Abstract
Non-syndromic low frequency sensorineural hearing loss (LFSNHL) affecting only 2000 Hz and below is an unusual type of hearing loss that worsens over time without progressing to profound deafness. This type of LFSNHL may be associated with mild tinnitus but is not associated with vertigo. We have previously reported two families with autosomal dominant LFSNHL linked to adjacent but non-overlapping loci on 4p16, DFNA6 and DFNA14. However, further study revealed that an individual with LFSNHL in the DFNA6 family who had a recombination event that excluded the DFNA14 candidate region was actually a phenocopy, and consequently, DFNA6 and DFNA14 are allelic. LFSNHL appears to be genetically nearly homogeneous, as only one LFSNHL family is known to map to a different chromosome (DFNA1). The DFNA6/14 critical region includes WFS1, the gene responsible for Wolfram syndrome, an autosomal recessive disorder characterized by diabetes mellitus and optic atrophy, and often, deafness. Herein we report five different heterozygous missense mutations (T699M, A716T, V779M, L829P, G831D) in the WFS1 gene found in six LFSNHL families. Mutations in WFS1 were identified in all LFSNHL families tested, with A716T arising independently in two families. None of the mutations was found in at least 220 control chromosomes with the exception of V779M, which was identified in 1/336 controls. This frequency is consistent with the prevalence of heterozygous carriers for Wolfram syndrome estimated at 0.3–1%. An increased risk of sensorineural hearing loss has been reported in such carriers. Therefore, we conclude that mutations in WFS1 are a common cause of LFSNHL.
INTRODUCTION
Non-syndromic sensorineural hearing loss affecting high frequencies is overall a common disorder known to be genetically heterogeneous. In contrast, low frequency sensorineural hearing loss (LFSNHL) is an unusual type of hearing loss in which frequencies at 2000 Hz and below are predominantly affected. Many patients with LFSNHL have tinnitus which is not particularly bothersome, but there are otherwise no associated features such as vertigo (l–3). Because high frequency hearing is generally preserved, LFSNHL patients retain excellent understanding of speech, although presbycusis or noise exposure may cause high frequency loss later in life. Consequently, LFSNHL is often asymptomatic, and many patients choose not to wear hearing aids. Although over 70 loci have been mapped in various types of non-syndromic sensorineural hearing loss, only a few (DFNA1, DFNA6 and DFNA14) are associated with LFSNHL. LFSNHL linked to DFNAl is caused by mutations in D1APH1, the homolog of Drosophila diaphanous (4). In contrast to LFSNHL associated with DFNA6/14, all affected individuals in the DFNAl family become profoundly deaf by the fourth decade of life (4,5).
DFNA6 was mapped in a large kindred from the United States (Family 59) with autosomal dominant LFSNHL (1,3), and DFNA14 was mapped in a family (Family T) of similar phenotype from The Netherlands (2.6). Previously, we reported that DFNA6 and DFNA14 map to adjacent but nonoverlapping regions on 4pl6, separated by 1.3 cM (6). We have subsequently identified two additional families withLFSNHL linked to DFNA6/14 (unpublished data). Thus, with the exception of one DFNA1 family, all described LFSNHL families are linked to 4p16.
WFS1, the gene responsible for Wolfram syndrome type 1, is located on 4p 16 near marker D4S431 and is thus, a possible candidate gene (7,8). Wolfram syndrome is also known as DIDMOAD to denote the association of diabetes insipidus, diabetes mellitus, optic atrophy and deafness (9). Juvenile- onset diabetes mellitus and bilateral progressive optic atrophy are the minimal diagnostic criteria for Wolfram syndrome (10). Other features include renal tract abnormalities, ataxia, peripheral neuropathy, mental retardation and psychiatric illness (11,12).
Hearing loss associated with Wolfram syndrome is typically a high frequency sensorineural hearing loss, although low frequencies may become affected as well (13,14). Hearing loss confined to the low frequencies is not described in association with Wolfram syndrome, although many reports do not specify the frequencies affected. The age of onset is typically between 5 and 15 years, similar to the age of onset of hearing loss associated with DFNA6 and DFNA14 (11).
It has been suggested that heterozygous carriers in Wolfram syndrome families are predisposed to hearing loss (15). Such carriers may be fairly common in the general population, with an estimated prevalence between 0.28 and 1% (9,11). Thus, we investigated whether or not heterozygous mutations in WFS1 cause non-syndromic LFSNHL in families linked to DFNA6/14. In addition, because of the near genetic homogeneity for LFSNHL, we also studied probands with LFSNHL from small families w’ithout a priori evidence of linkage to 4pl6.
RESULTS
DFNA6 and DFNA14 are allelic
Considering that LFSNHL is relatively rare, we found it perplexing that two families with very similar phenotypes were found to map to adjacent but non-overlapping loci. To investigate this further, we expanded the pedigree of Family 59. The audiogram of a newly ascertained individual with LFSNHL typical of the family is shown (Fig. 1). The haplotype analysis of key individuals in Family 59 for markers on 4p 16 is presented in Figure 2. A key recombination event, shared by II:1, II:3 and III:1, occurs between D4S827 and D4S431, defining a centro-meric boundary. III:7, who was not available for the previous study, is recombinant at D4S2354 and telomeric, phase unknown for D4S827 and D4S431, and non-recombinant for D4S2366 and centromeric markers. These recombinations define a candidate region of 600 kb, between D4S2354 and D4S431, overlapping the DFNA14 interval.
Figure 1.
Pure tone audiogram of 24-year-old female (111:7) from Family 59 with LFSNHL typical of DFNA6/14 phenotype. Circles, air conduction right ear; crosses, air conduction left ear. Masked bone conduction curves follow air conduction curves (not shown).
Figure 2.
Haplotype analysis of key individuals in Family 59 for markers on 4pl6 (telomeric to centromeric). Shaded symbols indicate those affected with LFSNHL. Open symbols indicate normal hearing. Bars indicate haplotypes with solid black bars denoting haplotype shared by affected family members. Solid black line in haplotype indicates indeterminate phase. Centromeric boundary is defined by a recombination event between D4S827 and D4S431 (II:1,II:3,III:1). The telomeric boundary is defined by a recombination event between D4S2354 and D4S827 (III:7). III:4 is shaded consistent with her affection status prior to linkage analysis, but III:4 does not share the haplotype common to the other affected individuals at D4S432 and below. Note that III:4 and her unaffected sister III:5 share the same maternal haplotype.
These findings contradicted previous results (3) based on data from individual 111:4 (Fig. 2). Because III:4 was classified as affected prior to linkage analysis, the recombination event between D4S412 and D4S432 places the DFNA6 interval telomeric to D4S432. The telomeric boundary of DFNA 14, D4S3023, is 1.3 cM proximal to D4S432, and thus, the results from III:4 implied that DFNA6 and DFNA14 did not overlap (6). Ascertainment of III:7, who is unambiguously affected, implied that III:4 may not be truly affected, since the recombination events would predict mutually exclusive intervals. Therefore, even though LFSNHL is rare and the chance of a phenocopy unlikely, we concluded that 111:4 is a phenocopy and that DFNA6 is allelic with DFNA 14. Since penetrance is complete (3), the recombination event in unaffected individual 111:5 supports this interpretation (Fig. 2).
Identification of other LFSNHL families
By combining the databases from the University of Michigan, The Rockefeller University and the University of Antwerp, additional families with non-syndromic LFSNHL were ascertained (Fig. 3). Family W (Fig. 3A), a Dutch family with autosomal dominant LFSNHL, segregates a common haplotype at 4p 16 (data not shown). Family 35, of Irish descent, has autosomal dominant LFNSHL linked to 4p 16, with a multipoint LOD score of 4.2 (Fig. 3B). The proband of Family 19 (Fig. 3C) has moderately severe bilateral LFSNHL with documented progression. Audiologic evaluation confirmed LFSNHL in the proband’s aunt and daughter, who was not aware of her hearing loss until she participated in this study. The proband of Family 21 is a child with unilateral LFSNHL and both parents are unaffected (Fig. 3D).
Figure 3.
Pedigrees of newly ascertained LFSNHL families. Shaded symbols indicate those affected with LFSNHL. Open symbols indicate normal hearing. (A) Family W. autosomal dominant LFSNHL. (B) Family 35, autosomal dominant LFSNHL. (C) Family 19. (D) Family 21. Arrow denotes proband.?, audiometric data not available.
Mutations in WFS1 are responsible for DFNA6/14
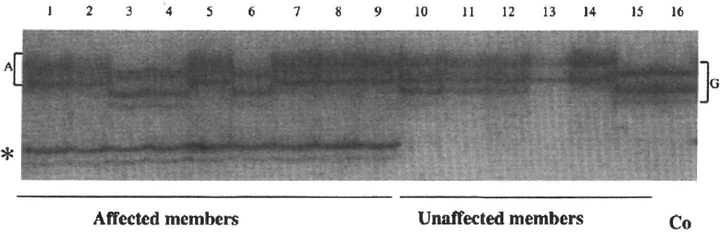
Because the candidate interval between D4S2354 and D4S431 contains the WFSI gene, we analyzed this gene for mutations in the LFSNHL families. SSCP analysis of the WFSI gene in Family 59 revealed a variant in exon 8 common to all affected family members but not seen in unaffected family members or controls (Fig. 4). Sequence analysis of exon 8 in Family 59 revealed a heterozygous, non-conservative missense mutation, 2656T->C (L829P). segregating with hearing loss (Fig. 5A and B; Table 1). Sequence analysis of the entire WFSI gene for individual 111:4 revealed six previously reported polymorphic variants (I333V, V395V, R456H, N500N, K811K, S855S) in the coding sequence as well as three intronic variants (IVS4 – 9a4g, IVS6 + 39A->G, IVS6 + 56C^T. with respect to GenBank ID AC004689.5). The absence of the L829P mutation as well as any other pathologic changes in WFSI confirms that 111:4 is a phenocopy.
Figure 4.
SSCP analysis of WFS1 exon 8–7 in Family 59. Lanes 1–9, affected family members; lanes 10–15, unaffected family members; lane 16, normal control. Note variant indicated by asterisk common to all affected family members but not seen in unaffected family members or control. Note also two distinct SSCP patterns in upper bands associated with the single nucleotide polymorphism 2735G→A (accession no. AF084481). Lane 1 shows the typical pattern associated with 2735G→A, whereas lane 16 shows the typical pattern associated with the wild-type
Figure 5.
Chromatograms demonstrating mutations in five families with LFSNHL. All chromatograms are from sequencing of forward strand. (A) Sequence from control showing wild-type nucleotide T at position 2656. The wild-type amino acid 829 is leucine. (B) Sequence of an affected individual from Family 59, showing the 2656T→C mutation. Note overlapping peaks (asterisk) indicating heterozygous mutation in comparison to the single peak in control (asterisk) shown in Figure 4A. The wild-type codon (CTG) is shown above the mutated codon (CCG). (C) Sequence of an affected individual from Family 19. Note overlapping peaks (asterisk) demonstrating the heterozygous mutation 2662G→A. (D) Sequence analysis of Family 21 demonstrating overlapping peaks (asterisk) indicative of the 2505G→A mutation. (E) Sequence analysis of an affected individual from Family T showing overlapping peaks (asterisk) indicative of the 2266C→T mutation. (F) Sequence analysis of an affected individual from Family W revealing overlapping peaks (asterisk) indicative of the 2316G→A mutation. An identical sequence change was found in Family 35 (data not shown).
Table 1.
Mutations in WFSI found in six LFSNHL families
| Family | Nationality | Nucleotide changea | WFS1 exon PCR primers | Amino acid change | Frequency in control chromosomes |
|---|---|---|---|---|---|
| T | The Netherlands | 2266C→T | 8–6 | T699M | 0/220 |
| W | The Netherlands | 2316G→A | 8–6 | A716T | 0/280 |
| 35 | United States (Irish) | 2316G→A | 8–6 | A716T | 0/280 |
| 21 | United States | 2505G→-A | 8–6 | V779M | 1/336 |
| 59 | United States | 2656T→C | 8–7 | L829P | 0/360 |
| 19 | United States | 2662G→.A | 8–7 | G831D | 0/360 |
Nucleotide numbering based on GenBank accession no. AF084481.
We then sequenced the entire WFSI gene in two Dutch LFSNHL families (Families W and T). Sequence analysis revealed mutations 2266C→T (T699M) segregating with hearing loss in Family T (Fig. 5E) and 23I6G→A (A716T) in Family W (Fig. 5F). The A716T mutation was also found to segregate with LFSNHL in Family 35. Analysis of SNPs in exon 8 for Families W and 35 showed absence of linkage disequilibrium (Table 2), suggesting that this mutation arose independently in each kindred. SSCP and sequence analysis of controls and subjects revealed multiple other known synonymous and non-synonymous variants (8,16–19).
Table 2.
Analysis of single nucleotide polymorphisms in WFSI exon 8
WFSI mutations are a common cause of low frequency hearing loss
Next, we analyzed DNA from LFSNHL patients from families without a priori evidence of linkage to DFNA6/14. SSCP variants in exon 8 for Families 19 and 21 were sequenced. A heterozygous missense mutation 2662G→A (G831D) was found in the proband of Family 19 (Fig. 4C), her aunt and her daughter, and 2505G→A (V779M) was identified in the proband of Family 21 (Fig. 4D). All five novel mutations are heterozygous missense transitions.
At least 220 control chromosomes were assayed for each variant (Table 1). Only one mutation, V779M, was found in any control. V779M abolishes a restriction site for HpyCH4III, and one of 90 anonymous controls (20) was heterozygous for the mutation.
DISCUSSION
Autosomal dominant, non-syndromic LFSNHL associated with DFNA6/14 is delayed onset, completely penetrant and slowly progressive (1–3,6). Whereas aging, noise exposure or drug toxicity can cause high frequency sensorineural hearing loss, LFSNHL appears to most commonly have a genetic etiology. The same phenotype has been described in other families with autosomal dominant inheritance (21,22). A series of 41 Japanese patients with LFSNHL included 10 familial cases (23), and 33% of 39 Danish patients with LFSNHL had a positive family history of hearing loss (24). LFSNHL is genetically homogeneous with the exception of a single DFNA1 family with a more severe phenotype (4).
We have identified WFSI as the gene responsible for LFSNHL in autosomal dominant families as well as sporadic cases. Extensive clinical evaluation of the families prior to molecular genetic analysis confirmed a non-syndromic hearing loss without associated features segregating with the hearing loss. Whereas it was not possible to specifically exclude the features of all possible hearing loss syndromes in the subjects prior to gene identification, it is clear that the affected subjects in this study do not have Wolfram syndrome, since none report juvenile-onset diabetes mellitus or optic atrophy.
The coding sequence of WFSI is 3628 bp arranged in eight exons of which the first is non-coding (7,8). Most mutations are found to affect exon 8. by far the largest (2609 bp). Wolfram syndrome is genetically heterogeneous, as some patients with Wolfram syndrome lack mutations in WFSI (7). A second locus (WFS2) has been mapped to 4q22–24 (25), which might be a candidate locus for other deafness genes. Because of genetic heterogeneity and the frequency of non- synonymous variants in the general population, the significance of missense mutations may be difficult to interpret.
A significant proportion of Wolfram syndrome patients are homozygous for WFSI null alleles (16,17). Several Wolfram syndrome patients have been found to be compound heterozygotes for a null allele and a missense mutation (7,8,16). Homozygous missense mutations in WFSI have rarely been reported in Wolfram syndrome patients. Known examples include two affected siblings homozygous for P724L (8), and one patient homozygous for P885L, five amino acids from the C-terminus (16).
Whereas most features of Wolfram syndrome are seen only in homozygotes, heterozygous carriers of Wolfram syndrome have an increased incidence of psychiatric illness, including severe depression, suicide attempts, psychosis, dementia and violent behavior (12,26,27). Several missense mutations in WFSI have been identified in patients with isolated psychiatric disorders but not in normal controls (17). Whereas LFSNHL patients in this study were not specifically evaluated for psychiatric illness, psychological testing and a comprehensive self-report questionnaire excluded psychiatric symptoms in the majority of Family 59 (1).
The five mutations presented here have not been previously described in the literature or in controls with one exception. In our study, V779M was found in 1/336 control chromosomes. Because the positive control is anonymous, no phenotypic information such as hearing status is available (20). Given the expected frequency of Wolfram syndrome carriers in the general population (9,11), finding a mutation at a very low frequency rate among controls is not unexpected. Such carriers have been shown to have an increased risk of hearing loss, although the hearing loss did not segregate as a Mendelian trait in the family studied (15). Detailed audiometric and psychiatric evaluation of heterozygous carriers of Wolfram syndrome would be useful to draw genotype-phenotype correlations.
WFSI encodes a protein of 890 amino acids, wolframin, with nine putative transmembrane domains. Wolframin lacks homology to other known proteins, and its function is currently unknown (7,8). A role in protein sorting or trafficking is hypothesized, as wolframin has been localized to the endoplasmic reticulum in cultured cells (28). WFSI is ubiquitously expressed, but brain expression is concentrated in certain populations of neurons, including the ventral cochlear nucleus and inferior colliculus. Whereas WFSI expression in the cochlea would be expected, given that mutations in WFSI cause both non-syndromic LFSNHL as well as hearing loss associated with Wolfram syndrome, the expression pattern of WFSI in the peripheral auditory system is not currently known. Neuropathologic studies of Wolfram syndrome revealed loss of nerve fibers in the cochlear nerves as well as mild neuronal loss and gliosis in the auditory brainstem (29). Further studies are necessary to determine why the mutations described herein selectively affect low frequency hearing, whereas the hearing loss associated with Wolfram syndrome has been reported to affect high frequencies.
Whereas mutations that cause Wolfram syndrome in the recessive state may occur nearly anywhere in the protein, all mutations in LFSNHL families were found in the portion of exon 8 that encodes the C-terminal domain. None of the mutations that cause hearing loss involves the N-terminal domain or any of the nine putative transmembrane domains. Current models of the wolframin protein predict a membrane topology in which the C-terminus is located on the cytoplasmic side of the endoplasmic reticulum membrane (16). Since the function of the wolframin protein remains completely unknown, the mechanism by which these mutations cause isolated low frequency hearing loss is yet unknown. However, we would predict that replacing a leucine with a proline would severely disrupt any secondary structure of the protein, and replacing the smallest amino acid, glycine, with a charged residue like aspartic acid would likewise be deleterious. All five of the amino acids affected by these mutations are conserved in human, rat and mouse, and both 829Leu and 799Val are conserved in Drosophila. These mutations might act in a dominant-negative fashion by interfering with or enhancing the specific function of the C-terminal domain, such as protein phosphorylation or protein-protein interactions.
The A716T mutation was found in both Family W and Family 35 and likely arose independently, since the alleles carrying the mutations have different nucleotides at three nearby SNPs. Three mutations (A716T, V779M, T699M) occurred in CpG nucleotides, which are known to be highly mutable in the human genome (30). It is unknown whether or not these mutations found in LFSNHL patients would cause the complete Wolfram syndrome in the homozygous state.
Despite their family history, many family members were not aware of their hearing loss until noise exposure or aging caused loss of hearing in the high frequencies as well. Because high frequencies important for speech discrimination are preserved, isolated LFSNHL may be otherwise asymptomatic. LFSNHL is difficult to diagnose by routine newborn screening methods, as typical screening protocols utilize stimuli of 2000 Hz and above. For these reasons, we suspect that LFSNHL is likely more frequent in the general population than currently recognized.
We have defined a subset of non-syndromic sensorineural hearing loss affecting the low frequencies without progression to profound deafness in which all individuals studied were found to have WFS1 mutations. Thus, we conclude that mutations in WFS1 are a common cause of low frequency sensorineural hearing loss.
MATERIALS AND METHODS
Patients
Four families with autosomal dominant non-syndromic LFSNHL had a priori evidence of linkage to 4pl6: Families 59 (1,3) and 35 (unpublished data) from the United States, and Families T (2,6) and W (unpublished data) from The Netherlands. Probands with LFSNHL from two additional families (Families 19 and 21) were identified without previous evidence of linkage to 4pl6. The Institutional Review Board of each institution approved these studies and written informed consent was obtained from each participant.
Genetic mapping
Patient DNA was prepared using standard methodology from peripheral lymphocytes or buccal epithelial cells (PureGene, Gentra). For fine mapping in Family 59, we genotyped polymorphic markers on 4p 16.3–4p 16.1: D4S169, D4S227, D4S43, D4S127, D4S412, D4S432, D4S3023. D4S2285, D4S2354, D4S827, D4S2354, D4S431, D4S2366, D4S2935, D4S394 and D4S3009. Marker locations were specified according to databases at the Whitehead Institute for Genomic Research, the National Center of Biotechnology Information and the Genome Database web sites. Polymorphisms were amplified by PCR with [γ−32P]ATP-labeled oligonucleotide primers. PCR reactions consisted of 20 ng of DNA template, 1 μl of l × PCR buffer II (Perkin Elmer), 0.8 μl each of 10 mM of each dNTP, 0.125 μal of 20 μM each forward and reverse primer, 0.6 μtl of 25 mM MgCl2 and 0.05 μl (0.25 U) of AmpliTaq Gold® DNA polymerase (Perkin Elmer), in a final volume of 10 μal. Conditions used for PCR were 93°C for 3 min, then 35 cycles of 93°C for 1 min, 58–62°C for 30s, 72°C for 30 s and 72°C for 10 min. An aliquot of 1 ^tl of each reaction was mixed with 2 μl of stop formamide buffer (87% formamide, 20 mM EDTA pH 8.0, 0.05% bromphenol blue, 0.05% xylene cyanol), denaturated for 5 min at 98°C and chilled on ice. An aliquot of 1 μl of each sample was loaded on a 6% polyacrylamide gel containing 42% urea and run at 60 W for 2–3 h. Gels were dried and exposed to Kodak X-OMAT film at room temperature overnight.
SSCP analysis
For Families 59, 19 and 21, all coding exons (2–8) of WFS1 were screened for variants by SSCP. Thirteen DNA fragments corresponding to each of exons 2–7 and to seven parts of exon 8 (8–1 through 8–7) were generated by PCR amplification of genomic DNA using available sequences of primers (7). PCR products were analyzed by SSCP using non-denaturing MDE gels (BioWhittaker Molecular Applications) overnight at 6 W and 25°C. PCR products that displayed SSCP patterns different from control samples were then sequenced.
Mutation analysis
The forward and reverse primers previously described were used for sequencing of both strands of the corresponding PCR fragments. Samples from Families 59, 19, 21 and 35 were sequenced with an ABI 3700 automated DNA sequencer (Applied Biosystems) at the University of Michigan DNA Sequencing Core. For Families W and T, the entire coding sequence of the WFS1 gene was sequenced using primers previously described (7) with an ABI3100 automated DNA sequencer. DNAs from Dutch controls were screened for the T699M and A716T mutations using the Snapshot kit on an automatic sequencer ABI3100.
For Families 59, 19 and 21, control samples consisted of 90 ethnically diverse samples representative of the US population of unknown hearing status from a commercially available panel (20) and 80–90 DNAs from Caucasians. Restriction digest assays were developed for each mutation to confirm the sequence, to test for segregation of the mutation with the hearing loss and to assay controls. Restriction enzymes used were HpaII (Promega, Madison, WI), HpyCH4III (New England Biolabs, Beverly, MA), and AvaII (New England Biolabs). For each assay, the relevant fragment of WFS1 exon 8 was amplified using PCR and incubated with the enzyme at 37°C for 1 h. The products were analyzed by electrophoresis on 2% agarose gels, stained with ethidium bromide and viewed under UV light.
The L829P mutation creates a unique HpaII site in exon 8–7. After digestion with HpaII, the wild-type allele yields one band of 300 bp and the mutated allele yields two bands (230, 70 bp). The V779M mutation abolishes the HpyCH4III site in exon 8–6. Digestion with HpyCH4III yields two bands (212 and 144 bp) for the wild-type allele, whereas the DNA of the mutated allele remains uncut (356 bp). The G831D mutation creates a new AvaII site in exon 8–7. After digestion with AvaII, the wild-type allele has two bands (261 and 39 bp), whereas the mutated allele yields three bands of 188, 73 and 39 bp.
Linkage disequilibrium
Sequence data from exon 8 from Family 35 and Family W were examined for the presence of known SNPs and compared to a reference sequence (AF084481).
ACKNOWLEDGEMENTS
We thank all the families for their participation and Edward Wilcox, Michael Hortsch, Margaret Lomax, Susan Barker, Pamella McMillan and Jay Hall. This work was supported by NIH grants DC00161 (M.M.L.), DC02982 (M.B.), DC03594 (S.M.L.), the University of Michigan Biomedical Research Council (M.B.), the University of Antwerp and the Flemish FWO (G.V.C.), the Starr Center for Human Genetics (S.M.L.) and the American Hearing Research Foundation (S.M.L.).
REFERENCES
- 1.Vanderbilt Hereditary Deafness Study Group (1968) Dominantly inherited low-frequency hearing loss. Arch. Otolaryngol, 88, 242–250. [PubMed] [Google Scholar]
- 2.Kunst H, Marres H. Huygen P, Van Camp G, Joosten F and Cremers C. (1999) Autosomal dominant non-syndromal low-frequency sensorineural hearing impairment linked to chromosome 4p 16 (DFNA14): statistical analysis of hearing threshold in relation to age and evaluation of vestibulo-ocular functions. Audiology, 38, 165–173. [DOI] [PubMed] [Google Scholar]
- 3.Lesperance MM, Hall JW, Bess FH, Fukushima K, Jain PK, Ploplis B, San Agustin TB, Skarka H, Smith RJ, Wills M et al. (1995) A gene for autosomal dominant nonsyndromic hereditary hearing impairment maps to 4p 16.3. Hum. Mol. Genet, 4, 1967–1972. [DOI] [PubMed] [Google Scholar]
- 4.Lynch ED, Lee MK, Morrow JE, Welcsh PL, Leon PE and King MC (1997) Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science, 278 1315–1318. [PubMed] [Google Scholar]
- 5.Leon PE, Raventos H, Lynch E, Morrow J and King MC (1992) The gene for an inherited form of deafness maps to chromosome 5q31. Proc. Natl Acad. Sci. USA, 89, 5181–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camp Van, Kunst H, Flothmann K, McGuirt W, Wauters J, Marres H. Verstreken M, Bespalova IN, Burmeister M, Van de Heyning PH et al. (1999) A gene for autosomal dominant hearing impairment (DFNA14) maps to a region on chromosome 4p 16.3 that does not overlap the DFNA6 locus. J. Med. Genet, 36, 532–536. [PMC free article] [PubMed] [Google Scholar]
- 7.Strom TM, Hortnagel K, Hofmann S. Gekeler F, Scharfe C, Rabl W, Gerbitz KD and Meitinger T. (1998) Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum. Mol. Genet, 7, 2021–2028. [DOI] [PubMed] [Google Scholar]
- 8.Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, Mueckler M. Marshall H, Donis-Keller H, Crock P et al. (1998) A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat. Genet, 20, 143–148. [DOI] [PubMed] [Google Scholar]
- 9.Fraser FC and Gunn T (1977) Diabetes mellitus, diabetes insipidus, and optic atrophy: an autosomal recessive syndrome? J. Med. Genet. 14 190–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfram DJ and Wagener HP. (1938) Diabetes mellitus and simple optic atrophy among siblings: report of four cases. Mayo Clin. Proc, 13, 715–718. [Google Scholar]
- 11.Barrett TG, Bundey SE and Macleod AF (1995) Neurodegeneration and diabetes: UK nationwide study of Wolfram (DIDMOAD) syndrome. Lancet. 346, 1458–1463. [DOI] [PubMed] [Google Scholar]
- 12.Swift RG, Sadler DB and Swift M (1990) Psychiatric findings in Wolfram syndrome homozygotes. Lancet, 336 667–669. [DOI] [PubMed] [Google Scholar]
- 13.Cremers CW, Wijdeveld PG and Pinckers AJ (1977) Juvenile diabetes mellitus, optic atrophy, hearing loss, diabetes insipidus, atonia of the urinary tract and bladder, and other abnormalities (Wolfram syndrome). A review of 88 cases from the literature with personal observations on 3 new patients. A eta Paediatr. Stand, 264, (suppl.), 1–16. [DOI] [PubMed] [Google Scholar]
- 14.Higashi K (1991) Otologic findings of DIDMOAD syndrome. Am. J. Otol, 12, 57–60. [PubMed] [Google Scholar]
- 15.Ohata T, Koizumi A, Kayo T, Shoji Y, Watanabe A, Monoh K, Higashi K, Ito S, Ogawa O, Wada Y and Takada G (1998) Evidence of an increased risk of hearing loss in heterozygous carriers in a Wolfram syndrome family. Hum. Genet, 103, 470–474. [DOI] [PubMed] [Google Scholar]
- 16.Hardy C, Khanim F. Torres R, Scott-Brown M, Seller A, Poulton J, Collier D, Kirk J, Polymeropoulos M, Latif F and Barrett T. (1999) Clinical and molecular genetic analysis of 19 Wolfram syndrome kindreds demonstrating a wide spectrum of mutations in WFS1. Am. J. Hum. Genet, 65 1279–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres R, Leroy E, Hu X. Katrivanou A, Gourzis P, Papachatzopoulou A, Athanassiadou A, Beratis S, Collier D and Polymeropoulos MH. (2001) Mutation screening of the Wolfram syndrome gene in psychiatric patients. Mol. Psychiatry-, 6, 39–43. [DOI] [PubMed] [Google Scholar]
- 18.Khanim F, Kirk J, Latif F and Barrett TG (2001) WFS1/wolframin mutations, Wolfram syndrome, and associated diseases. Hum. Mutat, 17, 357–367. [DOI] [PubMed] [Google Scholar]
- 19.Tessa A, Carbone I, Matteoli MC, Bruno C, Patrono C, Patera IP, De Luca F, Lorini R and Santorelli FM. (2001) Identification of novel WFS1 mutations in Italian children with Wolfram syndrome. Hum. Mutat, 17, 348–349. [DOI] [PubMed] [Google Scholar]
- 20.Collins FS, Brooks LD and Chakravarti A (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res, 8, 1229–1231. [DOI] [PubMed] [Google Scholar]
- 21.Konigsmark BW, Mengel M and Berlin CI (1971) Familial low frequency hearing loss. Laryngoscope, 81 759–771. [DOI] [PubMed] [Google Scholar]
- 22.Parving A, Johnsen NJ and Holm-Jensen S (1978) Dominantly inherited low-frequency hearing loss. Audiology. 17, 165–172. [DOI] [PubMed] [Google Scholar]
- 23.Iinuma T, Shitara T. Hoshino T and Kirikae I. (1967) Sensorineural hearing loss for low tones. Arch. Otolaryngol, 86 110–116. [DOI] [PubMed] [Google Scholar]
- 24.Parving A and Bak-Pedersen K (1978) Clinical findings and diagnostic problems in sensorineural low frequency hearing loss. Acta Otolaryngol., 85 184–190. [DOI] [PubMed] [Google Scholar]
- 25.El-Shanti H, Lidral AC, Jarrah N, Druhan L and Ajlouni K (2000) Homozygosity mapping identifies an additional locus for Wolfram syndrome on chromosome 4q. Am. J. Hum. Genet, 66, 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swift RG, Perkins DO, Chase CL, Sadler DB and Swift M (1991) Psychiatric disorders in 36 families with Wolfram syndrome. Am. J. Psychiatry 148, 775–779. [DOI] [PubMed] [Google Scholar]
- 27.Swift RG, Polymeropoulos MH, Torres R and Swift M (1998) Predisposition of Wolfram syndrome heterozygotes to psychiatric illness. Mol. Psychiatry, 3 86–91. [DOI] [PubMed] [Google Scholar]
- 28.Takeda K, Inoue H, Tanizawa Y, Matsuzaki Y, Oba J, Watanabe Y. Shinoda K and Oka Y. (2001) WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum. Mol. Genet, 10 477–484. [DOI] [PubMed] [Google Scholar]
- 29.Genis D, Davalos A, Molins A and Ferrer I. (1997) Wolfram syndrome: a neuropathological study. Acta Neuropathol. (Berl.) 93, 426–429. [DOI] [PubMed] [Google Scholar]
- 30.Cooper DN and Youssoufian H (1988) The CpG dinucleotide and human genetic disease. Hum. Genet, 78, 151–155. [DOI] [PubMed] [Google Scholar]