samples were aliquoted and exposed to a variety of temperature change regimens. As shown in Fig. (3A), samples exposed to a rapid H/F cycling process showed a significantly increased number of detectable miRNAs, with a total of 114 and 115 miRNAs detected in each sample, respectively (91 miRNAs were detected in both samples, and 23 and 24 miRNAs were detected in one sample but not the other). Compared to unprocessed serum, a substantial number of miRNAs is detected after H/F-cycling (138 targets total in the two H/F-cycled samples compared to 53 targets in unprocessed serum samples derived from the same serum aliquot) (Fig. 3B). Similar to miRNeasy RNA extraction method, reproducibility of quantifiable miRNA detection was high when comparing the two H/F-cycled samples (R2 value= 0.92 p-value<0.0001 – Fig. 3A) but not when comparing extracted H/F-cycled samples to unprocessed serum (Serum H/F-1 vs. serum-1: R2 value= 0.11, p-value=0.65; serum H/F-2 vs. serum-1: R2 value= -0.25; p-value=0.23; serum H/F-1 vs. serum-2: R2 value= 0.02; p-value=0.94; serum H/F-2 vs. serum-2: R2 value= -0.21; p-value=0.46 – Fig. 3B and S3). Importantly, as discussed below, miRNAs known to be present in microvesicles, miR-26a, miR-21-5p, miR-451a, miR-92a, let 7 family – reviewed in [25], are only present after the H/F cycle (Table S1 (739.6KB, pdf) ).
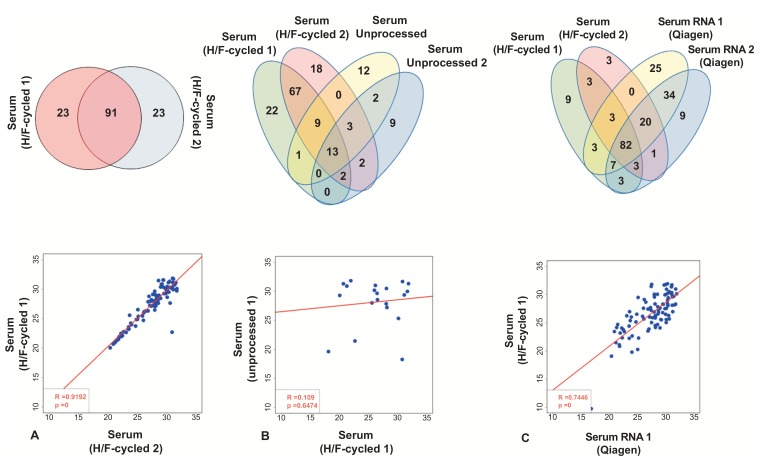
Fig. (3).
Heat/Freeze (H/F) cycle significantly improves miRNA detection. (A) Comparison of array results from two independent samples from the same subject after H/F process. Venn diagram between the two samples is shown in the top panel and regression analysis in the bottom panel (based on Ct values). (B) Comparison of array results from H/F and unprocessed serum samples. Venn diagram of two unprocessed samples and two H/F samples from the same subject is shown in the top panel. Regression analysis between one unprocessed serum sample and one H/F sample is shown in the bottom panel (based on Ct values). (C) Comparison of array results from a Qiagen RNA prep and Heat/Freeze (H/F) cycled serum. Top panel depicts miRNAs detected in all samples. Regression analysis between one RNA-extracted sample and one H/F processed serum sample (based on Ct values).
3.4. H/F-cycled Serum and Extracted RNA Samples Yield Comparable miRNA Profiles
To determine if the detection of miRNAs in H/F-cycled serum is comparable to extracted RNA, samples processed by both methodologies were compared. As shown in Fig. (3C), a total of 123 miRNAs were detected in H/F-cycled and RNA-extracted serum, with 82 miRNAs commonly detected in all 4 samples, 15 unique miRNA detected in H/F-cycled samples, and 70 unique miRNAs found only in RNA-extracted samples (Serum H/F-1 vs. RNA-1: R2 value= 0.74, p-value<0.0001; serum H/F-2 vs. RNA-1: R2 value= 0.51; p-value<0.0001; serum H/F-1 vs. RNA-2: R2 value= 0.69; p-value<0.0001; serum H/F-2 vs. RNA-2: R2 value= 0.57; p-value<0.0001 – Figs. 3C and S4 (739.6KB, pdf) ). Although a higher total number of miRNAs were detected in the miRNeasy-purified samples, these data indicate that the H/F-cycled method is highly reproducible while using significantly smaller sample volumes.
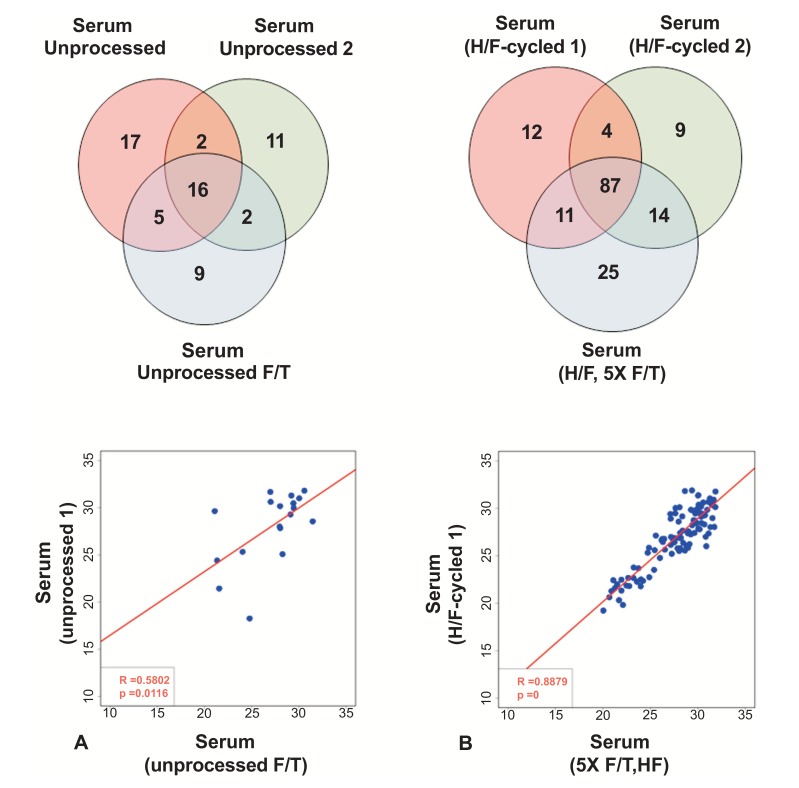
3.5. Repeated F/T Cycles Affect Detection of miRNAs in Unprocessed Serum but not in H/F-cycled Serum
We next investigated whether repeated F/T cycles affect miRNA detection in these samples. As shown in Fig. (4A) and S5, few miRNAs were detected in all 3 serum samples, and the correlation between measured miRNAs was low (Serum-1 vs. Serum F/T: R2 value= 0.58; p-value=0.012 and Serum-2 vs. Serum F/T: R2 value= 0.51; p-value=0.019). H/F-cycled serum, however, appears to overcome the deleterious effects of freeze/thaw cycles, perhaps a result of causing a more complete release of vesicle-encapsulated miRNAs. As shown in Fig. (4B), a serum sample that had been exposed to five F/T cycles to mimic repeated removal from storage was then processed with the H/F-cycle method and showed a similar miRNA profile as the original H/F-cycled sample: 87 miRNAs were commonly detected between all samples, and a significant correlation between expressed miRNAs was observed (Serum H/F-1 vs. Serum F/T+H/F: R2 value= 0.89; p-value<0.0001 and Serum H/F-2 vs. Serum F/T+H/F: R2 value= 0.81; p-value<0.0001 – Figs. 4B and S5 (739.6KB, pdf) ).
Fig. (4).
Repeated Freeze/Thaw (F/T) negatively impacts miRNA detection in unprocessed samples but improves it after Heat/Freeze (H/F) cycle. (A) Comparison of array results from two independent unprocessed samples and one F/T unprocessed samples. A 3 ml serum aliquot was defrosted on ice and frozen at -80oC a total of 5 times, followed by 3 H/F cycles and miRNA array. Venn diagram of the three samples is shown in the top panel and regression analysis of one unprocessed sample and one F/T samples is shown in the bottom panel (based on Ct values). (B) Comparison of array results from H/F samples before and after F/T. Venn diagram of all three samples from the same subject is shown in the top panel. Regression analysis between one H/F sample and one H/F plus 5 F/T cycles is shown in the bottom panel (based on Ct values).
3.6. H/F-cycling Improves Correlation between miRNA Profiles of Plasma and Serum
Because plasma is also known to contain extracellular vesicles [27], we hypothesized that H/F-cycling would improve the measurement of miRNAs in plasma samples. To test this hypothesis, two plasma samples were processed with the H/F-cycling method and analyzed with the Taqman array. As shown in Fig. (5A), 82 miRNAs were detected in both plasma samples, with only 25 and 26 unique miRNAs detected in samples 1 and 2, respectively. Moreover, the correlation between the two H/F-cycled plasma samples was similar to that observed for H/F-cycled serum samples (Plasma H/F-1 vs. Plasma H/F-2: R2 value= 0.95; p-value<0.0001). However, correlation between H/F-cycled plasma and standard plasma was low (Plasma H/F-1 vs. Plasma standard: R2 value= -0.24; p-value=0.28 and Plasma H/F-2 vs. Plasma standard: R2 value= -0.21; p-value=0.36 – data not shown), indicating that the H/F-cycling process had significantly altered the ability to detect miRNAs in these samples.
When comparing the Taqman miRNA array profiles of unprocessed plasma and serum (Fig. 1B above), little correlation between the two blood fractions was observed. However, when plasma and serum samples were processed with the H/F-cycling method, there was a significant increase in the number of targets detected in both sample types, with most targets found to be present in both plasma and serum fractions (Figs. 5B and S6 (739.6KB, pdf) ), and a high correlation between samples (Serum H/F-1 vs. Plasma H/F-1: R2 value= 0.92, p-value<0.0001; serum H/F-2 vs. plasma H/F-1: R2 value= 0.91; p-value<0.0001; serum H/F-1 vs. Plasma H/F-2: R2 value= 0.86; p-value<0.0001; serum H/F-2 vs. plasma H/F-2: R2 value= 0.84; p-value<0.0001).
3.7. Cohen Kappa Statistics Indicate Substantial Agreement between H/F and RNA-extracted Samples but not Between Unprocessed Samples
Cohen Kappa statistics were used to determine reliability of miRNA detection between duplicate samples. As shown in Table 1, substantial agreement was observed when comparing arrays using extracted RNA or H/F method. However, array comparisons using unprocessed serum or plasma resulted only in fair agreement.
Table 1.
Cohen Kappa statistics of various arrays. Agreement for the different values are: 0.01-0.2 – slight agreement; 0.21-0.4 – fair agreement; 0.41-0.6 – moderate agreement; 0.61-0.8 – substantial agreement; 0.81-0.99 – almost perfect agreement.
| Comparisons | Cohen Kappa |
|---|---|
| Serum-1 vs. Serum-2 | 0.34 |
| Serum vs. Plasma | 0.39 |
| RNA-1 vs. RNA-2 | 0.75 |
| Serum H/F-1 vs. Serum H/F-2 | 0.66 |
| Plasma H/F-1 vs. Plasma H/F-2 | 0.62 |
| Serum vs. Serum H/F | 0.18 |
| Serum H/F vs. Plasma H/F | 0.67 |
| Serum vs. RNA | 0.19 |
| Serum H/F vs. RNA | 0.62 |
4. DISCUSSION
As circulating miRNAs become more prevalent in the medical research and diagnostic fields, identification of optimal sample handling methods for the reliable detection of miRNAs in blood fractions will become more and more critical to establishing the utility of measuring miRNAs in these “liquid biopsies.” In the work presented here, a method for improving miRNA detection in plasma and serum is described. Detection of miRNAs after miRNA extraction can prove to be difficult when using samples from pediatric patients since, often, blood volumes obtained are relatively small. To circumvent this problem, guidelines from TaqMan array card suggest the use of 3 μl of serum or plasma without RNA extraction. As shown in Fig. (1) and Table 1, a limited number of miRNAs is identified using unprocessed serum.
To justify the use of unprocessed samples, the detection of circulating miRNAs would need to be comparable, at the very least, to the detection of miRNAs in samples that had been subjected to standard miRNA purification processes. A portion of circulating miRNAs are thought to be contained within membraned vesicles (EV, exosomes, etc) [14]. Unless these miRNAs are efficiently released before, or as part of, downstream measurement assays, they will likely go undetected, significantly impacting the miRNA profiles obtained from these samples. Given the low agreement and reduced number of miRNAs detected in unprocessed serum when compared to miRNeasy-processed serum (Table 1 and Fig. 2), we hypothesized that performing processing steps that would release miRNAs from vesicles would increase the number of targets detected in these assays.
The H/F-cycling method described here was developed with the intention of disrupting membraned vesicles in plasma and serum to release miRNAs and improve detection by RT-PCR. As clearly demonstrated with the Taqman miRNA array data presented here, the H/F-cycling method significantly increases both the number of targets detected and the reproducibility of results. It is not clear, however, that the H/F process does, in fact, disrupt vesicles and release miRNAs. However, studies have shown that some circulating miRNAs are only present in circulating vesicles. These include: (miR-26a, miR-21-5p, miR-451a, miR-92a, let 7 family – reviewed in [28]). Interestingly, we did not detect these miRNAs in unprocessed samples, suggesting that the H/F method results in the release of miRNAs from circulating vesicles. Nevertheless, until further work is done to specifically investigate how this process affects vesicles present in plasma and serum, it will remain a working hypothesis that release of miRNAs from microvesicles is the mechanism that improves detection of miRNAs in plasma and serum. Regardless, the H/F-cycling method represents a significant advancement for the reliable detection of miRNAs in blood fractions.
Independent of the methodology used, a subset of miRNAs in each sample comparison is unique to either one or the other sample being compared. A 3μl sampling of a patient sample consisting of 3μl of plasma or serum represents a small fraction of total patient serum. miRNAs that are in sufficiently low copy number within a sample may, or may not, be drawn up when pipetting small volumes for analysis. For high copy number targets, the likelihood of drawing copies of the miRNA into the analytical sample would be high, but for miRNAs with low copy numbers, there may be some randomness to the process that would result in low-abundance targets showing array-to-array detection variability. Similarly, high-throughput array detection methods historically require subsequent target validation steps to substantiate array data [29]. Though extremely valuable to the field, array formats have inherent platform variability, limiting their absolute reliability. Within the context of this study, variability introduced by either the randomness of sampling low copy number miRNAs from large volumes or inherent variability introduced by high-throughput detection array would both result in differences between miRNA profiles between the samples used in these experiments.
The identification of circulating miRNA biomarkers has relied on two distinct fractions of whole blood: plasma and serum. Though similar, these two fractions are distinctly different in composition. Namely, plasma is prepared without the significant activation of platelets and coagulation cascades. The result is a cell-free supernatant that is rich in fibrinogen and low in factors that are released by platelets upon activation. Serum, on the other hand, is created by the activation of coagulation cascades, causing a release of factors stored in platelet granules and the formation of a fibrin clot that is removed upon centrifugation. Some reports indicate that there are differences in the detection of miRNAs from plasma and serum [30, 31]. As shown in Fig. (1B), major differences are noted when using unprocessed plasma and serum, and the H/F-cycling method increases the number of miRNAs in both samples (Fig. 5 and S5). However, it is important to note that plasma used in the current study may contain platelets, and platelet miRNAs may be present. Interestingly, studies have shown that the same miRNAs thought to be enriched in platelets are also exclusively present in exosomes (reviewed in [28, 32]), and although several of these miRNAs are detected only after the H/F cycle (Supplement Table), determining the origin of these miRNAs is beyond the scope of this study. Importantly, these data suggest some differences commonly observed between platelet-rich plasma and serum may stem from inefficient handling and/or detection steps, and may contribute to differences in the actual miRNA content of plasma compared to serum.
CONCLUSION
What remains clear from the data presented in this report is that the H/F-cycling of plasma and serum is a reliable method to detect miRNAs from small volume samples. In addition to increasing the number of targets detected to levels that compare with common column-based miRNA purification strategies, H/F-cycling avoids exposing plasma and serum samples to conditions (precipitation or flow-through) that may selectively alter miRNA content. Similar to these studies, Breitbach et al. also showed that detection of cell-free DNA from unprocessed plasma samples results in more robust detection of DNA than initial DNA extraction [33], which they showed was due to loss of DNA in the flow-through of purification columns. The H/F-cycling of plasma and serum also avoids the use of denaturants and/or eluents that may inhibit downstream detection assays. Finally, the H/F-cycled samples require only a fraction of the plasma or serum inputs, with only 3ul of plasma or serum being sufficient to detect roughly the same number of miRNA targets. In summary, our results indicate that H/F processing of serum and plasma samples can be a good alternative to RNA extraction in samples in which large volumes are not available.
Ethics Approval and Consent to Participate
Blood collection was performed using standard phlebotomy techniques in accordance with an approved IRB protocol at the University of Colorado Denver.
Human and Animal Rights
No animal were used in this study. The reported experiments on human were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2008.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
This work was supported by the Jack Cooper Millisor Chair in Pediatric Heart Disease [to S.M.]; National Institutes of Health [HL126928 to S.M., HL107715 to B.S., HL119533 to C.S.]; and the Academic Industry Accelerator [AWD-162104 to C.S.]. Funding for open access charge: Academic Industry Accelerator [AWD-162104].
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
conflict of interest
Carmen Sucharov: Scientific founder and shareholder at miRagen, Inc. Brian Stauffer: Research support from Forest Laboratories, Inc. Carmen Sucharov, Shelley Miyamoto and Brian Stauffer: Scientific founders and shareholders at CoramiR, Inc. Peter Mariner: CEO at CoramiR, Inc. No potential conflicts of interest have been identified.
REFERENCES
- 1.Ghai V., Wang K. Recent progress toward the use of circulating microRNAs as clinical biomarkers. Arch. Toxicol. 2016;90(12):2959–2978. doi: 10.1007/s00204-016-1828-2. [DOI] [PubMed] [Google Scholar]
- 2.Cortez M.A., Calin G.A. MicroRNA identification in plasma and serum: A new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 2009;9(6):703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 3.Montani F., Bianchi F. Circulating cancer biomarkers: The macro-revolution of the microRNA. EBioMed. 2016;5:4–6. doi: 10.1016/j.ebiom.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono S., Lam S., Nagahara M., Hoon D.S. Circulating microRNA biomarkers as liquid biopsy for cancer patients: Pros and cons of current assays. J. Clin. Med. 2015;4(10):1890–1907. doi: 10.3390/jcm4101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuzaki J., Ochiya T. Circulating microRNAs and extracellular vesicles as potential cancer biomarkers: A systematic review. Int. J. Clin. Oncol. 2017;22(3):413–420. doi: 10.1007/s10147-017-1104-3. [DOI] [PubMed] [Google Scholar]
- 6.Correia C.N., Nalpas N.C., McLoughlin K.E., et al. Circulating microRNAs as potential biomarkers of infectious disease. Front. Immunol. 2017;8:118. doi: 10.3389/fimmu.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Q., Yang X. Circulating microRNA and long noncoding RNA as biomarkers of cardiovascular diseases. J. Cell. Physiol. 2016;231(4):751–755. doi: 10.1002/jcp.25174. [DOI] [PubMed] [Google Scholar]
- 8.Navickas R., Gal D., Laucevicius A., Taparauskaite A., Zdanyte M., Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: A systematic review. Cardiovasc. Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto S.D., Karimpour-Fard A., Peterson V., et al. Circulating microRNA as a biomarker for recovery in pediatric dilated cardiomyopathy. J. Heart Lung Transplant. 2015;34(5):724–733. doi: 10.1016/j.healun.2015.01.979. [DOI] [PubMed] [Google Scholar]
- 10.Zi Y., Yin Z., Xiao W., et al. Circulating microRNA as potential source for neurodegenerative diseases biomarkers. Mol. Neurobiol. 2015;52(3):1494–1503. doi: 10.1007/s12035-014-8944-x. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S., Reddy P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta. 2016;1862(9):1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kappel A., Keller A. miRNA assays in the clinical laboratory: Workflow, detection technologies and automation aspects. Clin. Chem. Lab. Med. 2017;55(5):636–647. doi: 10.1515/cclm-2016-0467. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moldovan L., Batte K.E., Trgovcich J., Wisler J., Marsh C.B., Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 2014;18(3):371–390. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrustincova A., Votavova H., Dostalova Merkerova M. Circulating microRNAs: Methodological aspects in detection of these biomarkers. Folia Biol. (Praha) 2015;61(6):203–218. doi: 10.14712/fb2015061060203. [DOI] [PubMed] [Google Scholar]
- 16.Farina N.H., Wood M.E., Perrapato S.D., et al. Standardizing analysis of circulating microRNA: clinical and biological relevance. J. Cell. Biochem. 2014;115(5):805–811. doi: 10.1002/jcb.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamam R., Hamam D., Alsaleh K.A., et al. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8(9):e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroh E.M., Parkin R.K., Mitchell P.S., Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marzi M.J., Montani F., Carletti R.M., et al. Optimization and standardization of circulating microrna detection for clinical application: The miR-Test case. Clin. Chem. 2016;62(5):743–754. doi: 10.1373/clinchem.2015.251942. [DOI] [PubMed] [Google Scholar]
- 20.McAlexander M.A., Phillips M.J., Witwer K.W. Comparison of methods for mirna extraction from plasma and quantitative recovery of rna from cerebrospinal fluid. Front. Genet. 2013;4:83. doi: 10.3389/fgene.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moret I., Sanchez-Izquierdo D., Iborra M., et al. Assessing an improved protocol for plasma microRNA extraction. PLoS One. 2013;8(12):e82753. doi: 10.1371/journal.pone.0082753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Kowdley K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012;431(1):69–75. doi: 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y.K., Yeo J., Kim B., Ha M., Kim V.N. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell. 2012;46(6):893–895. doi: 10.1016/j.molcel.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Podolska A., Kaczkowski B., Litman T., Fredholm M., Cirera S. How the RNA isolation method can affect microRNA microarray results. Acta Biochim. Pol. 2011;58(4):535–540. [PubMed] [Google Scholar]
- 25.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 2012;113(5):1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 26.Viera A.J., Garrett J.M. Understanding interobserver agreement: The kappa statistic. Fam. Med. 2005;37(5):360–363. [PubMed] [Google Scholar]
- 27.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75(2):193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleshner M., Crane C.R. Exosomes, DAMPs and miRNA: Features of stress physiology and immune homeostasis. Trends Immunol. 2017;38(10):768–776. doi: 10.1016/j.it.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajeevan M.S., Vernon S.D., Taysavang N., Unger E.R. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J. Mol. Diagn. 2001;3(1):26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Yuan Y., Cho J.H., McClarty S., Baxter D., Galas D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7(7):e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willeit P., Zampetaki A., Dudek K., et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ. Res. 2013;112(4):595–600. doi: 10.1161/CIRCRESAHA.111.300539. [DOI] [PubMed] [Google Scholar]
- 32.Sunderland N., Skroblin P., Barwari T., et al. MicroRNA biomarkers and platelet reactivity: the clot thickens. Circ. Res. 2017;120(2):418–435. doi: 10.1161/CIRCRESAHA.116.309303. [DOI] [PubMed] [Google Scholar]
- 33.Breitbach S., Tug S., Helmig S., et al. Direct quantification of cell-free, circulating DNA from unpurified plasma. PLoS One. 2014;9(3):e87838. doi: 10.1371/journal.pone.0087838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.