Abstract
Objectives
Characterization of the pharmacokinetic properties of the enantiomers of primaquine and carboxyprimaquine following administration of racemic primaquine given alone and in combination with commonly used antimalarial drugs.
Methods
Enantiomeric pharmacokinetics were evaluated in 49 healthy adult volunteers enrolled in three randomized cross-over studies in which a single dose of primaquine was given alone and then, after a suitable washout period, in combination with chloroquine, dihydroartemisinin/piperaquine or pyronaridine/artesunate. Non-linear mixed-effects modelling was used to characterize pharmacokinetics and assess the impact of drug–drug interactions.
Results
The volume of distribution of racemic primaquine was decreased by a median (95% CI) of 22.0% (2.24%–39.9%), 24.0% (15.0%–31.5%) and 25.7% (20.3%–31.1%) when co-administered with chloroquine, dihydroartemisinin/piperaquine and pyronaridine/artesunate, respectively. The oral clearance of primaquine was decreased by a median of 19.1% (14.5%–22.8%) when co-administered with pyronaridine/artesunate. These interactions were enantiospecific with a relatively higher effect on (+)-S-primaquine than on (−)-R-primaquine. No drug–drug interaction effects were seen on the pharmacokinetics of either carboxyprimaquine enantiomer.
Conclusions
Population pharmacokinetic models characterizing the enantiospecific properties of primaquine were developed successfully. Exposure to primaquine, particularly to the (+)-S-primaquine but not the carboxy metabolites, increased by up to 30% when co-administered with commonly used antimalarial drugs. A better mechanistic understanding of primaquine metabolism is required for assessment of its efficacy and haematological toxicity in humans.
Introduction
The 8-aminoquinoline primaquine is the only approved drug active against the dormant liver-stage malaria parasites (i.e. hypnozoites) of Plasmodium vivax and Plasmodium ovale. Primaquine also has significant asexual-stage activity against these parasites. It is used for the radical treatment of P. vivax and P. ovale malaria together with blood-stage schizontocidal drugs such as chloroquine or artemisinin-based combination therapies (ACTs). The WHO recommends 0.25–0.5 mg of primaquine base/kg body weight once daily for 14 days for the treatment of P. vivax and P. ovale malaria in children and adults. Primaquine causes dose-dependent haemolysis in people with glucose-6-phosphate dehydrogenase (G6PD) deficiency. This enzymopathy is common in tropical areas, with prevalences between 3% and 30%. To mitigate the haemolytic risk in individuals with milder variants of G6PD deficiency, once-weekly primaquine (0.75 mg base/kg) for 8 weeks is recommended.1 Primaquine is also an effective gametocytocidal drug, eliminating mature Plasmodium falciparum gametocytes, and thereby reducing transmission of malaria from humans to mosquitoes. A single low dose of primaquine (0.25 mg base/kg) is considered to be as effective as the previously recommended dose of 0.75 mg base/kg in blocking transmission, with a very low risk of haemolysis in G6PD-deficient individuals.2–4 Recent studies confirm that the addition of a single low dose of primaquine (0.25 mg base/kg) to artemether/lumefantrine and dihydroartemisinin/piperaquine treatments in P. falciparum malaria reduced gametocytaemia and transmission of malaria to mosquitoes compared with the ACT treatment alone.5,6
Primaquine phosphate formulations used clinically are racemic (50:50) mixtures of (+)-S- and (−)-R-enantiomers. The drug is almost completely absorbed after oral administration (96% bioavailability)7 and is readily metabolized in the liver. The major metabolite, carboxyprimaquine, is formed principally by hepatic monoamine oxidase A (MAO-A).8,9 Carboxyprimaquine is believed to be pharmacologically inactive and non-toxic.10,11 In contrast, reactive ring-hydroxylated primaquine metabolites, generated through cytochrome P450 (CYP) (mainly 2D6)-mediated metabolism, are thought to mediate the anti-hypnozoite and gametocytocidal activities and also the haemolytic toxicity in G6PD-deficient individuals.12–16
There is evidence to suggest that the metabolism of primaquine is enantioselective. In vitro metabolism studies using rat liver fractions and human hepatocytes showed that (−)-R-primaquine was preferentially converted into (−)-R-carboxyprimaquine.17–19 Studies of CYP2D6-mediated metabolism in vitro revealed that (+)-S-primaquine preferentially generated 2- and 5-hydroxyprimaquine while (−)-R-primaquine preferentially generated 3- and 4-hydroxyprimaquine.20 However, the relative contributions of each of the hydroxylated primaquine metabolites to antimalarial activity and haemolytic toxicity are still unclear. The first evidence of different pharmacokinetic profiles of primaquine enantiomers in humans (n = 6) after racemic primaquine administration showed higher exposure to (+)-S-primaquine than to (−)-R-primaquine, and only (−)-R-carboxyprimaquine was detected in plasma.21
Despite its extensive use as a partner drug with other antimal-arials, there are few studies of primaquine drug–drug interactions. Non-compartmental pharmacokinetic analyses assessing interactions between primaquine and chloroquine, dihydroartemisinin/piperaquine (Eurartesim®) and pyronaridine/artesunate (Pyramax®) all suggested higher primaquine exposures when administered in combination compared with when primaquine was administered alone.22–24 Enantiospecific pharmacokinetic interactions were not addressed in these studies. The objective of this study was to characterize the pharmacokinetic properties of racemic and enantiomeric primaquine and carboxyprimaquine, following administration of racemic primaquine, and to investigate interactions with the three blood-stage antimalarial drug combinations using non-linear mixed-effects population pharmacokinetic modelling.
Methods
Study design and participants
Data from three individual studies investigating potential pharmacokinetic drug–drug interactions of racemic primaquine when co-administered with chloroquine (Study I, Clinicaltrials.gov identifier NCT01218932), dihydroartemisinin/piperaquine (Study II, Clinicaltrials.gov identifier NCT01525511) and pyronaridine/artesunate (Study III, Clinicaltrials.gov identifier NCT01552330) were pooled to develop population pharmacokinetic models of primaquine and carboxyprimaquine, and their enantiomers. These studies were randomized, open-label, two-arm, three-period, single-dose cross-over studies in healthy G6PD-normal Thai adult volunteers conducted at the Clinical Therapeutics Unit, Mahidol-Oxford Tropical Medicine Research Unit, Bangkok, Thailand. Complete details of study design, inclusion criteria, and non-compartmental analysis results have been reported in full elsewhere.22–24
In each study, 16–17 volunteers were randomized to receive primaquine alone, primaquine combined with co-administered drug, and co-administered drug alone. The dose of each drug was as follows: 30 mg of primaquine; 600 mg of chloroquine; 120 mg of dihydroartemisinin plus 960 mg of piperaquine tetraphosphate; and 540 mg of pyronaridine tetraphosphate plus 180 mg of artesunate. Washout was 7 days after primaquine alone and 8 weeks after chloroquine-, piperaquine- and pyronaridine-containing regimens. The study drugs were administered as a single oral dose 30 min after a light standard meal followed by 4 h of fasting after dosing.
Ethics
All studies were conducted in accordance with good clinical practice and the guiding principles of the Declaration of Helsinki and were approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (Study I, MUTM 2010-028-01; Study II, MUTM 2012-009-01; Study III, MUTM 2012-013-01) and the Oxford Tropical Research Ethics Committee, University of Oxford (Study I, OXTREC 39-10; Study II, OXTREC 58-11; Study III, OXTREC 04-12). Signed and dated written informed consent was obtained from each participant prior to participation in the study.
Drug analysis
Blood samples for plasma drug concentration measurements were collected pre-dose and at 0.25, 0.5, 1, 1.5, 2, 3, 4, 6, 8, 10, 12 and 24 h and on day 3 (48–54 h) post-dose. Additional blood samples were collected on days 4, 7, 11, 15, 22, 36 and 42 when primaquine was combined with co-administered drugs. The pharmacokinetic properties of the co-administered antimalarial drugs have been reported previously.22–25
Determination of racemic and enantiomeric primaquine and carboxyprimaquine concentrations in plasma samples was performed using an LC–MS/MS method that was validated according to US FDA guidelines (W. Hanpithakpong, N. P. Day, N. J. White and J. Tarning, unpublished data). Racemic primaquine and carboxyprimaquine were provided by the Worldwide Antimalarial Resistance Network (WWARN). Enantiomeric primaquine and carboxyprimaquine and their stable isotope-labelled internal standards (6-trideuteromethoxyprimaquine diphosphate and 6-trideuteromethoxycarboxyprimaquine) were obtained from the National Centre for Natural Products Research (University of Mississippi, MS, USA). Plasma samples were processed by protein precipitation followed by phospholipid removal by solid-phase extraction. Racemic separations were performed using a Hypersil GOLD column [100 mm × 4.6 mm, internal diameter (ID) 3 μm; Thermo Scientific, Fremont, CA, USA] with 10 mM ammonium acetate/acetonitrile (50:50 v/v) at a flow rate of 0.5 μL/min in isocratic elution mode for 8 min. Enantiomeric separations were achieved using a CHIRALCEL OD-3R column (150 mm × 2.1 mm, ID 3 μm; Chiral Technologies Inc., PA, USA) with 20 mM ammonium formate with 0.1% acetic acid/acetonitrile (75:25 v/v) and methanol/acetonitrile (75:25 v/v) at a flow rate of 1.0 mL/min in gradient elution mode for 26 min.
Detection was performed using an API 5000 triple-quadrupole system (Applied Biosystems/MDS Sciex, Foster City, CA, USA) operated in the positive electrospray ionization mode. Quantification was performed using multiple reaction monitoring of the transitions at m/z 260–175 and 263–86 for primaquine and its internal standard, respectively, and at m/z 275–175 and 278–178 for carboxyprimaquine and its internal standard, respectively. The within-day and between-day precisions of racemic and enantiomeric primaquine and carboxyprimaquine were <9% at all tested quality control concentrations and <15% at the lower limit of quantification (LLOQ). The LLOQ was 1.14 and 4.88 ng/mL for racemic primaquine and carboxyprimaquine, respectively, and 0.571 and 2.44 ng/mL for enantiomeric primaquine and carboxyprimaquine, respectively.
Population pharmacokinetic analysis
Concentrations of racemic and enantiomeric primaquine and carboxyprimaquine were converted into equivalent molar units and transformed to their natural logarithms before analysis. Population pharmacokinetic models were developed using non-linear mixed-effects modelling in NONMEM version 7.3 (ICON Development Solutions, Ellicott City, MD, USA).26 The first-order conditional estimation method with interactions (FOCE-I) and subroutine ADVAN5 TRANS1 were used throughout the study. Data post-processing was performed using Perl-speaks-NONMEM (PsN)27,28 version 4.6.0 and Xpose29 version 4.0 in the programming language R version 3.3.0.30
Racemic and enantiomeric entities were modelled separately. Racemic primaquine and carboxyprimaquine concentrations were available for plasma samples from Study I, and enantiomeric concentrations were available for plasma samples from Study II and Study III. Therefore, the sum of the concentrations of each enantiomer was used in the racemic models.
A simultaneous parent–metabolite model was developed to describe the pharmacokinetics of racemic and enantiomeric primaquine and carboxyprimaquine. Complete in vivo conversion of primaquine to carboxyprimaquine was assumed to maintain structural identifiability. Enantiomeric primaquine and carboxyprimaquine were modelled assuming a complete conversion of (−)-R-primaquine to (−)-R-carboxyprimaquine and likewise for the (+)-S-enantiomers.
The structural base models were parameterized as primaquine first-order absorption rate constant (ka), relative oral bioavailability (F), elimination clearance of primaquine (CL/FPRQ) and carboxyprimaquine (CL/FCPRQ), and volume of distribution of primaquine (V/FPRQ) and carboxyprimaquine (V/FCPRQ). Since intravenous data were not available, F was fixed to unity for the population but inter-individual variability (IIV) was allowed. Different distribution models (one-, two- and three-compartment) and absorption models (zero-order, first-order absorption with and without lag time and a flexible transit compartment model31) were evaluated. For the transit compartment absorption model, the number of transit compartments was determined by stepwise addition. Mean absorption transit time (MTT) was estimated as the number of transit compartments divided by the transit rate constant (ktr). A model incorporating a first-pass effect was evaluated where an estimated fraction of the absorbed oral primaquine was converted into carboxyprimaquine during the first-pass hepatic metabolism (Fm), while the remaining fraction of primaquine (1 − Fm) was absorbed unchanged. IIV and inter-occasion variability (IOV) were implemented exponentially, and assumed to be independent and normally distributed around zero with variance ω2. Primaquine taken alone and when co-administered with other antimalarial drugs were regarded as separate occasions in the same individual. Residual variability was modelled as separate additive errors for each entity on log-transformed data, which essentially correspond to exponential residual errors on the normal scale. The level 2 (L2) data item in NONMEM was used to account for potential correlations between observations (i.e. residual errors) of parent drug and its metabolite when their measurements were from the same plasma samples.
The best structural base models were used subsequently for covariate model building. Body weight, centred on the median value of the population, was implemented a priori using allometric scaling with a fixed exponent of 0.75 on clearance parameters and 1 on volume parameters.32 Possible drug–drug interactions were modelled as proportional effects on each parameter. Other covariates, including age, gender and baseline haematological and serum chemistry measurements, were screened based on physiological plausibility and statistical significance (P < 0.05, using Pearson’s, Spearman’s and Kendall’s parameter–covariate correlations) and evaluated formally as linear, exponential or power relationships using a stepwise covariate model with forward addition (P < 0.05) and backward deletion (P < 0.001). In addition, a full covariate model was constructed separately to estimate the impact of each co-administration by including the covariate simultaneously on all parameters, except F, in order to retain model identifiability. The 95% CI of the estimated covariate effect was obtained by bootstrapping the full model (n = 1000). A significant drug–drug interaction effect was defined as a difference of at least 20% in parameter estimates. Secondary pharmacokinetic parameters, including plasma Cmax, plasma Tmax, terminal elimination t1/2 and total plasma AUC, were calculated from the individual post hoc empirical Bayes estimates (EBEs) of primary parameters from the final models.
The objective function value (OFV; equal to −2 log-likelihood) was used to discriminate between two competing hierarchical models. A decrease in OFV (ΔOFV) of 3.84, 6.63 and 10.8 was considered a significant improvement in model fit at P < 0.05, P < 0.01 and P < 0.001, respectively, with one additional parameter. Physiological plausibility, precision of parameter estimates and goodness-of-fit (GOF) were employed to guide model selection. Shrinkage in random effects was calculated to assess the reliability of individual parameter estimates (EBEs) and GOF diagnostics.33 Non-parametric bootstrapping (n = 1000), stratified by study, was used to calculate CIs and relative standard errors (RSEs) of parameter estimates of the final models. The predictive performance of the model was examined by using simulation-based diagnostics (numerical and visual predictive checks).34
Results
A total of 49 healthy volunteers (32 females and 17 males) from three separate studies were included in this population pharmacokinetic analysis. The median (range) age was 32 years (20–51 years) and the median weight was 60.2 kg (46.8–71.4 kg). Full demographic data and non-compartmental pharmacokinetic analyses assessing drug–drug interactions between primaquine and chloroquine, dihydroartemisinin/piperaquine and pyronaridine/artesunate have been published in full elsewhere.22–24 There were no serious adverse events. The collected data consisted of 1274 observations for each of the racemic plasma concentrations of primaquine and carboxyprimaquine and 830 observations for each enantiomeric primaquine and carboxyprimaquine plasma concentration.
Pharmacokinetics of racemic and enantiomeric primaquine and carboxyprimaquine
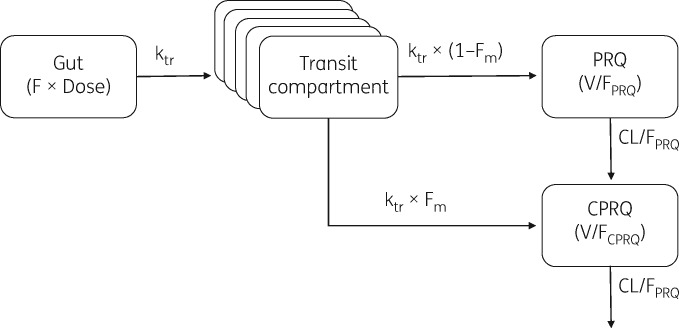
The disposition pharmacokinetics of primaquine and carboxyprimaquine, in racemic and enantiomeric models, were both best described by a one-compartment model with first-order absorption and elimination. A two-compartment model for primaquine did not improve the model fit significantly (P > 0.05). A two-compartment disposition model for carboxyprimaquine improved the model fit significantly (ΔOFV of −84.7) for the racemic model but V/F of the central compartment and its IIV were estimated with poor precision (RSE >100%). For enantiomeric models, the sampling periods were too short to allow precise determination of a potential peripheral compartment. The one-compartment disposition model for carboxyprimaquine showed no systematic bias in the racemic or enantiomeric models and was therefore carried forward. Oral absorption was best described by a transit-compartment model, with ka set as equal to ktr for model stability. The number of transit compartments was fixed to five for the racemic and (−)-R-enantiomer models and to six for the (+)-S-enantiomer model. Incorporation of hepatic first-pass metabolism showed a significant improvement (P < 0.05) in model fit (ΔOFV of −260, −11.1 and −42.5 for racemic, (+)-S- and (−)-R-enantiomer models, respectively). A schematic representation of the final structural model is depicted in Figure 1. Only a small number of observations were below the LLOQ (<5% of total observations) and therefore excluded from the analysis. A relatively larger fraction of observations were measured to be below the LLOQ in the terminal elimination phase (28.9% of primaquine concentrations at 48–54 h after dosing, and 18.8% of carboxyprimaquine concentrations at 144 h after dosing) but the exclusion of these LLOQ data did not show any systemic bias in simulation-based diagnostics [i.e. categorical visual predictive checks (VPCs), data not shown]. Adding IOV in MTT between occasions when primaquine was taken alone and when it was co-administered with other antimalarial drugs improved the model fit significantly (ΔOFV of −1330, −973 and −1070 for racemic, (+)-S- and (−)-R-enantiomer models, respectively). Additional IOV in Fm also resulted in significantly improved models (ΔOFV of −295, −155 and −45.8 for racemic, (+)-S- and (−)-R-enantiomer models, respectively). Addition of correlation between parent and metabolite residual errors (i.e. L2 data item) improved the model fit significantly (ΔOFV of 1160, 83.2 and 266 for racemic, (+)-S- and (−)-R-enantiomer models, respectively).
Figure 1.
Structural representation of the final population pharmacokinetic model for racemic and enantiomeric primaquine (PRQ) and carboxyprimaquine (CPRQ).
The mean V/FPRQ of (+)-S-primaquine (126 L) was lower than that of (−)-R-primaquine (160 L). The mean CL/FPRQ of (+)-S-primaquine (12.5 L/h) was ∼2-fold lower than that of (−)-R-primaquine (23.1 L/h). The metabolite/parent AUC ratio (AUCCPRQ/AUCPRQ) was 35.2, 1.98 and 109 for racemic, (+)-S- and (−)-R-primaquine, respectively.
Body weight was implemented a priori as an allometric function on volume and clearance parameters and resulted in an improvement in the population models (ΔOFV of −12.9, −4.92 and −21.1 for racemic, (+)-S- and (−)-R-enantiomer models, respectively). Using a stepwise covariate modelling approach, we found that co-administration of all three commonly used antimalarial treatments decreased V/FPRQ significantly, and that co-administration of primaquine with pyronaridine/artesunate significantly decreased CL/FPRQ. Furthermore, the drug–drug interactions did not affect (+)-S- and (−)-R-primaquine equally (Table 1). No other covariates had significant effects on model parameters.
Table 1.
Population pharmacokinetic parameter estimates from the final population pharmacokinetic models
| Racemic |
(+)-S-enantiomer |
(−)-R-enantiomer |
||||
|---|---|---|---|---|---|---|
| Parameter | estimatea (%RSEb) | 95% CIb | estimatea (%RSEb) | 95% CIb | estimatea (%RSEb) | 95% CIb |
| Fixed effects | ||||||
| F (%) | 100 fixed | NA | 100 fixed | NA | 100 fixed | NA |
| MTT (h) | 0.893 (5.47) | 0.796–0.986 | 0.812 (5.31) | 0.734–0.901 | 0.833 (5.61) | 0.743–0.927 |
| No. of transit compartments | 5 fixed | NA | 6 fixed | NA | 5 fixed | NA |
| CL/FPRQ (L/h) | 16.0 (5.31) | 14.4–17.8 | 12.5 (5.58) | 11.2–14.0 | 23.1 (5.94) | 20.7 –26.1 |
| V/FPRQ (L) | 156 (5.64) | 139–173 | 129 (5.48) | 117–145 | 160 (7.27) | 139–184 |
| Fm (%) | 0.360 (4.83) | 0.327–0.394 | 0.298 (6.77) | 0.255–0.335 | 0.460 (5.08) | 0.416–0.506 |
| CL/FCPRQ (L/h) | 0.729 (3.49) | 0.680–0.780 | 8.11 (7.14) | 6.96–9.27 | 0.377 (4.62) | 0.343–0.411 |
| V/FCPRQ (L) | 16.5 (2.70) | 15.7–17.5 | 159 (5.45) | 142–175 | 10.5 (1.98) | 10.1–11.0 |
| Drug–drug interaction effects (% decrease) | ||||||
| chloroquine | ||||||
| V/FPRQ | 13.8 (15.8) | 9.62–17.9 | NA | NA | NA | NA |
| dihydroartemisinin/piperaquine | ||||||
| V/FPRQ | 15.0 (21.9) | 8.34–20.8 | 16.0 (18.2) | 10.8–22.1 | 22.3 (13.6) | 15.8–27.6 |
| pyronaridine/artesunate | ||||||
| CL/FPRQ | 18.0 (19.1) | 10.5–23.7 | 19.5 (19.6) | 11.6–26.5 | NA | NA |
| V/FPRQ | 24.3 (18.0) | 14.8–31.8 | 27.5 (15.7) | 18.4–35.1 | 10.2 (25.2) | 4.79–14.8 |
| Random effects [%CV (%RSE)] | ||||||
| IIV F | 15.8 (11.3) | 12.0–18.9 | 26.7 (14.3) | 18.5–34.0 | 13.8 (11.2) | 10.4–16.4 |
| IIV CL/FPRQ | 16.0 (16.3) | 11.0–21.5 | NA | NA | NA | NA |
| IIV V/FPRQ | 15.2 (24.9) | 7.52–22.5 | 19.0 (13.4) | 13.3–23.2 | 9.90 (17.4) | 6.26–12.6 |
| IIV CL/FCPRQ | 19.1 (9.99) | 15.2–22.6 | 25.9 (15.1) | 17.8–33.4 | 24.5 (13.7) | 17.8–31.0 |
| IOV MTT | 51.8 (6.05) | 44.9–58.5 | 44.1 (6.95) | 37.2–50.2 | 48.3 (6.47) | 41.1–54.6 |
| IOV Fm | 39.6 (8.37) | 31.8–46.1 | 53.4 (11.2) | 40.4–65.9 | 41.7 (10.6) | 32.1–50.8 |
| RUVPRQ | 42.6 (6.17) | 37.0–48.4 | 26.5 (8.20) | 22.3–31.0 | 37.4 (7.26) | 32.1–43.5 |
| RUVCPRQ | 21.4 (8.33) | 18.1–25.2 | 17.5 (11.1) | 13.7–21.4 | 16.8 (7.34) | 14.3–19.3 |
| RUVPRQ,CPRQ | 27.2 (8.83) | 22.1–31.6 | 14.1 (11.8) | 10.7–17.1 | 20.6 (11.8) | 15.9–25.3 |
RUV, additive residual variability; NA, data not available.
Population mean, IIV and IOV were computed from NONMEM. IIV and IOV are presented as [exp(estimate) − 1]1/2 × 100.
RSE and 95% CI were computed using the non-parametric bootstrap method of the final pharmacokinetic models (n = 1000).
All secondary parameters of primaquine and carboxyprimaquine calculated from final population pharmacokinetic models are presented in Table 2. Interactions with all three tested antimalarial treatments resulted in a significantly higher Cmax and a significantly shorter t1/2 of racemic primaquine. A significant increase in AUC of racemic primaquine and a decrease in AUCCPRQ/AUCPRQ were observed when primaquine was co-administered with pyronaridine/artesunate, but not with the other antimalarials.
Table 2.
Secondary parameters of primaquine and carboxyprimaquine from the final population pharmacokinetic models
| Secondary parameters, median (range) | Primaquine alone | Primaquine co-administered with antimalarial drugs |
||
|---|---|---|---|---|
| chloroquine | dihydroartemisinin/ piperaquine | pyronaridine/artesunate | ||
| Racemic | ||||
| Cmax PRQ (ng/mL) | 113 (48.7–219) | 120 (71.7–197)** | 131 (56.4–219)** | 144 (94.5–292)** |
| Tmax PRQ (h) | 1.96 (0.721–3.67) | 1.73 (0.745–3.33)** | 1.31 (0.784–2.97) | 1.35 (0.744–2.51) |
| t1/2 PRQ (h) | 6.66 (4.51–9.02) | 6.11 (4.60–7.77)** | 5.88 (4.51–7.10)** | 5.74 (4.16–7.20)** |
| AUC0–48 PRQ (ng·h/mL) | 1190 (629–2680) | 1170 (745–2300) | 1160 (506–2280) | 1370 (648–2900)* |
| AUC0–∞ PRQ (ng·h/mL) | 1200 (638–2730) | 1180 (750–2340) | 1170 (510–2300) | 1380 (648–2910)* |
| Cmax CPRQ (ng/mL) | 1040 (725–1550) | 1070 (865–1560)** | 1060 (798–1440)** | 1220 (781–1630)** |
| Tmax CPRQ (h) | 10.5 (5.62–15.7) | 9.84 (7.54–14.6)* | 9.83 (5.62–15.0) | 8.87 (7.06–11.2)** |
| t1/2 CPRQ (h) | 15.6 (10.2–23.2) | 16.6 (10.2–20.8) | 15.3 (12.6–23.2) | 13.8 (12.2–19.2) |
| AUC0–48 CPRQ (ng·h/mL) | 35 800 (21 600–55 400) | 33 500 (24 500–49 200)** | 33 600 (22 000–50 400)* | 34 600 (20 600–53 800)** |
| AUC0–∞ CPRQ (ng·h/mL) | 43 700 (24 500–73 100) | 45 400 (28 500–63 600)** | 41 700 (24 900–64 500)** | 45 700 (24 500–73 300)** |
| AUCCPRQ/AUCPRQ | 35.2 (13.7–66.2) | 35.9 (14.4–56.5) | 36.8 (24.4–64.2) | 32.9 (16.0–41.8)* |
| (+)-S-enantiomer | ||||
| Cmax PRQ (ng/mL) | 77.9 (36.1–145) | NA | 89.5 (36.1–148)** | 106 (60.1–198)** |
| Tmax PRQ (h) | 1.52 (0.869–3.00) | NA | 1.37 (0.816–2.77) | 1.35 (0.718–2.53) |
| t1/2 PRQ (h) | 7.07 (4.53–9.33) | NA | 6.21 (4.87–7.84)** | 6.03 (4.08–8.38)** |
| AUC0–48 PRQ (ng·h/mL) | 870 (385–1960) | NA | 886 (372–1810) | 1000 (402–2110)** |
| AUC0–∞ PRQ (ng·h/mL) | 878 (386–2020) | NA | 890 (372–1840) | 1010 (403–2130)** |
| Cmax CPRQ (ng/mL) | 51.2 (28.1–86.6) | NA | 55.8 (39.9–93.6)** | 52.7 (30.4–84.0)** |
| Tmax CPRQ (h) | 10.7 (5.21–18.7) | NA | 9.59 (5.21–17.7)** | 10.0 (5.79–13.0)** |
| t1/2 CPRQ (h) | 13.0 (9.31–25.5) | NA | 12.8 (8.78–25.5) | 13.0 (9.31–22.0) |
| AUC0–48 CPRQ (ng·h/mL) | 1660 (753–3120) | NA | 1710 (1030–3240)** | 1690 (760–3130)** |
| AUC0–∞ CPRQ (ng·h/mL) | 1980 (816–4340) | NA | 1990 (1080–4290)** | 1990 (816–4360)** |
| AUCCPRQ/AUCPRQ | 1.98 (1.35–4.40) | NA | 2.16 (1.31–4.44) | 1.86 (1.21–3.14)** |
| (−)-R-enantiomer | ||||
| Cmax PRQ (ng/mL) | 46.1 (22.6–77.5) | NA | 54.1 (22.6–77.5)** | 57.2 (43.3–101)* |
| Tmax PRQ (h) | 1.49 (0.851–3.13) | NA | 1.25 (0.762–2.87) | 1.41 (1.05–2.17) |
| t1/2 PRQ (h) | 4.66 (3.81–5.81) | NA | 3.84 (3.36–4.56)** | 4.26 (3.81–5.07) |
| AUC0–48 PRQ (ng·h/mL) | 349 (193–600) | NA | 328 (151–481) | 383 (246–660) |
| AUC0–∞ PRQ (ng·h/mL) | 350 (193–600) | NA | 328 (151–481) | 384 (246–660) |
| Cmax CPRQ (ng/mL) | 989 (671–1460) | NA | 983 (771–1320)** | 1120 (719–1480)** |
| Tmax CPRQ (h) | 8.70 (5.69–13.9) | NA | 8.23 (5.16–12.5)* | 8.19 (6.77–10.1)** |
| t1/2 CPRQ (h) | 18.0 (14.5–37.9) | NA | 17.9 (14.5–37.9) | 17.2 (14.8–25.3) |
| AUC0–48 CPRQ (ng·h/mL) | 33 100 (19 800–51 400) | NA | 30 800 (20 000–46 100)** | 33 600 (20 400–52 200) |
| AUC0–∞ CPRQ (ng·h/mL) | 42 100 (22 800–74 200) | NA | 40 900 (22 800–66 600)** | 45 000 (23 400–74 200) |
| AUCCPRQ/AUCPRQ | 109 (72.2–200) | NA | 130 (74.4–254) | 113 (68.7–180) |
Secondary parameter estimates were calculated from the individual empirical Bayes post hoc estimates of the primary pharmacokinetic parameters.
AUCCPRQ/AUCPRQ, metabolite/parent drug AUC ratio; NA, data not available.
P < 0.05
P < 0.01 by Wilcoxon signed rank test.
Co-administration with dihydroartemisinin/piperaquine and pyronaridine/artesunate resulted in a significantly higher Cmax of both (+)-S-primaquine and (−)-R-primaquine, and a significantly shorter t1/2 of (+)-S-primaquine. Only co-administration of dihydroartemisinin/piperaquine resulted in a significantly shorter t1/2 of (−)-R-primaquine. The AUC of (+)-S-primaquine was increased significantly by co-administration with pyronaridine/artesunate, which had led to a decrease in AUCCPRQ/AUCPRQ.
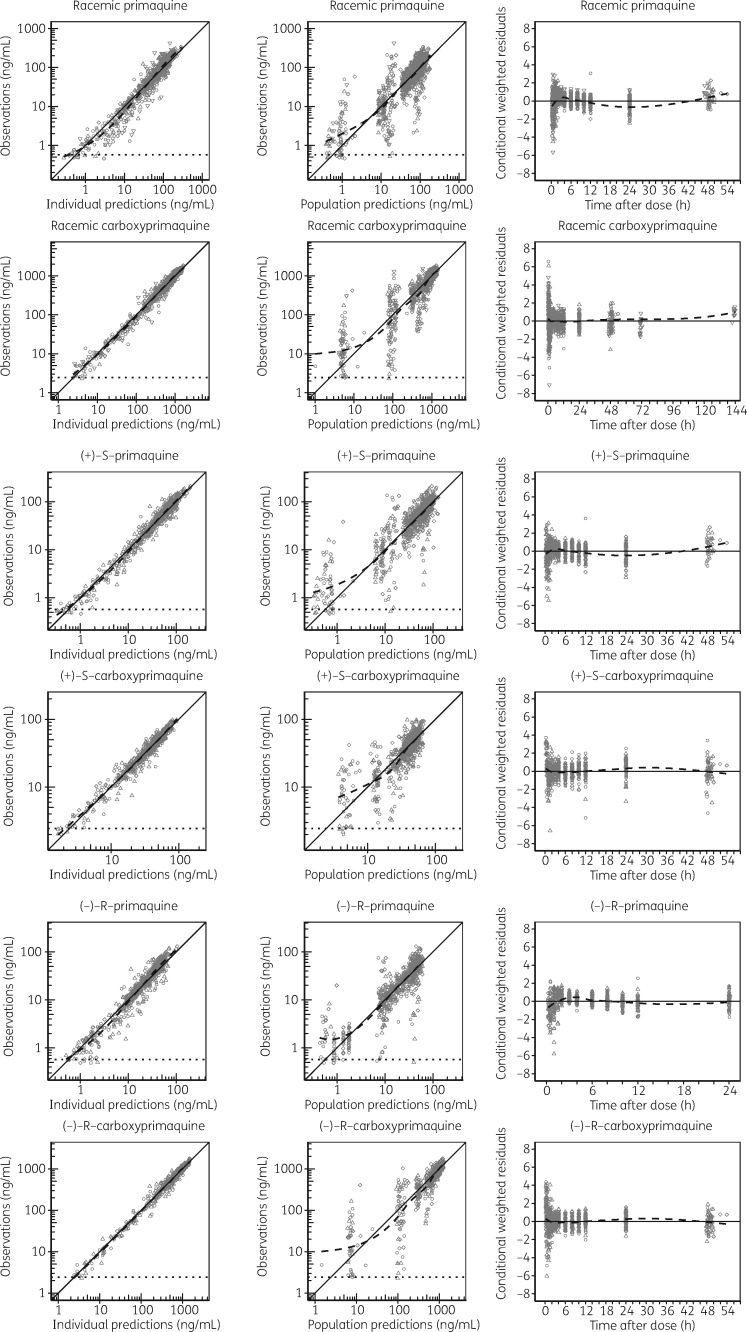
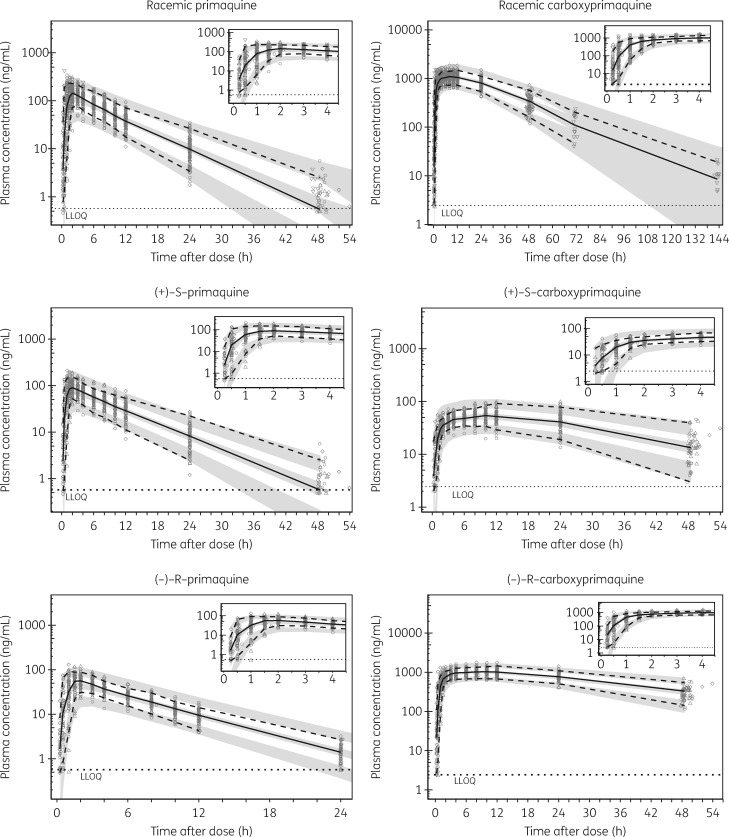
In all three models, IIV and IOV estimates <10% were fixed to zero variability and omitting them did not affect the overall model fits. The final pharmacokinetic model parameters were estimated with good precision (Table 1). Relatively low η shrinkages (<15%) were observed for all of the included random effects except IIV on V/FPRQ (24.6%) in the racemic model. The ε shrinkages were generally low (<10%) for both primaquine and carboxyprimaquine in racemic and enantiomeric models. GOF plots showed no major model misspecification (Figure 2). VPCs of the final population models are shown in Figure 3. The final population parameter estimates with 95% CIs obtained from the bootstrap analysis are shown in Table 1.
Figure 2.
GOF diagnostics of the final population pharmacokinetic models of racemic and enantiomeric primaquine and carboxyprimaquine. The observed concentrations, population predictions and individual predictions are presented on a logarithmic (base 10) scale. Open circles, observed data for primaquine alone; downward triangles, observed data for primaquine with chloroquine; upward triangles, observed data for primaquine with dihydroartemisinin/piperaquine; diamonds, observed data for primaquine with pyronaridine/artesunate; broken lines, locally weighted least-squares regressions; solid lines, lines of identity; dotted horizontal lines, LLOQ.
Figure 3.
VPCs of the final population pharmacokinetic models describing racemic and enantiomeric primaquine and carboxyprimaquine. Open circles, observed data for primaquine alone; downward triangles, observed data for primaquine with chloroquine; upward triangles, observed data for primaquine with dihydroartemisinin/piperaquine; diamonds, observed data for primaquine with pyronaridine/artesunate; solid lines, 50th percentiles of the observed data; broken lines, 5th and 95th percentiles of the observed data; shaded areas, 95% CIs of simulated (n = 2000) 5th, 50th and 95th percentiles; dotted horizontal lines, LLOQ. Concentrations are displayed on a logarithmic (base 10) scale.
Drug–drug interactions
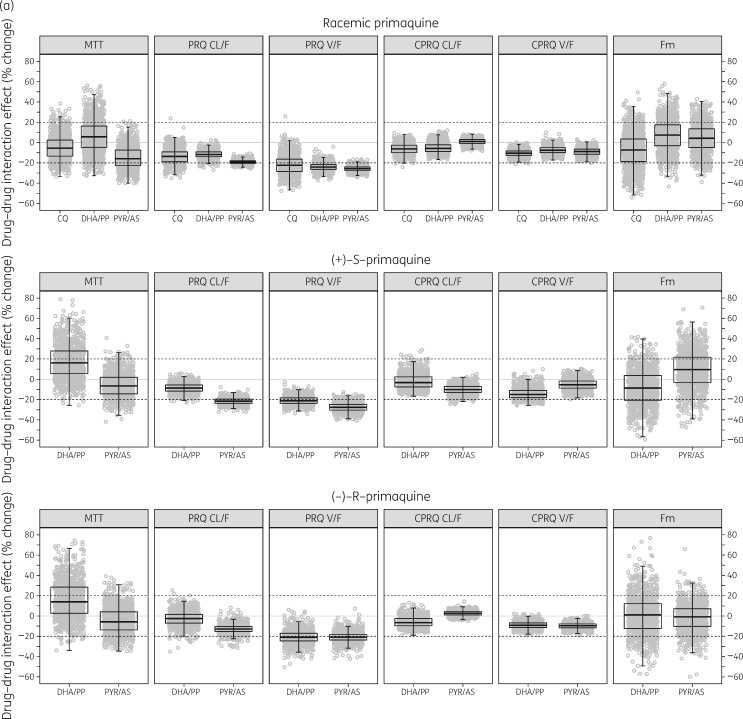
Co-administration of primaquine with other antimalarial treatments was found to affect the pharmacokinetic properties of primaquine significantly, but it did not alter the pharmacokinetics of carboxyprimaquine. The effects of each blood-stage antimalarial co-administration were evaluated further using a full covariate approach and bootstrap diagnostics, as shown in Figure 4(a and b).
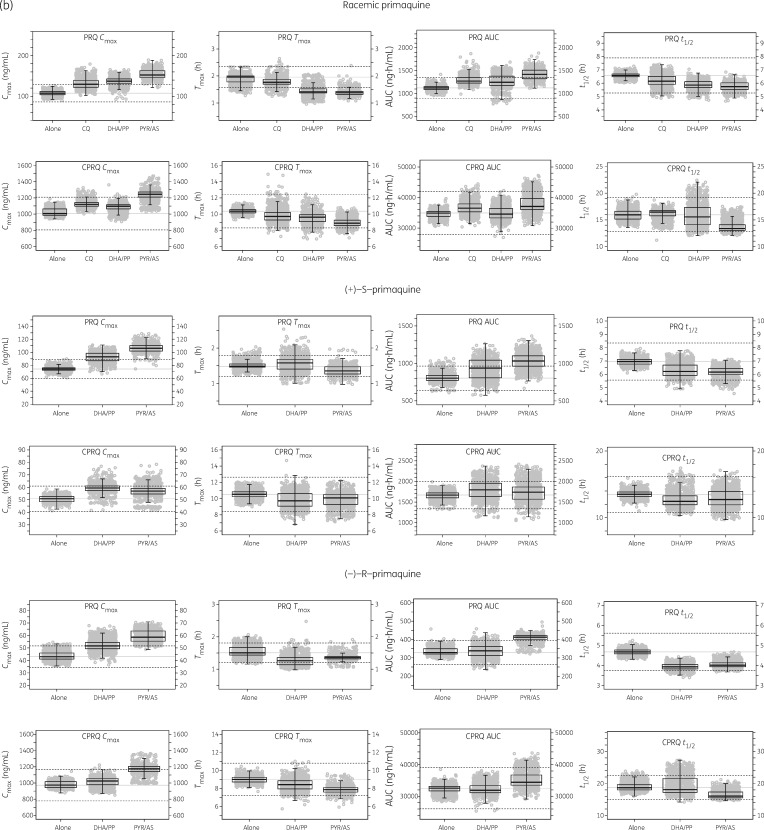
Figure 4.
Effect of co-administration of chloroquine (CQ), dihydroartemisinin/piperaquine (DHA/PP) or pyronaridine/artesunate (PYR/AS) when evaluated on (a) primary and (b) secondary pharmacokinetic parameters of primaquine and carboxyprimaquine using a full covariate approach. The dashed lines represent ±20% differences in parameter estimates caused by co-administration of antimalarial drugs.
The V/F of racemic primaquine was decreased by a median (95% CI) of 22.0% (2.24%–39.9%), 24.0% (15.0%–31.5%) and 25.7% (20.3%–31.1%) in the presence of chloroquine, dihydroartemisinin/piperaquine and pyronaridine/artesunate, respectively. The CL/F of racemic primaquine decreased by a median of 19.1% (14.5%–22.8%) in the presence of pyronaridine/artesunate. This resulted in a significant increase in Cmax and AUC of primaquine and carboxyprimaquine.
The V/F of (+)-S-primaquine was decreased by a median of 21.0% (12.0%–28.8%) and 27.6% (18.2%–35.9%) in the presence of dihydroartemisinin/piperaquine and pyronaridine/artesunate, respectively. The CL/F of (+)-S-primaquine was decreased by a median of 21.5% (14.9%–27.5%) in the presence of pyronaridine/artesunate.
The V/F of (−)-R-primaquine was decreased by a median of 20.8% (5.93%–34.8%) and 20.9% (9.61%–31.8%) in the presence of dihydroartemisinin/piperaquine and pyronaridine/artesunate, respectively. However the CL/F of (−)-R-primaquine was not significantly affected by the presence of dihydroartemisinin/piperaquine or pyronaridine/artesunate. It was not possible to evaluate the enantiospecific impact of chloroquine since only racemic concentrations were measured in this clinical study.
Discussion
This study described successfully the pharmacokinetic properties of racemic and enantiomeric primaquine and its major metabolite, carboxyprimaquine, after oral administration of primaquine racemate using a population pharmacokinetic modelling approach. We confirmed that the pharmacokinetics of primaquine, but not carboxyprimaquine, were altered when it was co-administered with chloroquine, dihydroartemisinin/piperaquine or pyronaridine/artesunate to healthy volunteers.
The structural model, comprising a series of transit absorption compartments followed by one disposition compartment for primaquine and one disposition compartment for carboxyprimaquine, was able to characterize adequately the plasma concentration profiles of a racemic mixture and both (+)-S- and (−)-R-enantiomers. Including pre-systemic metabolism of primaquine in the model improved the model fit significantly, consistent with a recent published pharmacokinetic study.35
Clearance and volume parameters were allometrically scaled by body weight, which has also been described in previously published pharmacokinetic models of primaquine.32,35
The data presented here show that the pharmacokinetic properties of primaquine are enantiospecific. (−)-R-primaquine was found to have relatively higher CL/F and V/F than (+)-S-primaquine, resulting in a 2-fold higher exposure to (+)-S-primaquine, consistent with previous reports.21,36 However, there were substantially different exposures to the carboxyprimaquine enantiomers, resulting in a 1000-fold higher exposure to (−)-R-carboxyprimaquine compared with (+)-S-carboxyprimaquine. Considering that MAO-A, based on in vitro recombinant enzyme assays, is responsible for the formation of carboxyprimaquine, this observation may suggest that (+)-S-primaquine would be a more important substrate than (−)-R-primaquine for formation of an active metabolite through the CYP2D6-mediated metabolism pathway (assuming approximately similar clearance and volume values for enantiomeric carboxyprimaquine). Chloroquine, dihydroartemisinin and pyronaridine have all been reported to inhibit CYP2D6 activity, though with different degrees of inhibition capacity.37–39 Consistent with these reports, we found that co-administration of chloroquine, dihydroartemisinin/piperaquine and pyronaridine/artesunate caused a small reduction in overall clearance, with pyronaridine/artesunate showing the largest inhibition (19.3%). The interaction effect was more pronounced on the metabolism of (+)-S-primaquine compared with that of the (−)-R-enantiomer. This finding is consistent with the earlier report of enantiomeric preference in the metabolism of primaquine.20
In addition, co-administration of these drugs also caused a decrease in V/FPRQ. This effect was more prominent with (+)-S-primaquine compared with (−)-R-primaquine. Possible explanations include tissue binding site competition or inhibition of transporter-mediated uptake into cellular compartments impeding tissue accumulation of primaquine. Although this interaction might cause reduced primaquine transport to infected hepatocytes and subsequently result in the decrease in active metabolite exposure by preventing metabolism, potentiation of the anti-relapse efficacy of primaquine by chloroquine was reported by Alving et al.40 Since primaquine is a substrate of the efflux transporter P-glycoprotein (P-gp), inhibition of P-gp on the surface of hepatocytes by co-administered drugs would be expected to retain primaquine in the liver.41
On the other hand, inhibition of the P-gp transporter in the luminal membrane of the small intestine might also promote primaquine absorption into enterocytes, which could explain the reduced MTT of primaquine when co-administered with the P-gp inhibitor pyronaridine. However, this increase in absorption was minimal as primaquine is normally well absorbed.7
Accurate prediction of potential drug–drug interactions of primaquine is challenging as it has been shown to interact with other commonly used antimalarial drugs through several drug transporters and the CYP2D6 enzyme pathway, all of which are potentially affected by genetic polymorphisms and therefore increase IIV pharmacokinetics. This suggests that the efficacy and safety of primaquine-containing combination therapies should be assessed further in individuals with these polymorphisms.
The first limitation of the study is that the active metabolite(s) of primaquine, responsible for its main pharmacodynamic effects, is unknown and there is still not a clear understanding of the enantioselective metabolism of primaquine. This limits the ability of pharmacokinetic models to predict antiparasitic efficacy and haematological toxicity due to drug–drug interactions. Second, it was not possible to evaluate the enantiospecific interaction effect mediated by co-administration with chloroquine since the enantiomeric plasma concentrations were not available. However, a relatively higher inhibitory effect of chloroquine on (+)-S-primaquine than on the (−)-R-enantiomer might be expected based on observations from previous in vitro studies using recombinant human CYP2D6 and MAO incubations.42 Chloroquine inhibited (+)-S-primaquine metabolism to form CYP2D6-dependent metabolites but it did not affect MAO-mediated metabolism. Lastly, this study was performed in healthy volunteers, who may not be representative for assessing the effects of acute drug–drug interactions in patients. Despite this, a previous study confirmed that co-administration of primaquine (30 mg of base daily for 14 days) with either dihydroartemisinin/piperaquine (standard daily dosing of 120 mg of dihydroartemisinin and 960 mg of piperaquine phosphate for 3 days) or pyronaridine/artesunate (single daily dosing of 540 mg of pyronaridine phosphate and 180 mg of artesunate for 3 days) did not compromise primaquine safety or efficacy for radical cure of vivax malaria in patients.43
Conclusions
We have successfully characterized the pharmacokinetic properties of racemic and enantiomeric primaquine and carboxyprimaquine using population pharmacokinetic modelling. The pharmacokinetic properties of the enantiomers showed substantial differences in the exposure to primaquine and carboxyprimaquine enantiomers. All evaluated antimalarial drugs showed a drug–drug interaction with primaquine, resulting in an ∼10%–30% increase in exposure to primaquine. This pharmacokinetic interaction was also enantiospecific. The developed pharmacokinetic models presented here can be used for further assessment of efficacy and safety of primaquine and other antimalarial drug combination therapies in future studies.
Acknowledgements
We thank the volunteers for their participation, and the personnel at the Clinical Therapeutics Unit for collecting the pharmacokinetic samples. The diligent staff of the Department of Clinical Pharmacology, Mahidol-Oxford Tropical Medicine Research Unit are acknowledged for conducting drug measurements. We are very grateful to Prof Larry Walker (National Center for Natural Product Research, University of Mississippi) for the kind donation of internal standards.
Funding
This work was supported by the Wellcome Trust and was part of the Wellcome Trust Mahidol University-Oxford Tropical Medicine Research Programme. Study II and Study III were partially funded by the Medicines for Malaria Venture (MMV). This work was partly funded by the Bill & Melinda Gates Foundation. The funders had no part in the study design, implementation and analysis of the results or the decision to publish this manuscript.
Transparency declarations
None to declare.
References
- 1. WHO. Guidelines for the Treatment of Malaria—Third Edition 2015. http://www.who.int/malaria/publications/atoz/9789241549127/en/.
- 2. White N, Qiao L, Qi G. et al. Rationale for recommending a lower dose of primaquine as a Plasmodium falciparum gametocytocide in populations where G6PD deficiency is common. Malar J 2012; 11: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO. WHO Global Malaria Programme: Policy Brief on Single-Dose Primaquine as a Gametocytocide in Plasmodium falciparum Malaria 2015. http://www.who.int/entity/malaria/publications/atoz/who_htm_gmp_2015.1.pdf?ua=1.
- 4. Bancone G, Chowwiwat N, Somsakchaicharoen R. et al. Single low dose primaquine (0.25mg/kg) does not cause clinically significant haemolysis in G6PD deficient subjects. PLoS One 2016; 11: e0151898.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goncalves BP, Tiono AB, Ouedraogo A. et al. Single low dose primaquine to reduce gametocyte carriage and Plasmodium falciparum transmission after artemether-lumefantrine in children with asymptomatic infection: a randomised, double-blind, placebo-controlled trial. BMC Med 2016; 14: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dicko A, Brown JM, Diawara H. et al. Primaquine to reduce transmission of Plasmodium falciparum malaria in Mali: a single-blind, dose-ranging, adaptive randomised phase 2 trial. Lancet Infect Dis 2016; 16: 674–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mihaly GW, Ward SA, Edwards G. et al. Pharmacokinetics of primaquine in man. I. Studies of the absolute bioavailability and effects of dose size. Br J Clin Pharmacol 1985; 19: 745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pybus BS, Sousa JC, Jin X. et al. CYP450 phenotyping and accurate mass identification of metabolites of the 8-aminoquinoline, anti-malarial drug primaquine. Malar J 2012; 11: 259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Constantino L, Paixao P, Moreira R. et al. Metabolism of primaquine by liver homogenate fractions. Evidence for monoamine oxidase and cytochrome P450 involvement in the oxidative deamination of primaquine to carboxyprimaquine. Exp Toxicol Pathol 1999; 51: 299–303. [DOI] [PubMed] [Google Scholar]
- 10. Brossi A, Millet P, Landau I. et al. Antimalarial activity and inhibition of monoamine oxidases A and B by exo-erythrocytic antimalarials. Optical isomers of primaquine, N-acylated congeners, primaquine metabolites and 5-phenoxy-substituted analogues. FEBS Lett 1987; 214: 291–4. [DOI] [PubMed] [Google Scholar]
- 11. Bates MD, Meshnick SR, Sigler CI. et al. In vitro effects of primaquine and primaquine metabolites on exoerythrocytic stages of Plasmodium berghei. Am J Trop Med Hyg 1990; 42: 532–7. [DOI] [PubMed] [Google Scholar]
- 12. Ganesan S, Tekwani BL, Sahu R. et al. Cytochrome P(450)-dependent toxic effects of primaquine on human erythrocytes. Toxicol Appl Pharmacol 2009; 241: 14–22. [DOI] [PubMed] [Google Scholar]
- 13. Pybus BS, Marcsisin SR, Jin X. et al. The metabolism of primaquine to its active metabolite is dependent on CYP 2D6. Malar J 2013; 12: 212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bowman ZS, Oatis JE, Whelan JL. et al. Primaquine-induced hemolytic anemia: susceptibility of normal versus glutathione-depleted rat erythrocytes to 5-hydroxyprimaquine. J Pharmacol Exp Ther 2004; 309: 79–85. [DOI] [PubMed] [Google Scholar]
- 15. Potter BMJ, Xie LH, Vuong C. et al. Differential CYP 2D6 metabolism alters primaquine pharmacokinetics. Antimicrob Agents Chemother 2015; 59: 2380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bennett JW, Pybus BS, Yadava A. et al. Primaquine failure and cytochrome P-450 2D6 in Plasmodium vivax malaria. N Engl J Med 2013; 369: 1381–2. [DOI] [PubMed] [Google Scholar]
- 17. Baker JK, McChesney JD.. Differential metabolism of the enantiomers of primaquine. J Pharm Sci 1988; 77: 380–2. [DOI] [PubMed] [Google Scholar]
- 18. Bortocan R, Bonato PS.. Enantioselective analysis of primaquine and its metabolite carboxyprimaquine by capillary electrophoresis. Electrophoresis 2004; 25: 2848–53. [DOI] [PubMed] [Google Scholar]
- 19. Fasinu PS, Avula B, Tekwani BL. et al. Differential kinetic profiles and metabolism of primaquine enantiomers by human hepatocytes. Malar J 2016; 15: 224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fasinu PS, Tekwani BL, Nanayakkara NP. et al. Enantioselective metabolism of primaquine by human CYP2D6. Malar J 2014; 13: 507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tekwani BL, Avula B, Sahu R. et al. Enantioselective pharmacokinetics of primaquine in healthy human volunteers. Drug Metab Dispos 2015; 43: 571–7. [DOI] [PubMed] [Google Scholar]
- 22. Hanboonkunupakarn B, Ashley EA, Jittamala P. et al. Open-label crossover study of primaquine and dihydroartemisinin-piperaquine pharmacokinetics in healthy adult Thai subjects. Antimicrob Agents Chemother 2014; 58: 7340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jittamala P, Pukrittayakamee S, Ashley EA. et al. Pharmacokinetic interactions between primaquine and pyronaridine-artesunate in healthy adult Thai subjects. Antimicrob Agents Chemother 2015; 59: 505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pukrittayakamee S, Tarning J, Jittamala P. et al. Pharmacokinetic interactions between primaquine and chloroquine. Antimicrob Agents Chemother 2014; 58: 3354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chotsiri P, Wattanakul T, Hoglund RM. et al. Population pharmacokinetics and electrocardiographic effects of dihydroartemisinin-piperaquine in healthy volunteers. Br J Clin Pharmacol 2017; 83: 2752–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beal S, Sheiner LB, Boeckmann A. et al. NONMEM User’s Guides (1989-2009). Ellicott City, MD, USA: Icon Development Solutions, 2009. [Google Scholar]
- 27. Lindbom L, Pihlgren P, Jonsson EN.. PsN-Toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed 2005; 80: 241–57. [DOI] [PubMed] [Google Scholar]
- 28. Lindbom L, Ribbing J, Jonsson EN.. Perl-speaks-NONMEM (PsN)—a Perl module for NONMEM related programming. Comput Methods Programs Biomed 2004; 75: 85–94. [DOI] [PubMed] [Google Scholar]
- 29. Jonsson EN, Karlsson MO.. Xpose—an S-PLUS based population pharmacokinetic/pharmacodynamic model building aid for NONMEM. Comput Methods Programs Biomed 1999; 58: 51–64. [DOI] [PubMed] [Google Scholar]
- 30. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2014. http://www.R-project.org/. [Google Scholar]
- 31. Savic RM, Jonker DM, Kerbusch T. et al. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn 2007; 34: 711–26. [DOI] [PubMed] [Google Scholar]
- 32. Moore BR, Salman S, Benjamin J. et al. Pharmacokinetic properties of single-dose primaquine in Papua New Guinean children: feasibility of abbreviated high-dose regimens for radical cure of vivax malaria. Antimicrob Agents Chemother 2014; 58: 432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Savic RM, Karlsson MO.. Importance of shrinkage in empirical Bayes estimates for diagnostics: problems and solutions. AAPS J 2009; 11: 558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bergstrand M, Hooker AC, Wallin JE. et al. Prediction-corrected visual predictive checks for diagnosing nonlinear mixed-effects models. AAPS J 2011; 13: 143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goncalves BP, Pett H, Tiono AB. et al. Age, weight, and CYP2D6 genotype are major determinants of primaquine pharmacokinetics in African children. Antimicrob Agents Chemother 2017; 61: pii: e02590–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saunders D, Vanachayangkul P, Imerbsin R. et al. Pharmacokinetics and pharmacodynamics of (+)-primaquine and (−)-primaquine enantiomers in rhesus macaques (Macaca mulatta). Antimicrob Agents Chemother 2014; 58: 7283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morris CA, Pokorny R, Lopez-Lazaro L. et al. Pharmacokinetic interaction between pyronaridine-artesunate and metoprolol. Antimicrob Agents Chemother 2014; 58: 5900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lancaster DL, Adio RA, Tai KK. et al. Inhibition of metoprolol metabolism by chloroquine and other antimalarial drugs. J Pharm Pharmacol 1990; 42: 267–71. [DOI] [PubMed] [Google Scholar]
- 39. Asimus S, Elsherbiny D, Hai TN. et al. Artemisinin antimalarials moderately affect cytochrome P450 enzyme activity in healthy subjects. Fundam Clin Pharmacol 2007; 21: 307–16. [DOI] [PubMed] [Google Scholar]
- 40. Alving AS, Arnold J, Hockwald RS. et al. Potentiation of the curative action of primaquine in vivax malaria by quinine and chloroquine. J Lab Clin Med 1955; 46: 301–6. [PubMed] [Google Scholar]
- 41. Jin X, Luong TL, Reese N. et al. Comparison of MDCK-MDR1 and Caco-2 cell based permeability assays for anti-malarial drug screening and drug investigations. J Pharmacol Toxicol Methods 2014; 70: 188–94. [DOI] [PubMed] [Google Scholar]
- 42. Fasinu PS, Tekwani BL, Avula B. et al. Pathway-specific inhibition of primaquine metabolism by chloroquine/quinine. Malar J 2016; 15: 466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nelwan EJ, Ekawati LL, Tjahjono B. et al. Randomized trial of primaquine hypnozoitocidal efficacy when administered with artemisinin-combined blood schizontocides for radical cure of Plasmodium vivax in Indonesia. BMC Med 2015; 13: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]