Abstract
Purpose
Warfarin is an oral anticoagulant associated with adverse reaction to drugs due to wide inter- and intra-individual dosage variability. Warfarin dosage has been related to non-genetic and genetic factors. CYP2C9 and VKORC1 gene polymorphisms affect warfarin metabolism and dosage. Due to the central role of populations’ ethnical and genetic origin on warfarin dosage variability, novel algorithms for Latin American subgroups are necessary to establish safe anticoagulation therapy.
Patients and methods
We genotyped CYP2C9*2 (c.430C > T), CYP2C9*3 (c.1075A > C), CYP4F2 (c.1297G > A), and VKORC1 (−1639 G > A) polymorphisms in 152 Colombian patients who received warfarin. We evaluated the impact on the variability of patients’ warfarin dose requirements. Multiple linear regression analysis, using genetic and non-genetic variables, was used for creating an algorithm for optimal warfarin maintenance dose.
Results
Median weekly prescribed warfarin dosage was significantly lower in patients having the VKORC1-1639 AA genotype and poor CYP2C9*2/*2,*2/*3 metabolizers than their wild-type counterparts. We found a 2.3-fold increase in mean dose for normal sensitivity patients (wild-type VKORC1/CYP2C9 genotypes) compared to the other groups (moderate and high sensitivity); 31.5% of the patients in our study group had warfarin sensitivity-related genotypes. The estimated regression equation accounted for 44.4% of overall variability in regard to warfarin maintenance dose. The algorithm was validated, giving 45.9% correlation (R2=0.459).
Conclusion
Our results describe and validate the first algorithm for predicting warfarin maintenance in a Colombian mestizo population and have contributed toward the understanding of pharmacogenetics in a Latin American population subgroup.
Keywords: genetic polymorphism, adverse drug reaction, gene frequency, anticoagulants
Video abstract
Introduction
Warfarin is an oral anticoagulant which is prescribed worldwide for the prophylaxis and treatment of several diseases such as deep venous thrombosis, pulmonary embolism, and ischemic cerebrovascular events.1–3 Although warfarin has high clinical efficacy, it has been associated with different bleeding-related hospital readmission rates.4,5 The US Food and Drug Administration (FDA) has stated that this complication has been partly due to wide inter- and intra-individual dosage variability, thereby contributing to increased incidence of adverse effects and complications, some of which can be life-threatening.1 Indeed, routine high-dose usage can lead to severe bleeding episodes.
Warfarin doses are usually established empirically and are adjusted by dose titration in line with the international normalized ratio (INR) for monitoring patients’ anticoagulant response.2,6,7 Chemically, warfarin is a racemic mixture of R- and S-enantiomers acting on the coagulation cascade by inhibiting VKORC1, which prevents clotting factor carboxylation. The S-enantiomer is metabolized by the CYP2C9 enzyme. When compared with R-enantiomer, S-warfarin has greater anticoagulant activity.6 The CYP4F2 enzyme participates in the same molecular cascade in vitamin K catabolism.7
Warfarin dosage has been related to non-genetic factors such as alcohol consumption, age, gender, and advanced liver or kidney disease.8 Genetic factors such as CYP2C9 and VKORC1 gene polymorphisms (eg, CYP2C9*2, rs1799853; CYP2C9*3, rs1057910; VKORC1 rs9923231) have been related to warfarin metabolism and dosage.2,3,6,9–12
In vitro studies have indicated that CYP2C9*2 impairs S-warfarin metabolism by 30% and *3 alleles by 90%.13 The rs9923231-VKORC1 polymorphism has been related to VKORC1 protein expression modifications and changes in warfarin sensitivity,14 while the CYP4F2 rs2108622 sequence variant has been linked to enzymatic activity modification and warfarin dose variability.15
The FDA and Clinical Pharmacogenetic Implementation Consortium (CPIC) have recommended genotyping these pharmacogenes when prescribing an initial dose of warfarin for more than 10 years now.1,16 Depending on CYP2C9 and VKORC1 alleles and non-genetic factors, some pharmaco-genetic algorithms have been described for establishing an adequate prediction for warfarin dosage in different populations, such as Asian Americans, Hispanics, Caucasians, and African-Americans.17–19
Latin Americans account for the largest recently admixed population in the world.20 México, Perú, Ecuador, and Guatemala have a greater Amerindian influence than Argentina, Puerto Rico, Costa Rica, Nicaragua, and Uruguay. These countries have ethnic influence mainly of European populations. For Colombia, high inter-population variability has been recorded.21 Colombia has ~50 million inhabitants; 49% of the population is mestizo (European and Amerindian ancestries), 37% has European origin, 10% has African ancestry, and 3.4% is Amerindian.22
Due to the central role of populations’ ethnical and genetic origin on warfarin dosage variability, novel algorithms for Latin American subgroups are necessary to establish safe anticoagulation therapy protocols. The present study has thus assessed VKORC1, CYP2C9, and CYP4F2 polymorphism impact on the variability of Colombian patients’ warfarin dose requirements and determined the frequency of individuals having sensitivity to this drug. Our results indicated that median weekly prescribed warfarin dosage was significantly lower in patients having the VKORC1-1639 AA genotype and poor CYP2C9*2/*2,*2/*3 metabolizers than their wild-type counterparts. Regarding warfarin sensitivity, we found a 2.3-fold increase in mean dose for normal sensitivity patients (wild-type VKORC1/CYP2C9 genotypes) compared to the other groups (moderate and high sensitivity); 31.5% of the patients in our study group had warfarin sensitivity-related genotypes.
We thus proposed and validated a dosage prediction algorithm for anticoagulated patients having stable INR. We evaluated the contribution of genetic and non-genetic factors to dosage variability. The estimated regression equation accounted for 44.4% of total variability in regard to warfarin maintenance dose. Our study gives the first description and validation of an algorithm for predicting warfarin dosing maintenance for a cohort of the Colombian patients and provides a starting point for future initiatives involving increased patient panel size. The results presented here should be used and monitored by doctors working in public and private medical institutions.
Patients and methods
Sampling and data collection
Patients were divided into two groups (Generation-G and Validation-V) for the study. Cohort G included 152 patients who were attending the Rosario University’s Teaching Hospital (Méderi), a third-level hospital in Bogotá (Colombia). These patients were already using warfarin and had at least three consecutive INR measurements (2.0–3.0). All eligible individuals were asked to participate in the study and those who agreed to do so signed an informed consent form. Several variables were recorded for each participant, such as demographic information, gender, anthropometrical measurements (weight, height, BMI, warfarin dose, INR measurements, and concomitant drug use (eg, amiodarone or statins) which theoretically could interact with warfarin (Table 1).
Table 1.
Patients’ characteristics in the G and V groups
| Parameters | G group | V group |
|---|---|---|
|
| ||
| Total (n=152 patients) | Total (n=87 patients) | |
|
| ||
| Age (years) | 62.67±15.34 | 62.29±12.36 |
| Gender (male: female) | 85:67 | 46:41 |
| Height (cm) | 162±0.08 | 161±0.09 |
| Weight (kg) | 69.21±13.39 | 72.1±13.54 |
| Body mass index (kg/m2) | 25.62±6.04 | 27.65±4.86 |
| Mean weekly warfarin dose (mg) | 32.02±11.68 | 29.1±12.46 |
| Diagnosis, n (%) | ||
| Heart disease | 53 (34.86%) | 48 (55.2%) |
| Deep venous thrombosis | 48 (31.57%) | 27 (31.0%) |
| Pulmonary thromboembolism | 15 (9.86%) | 13 (14.9%) |
| Cerebrovascular disease | 7 (3.94%) | 11 (12.6%) |
| Antiphospholipid syndrome | 3 (1.97%) | 3 (3.4%) |
| Others | 26 (17.10%) | 5 (5.7%) |
Subjects having INR values outside the therapeutic range and individuals suffering from renal or hepatic disease were excluded from the study.
All the experimental procedures followed in this study were approved by the Universidad del Rosario’s Ethics Committee, and the study was conducted in line with the Declaration of Helsinki (approval date: April 13, 2010; institutional review board reference DVG-088 and ABN062).
Cohort V (validation) (n=87) consisted of patients attending the Military Hospital in Bogotá (Colombia). Data on gender, weight, height, BMI, mean weekly warfarin dose, and age were collected for Cohort V. The characteristics of patients belonging to groups G and V were similar (Table 1). V-group data were only used for validating the warfarin dosage algorithm.
CYP2C9, CYP4F2, and VKORC1 genotyping
Genomic DNA was obtained from patients’ blood samples using the conventional salting-out procedure. Genomic regions encompassing CYP2C9*2 (c.430C>T), CYP2C9*3 (c.1075A>C), CYP4F2 (c.1297G>A), and VKORC1 (−1639G>A) polymorphisms were amplified by PCR, purified, and directly sequenced. Primer sequences and PCR conditions are available upon request. Wild-type sequences are those listed in the Ensembl database (https://www.ensembl.org/index.html): ENST00000260682.7 (CYP2C9), ENST00000221700.10 (CYP4F2), and ENST00000394975.2 (VKORC1). The details of the protocol can be obtained at protocols.io: http://dx.doi.org/10.17504/protocols.io.pbedije
Data analysis
CYP2C9, CYP4F2, and VKCORC1 variant allele and genotype frequencies were determined. Hardy–Weinberg equilibrium (HWE) was evaluated using a chi-squared test. Combined CYP2C9 and VKCORC1 genotype frequency was also established. The CYP4F2 was not considered in the combined genotypic analysis as it was not in HWE. To select genetic and non-genetic factors for the multiple linear regression analysis, we performed a univariate analysis for each variable (eg, age, height, weight, gender, BMI, amiarodone, VKORC1 and CYP2C9 genotypes), and those with a significant P-value were included. Multiple linear regression analysis, using genetic and non-genetic variables, was used for creating an algorithm for each patient’s optimal warfarin maintenance dose. This algorithm was then assessed and validated using the V cohort. Correlation analysis was used to determine correlation between warfarin actual and predicted (based on the algorithm) maintenance dose.
By using the clinical and genetic data of the G group, we evaluated the predictive power of some previously reported algorithms.
Those algorithms were considered according to the following features: 1) evaluation of CYP2C9*2, CYP2C9*3, and VKORC1-1639G>A (or VKORC1 1173C>T, in this case we imputed its value); 2) equations to predict stable warfarin dosing; 3) ethnicity; and 4) equations containing clinical parameters similar to our study. Three algorithms from different studies that met our inclusion criteria were finally selected from the literature: Sconce et al,23 International Warfarin Pharmacogenetics Consortium IWPC,24 and Perini et al.25 The statistical test was performed considering a P-value <0.05 as statically significant (95% CI) and a statistical power of 0.8.
Results
Demographic and clinical characteristics
Patients’ characteristics of G group are summarized in Table 1. The patients’ median age was 62.7 years (range: 26–88); 67 were females and 85 were males. The main medical indications for anticoagulation therapy were the presence of heart disease (34.9%) and deep vein thrombosis (31.6%). The study cohort’s mean weekly warfarin dosage was 32.02±11.68 mg/week; 40.1% of our patients suffered adverse drug reactions (ADRs).
CYP2C9, CYP4F2, and VKORC1 variant genotype frequency
CYP4F2 CC genotype frequency was 50.65%, CYP4F2 CT 34.21%, and CYP4F2 TT 15.13%, while VKORC1 GG genotype frequency was 32.2%, VKORC1 GA 47.6%, and VKORC1 AA 20.4% (Table 2). CYP2C9 genotype frequencies were *1/*1 80.92%, *1/*2 11.11%, *1/*3 5.92%, *2/*2 1.31%, and *2/*3 0.65%; CYP2C9*3 homozygous mutant alleles were not identified (Table 2). The CYP2C9 genotype-based predicted metabolizer frequencies were normal metabolizers (*1/*1) 81.01%, intermediate metabolizers (*1/*2, *1/*3) 17.03%, and poor metabolizers (*2/*2, *2/*3) 1.96% (Table 3). VKORC1 (P=0.38) and CYP2C9 (P=0.99) allele frequencies had HWE compared to the CYP4F2 variant which was not in HWE (P=0.02), which was thereby excluded from further analysis and from the algorithm.
Table 2.
Genotype frequencies of VKORC1, CYP2C9, and CYP4F2 variants in the study group
| Gene | Genotype | Genotype frequency |
|---|---|---|
|
| ||
| VKORC1 | GG | 32.2% (n=49) |
| GA | 47.6% (n=72) | |
| AA | 20.4% (n=31) | |
| CYP2C9 | *1/*1 | 80.92% (n=123) |
| *1/*2 | 11.11% (n=17) | |
| *1/*3 | 5.92% (n=9) | |
| *2/*2 | 1.31% (n=2) | |
| *2/*3 | 0.65% (n=1) | |
| CYP4F2 | CC | 50.65% (n=77) |
| CT | 34.21% (n=52) | |
| TT | 15.13% (n=23) | |
Table 3.
Genotype frequencies within warfarin metabolism and sensitivity groups
| Warfarin metabolism (CYP2C9)
| ||
|---|---|---|
| Metabolism | Genotype | Genotype frequency |
|
| ||
| Normal | *1/*1 | 81.01% (n= 123) |
| Intermediate | *1/*2, *1/*3 | 17.03% (n=26) |
| Poor | *2/*2 *2/*3 | 1.96% (n=3) |
|
Warfarin sensitivity (VKORCI/CYP2C9) | ||
| Sensitivity | Genotype | Genotype frequency |
| Normal | GG/*1*1, GG/*1*2, GA/*1*1 | 68.4% (n= 104) |
| Moderate | GG/*1*3, GG/*2*2, GG/*2*3, GA/*1*2, GA/*1*3, GA/2*2, AA/*1*1, AA/*1*2 | 28.9% (n=44) |
| High | GG/*3*3, GA/*2*3, GA/*3*3, AA/*1*3, AA/*2*2, AA/*2*3, AA/*3*3 | 2.6% (n=4) |
Assessing CYP2C9 and VKORC1 genotype combination regarding warfarin sensitivity and dosing
The weekly prescribed median warfarin dosage was significantly higher (1.7 times) in patients with VKORC1 GG genotypes (39.33 mg/week) compared to VKORC1 GA/AA genotype (22.95 mg/week) (P<0.0001). We found a statistically significant difference (1.6 times) regarding weekly median warfarin dose between normal (33.14 mg/week) and poor (20 mg/week) CYP2C9 metabolizers (P<0.001) (Table 4).
Table 4.
Effect of CYP2C9 and VKORC1 on mean dose (mg/week), according to genotype and sensitivity group
| Gene | Genotype | Mean dose (mg/week) | P-value | |
|---|---|---|---|---|
|
| ||||
| VKORCI | GG | 39.33 (mg/week) | 0.0001 | |
| AA | 22.95 (mg/week) | |||
| CYP2C9 | *l/*l | 33.14 (mg/week) | 0.001 | |
| *2/*2 *2/*3 | 20 (mg/week) | |||
|
| ||||
| Sensitivity groups | Mean dose (mg/week) | P-value | Adverse drug reactions | P-value |
|
| ||||
| Normal (n=104) | 35.53 (mg/week) | 0.001 | 35.6% (n=37) | 0.002 |
| Moderate (n=44) | 25.21 (mg/week) | 65.9% (n=29) | ||
| High (n=4) | 15.6 (mg/week) | 75% (n=3) | ||
We established three groups regarding predicted warfarin sensitivity (normal, moderate, and high), depending on CYP2C9 and VKCORC1 combined genotypes. The normal sensitivity group included GG/*1*1, GG/*1*2, and GA/*1*1 (VKORC1/CYP2C9) genotypes, accounting for 68.4% of the patients (G group). The moderate sensitivity group consisted of 28.9% of patients and included the GG/*1*3, GG/*2*2, GG/*2*3, GA/*1*2, GA/*1*3, GA/2*2, AA/*1*1, and AA/*1*2 genotypes (G group). The GG/*3*3, GA/*2*3, GA/*3*3, AA/*1*3, AA/*2*2, AA/*2*3, and AA/*3*3 genotypes formed the high sensitivity group (2.6% of the patients) (G group) (Table 3).
The required average dose according to combined genotypes and warfarin sensitivity was 35.5 mg/week for the normal sensitivity group, 25.2 mg/week for moderate sensitivity patients, and 15.6 mg/week for high sensitivity genotypes. The median doses for the three groups displayed statistically significant differences (P<0.001) (Table 4). We found that 75% of patients had adverse reactions in the high warfarin sensitivity groups, 65.9% in the moderate group, and 35.6% in the normal group (P=0.002).
Proposing and validating a warfarin-dosing algorithm
Multivariate stepwise linear regression analysis was used for selecting the factors involved in the warfarin maintenance dosage. The study revealed that only age, gender, and VKORC1 and CYP2C9 genotypes had a relevant influence on warfarin dosage variability. The weight, height, BMI, amiodarone treatment, and CYP4F2 genotype did not reach a statistical significance (P>0.05). The following dosing algorithm for Colombian patients was thus proposed: square root of weekly dose (mg) = 9.672 – (0.02*age) – (0.404*gender) – (0.794*VKORC1) – (0.607*CYP2C9); age was defined in years and gender was coded as 1 for men and 2 for women. VKORC1 was coded as 1 for GG, 2 for GA, and 3 for AA, while CYP2C9 was coded as 1 for *1/*1, 2 for *1/*2, *1/*3, or *2/*2, and 3 for 2*/3*.
Multivariate linear regression estimated 44.4% (R2=0.444) inter-individual variability in regard to maintenance warfarin dosage. VKORC1 explained 26% of the model’s genetic factors and CYP2C9 4%. The algorithm was validated on the V group (n=87), giving 45.9% correlation (R2=0.459) between predicted dose (using the proposed model) and that used in V-group patients (P<0.0001) (Figure 1).
Figure 1.
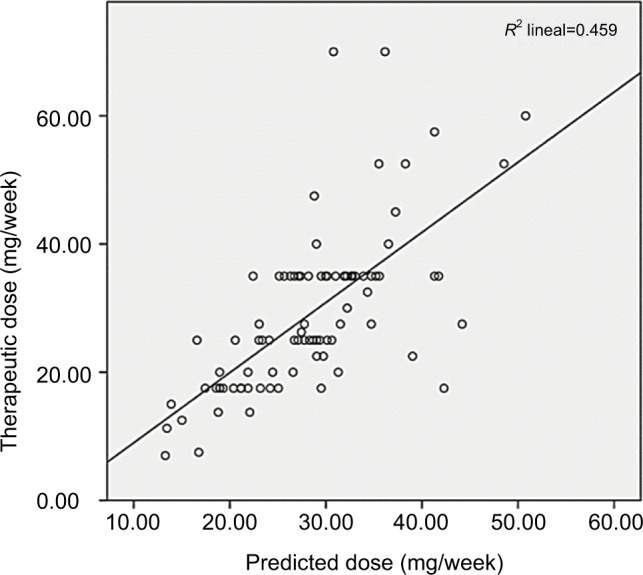
Validation of the proposed warfarin dosing algorithm in the validation group.
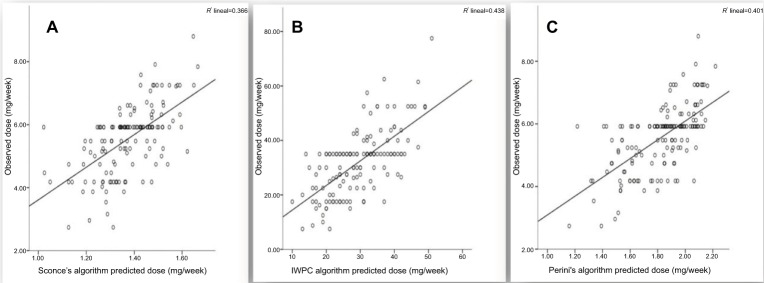
The correlation between warfarin predicted and observed doses, in the G group, with the algorithms by Sconce et al,23 Perini et al,25 and IWPC24 indicated R2 values of 36,6%, 40.1%, and 43.8%, respectively (Figure 2).
Figure 2.
Predictive power of reported algorithms in the G group.
Notes: (A) Predicted dose by Sconce’s algorithm, (B) predicted dose by IWPC algorithm, and (C) predicted dose by Perini’s algorithm.
Abbreviation: IWPC, International Warfarin Pharmacogenetics Consortium.
Discussion
Warfarin is a frequently used drug with a narrow therapeutic index and considerable inter-individual variability regarding the accurate dose required to ensure safe anticoagulation. Complications arising from inadequate warfarin dose are frequently reported to the FDA.9,26 An initial dose is usually prescribed empirically and then adjusted until stable INR is attained; patients have an increased risk of bleeding or suffering from thromboembolism during this lapse of time.1 A suitable INR value was reached in our patients using a 7.5–77.5 mg/weekly warfarin dose range, which evoked high inter-individual variability (10.3 mg/week). Considerable ranges of variability have been described in other populations (1–20 mg/day), which has been shown to be strongly related to inter-individual differences regarding genetic variants.1,27
It has been widely accepted that genotyping CYP2C9 and VKORC1 variants must be the first pharmacogenetic step for initial warfarin dosage prescription.28,29 Regarding these genes, we have identified a relationship between the weekly warfarin maintenance dose and the VKORC1-1639G>A, CYP2C9*2, and CYP2C9*3 variants. The VKORC1-1639 A allele has been associated with lower daily warfarin dose requirement.14,30–33 The warfarin dose required by our patients carrying VKORC1 GA or AA genotypes to reach a stable INR was significantly lower than that for those having the GG counterpart (P<0.0001). Most of our patients (68%) had been initially overdosed with warfarin as they carried at least one A-VKORC1 allele. Our results are consistent with the foregoing data of patients of different ethnic origins in which the mean warfarin dose (mg/week) in patients with GG genotype is significantly higher compared with that with an AA genotype (African-Americans: 45 vs 35; Hispanic-Americans: 42.5 vs 16.1; Asian: 34.3 vs 21.7, and Caucasians: 31.7 vs 15.6).23,34 Wang et al, conducted a functional study of VKORC1-1639G>A polymorphism and demonstrated that the −1639A allele was associated with twofold lower mRNA levels in human liver than the −1639G allele. The VKORC1-1639 G>A substitution creates a suppressor E-box binding site which attracts repressive E-box binding proteins.35 Moreover, this variant has been related to a quantitative change in VKORC1 expression.36 This finding is consistent with a reduced warfarin maintenance dose in patients carrying the VKORC1-1639A allele.35
The VKORC1-1639GA frequencies identified in our study were significantly different to those reported for African-American and Asian populations.37–40 VKORC1-AA genotype frequency is substantially higher in Asians (55%) compared to that in our population (20.4%) which has been partly associated with Asian patients’ reduced warfarin dose requirement.41 Conversely, this variant occurs less frequently in individuals having African ancestry (2%), partially accounting for this population’s higher warfarin dose requirements.42,43 As in our data, VKORC1-1639A homozygotes alleles in other populations (Caucasian and Hispanic) represent about 18% of allele frequency.44
Regarding CYP2C9, our results indicated that around 81% of Colombian patients carried homozygous CYP2C9*1/*1 wild-type alleles, suggesting that they were normal metabolizers. It has been stated that individuals inheriting one or two CYP2C9*2 or CYP2C9*3 copies require lower warfarin doses to reach therapeutic anticoagulation levels compared to CYP2C9 patients carrying the homozygous wild-type version (*1/*1).1 Similarly, our data showed that patients who are poor metabolizers needed a significantly lower warfarin dose to reach stable INR compared to patients carrying a wild-type genotype (P<0.001).
Concerning our G group, 19% of patients carried one or two copies of CYP2C9*2 or CYP2C9*3. It has been established that individuals carrying these genotypes are at a higher risk of bleeding during warfarin therapy compared to those carrying the CYP2C9*1 allele and require more time to achieve a stable INR.45 Such data highlighted the need for careful initial dosing of our patients carrying one or more CYP2C9 allele variants to reduce the total time needed for them to achieve therapeutic anticoagulation.
Other populations (eg, Caucasian, Middle Eastern, South Asian) have displayed CYP2C9 genetic profiles similar to those from our patients. However, African, African-American, and East Asian patients have shown different genetic profiles (0%–0.05% CYP2C9*2/*2, or *2/*3).1 African and Asian patients thus do not suffer the effect of these variants. Additional variants are associated with African-Americans’ warfarin dose requirements (eg, CYP2C9*5, *6,*8, and *11 polymorphisms) which are rare in non-African ancestry populations.45 This scenario reinforces the importance of systematic dose-guided genotyping in warfarin users.
CYP2C9 and VKORC1 combined genotypes were used to assess warfarin sensitivity in the study group (G). We determined that moderate sensitivity patients and high sensitivity patients required 1.4 times and 2.3 times fewer amounts of warfarin than the recommended initial dose (35 mg/week), respectively; 31.5% of our patients were likely to be sensitive to warfarin treatment. Previous studies have reported that combined CYP2C9 and VKORC1 functioning was essential for correct warfarin clearance which has led to determining that warfarin dosage requirement should be adjusted depending on these genes’ specific polymorphisms.46
ADR frequency was almost twice as high in our inter-mediate (75%) or high (65.9%) warfarin sensitivity groups as in patients having a normal sensitivity (35.6%), thereby highlighting the importance of genetic factors in this drug’s safety profile.
It has been reported that patients who are treated depending on their specific warfarin sensitivity genotypes reach a target stable INR earlier than patients where genetic information is lacking.47 Furthermore, patients who received genotype-guided warfarin dosing experienced fewer episodes of bleeding. These findings highlighted the importance of incorporating genomic variant analysis related to adverse effects occurring in patients who are warfarin sensitive.
Warfarin’s narrow therapeutic index and differential ethnical responses to its administration have led to the description of dose prediction algorithms incorporating genetic and non-genetic factors.8
Our study is the first to validate an algorithm in Colombian patients who are broadly considered a mestizo population.48 We found that age, gender, and polymorphisms in CYP2C9 and VKORC1 influenced warfarin dose in this sample of Colombian patients, accounting for 44.4% of warfarin dose variability required to reach a 2–3 INR. In our algorithm, the CYP2C9 genetic variant explained 4% of warfarin maintenance dose variability. VKORC1 explained 26% of such variability, while non-genetic variants accounted for 14.4%. Contrary to that reporter by others, concurrent medication such as amiodarone was not significantly associated to warfarin dose and therefore were excluded from our algorithm.49 It has been shown that by incorporating other drugs (statins, amlodipine, and diuretics) as predictor variables in algorithms, an improvement of about 5% can be achieved.25,49,50 Interestingly, other models have described that non-genetic factors have an even higher contribution (30%–40%) to warfarin dosing than the genetic factors.51–53 This might explain, at least in part, the efficiency differences on algorithms which analyze common genes in ethnically similar populations. In our study, only two out of nine non-genetic variables were included in the final algorithm (P<0.05 in the univariate analysis). Their contribution was similar to that reported by Perini et al, in Brazilian patients (13%),25 but different from that reported by other studies (30%–41%).51,52 We can infer that the inclusion of a greater number of non-genetic variables may impact on predictive analyses of ideal warfarin doses.
Regarding genetic variables, our results are in accordance with the literature. CYP2C9 and VKORC1 genotypes contribute to the inter-individual variability of warfarin dosing in European population (21%),23 Latin American population (38%),25 and Asian population (22%).52 The VKORC1 rs9923231 frequency is a major determinant of the differences observed across populations. In African population, the polymorphisms in VKORC1 and CYP2C9 explain only 11% of the warfarin dose variation.54–56 The incorporation of additional polymorphisms (eg, CALU, rs339097, CYP2C9 *8, *5, *6, and *11) could reclassify the predicted metabolic phenotypes of almost 15% of African-Americans.57 We estimate that in our mestizo patients, CYP2C9*2, CYP2C9*3, and VKORC1-1631G>A single nucleotide polymorphisms are the major genetic factors responsible for inter-individual variation in warfarin dosing.
Our algorithm generated a similar R2 value to that reported by others, including the IWPC algorithm (27 to 51%).8,58–60 Multivariate analysis in several populations has led to specific warfarin dosing algorithms being proposed which have explained 27%–67% of dosing variability.17–19,50,51,55,61–65 For instance, Limdi et al proposed that algorithms explained 40%–60% of inter-individual warfarin dose variation,66 and when these algorithms were used with individuals having Caucasian and Asian origins, they explained ~50% of warfarin dose variability; however, when used with African-Americans, they could only explain 30% of such differences.67 Interestingly, expanded genetic algorithms used for African-Americans populations, which incorporate CYP2C9 (*6 and *8), CYP4F2*3, and CALU (p.R4Q), improved the R2 value to 41%.51 In Latin America, several warfarin dosage algorithms have been described, but they are restricted to some countries/populations (eg, Brazil, Mexican American, and Puerto Rico).25,34,49,50,68 Our R2 value was similar to that of two studies performed in Brazilian (40%) and Puerto Rican patients (48%),49,69 but lower than those reported by other studies (from 51% to 70%).25,34,50,61,68 Some authors have recognized that in ethnically admixed populations, lower R2 values are obtained in warfarin dosing algorithms respect to homogeneous ancestry populations.49 It should be noted that one of the Latin American studies that has published the highest value of R2 (63.3%) included southern Brazilian patients of European ancestry.50 Other studies in Mexican American, Caribbean Latino, and Brazilian populations have developed algorithms that explain 68%, 70%, 63% of the inter-individual variability in warfarin dosing with the inclusion of CYP4F2*3, F2, CYP2C9*5, CYP2C9*6, CYP2C9*8, CYP2C9*1, and NQO1*2 genotypes plus VKORC1 haplotype and several concurrent medications.34,50,68 Taken together, those variables add about 11% to the proposed models. This might have been due to the contribution of additional polymorphisms in genes associated with warfarin metabolism. For instance, the NQO1*2 allele has displayed an increased minor allele frequency in Latin American patients, compared to other populations, and has been associated with increased warfarin dose requirements.34 The addition of CYP4F2*3 and NQO1*2 explained about 68% of warfarin dose variability in Latin American populations compared with 58% in algorithms that do not include these variants.8 These results strongly argued in favor of the clinical utility of incorporating population-specific genetic variants to those of warfarin dosing prediction. Although the CYP4F2*3 variant has been show to contribute to warfarin dose variability, our study displayed a deviation from HWE in the G group which led to exclude it from the prediction algorithm. Despite the HWE deviation, this variant has been reported in patients with acute coronary syndrome. However, only 5% of our cohort exhibited this phenotype. We suggest that an expanded population analysis with CYP4F2*3 would be necessary to verify our result.
Individuals having European ancestry have been widely studied to date regarding warfarin pharmacogenetics. For instance, the European Pharmacogenetics of Anticoagulation Therapy (EU-PACT) trial studied 455 patients (227 with genotype-guided dosing and 228 controls). Significant differences were found between groups regarding the average time to reach the therapeutic dose and INR. In the genotype-guided patients over-anticoagulation was significantly reduced.17 The Clarification of Optimal Anticoagulation through Genetics (COAG) trial adopted similar approaches in diverse populations including African-American participants. By contrast with the EU-PCT, this clinical trial revealed no improvement regarding the time taken to achieve the therapeutic dose using a pharmacogenetic algorithm. The COAG trial concluded that additional molecular polymorphisms related with warfarin dose in African-Americans should be included in pharmacogenetic algorithms.70 Despite inconsistencies in the EU-PACT and COAG trial outcomes, the CPIC included separate recommendations for ancestry-based genotype-guided therapy.1
Validation of the warfarin dosing algorithm in a different group of patients within the present study (V group) displayed a similar correlation (R2=45.9%) (P<0.0001) to that identified for the G group. We thus consider that the proposed model might be systematically used on Colombian patients carrying a similar VKORC1 and CYP2C9 genetic background to the population involved here. It is worth noting that another study on Colombian individuals reported a Hispanic ancestry-related genetic isolate (the self-designated “Paisa” community) explaining 38.3% of warfarin variability. As in our study, it concluded that VKORC1-1639 G>A was the main genetic determinant for such variability (11.2%).71
Compared with ours, the algorithm derived by Sconce et al23 (European population) showed a lower correlation (R2=36.6%) and our pharmacogenetics algorithm gave a better “ideal dose” estimates (R2=44.4% in G group and R2=45.9% in V group) in relation to the algorithms proposed by IWPC24 (R2=43.8%) and Perini et al25 (R2=40.1%) (Figure 2). Those models that involved ethnically non-homogeneous populations (IWPC and Perini) produced similar accuracy to that observed in our model. In accordance with Ramos et al,61 the predictive power of models for ideal warfarin dose may be dependent on the ethnic origin of the population, and as postulated by Suarez-Kurtz et al,72 all pharmacogenomics is highly sensitive to within-population diversity.
Bottom et al50 developed an algorithm with high predictive power (63%) for the Brazilian population of European ancestry, by evaluating patients with models obtained from an admixed Brazilian cohort (eg, white, brown, and black population);25 an inferior performance, attributed to the differences in the ancestry between patients, was observed.50 In addition, Suarez-Kurtz57 reported that the predictive power of the Bottom’s algorithm was higher in Brazilian white patients (R2=0.50) compared with that from brown and black individuals (R2=0.40). Taken together, our results permit to establish that novel algorithms for warfarin treatment, specifically designed for Colombian individuals, should predict dosing more accurately than guidelines used for other populations. However, we consider that for Latin American populations, the inclusion of other genes such as CYP4F2*3, NQO1*2, F2 rs5896 and non-genetic variables (dose-adjusted INR, admixture index) can be crucial to obtain a model with better predictive power, such as described by Duconge (70%),68 Ramos (67%),61 Bress (68%),34 and Botton (63%).50
Limitations
We consider that the main limitation of our study was the small sample size. In addition, the evaluation of only two CYP2C9 single nucleotide polymorphisms and the exclusion of the CYP4F2 gene might be related to inaccurate R2 calculations. Further studies on an expanded Colombian population are therefore necessary.
Conclusion
Taken together, our results describe the first validated algorithm for predicting warfarin maintenance in a Colombian population and have contributed toward the understanding of pharmacogenetics in a Latin American population subgroup.
Acknowledgments
The present study was supported by the Universidad del Rosario (Grant CS/Genetics/ABN062-2018 and DVG088-FIUR-2010). Laissue’s lab is supported by the Universidad del Rosario.
Footnotes
Author contributions
JMG: substantial contribution to concept and design, revising the article for important intellectual content, analysis and interpretation of data; CMR: substantial contribution to concept and design; NCC: substantial contribution to analysis and interpretation of data; CA: substantial contribution to acquisition of data; CC-O: substantial contribution to conception and design; NP: substantial contribution to acquisition of data; RAC: substantial contribution to analysis and interpretation of data; DD: substantial contribution to analysis of data, revising the article; PL: revising the article critically for important intellectual content, final approval of the version to be published; DJF: substantial contribution to concept and design, revising the article and final approval of the version to be published. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther. 2017;102(3):397–404. doi: 10.1002/cpt.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuteja S, Limdi N. Pharmacogenetics in Cardiovascular Medicine. Curr Genet Med Rep. 2016;4(3):119–129. doi: 10.1007/s40142-016-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi F, Mcginnis R, Bourgeois S, et al. A genomewide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rojas-Velandia C, Ruiz-Garzón J, Moscoso-Alcina JC, et al. Characterization of adverse drug reactions causing admission to an intensive care unit. Br J Clin Pharmacol. 2017;83(5):1134–1140. doi: 10.1111/bcp.13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shehab N, Lovegrove MC, Geller AI, Rose KO, Weidle NJ, Budnitz DS. US Emergency Department Visits for Outpatient Adverse Drug Events, 2013-2014. JAMA. 2016;316(20):2115–2125. doi: 10.1001/jama.2016.16201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari M, Pengo V, Barolo M, Bezzo F, Padrini R. Assessing the relative potency of (S)- and (R)-warfarin with a new PK-PD model, in relation to VKORC1 genotypes. Eur J Clin Pharmacol. 2017;73(6):699–707. doi: 10.1007/s00228-017-2248-9. [DOI] [PubMed] [Google Scholar]
- 7.Mcdonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol. 2009;75(6):1337–1346. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaye JB, Schultz LE, Steiner HE, Kittles RA, Cavallari LH, Karnes JH. Warfarin Pharmacogenomics in Diverse Populations. Pharmacotherapy. 2017;37(9):1150–1163. doi: 10.1002/phar.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin T, Miyata T. Warfarin dose and the pharmacogenomics of CYP2C9 and VKORC1 – rationale and perspectives. Thromb Res. 2007;120(1):1–10. doi: 10.1016/j.thromres.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Quinn AL, Liko I, Lee JC. Clinical effect of CYP2C9*5/*6 genotype on a patient’s warfarin dose requirement. Pharmacogenomics. 2017;18(11):1051–1057. doi: 10.2217/pgs-2017-0059. [DOI] [PubMed] [Google Scholar]
- 11.Wakamiya T, Hokosaki T, Tsujimoto S, et al. Effect of VKORC1, CYP2C9, CFP4F2, and GGCX Gene Polymorphisms on Warfarin Dose in Japanese Pediatric Patients. Mol Diagn Ther. 2016;20(4):393–400. doi: 10.1007/s40291-016-0212-5. [DOI] [PubMed] [Google Scholar]
- 12.Mili FD, Allen T, Wadell PW, et al. VKORC1-1639A allele influences warfarin maintenance dosage among Blacks receiving warfarin anticoagulation: a retrospective cohort study. Future Cardiol. 2018;14(1):15–26. doi: 10.2217/fca-2017-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12(3):251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 14.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 15.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111(8):4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahtani KR, Heneghan CJ, Nunan D, et al. Optimal loading dose of warfarin for the initiation of oral anticoagulation. Cochrane Database Syst Rev. 2012;12:CD008685. doi: 10.1002/14651858.CD008685.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 18.Alzubiedi S, Saleh MI. Pharmacogenetic-guided Warfarin Dosing Algorithm in African-Americans. J Cardiovasc Pharmacol. 2016;67(1):86–92. doi: 10.1097/FJC.0000000000000317. [DOI] [PubMed] [Google Scholar]
- 19.Lin M, Yu L, Qiu H, Wang Q, Zhang J, Song H. Verification of five pharmacogenomics-based warfarin administration models. Indian J Pharmacol. 2016;48(3):258–263. doi: 10.4103/0253-7613.182876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adhikari K, Mendoza-Revilla J, Chacón-Duque JC, Fuentes-Guajardo M, Ruiz-Linares A. Admixture in Latin America. Curr Opin Genet Dev. 2016;41:106–114. doi: 10.1016/j.gde.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Salzano FM, Sans M. Interethnic admixture and the evolution of Latin American populations. Genet Mol Biol. 2014;37(1 Suppl):151–170. doi: 10.1590/s1415-47572014000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Castro M, Restrepo CM. Genetics and genomic medicine in Colom-bia. Mol Genet Genomic Med. 2015;3(2):84–91. doi: 10.1002/mgg3.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106(7):2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 24.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perini JA, Struchiner CJ, Silva-Assunção E, et al. Pharmacogenetics of warfarin: development of a dosing algorithm for brazilian patients. Clin Pharmacol Ther. 2008;84(6):722–728. doi: 10.1038/clpt.2008.166. [DOI] [PubMed] [Google Scholar]
- 26.Agaba P, Kildow BJ, Dhotar H, Seyler TM, Bolognesi M. Comparison of postoperative complications after total hip arthroplasty among patients receiving aspirin, enoxaparin, warfarin, and factor Xa inhibitors. J Orthop. 2017;14(4):537–543. doi: 10.1016/j.jor.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Spall HG, Wallentin L, Yusuf S, et al. Variation in warfarin dose adjustment practice is responsible for differences in the quality of anticoagulation control between centers and countries: an analysis of patients receiving warfarin in the randomized evaluation of long-term anticoagulation therapy (RE-LY) trial. Circulation. 2012;126(19):2309–2316. doi: 10.1161/CIRCULATIONAHA.112.101808. [DOI] [PubMed] [Google Scholar]
- 28.Shaw K, Amstutz U, Kim RB, et al. Clinical Practice Recommendations on Genetic Testing of CYP2C9 and VKORC1 Variants in Warfarin Therapy. Ther Drug Monit. 2015;37(4):428–436. doi: 10.1097/FTD.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 29.Nutescu EA, Drozda K, Bress AP, et al. Feasibility of implementing a comprehensive warfarin pharmacogenetics service. Pharmacotherapy. 2013;33(11):1156–1164. doi: 10.1002/phar.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt MT, Wolfson SK, Kuller LH. Lower extremity arterial disease and the aging process: a review. J Clin Epidemiol. 1992;45(5):529–542. doi: 10.1016/0895-4356(92)90102-s. [DOI] [PubMed] [Google Scholar]
- 31.Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14(13):1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 32.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bress A, Patel SR, Perera MA, Campbell RT, Kittles RA, Cavallari LH. Effect of NQO1 and CYP4F2 genotypes on warfarin dose requirements in Hispanic-Americans and African-Americans. Pharmacogenomics. 2012;13(16):1925–1935. doi: 10.2217/pgs.12.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112(4):1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathore SS, Agarwal SK, Pande S, Mittal T, Mittal B. The impact of VKORC1-1639 G>A polymorphism on the maintenance dose of oral anticoagulants for thromboembolic prophylaxis in North India: A pilot study. Indian J Hum Genet. 2011;17(Suppl 1):S54–S57. doi: 10.4103/0971-6866.80360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dickmann LJ, Rettie AE, Kneller MB, et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol. 2001;60(2):382–387. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- 38.Cen HJ, Zeng WT, Leng XY, et al. CYP4F2 rs2108622: a minor significant genetic factor of warfarin dose in Han Chinese patients with mechanical heart valve replacement. Br J Clin Pharmacol. 2010;70(2):234–240. doi: 10.1111/j.1365-2125.2010.03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi JR, Kim JO, Kang DR, et al. Proposal of pharmacogenetics-based warfarin dosing algorithm in Korean patients. J Hum Genet. 2011;56(4):290–295. doi: 10.1038/jhg.2011.4. [DOI] [PubMed] [Google Scholar]
- 40.Yoshizawa M, Hayashi H, Tashiro Y, et al. Effect of VKORC1-1639 G>A polymorphism, body weight, age, and serum albumin alterations on warfarin response in Japanese patients. Thromb Res. 2009;124(2):161–166. doi: 10.1016/j.thromres.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Dang MT, Hambleton J, Kayser SR. The influence of ethnicity on warfarin dosage requirement. Ann Pharmacother. 2005;39(6):1008–1012. doi: 10.1345/aph.1E566. [DOI] [PubMed] [Google Scholar]
- 42.Limdi NA, Beasley TM, Crowley MR, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;9(10):1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perera MA, Gamazon E, Cavallari LH, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89(3):408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11(6):781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavallari LH. Time to revisit warfarin pharmacogenetics. Future Cardiol. 2017;13(6):511–513. doi: 10.2217/fca-2017-0061. [DOI] [PubMed] [Google Scholar]
- 46.Sangviroon A, Panomvana D, Tassaneeyakul W, Namchaisiri J. Pharmacokinetic and pharmacodynamic variation associated with VKORC1 and CYP2C9 polymorphisms in Thai patients taking warfarin. Drug Metab Pharmacokinet. 2010;25(6):531–538. doi: 10.2133/dmpk.dmpk-10-rg-059. [DOI] [PubMed] [Google Scholar]
- 47.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83(3):460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 48.Yunis JJ, Acevedo LE, Campo DS, Yunis EJ. Geno-geographic origin of Y-specific STR haplotypes in a sample of Caucasian-Mestizo and African-descent male individuals from Colombia. Biomedica. 2013;33(3):459–467. doi: 10.7705/biomedica.v33i3.807. [DOI] [PubMed] [Google Scholar]
- 49.Santos PC, Marcatto LR, Duarte NE, et al. Development of a pharmacogenetic-based warfarin dosing algorithm and its performance in Brazilian patients: highlighting the importance of population-specific calibration. Pharmacogenomics. 2015;16(8):865–876. doi: 10.2217/pgs.15.48. [DOI] [PubMed] [Google Scholar]
- 50.Botton MR, Bandinelli E, Rohde LE, Amon LC, Hutz MH. Influence of genetic, biological and pharmacological factors on warfarin dose in a Southern Brazilian population of European ancestry. Br J Clin Pharmacol. 2011;72(3):442–450. doi: 10.1111/j.1365-2125.2011.03942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez AH, Shi Y, Schildcrout JS, et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13(4):407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho HJ, On YK, Bang OY, et al. Development and comparison of a warfarin-dosing algorithm for Korean patients with atrial fibrillation. Clin Ther. 2011;33(10):1371–1380. doi: 10.1016/j.clinthera.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Jiang NX, Ge JW, Xian YQ, et al. Clinical application of a new warfarin-dosing regimen based on the CYP2C9 and VKORC1 genotypes in atrial fibrillation patients. Biomed Rep. 2016;4(4):453–458. doi: 10.3892/br.2016.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Momary KM, Shapiro NL, Viana MA, Nutescu EA, Helgason CM, Cavallari LH. Factors influencing warfarin dose requirements in African-Americans. Pharmacogenomics. 2007;8(11):1535–1544. doi: 10.2217/14622416.8.11.1535. [DOI] [PubMed] [Google Scholar]
- 55.Hernandez W, Gamazon ER, Aquino-Michaels K, et al. Ethnicity-specific pharmacogenetics: the case of warfarin in African Americans. Pharmacogenomics J. 2014;14(3):223–228. doi: 10.1038/tpj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Limdi NA, Brown TM, Yan Q, et al. Race influences warfarin dose changes associated with genetic factors. Blood. 2015;126(4):539–545. doi: 10.1182/blood-2015-02-627042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suarez-Kurtz G. Population diversity and the performance of warfarin dosing algorithms. Br J Clin Pharmacol. 2011;72(3):451–453. doi: 10.1111/j.1365-2125.2011.04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bodin L, Verstuyft C, Tregouet DA, et al. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood. 2005;106(1):135–140. doi: 10.1182/blood-2005-01-0341. [DOI] [PubMed] [Google Scholar]
- 59.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5(4):262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 60.Veenstra DL, You JH, Rieder MJ, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15(10):687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 61.Ramos AS, Seip RL, Rivera-Miranda G, et al. Development of a pharmacogenetic-guided warfarin dosing algorithm for Puerto Rican patients. Pharmacogenomics. 2012;13(16):1937–1950. doi: 10.2217/pgs.12.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho SM, Lee KY, Choi JR, Lee KA. Development and Comparison of Warfarin Dosing Algorithms in Stroke Patients. Yonsei Med J. 2016;57(3):635–640. doi: 10.3349/ymj.2016.57.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao L, Chen C, Li B, et al. Verification of pharmacogenetics-based warfarin dosing algorithms in Han-Chinese patients undertaking mechanic heart valve replacement. PLoS One. 2014;9(4):e94573. doi: 10.1371/journal.pone.0094573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei X, Guo Y, Sun J, et al. Accuracy assessment of pharmacogenetic algorithms for warfarin dose prediction in Chinese patients. Am J Hematol. 2012;87(5):541–544. doi: 10.1002/ajh.23151. [DOI] [PubMed] [Google Scholar]
- 65.Borobia AM, Lubomirov R, Ramírez E, et al. An acenocoumarol dosing algorithm using clinical and pharmacogenetic data in Spanish patients with thromboembolic disease. PLoS One. 2012;7(7):e41360. doi: 10.1371/journal.pone.0041360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28(9):1084–1097. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eby C. Warfarin pharmacogenetics: does more accurate dosing benefit patients? Semin Thromb Hemost. 2012;38(7):661–666. doi: 10.1055/s-0032-1326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duconge J, Ramos AS, Claudio-Campos K, et al. A Novel Admixture-Based Pharmacogenetic Approach to Refine Warfarin Dosing in Caribbean Hispanics. PLoS One. 2016;11(1):e0145480. doi: 10.1371/journal.pone.0145480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bosch LA. A Proposal for an Individualized Pharmacogenetic-Guided Warfarin Dosage Regimen for Puerto Rican Patients Commencing Anticoagulation Therapy. J Pharmacogenomics Pharmacoproteomics. 2014;5(1) doi: 10.4172/2153-0645.T-001. pii:T-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palacio L, Falla D, Tobon I, et al. Pharmacogenetic impact of VKORC1 and CYP2C9 allelic variants on warfarin dose requirements in a hispanic population isolate. Clin Appl Thromb Hemost. 2010;16(1):83–90. doi: 10.1177/1076029608330472. [DOI] [PubMed] [Google Scholar]
- 72.Suarez-Kurtz G, Pena SD, Hutz MH. Application of the F(ST) statistics to explore pharmacogenomic diversity in the Brazilian population. Pharmacogenomics. 2012;13(7):771–777. doi: 10.2217/pgs.12.39. [DOI] [PubMed] [Google Scholar]