Abstract
Rapid evolution in annual plants can be quantified by comparing phenotypic and genetic changes between past and contemporary individuals from the same populations over several generations. Such knowledge will help understand the response of plants to rapid environmental shifts, such as the ones imposed by global climate change. To that end, we undertook a resurrection approach in Spanish populations of the annual plant Arabidopsis thaliana that were sampled twice over a decade. Annual weather records were compared to their historical records to extract patterns of climatic shifts over time. We evaluated the differences between samplings in flowering time, a key life-history trait with adaptive significance, with a field experiment. We also estimated genetic diversity and differentiation based on neutral nuclear markers and nucleotide diversity in candidate flowering time (FRI and FLC) and seed dormancy (DOG1) genes. The role of genetic drift was estimated by computing effective population sizes with the temporal method. Overall, two climatic scenarios were detected: intense warming with increased precipitation and moderate warming with decreased precipitation. The average flowering time varied little between samplings. Instead, within-population variation in flowering time exhibited a decreasing trend over time. Substantial temporal changes in genetic diversity and differentiation were observed with both nuclear microsatellites and candidate genes in all populations, which were interpreted as the result of natural demographic fluctuations. We conclude that drought stress caused by moderate warming with decreased precipitation may have the potential to reduce within-population variation in key life-cycle traits, perhaps as a result of stabilizing selection on them, and to constrain the genetic differentiation over time. Besides, the demographic behaviour of populations probably accounts for the substantial temporal patterns of genetic variation, while keeping rather constant those of phenotypic variation.
Keywords: Arabidopsis thaliana, broad sense heritability, field experiments, flowering time, microsatellite genotyping, precipitation, resurrection approach, warming
Plant populations vary over time as a result of the effects of environmental variation on life-cycle traits and genetic diversity. It is important to quantify how much populations actually change over time to better understand the response of plants to rapid environmental shifts. Our resurrection approach indicated that populations of the annual plant Arabidopsis thaliana exhibited substantial genetic changes over just a decade. Interestingly, such changes seemed to be mediated by the combination of the extent of warming and precipitation. Nevertheless, populations remained viable over time suggesting that plants may possess the means to cope with global climate change.
Introduction
All plant populations exhibit to some extent spatio-temporal phenotypic and genetic changes as a result of environmental variation. The quantification of the pace and intensity of such phenotypic and genetic changes, as well as their impact on fitness and population persistence, is a particularly pressing issue to understand the evolutionary potential of plants in rapidly changing environments, such as the ones currently posed by global climate change (Shaw and Etterson 2012; Kopp and Matuszewski 2014). The dominant view of modern population genetics is that adaptation to changing environments is chiefly driven by the random genetic drift and selection on consistently beneficial or deleterious mutations (Messer et al. 2016). This paradigm implies a slow evolutionary process because common mutations are expected to have only small effects on fitness (Messer et al. 2016). However, important phenotypic changes, interpreted as events of rapid or contemporary evolution, have been observed in plant populations over relatively short periods of time for different plant traits, such as flowering time (Franks et al. 2007) and self-fertilization rates (Bodbyl Roels and Kelly 2011), as well as processes, such as the invasiveness of exotic species in recipient communities (Maron et al. 2004) or the dynamics of plant–pathogen interactions (Gilbert and Parker 2010). Overall, it is increasingly accepted that the standard genetic model might be insufficient to describe the rapid phenotypic adaptive changes observed in natural populations (Messer et al. 2016). Further work is clearly needed to quantify the timescales of genetic and phenotypic variation in plant populations and to infer their ecological and evolutionary consequences in a context of rapid environmental changes.
The resurrection approach represents a powerful, albeit time-consuming, means to quantify the extent of phenotypic and genetic variation in plant populations over time. In particular, the resurrection approach compares past and contemporary performance under common conditions of individuals, i.e. the ancestors and their descendants, whose seeds were collected from the same populations at different points in time encompassing several generations (Bennington et al. 1991; McGraw et al. 1991; Davis et al. 2005; Franks et al. 2007, 2008, 2018; Anderson et al. 2012; Franks and Hoffmann 2012; Nevo et al. 2012; Fukano et al. 2013; Gomulkiewicz and Shaw 2013; Sultan et al. 2013; Bustos-Segura et al. 2014; Van Dijk and Hautekèete 2014; Thomann et al. 2015; Welt et al. 2015; Etterson et al. 2016; Horgan-Kobelski et al. 2016; Kuester et al. 2016; O’Hara et al. 2016; Frachon et al. 2017). An exceptional value of the resurrection approach is that phenotypic differences in fitness-related traits between ancestors and descendants may be attributed to rapid evolution, i.e. fast genetically based evolutionary shifts imposed by environmental changes (Franks et al. 2007), which has been detected in annual plants over a few generations (Maron et al. 2004; Franks et al. 2007; Rhoné et al. 2010; Franks and Hoffmann 2012; Sultan et al. 2013; Etterson et al. 2016; Frachon et al. 2017). In order to enhance its impact and value, resurrection studies should address phenotypic variation in fitness-related traits along with neutral and functional genetic variation across generations. Whilst neutral genetic variation can provide hints on the demographic behaviour that populations experienced over time, functional genetic variation can provide insight into the genetic basis of variation in phenotypic traits.
Beyond the interest of the resurrection approach for evolutionary biology, resurrection experiments may also become extremely valuable in quantifying the responses of plant populations to climate change (Anderson et al. 2012; Franks et al. 2014; Etterson et al. 2016). Many climate change studies examine models predicting distribution range changes for a wide array of plants in climate change scenarios (Pearson and Dawson 2003; Beaumont et al. 2008; Bellard et al. 2012; Parmesan and Hanley 2015). Besides, there exists a large body of literature reporting generalized advances in plant phenology with a warming climate using existing long-term observational data sets (Peñuelas and Filella 2001; Menzel et al. 2006; Parmesan 2007; Parmesan and Hanley 2015), as well as estimating the effects of experimental warming on plant performance for various species in different biomes (Lloret et al. 2004; Walker et al. 2006; Wolkovich et al. 2012; Peñuelas et al. 2013). However, one important piece is still missing to fully comprehend the effects of climate change on plant populations and communities: the microevolutionary consequences of climate change, i.e. the genetic changes driven by selection imposed by environmental changes (Holt 1990; Karell et al. 2011). In the long term, the resurrection approach can supply this missing piece of knowledge by quantifying phenotypic and genetic variation across generations. Furthermore, if phenotypic variation in fitness-related traits between ancestors and descendants can be associated to variation in putative agents of natural selection that occurred between the points in time when seeds were collected, we will have the means to parameterize process-based models with functions affecting fitness under short- and mid-term climate change scenarios.
The main goal of this study was to quantify the extent of phenotypic and genetic temporal change in natural populations of the annual plant Arabidopsis thaliana by means of a resurrection approach. To this end, we used seeds from four well-known Spanish A. thaliana populations sampled in 2003–04 (Picó et al. 2008) and resampled in 2012–13 specifically for this study. Overall, these four populations are a good illustration of the Mediterranean therophyte community with a great diversity of annual plants including A. thaliana. We carried out a resurrection approach on A. thaliana through three specific objectives. First, we conducted a series of field experiments to estimate temporal variation in flowering time, a key developmental trait with adaptive significance affecting fitness in A. thaliana (Weinig et al. 2002; Korves et al. 2007; Ågren and Schemske 2012; Manzano-Piedras et al. 2014; Exposito-Alonso et al. 2018a). Besides, experiments were also performed to detect hidden genetic variation in flowering time in study populations over time. Second, we analysed the amount and spatio-temporal distribution of genetic variation by genotyping populations from the two samplings with neutral microsatellite loci, which provided hints on the demographic behaviour of A. thaliana populations over the study period (e.g. effective population size). Third, we quantified the spatio-temporal patterns of nucleotide diversity in well-known genes affecting life-history traits in populations from the two samplings. In particular, we sequenced FRI and FLC, two genes involved in the vernalization pathway for flowering (Koornneef et al. 1998; He et al. 2003; Kim et al. 2009; Kim and Sung 2014; Méndez-Vigo et al. 2016), and DOG1, a seed specific gene affecting seed dormancy (Alonso-Blanco et al. 2003; Bentsink et al. 2006, 2010; Kronholm et al. 2012; Chiang et al. 2013; Vidigal et al. 2016). Importantly, we overcame one of the main caveats of the resurrection approach, i.e. aging effects on stored seed collections, by simultaneously bulking up seeds from each population and sampling to remove aging and environmental maternal effects on stored old and field-collected contemporary seeds, respectively, prior to experiments. We discuss the results in the context of the added value and implications of the resurrection approach for evolutionary studies in plants in a rapidly changing world.
Methods
Source populations
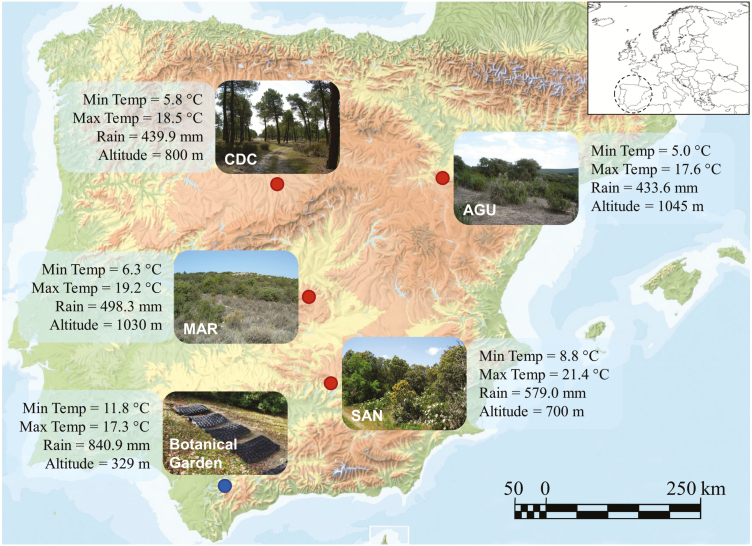
A total of four Spanish A. thaliana populations were selected for this study (Fig. 1): Aguarón (AGU; 41.32°N, 1.34°W, 1045 m a.s.l., Zaragoza province), Ciruelos de Coca (CDC; 41.21°N, 4.55°W, 800 m a.s.l., Segovia province), Marjaliza (MAR; 39.58°N, 3.93°W, 1030 m a.s.l., Toledo province) and Santa Elena (SAN; 38.33°N, 3.51°W, 700 m a.s.l., Jaén province). In these locations, A. thaliana occurs in typically Mediterranean environments (Fig. 1), such as forests and scrublands dominated by evergreen Quercus species (AGU, MAR and SAN) and sandy open sites and edges of Pinus pinaster forests (CDC). In all populations, A. thaliana occurred in patches of different size and density in the two samplings. Populations were also rather similar in terms of general weather conditions (Fig. 1), based on data available from different digital geographical databases used previously to characterize the Iberian collection of A. thaliana populations (Marcer et al. 2016). The four populations, on average separated by 267 km (range = 143.6–377.0 km), are genetically differentiated from each other (Picó et al. 2008; Méndez-Vigo et al. 2013).
Figure 1.
Map of Spain with the four Arabidopsis thaliana populations of study (red dots). A photograph of each population along with environmental characteristics of populations, such as average annual minimum temperature, average annual maximum temperature, total annual precipitation and altitude, are given. The location of the El Castillejo Botanical Garden where field experiments were conducted is also indicated (blue dot) along with a photograph of experimental blocks and the environmental characteristics of the experimental facility.
All populations were sampled twice: AGU in 2004 and 2012 (spanning 8 years), CDC in 2003 and 2012 (9 years), MAR in 2003 and 2012 (9 years) and SAN in 2003 and 2013 (10 years). In the first and second samplings, we collected seeds from 20–32 and 60–70 individuals per population, respectively. In MAR, the individuals sampled in the second sampling were collected in 2012 (N = 29) and in 2013 (N = 41), but for the sake of simplicity we did not make any distinction between them. Important for this study, the two samplings were carried out around a GPS location recorded in the first sampling characterizing a representative abundant patch within each population. To the extent possible, we took the precaution to sample A. thaliana individuals separated from each other by at least 1 m within the same area around the GPS location (~50 × 50 m2) in the two samplings.
Field-collected seeds from the first sampling were multiplied by the single seed descent method in a glasshouse from the Centro Nacional de Biotecnología (CNB-CSIC) of Madrid a few months after collection. Multiplied seeds were stored in dry conditions in cellophane bags at room temperature in darkness, storing conditions that can preserve A. thaliana seeds for years. Collected seeds from the first sampling and field-collected seeds from the second sampling were multiplied again in the same manner in a single experiment in 2014 to obtain the final collection of seeds from individuals of the first (2003–04) and second (2012–13) sampling from each population.
Environmental data
For each A. thaliana population, weather records were obtained from the Agencia Estatal de Meteorología of Spain (AEMET), including daily records of minimum and maximum temperatures and precipitation from the nearest automatic meteorological stations. We used data from 1–3 meteorological stations per population depending on their geographical distance to populations, as well as on the degree of data completeness for the study period. On average, meteorological stations were located at 28.9 km (range = 2.4–48.8 km) far from A. thaliana populations. When more than one meteorological station was used per population, i.e. MAR and SAN, we computed daily mean values across meteorological stations for minimum temperature, maximum temperature and precipitation. For each population, we also obtained historical climate data on average annual minimum temperature, average annual maximum temperature and total annual precipitation. Historical data were obtained from the Digital Climatic Atlas from the Iberian Peninsula, based on a spatial interpolation using records from a total of 2285 meteorological stations across Portugal and Spain during the period 1951–99.
Aerial orthophotographs were retrieved from different public administrations in Spain (i.e. Plan Nacional de Ortofotografía Aérea, Sistema de Información Territorial de Aragón, Portal de Mapas de la Junta de Comunidades de Castilla-La Mancha and Infraestructura de Datos Espaciales de Andalucía) and used to detect major landscape changes in A. thaliana populations over the study period [seeSupporting Information—Fig. S1]. AGU, MAR and SAN did not show any significant major natural disturbance, e.g. fires or landslides, between samplings. In contrast, CDC exhibited some partial changes in tree density [seeSupporting Information—Fig. S1]. Unlike the other populations, CDC occupies sandy open sites and edges of P. pinaster forests, which have been exploited for centuries for resin extraction. Nonetheless, A. thaliana distribution seemed not to be affected by this partial modification, probably because A. thaliana only occurred along P. pinaster forest edges. However, we ignore whether other biotic factors, e.g. herbivores and pathogens, have affected these populations over the time period of study. We can at least ensure that we did not detect any specific damage during the two samplings or in visits conducted between samplings to some of the populations, such as CDC and MAR (C. Alonso-Blanco and F. X. Picó, pers. obs.).
Field experiments
We conducted a total of three field experiments using 15 haphazardly chosen individuals per population (CDC, MAR and SAN) and sampling (3 populations × 15 individuals × 2 samplings = 90 individuals). Due to technical problems, individuals from AGU from the 2012 sampling could not be multiplied and this population was excluded from the experiments. In addition, CDC and MAR from the first sampling were eventually represented by 14 individuals. Experiments were conducted at El Castillejo Botanical Garden of Sierra de Grazalema Natural Park in south-west Spain (Fig. 1) following the same protocols used in previous successful experiments with Iberian A. thaliana accessions at the same facility (Méndez-Vigo et al. 2013; Manzano-Piedras et al. 2014; Exposito-Alonso et al. 2018a). In short, all individuals were replicated six times in all experiments (3 experiments × 88 individuals × 6 replicates = 1584 experimental units). Each replicate contained 60 filled seeds (1584 experimental units × 60 seeds = 95040 seeds) that were sown in square plastic pots (12 × 12 × 12 cm3) filled with standard soil mixture (Abonos Naturales Cejudo Baena S.L., Utrera, Spain). Previously, batches of 60 seeds were prepared 1 month before each sowing, and stored in 1.5-mL plastic tubes at room temperature in darkness until the sowing day.
Experiments were designed to quantify flowering time in field conditions of A. thaliana individuals in three different scenarios determined by sowing dates. Hence, all individuals from each population and sampling were allowed to germinate in mid autumn (sowing date: 7 October 2014), late autumn (sowing date: 5 December 2014) and mid winter (sowing date: 27 January 2015). Such sowing schedule reproduced quite well the germination behaviour observed in natural populations from different Iberian environments (Montesinos et al. 2009; Picó 2012). Our goal was to force plants from the two samplings to complete the life cycle in progressively shorter periods of time as a means to express hidden genetic variation in flowering time (see Le Rouzic and Carlborg 2008). Although late spring germination is also observed in natural populations from Mediterranean environments, sowing in spring reduces the vegetative phase in a way that A. thaliana cannot complete the life cycle at El Castillejo Botanical Garden (F. X. Picó, unpubl. data). Flowering time in experimental field conditions was estimated as the number of days between 15 days after sowing, when most seeds had already germinated in all experiments, and flowering date. Flowering date was given at the pot level when the majority of plants in the pot, which were sisters and showed homogeneous flowering behaviour, had the first flower open. In these experiments, we only focused on flowering time because this trait showed the largest quantitative genetic differentiation among populations when compared with other life-cycle traits, e.g. recruitment and fecundity (Méndez-Vigo et al. 2013). Besides, flowering time estimated for A. thaliana in field conditions strongly correlates with fitness, estimated as the product between survivorship and fecundity (Exposito-Alonso et al. 2018a).
Statistical analyses
For each experiment, general linear models were used to test the effect of sampling (fixed factor: 2003–04 and 2012–13), population (random factor: CDC, MAR and SAN) and individuals nested within population (random factor: 14–15 individuals per population) on flowering time. We inspected the distribution of the residuals to check that the assumptions of the analyses were met. For each population, sampling and experiment, we estimated the broad sense heritability of flowering time as h2 = VG/(VG + VE), where VG is the estimated among-individual variance component and VE is the residual variance (Le Corre 2005). The 95 % confidence intervals (CIs) for h2 values were computed with the (co)variances method using restricted maximum likelihood (REML) variance components (Lynch and Walsh 1998). Analyses were performed with SPSS v.23 statistical software (IBM, Chicago, IL, USA).
Genetic analyses
A total of 226 A. thaliana individuals were collected in the second sampling (2012–13). They were genotyped for 12 nuclear microsatellites (see DNA extraction and marker genotyping protocols in Picó et al. 2008; Méndez-Vigo et al. 2011). Individuals eventually used from the second sampling were distributed as follows: 53 from AGU, 58 from CDC, 70 from MAR and 45 from SAN. Microsatellite data from the second sampling and microsatellite data obtained from the first one (2003–04; Picó et al. 2008) were joined together. Microsatellite data obtained from the first sampling included a total of 103 individuals: 20, 32, 30 and 21 individuals from AGU, CDC, MAR and SAN, respectively. Hence, the overall microsatellite data set of our resurrection approach totalled 329 individuals. Ten individuals from the second sampling were genotyped twice for all microsatellites. This was used to estimate an average genotyping error rate of 0.021 per locus, similar in magnitude to the error obtained from the existing data set from the first sampling, i.e. 0.047 per locus (Picó et al. 2008). In both cases, microsatellite genotyping errors were mostly due to allele dropout at heterozygous loci. Electropherograms obtained for the microsatellite genotyping from the second sampling were visually inspected and manually scored using GeneMapper v.4.1 software (Applied Biosystems, Foster City, CA, USA).
The number of individuals genotyped per population in the second sampling more than doubled those from the first one. In order to avoid sample size effects and subsequent bias in genetic parameters, we created 100 random subsamples of 20 individuals each, which was the minimum sample size used in this study, per population and sampling. Genetic diversity was estimated with GenAlEx v.6.5 (Peakall and Smouse 2012), including observed heterozygosity (HO) and mean gene diversity (HS). We also computed outcrossing rates (OR) as (1 − FIS)/(1 + FIS), where FIS is the inbreeding coefficient. For each population, genetic differentiation was also estimated by partitioning the genetic variance among samplings, among individuals within samplings and within individuals, calculating F-statistics via the analysis of molecular variance (AMOVA) with GenAlEx. Significance of F-statistics was estimated by performing 1000 permutations. Final values for genetic diversity and genetic differentiation parameters were obtained by calculating mean values across the 100 random subsamples of equal size. Finally, we computed the number and frequency of non-redundant multilocus genotypes (NG) per population and sampling, which provided a direct measure of unique combinations of alleles per population and sampling. We used NG to perform principal coordinate analysis (PCoA) with GenAlEx for visualizing the relationship among non-redundant multilocus genotypes, revealing the genetic structure of our study system in space and time.
Given that we genotyped the same populations in two different points in time, we estimated the effective population size (Ne) using the temporal method of Nei and Tajima (1981) and Waples (1989), as implemented in NeEstimator v.2 (Do et al. 2014). The Ne parameter is computed by relating the observed amount of temporal change in allele frequency to that expected under pure genetic drift. The comparison of Ne values among populations allows the inference of the extent of genetic drift in them, assuming that the effect of systematic forces, i.e. mutation, selection and migration, was low and that sampling effects were similar in all populations (Wang 2005). We used plan II, i.e. sampling before reproduction and not replaced, to estimate the standardized variance in the temporal change of allele frequency, which is reciprocally proportional to Ne, by using all individuals from the two samplings per population and without restrictions on the lowest allele frequency. The 95 % CIs for Ne were estimated by the Jackknife method on loci.
Two flowering time genes (FRI and FLC) and one seed dormancy gene (DOG1) were sequenced in 21–23 haphazardly chosen individuals per population of the second sampling. We used existing sequence data on these genes from 9–10 individuals of the same populations from previous works corresponding to the first sampling (2003–04; Kronholm et al. 2012; Méndez-Vigo et al. 2013). These genes, known to account for variation in flowering time and seed dormancy in A. thaliana, were previously analysed in Iberian A. thaliana accessions and populations (Méndez-Vigo et al. 2011, 2013; Kronholm et al. 2012), allowing the identification of the gene regions concentrating the highest nucleotide diversity of interest. Thus, we sequenced the complete FRI gene (3.5 kb; Méndez-Vigo et al. 2011, 2013), a fragment corresponding to a 0.70-kb segment of FLC intron 1 (Méndez-Vigo et al. 2011, 2013) and a fragment corresponding to a 0.39-kb segment of DOG1 exon 1 (Kronholm et al. 2012). In all cases, between one and seven overlapping fragments of 0.5–0.7 kb were PCR-amplified using described primers (Méndez-Vigo et al. 2011, 2013; Kronholm et al. 2012). PCR products were sequenced using an ABI PRISM 3700 DNA analyser (Applied Biosystems, Foster City, CA, USA). DNA sequences were aligned using DNASTAR v.8.0 (Lasergene, Madison, WI, USA). Alignments were inspected and edited by hand with GENEDOC v.2.7.0 (Nicholas et al. 1997). Nucleotide diversity was estimated with DnaSP v.5 (Librado and Rozas 2009). Polymorphisms were used to estimate the number and frequency of non-redundant multilocus haplotypes (NH) per gene, population and sampling. We did not use random subsamples of equal size to estimate gene parameters because variation in sample sizes was much lower in genes than in microsatellites. Besides, the ratio between the number of individuals from the first and second samplings used for gene sequencing was practically the same for all populations. GenBank accession numbers of DNA sequences generated in this study from the second sampling are MF142982–MF143073 for FRI, MF142894–MF142981 for FLC and MF142804–MF142893 for DOG1. Accession numbers of gene sequences from the first sampling are available elsewhere (Kronholm et al. 2012; Méndez-Vigo et al. 2013).
Results
Climatic trends
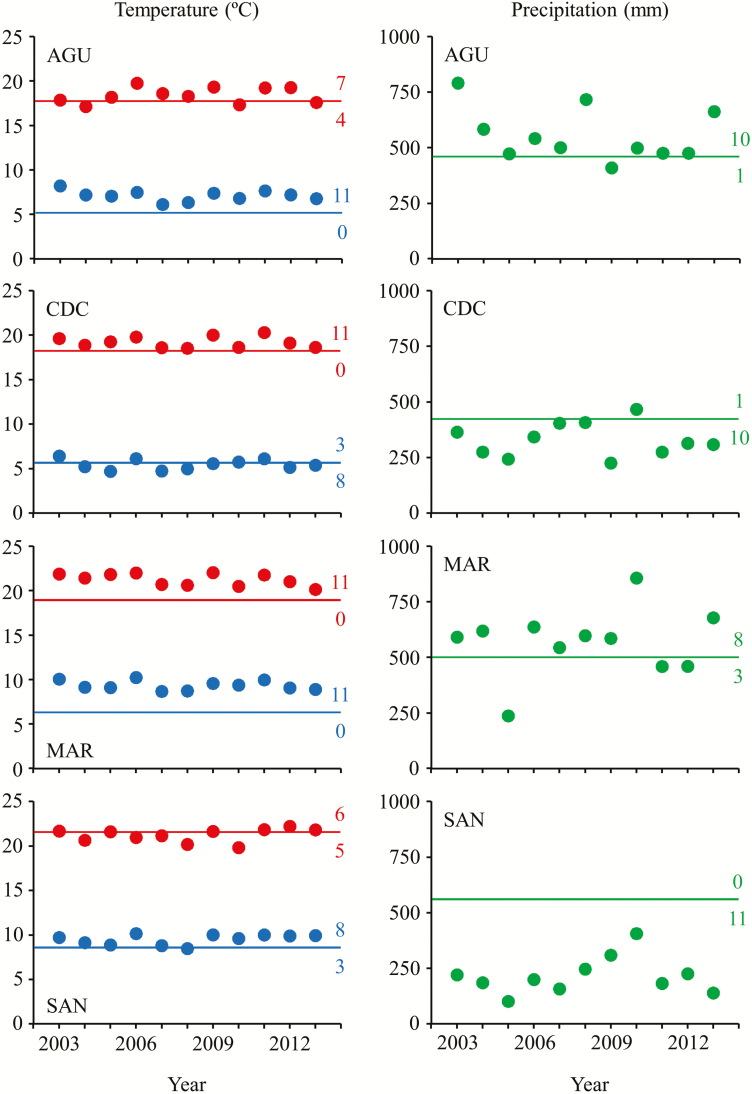
For each population, we plotted average annual minimum temperature, average annual maximum temperature and total annual precipitation recorded between 2003 and 2013 against the historical values obtained from the Digital Climatic Atlas from the Iberian Peninsula. Average annual minimum temperatures were clearly above the historical record in AGU, MAR and SAN (average ± SE range of average annual minimum temperature above the historical record = 0.52 ± 0.18–3.15 ± 0.17 °C), but not in CDC (average annual minimum temperature below the historical record = 0.33 ± 0.18 °C; Fig. 2). In the case of average annual maximum temperatures, there was a trend for annual values to surpass the historical record in AGU, CDC and MAR (average ± SE range of average annual maximum temperature above the historical record = 0.52 ± 0.28–2.06 ± 0.22 °C), but not in SAN (average annual maximum temperature below the historical record = 0.35 ± 0.24 °C; Fig. 2). Finally, substantial year-to-year variation in total annual precipitation was detected in all populations over the study period. CDC and SAN exhibited precipitation values below the historical record in practically all years (average ± SE range of total annual precipitation below the historical record = 111.89 ± 23.91–329.53 ± 26.59 mm), whereas AGU and MAR showed the opposite trend (average ± SE range of total annual precipitation above the historical record = 65.69 ± 48.80–86.71 ± 37.56 mm; Fig. 2).
Figure 2.
Average annual minimum temperatures (blue dots), average annual maximum temperatures (red dots) and annual total precipitation (green dots) for each Arabidopsis thaliana population for the period 2003–13. For each weather record, lines indicate the historical value corresponding to the period 1950–99, extracted from the Digital Climatic Atlas from the Iberian Peninsula. For the sake of clarity, the number of years above and below the historical records is also given.
Flowering time
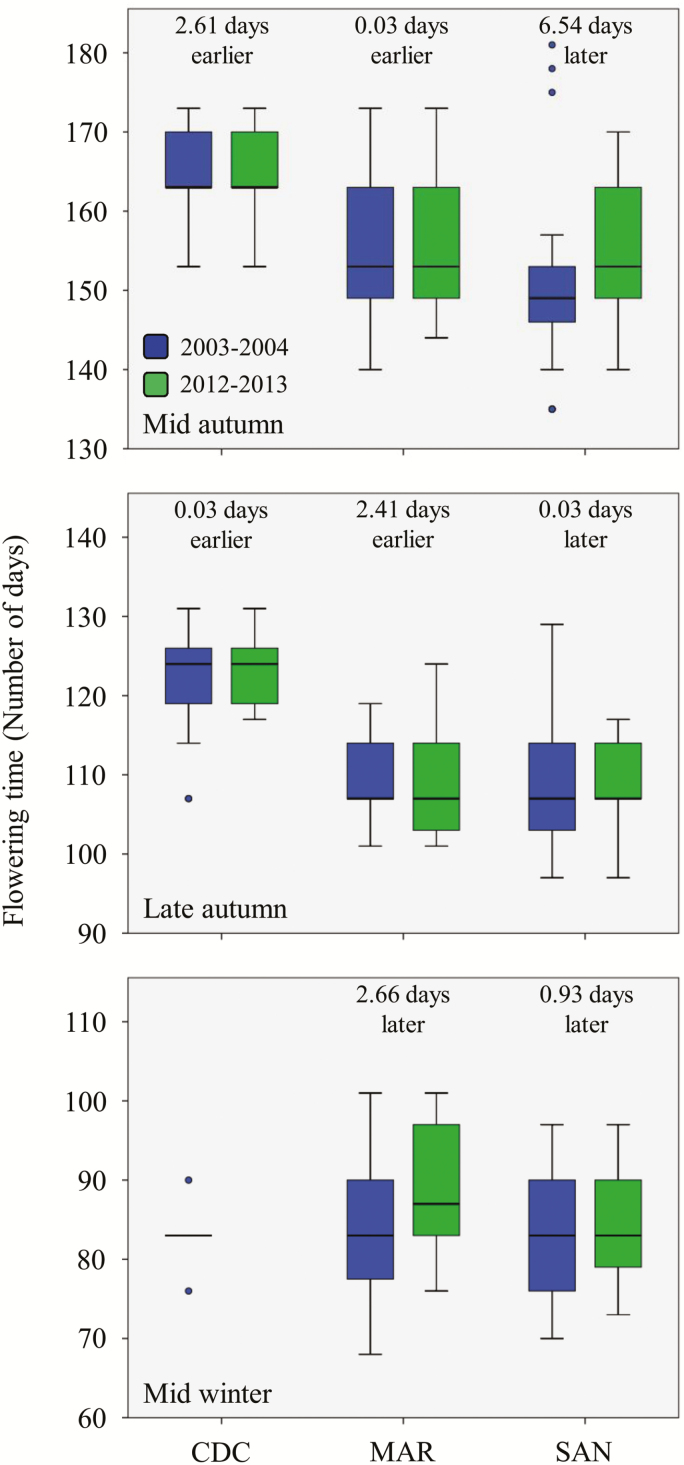
We conducted field experiments to quantify variation in flowering time of A. thaliana individuals between the two samplings. We carried out three sequential experiments, 2 months apart from each other, differing in sowing date, which forced all individuals to complete their life cycle in increasingly shorter periods of time. All individuals from all populations and samplings completed the life cycle in the mid autumn and late autumn sowing experiments. In the mid winter sowing experiment, all but one individual from CDC from the first sampling failed to complete the life cycle and died before reproduction (Fig. 3), only one individual from MAR from the first sampling did not reach maturity and all individuals from SAN from both samplings completed the life cycle.
Figure 3.
Summary statistics for flowering time. For each experiment and sampling, boxes show the lower and upper quartiles, whiskers are drawn down to the 10th percentile and up to the 90th, the line is the median of observations and hollow dots indicate outliers. For the sake of clarity, the mean difference in flowering time between samplings is also given. Only one CDC individual from the first sampling and none from the second sampling flowered in the mid winter experiment.
Overall, our analyses indicated that sampling did not significantly affect flowering time in any experiment (Table 1). The mean (±SE) for flowering time was 157.2 ± 0.5, 113.5 ± 0.3 and 84.5 ± 0.4 days for the mid autumn, late autumn and mid winter experiments, respectively. Population and individuals nested within population were significantly different in nearly all experiments (Table 1). CDC was the population with the latest flowering times in all experiments (mean flowering time ± SE = 164.5 ± 0.6 and 123.1 ± 0.3 days for the mid autumn and late autumn experiments, respectively), followed by MAR (156.0 ± 0.9, 109.0 ± 0.5 and 86.0 ± 0.7 days for the mid autumn, late autumn and mid winter experiments, respectively) and SAN (152.8 ± 0.8, 109.0 ± 0.4 and 83.1 ± 0.5 days for the mid autumn, late autumn and mid winter experiments, respectively).
Table 1.
General linear model testing the effect of sampling (2003–04 and 2012–13), population (CDC, MAR and SAN) and individuals nested within populations (14–15 individuals per population) on flowering time of Arabidopsis thaliana in three field experiments (sowing times in parenthesis). Degrees of freedom (d.f.), F-values and their significance are given for all factors and experiments. Significance: ***P < 0.0001; **P < 0.01; *P < 0.05; ns, non-significant.
| Factor | Experiment #1 (mid autumn) | Experiment #2 (late autumn) | Experiment #3 (mid winter) | |||
|---|---|---|---|---|---|---|
| d.f. | F-value | d.f. | F-value | d.f. | F-value | |
| Sampling (S) | 1 | 1.28ns | 1 | 0.70ns | 1 | 2.94ns |
| Population (P) | 2 | 5.27ns | 2 | 46.08** | 1 | 5.39* |
| Individual | 42 | 3.64*** | 42 | 5.73*** | 28 | 4.95*** |
| S × P | 2 | 13.49*** | 2 | 7.47** | 1 | 0.79ns |
| Error | 299 | 475 | 256 | |||
The interaction between sampling and population was also significant for the mid autumn and late autumn experiments (Table 1), indicating that the effect of sampling differed among populations. Differences between the two samplings for mean flowering time per population and experiment were small, ranging from a low of 0.03 to a high of 2.66 days, except for SAN in the mid autumn experiment with a difference of 6.54 days between samplings (Fig. 3). In CDC, individuals from the second sampling flowered earlier than individuals from the first sampling (Fig. 3). In the case of MAR, this population exhibited a similar behaviour, except for the mid winter experiment in which individuals from the second sampling flowered later than those from the first sampling (Fig. 3). Finally, SAN exhibited later flowering times for individuals from the second sampling than for individuals from the first sampling in all three experiments, particularly in the mid autumn experiment (Fig. 3).
Broad sense heritability (h2) values for flowering time for each population and experiment varied between 0.25 and 0.79 for individuals from the first sampling and between 0.00 and 0.65 for individuals from the second sampling (Table 2). Such a generalized decrease in h2 values for flowering time was mainly observed in SAN in all experiments, in MAR in the mid winter experiment and in CDC in the late autumn experiment (Table 2). Exceptions were CDC in the mid autumn experiment in which h2 values for flowering time slightly increased, and MAR in the two autumn experiments in which h2 values for flowering time remained rather similar (Table 2). SAN was the population with the most pronounced decrease in h2 values between samplings in all experiments (Table 2). In contrast, h2 values for flowering time in MAR showed the least difference between samplings, particularly in the mid and the late autumn experiments (Table 2).
Table 2.
Broad sense heritability (h2) values for flowering time in three Arabidopsis thaliana populations estimated in three field experiments with individuals collected from two samplings (2003–04 and 2012–13). The 95 % CIs for h2 values are given in parentheses. No individuals from CDC, except one from the first sampling, were able to complete the life cycle in the mid winter experiment.
| Population | Experiment | 2003–04 | 2012–13 |
|---|---|---|---|
| CDC | Mid autumn | 0.25 (0.14–0.33) | 0.39 (0.30–0.46) |
| CDC | Late autumn | 0.79 (0.73–0.82) | 0.08 (0.02–0.13) |
| CDC | Mid winter | – | – |
| MAR | Mid autumn | 0.67 (0.60–0.72) | 0.65 (0.57–0.69) |
| MAR | Late autumn | 0.61 (0.53–0.66) | 0.60 (0.53–0.65) |
| MAR | Mid winter | 0.75 (0.69–0.79) | 0.58 (0.50–0.63) |
| SAN | Mid autumn | 0.76 (0.71–0.80) | 0.33 (0.24–0.39) |
| SAN | Late autumn | 0.67 (0.61–0.72) | 0.00 (0.00–0.00) |
| SAN | Mid winter | 0.32 (0.24–0.39) | 0.19 (0.11–0.25) |
Genetic diversity
We used genetic data from a total of 329 A. thaliana individuals based on 12 nuclear microsatellite markers, including 103 individuals from the first sampling and 226 individuals from the second sampling, to assess temporal changes in genetic diversity, genetic differentiation and genetic structure in A. thaliana populations. After correcting for sample size differences between samplings and populations by random subsampling of equal size (N = 20), our results showed contrasting and pronounced patterns of temporal changes in genetic diversity parameters in A. thaliana populations. For example, the genetic diversity (HS) in AGU and MAR increased by 103.2 and 13.1 % between the two samplings, respectively (Table 3). In contrast, genetic diversity in CDC decreased by 19.8 %, whereas that in SAN remained fairly stable over time with a slight decrease of 7.9 % between the two samplings (Table 3). Observed heterozygosity (HO) and outcrossing rates (OR) substantially increased in all populations over the study period, except those in AGU that showed the opposite pattern (Table 3). Nonetheless, observed values for HO and OR in both samplings fell within the expected values for the highly self-fertilizing A. thaliana: HO varied between 0.004 and 0.089, and OR between 0.004 and 0.094 (Table 3).
Table 3.
Genetic diversity of Arabidopsis thaliana populations obtained from each sampling estimated from 12 nuclear microsatellite loci. The number of multilocus genotypes (NG) with the number of sampled individuals in parenthesis (N), observed heterozygosity (HO), outcrossing rates (OR) and mean gene diversity (HS) are given. HO, OR and HS are means (±SD) from 100 subsamples of 20 individuals each per population and sampling.
| Population | N G (N) | H O | O R | H S |
|---|---|---|---|---|
| AGU (2004) | 12 (20) | 0.046 ± 0.000 | 0.094 ± 0.000 | 0.282 ± 0.000 |
| AGU (2012) | 31 (53) | 0.021 ± 0.009 | 0.018 ± 0.008 | 0.573 ± 0.022 |
| CDC (2003) | 26 (32) | 0.021 ± 0.008 | 0.020 ± 0.008 | 0.602 ± 0.015 |
| CDC (2012) | 38 (58) | 0.066 ± 0.023 | 0.082 ± 0.038 | 0.483 ± 0.050 |
| MAR (2003) | 20 (30) | 0.026 ± 0.008 | 0.033 ± 0.014 | 0.540 ± 0.039 |
| MAR (2012) | 63 (70) | 0.089 ± 0.022 | 0.068 ± 0.018 | 0.611 ± 0.018 |
| SAN (2003) | 18 (21) | 0.004 ± 0.001 | 0.004 ± 0.001 | 0.671 ± 0.008 |
| SAN (2013) | 22 (45) | 0.019 ± 0.009 | 0.017 ± 0.009 | 0.618 ± 0.032 |
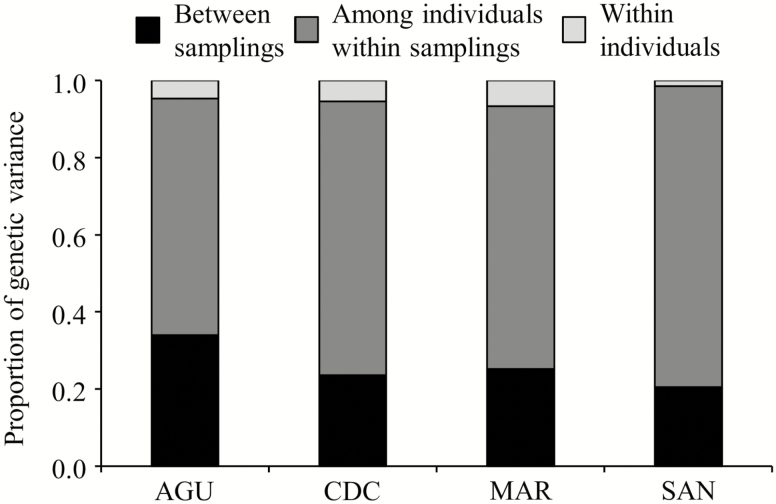
The AMOVA indicated that the extent of genetic differentiation between the two samplings was substantial in all populations, with values as high as 34.0 % for AGU, 23.6 % for CDC, 25.1 % for MAR and 19.9 % for SAN (Fig. 4). These results indicated that the highest temporal genetic differentiation values were observed in the northern and central populations (AGU, CDC and MAR), whereas the lowest value was recorded in the southernmost population (SAN). Genetic differentiation among individuals within samplings reached the highest values, ranging between 61.4 % (AGU) and 76.0 % (SAN) (Fig. 4). As expected for a highly self-fertilizing plant, genetic differentiation within individuals showed the lowest values (range = 1.4–6.7 %; Fig. 4). When taken all data together in a single AMOVA including sampling as the highest hierarchical level, we found that genetic differentiation between samplings was of 3.1 %, genetic differentiation among populations within samplings of 29.9 %, genetic differentiation among individuals within populations of 63.0 % and genetic differentiation within individuals of 4.0 %. All F-statistics were significant (P < 0.0001).
Figure 4.
Proportion of genetic variance obtained from AMOVA for each Arabidopsis thaliana population. Genetic variance is partitioned between samplings, among individuals within samplings and within individuals. Mean values from 100 analyses using subsamples of equal size are given. Standard deviations are not shown because they were very low.
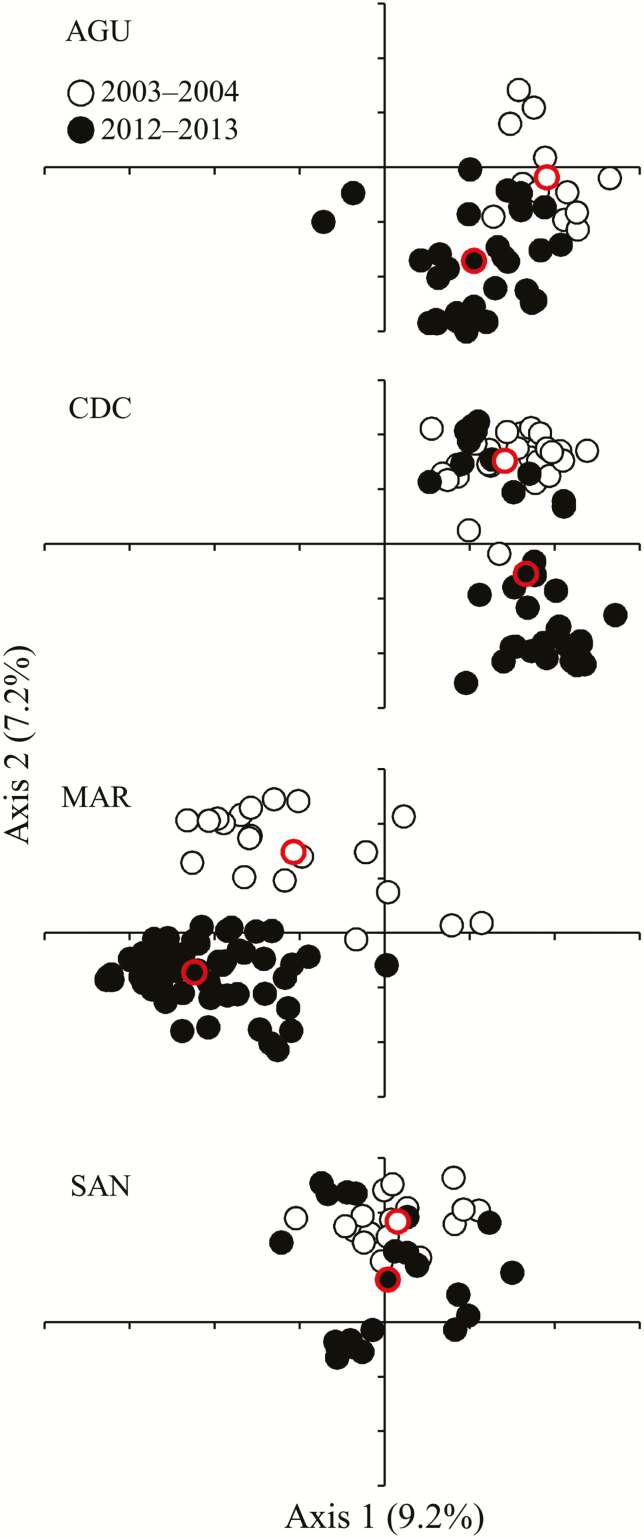
We found that 69.9 % of the genotyped A. thaliana individuals (230 of 329) were non-redundant multilocus genotypes (Table 3). Non-redundant multilocus genotypes represented by more than one individual (48 of 230; 20.9 %) included a minimum of two individuals and a maximum of 14 (mean ± SD = 3.1 ± 2.2 individuals per multilocus genotype). It must be noted that no identical multilocus genotype was found among populations or between samplings. All individuals with the same multilocus genotype came from the same population and sampling. Non-redundant multilocus genotypes from all populations and samplings were also used to explore how the genetic structure of A. thaliana populations varied between samplings by means of PCoA. The first three eigenvalues explained up to 22.4 % of the variation. The graphical representation of the first two axes, accounting for 16.4 % of the variation, indicated a clear trend for temporal structuring between multilocus genotypes from the two samplings in each A. thaliana population (Fig. 5). MAR exhibited the clearest pattern of temporal genetic structuring between samplings (distance between centroids for multilocus genotypes from the first and second samplings = 0.50), AGU and CDC exhibited intermediate overlap patterns between multilocus genotypes from the two samplings (0.36 and 0.41 for AGU and CDC, respectively) and SAN showed the highest overlap between multilocus genotypes from the two samplings (0.20; Fig. 5). Thus, SAN had the largest genetic similarities between multilocus genotypes detected in the two samplings (Fig. 5).
Figure 5.
Scatter plots displaying the first two eigenvectors estimated by PCoA of Arabidopsis thaliana non-redundant multilocus genotypes using 12 nuclear microsatellites. For the sake of clarity, populations are shown in different panels, although eigenvectors come from a unique analysis combining all data from all populations and samplings. Multilocus genotypes from the first sampling (2003–04; hollow dots) and multilocus genotypes from the second sampling (2012–13; filled dots) are indicated for each population. The centroids (dots with red contour) for multilocus genotypes from the two samplings and populations are also indicated.
We evaluated the extent of genetic drift in all populations by estimating effective population size (Ne) with the temporal method. AGU exhibited a Ne value of 10.8 (95 % CI = 7.3–15.3). The rest of populations had higher Ne estimates with values of 18.4 (12.8–25.4) for CDC, 20.5 (12.5–31.1) for MAR and 21.5 (16.0–28.3) for SAN. Hence, the effects of genetic drift in CDC, MAR and SAN were rather similar, whereas those in AGU were more intense. This result can be related to the fact that the lowest genetic diversity (HS) was in AGU in the 2004 sampling (Table 3).
We also sequenced the regions concentrating the highest nucleotide diversity in two flowering time genes (FRI and FLC) and one seed dormancy gene (DOG1) in individuals from the two samplings from each population. FRI was the gene with more polymorphisms in all populations (range = 2–20 single nucleotide polymorphisms [SNPs]) generating more multilocus haplotypes (Table 4). FLC and DOG1 exhibited less polymorphisms or even none at all (range = 0–4 SNPs and 0–8 SNPs for FLC and DOG1, respectively; Table 4). Temporal patterns of variation in silent nucleotide diversity were quite erratic in these genes: FRI decreased its nucleotide diversity in all populations except in AGU, FLC decreased it in AGU and CDC and increased it in MAR and DOG1 sharply decreased it in AGU but strongly increased it in MAR and SAN (Table 4).
Table 4.
Genetic diversity of Arabidopsis thaliana populations from each sampling estimated for two flowering genes (FRI and FLC) and one seed dormancy gene (DOG1). The number of multilocus haplotypes (NH) with the number of sampled individuals in parenthesis (N), the number of polymorphisms (NP) that were not in linkage disequilibrium, and silent (πsilent) and non-synonymous (πnon-syn) nucleotide diversity are given.
| Population | FRI | FLC | DOG1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N H (N) | NP | πsilent | πnon-syn | N H (N) | NP | πsilent | πnon-syn | N H (N) | NP | πsilent | πnon-syn | |
| AGU (2004) | 2 (9) | 5 | 0.00076 | 0.00055 | 4 (9) | 4 | 0.00454 | – | 3 (10) | 3 | 0.00595 | 0.00374 |
| AGU (2012) | 2 (23) | 5 | 0.00147 | 0.00042 | 2 (23) | 3 | 0.00080 | – | 2 (23) | 2 | 0 | 0.00292 |
| CDC (2003) | 5 (9) | 12 | 0.00204 | 0.00094 | 4 (9) | 3 | 0.00232 | – | 1 (10) | 0 | 0 | 0 |
| CDC (2012) | 3 (23) | 2 | 0.00012 | 0 | 3 (23) | 3 | 0.00217 | – | 1 (23) | 0 | 0 | 0 |
| MAR (2003) | 6 (9) | 12 | 0.00373 | 0.00153 | 3 (9) | 2 | 0.00107 | – | 3 (10) | 5 | 0.00395 | 0.00148 |
| MAR (2012) | 15 (23) | 16 | 0.00255 | 0.00158 | 5 (21) | 4 | 0.00256 | – | 4 (21) | 5 | 0.03650 | 0.00370 |
| SAN (2003) | 5 (9) | 13 | 0.00163 | 0.00248 | 1 (9) | 0 | 0 | – | 4 (10) | 7 | 0.00224 | 0.00560 |
| SAN (2013) | 9 (23) | 20 | 0.00149 | 0.00199 | 1 (21) | 0 | 0 | – | 7 (23) | 8 | 0.01108 | 0.00550 |
Discussion
The analysis of phenotypic and genetic variation between individuals of the same populations sampled at different times enables us to quantify how populations have actually changed across generations. Annual plants are particularly well suited for conducting the resurrection approach, as shown by recent illuminating examples on A. thaliana (Frachon et al. 2017), Brassica rapa (Franks et al. 2007, 2016; Franks and Weis 2008; Welt et al. 2015), Centaurea cyanus (Thomann et al. 2015), Datura stramonium (Bustos-Segura et al. 2014), Hordeum spontaneum (Nevo et al. 2012), Ipomoea purpurea (Kuester et al. 2016), Polygonum cespitosum (Sultan et al. 2013; Horgan-Kobelski et al. 2016) and Triticum dicoccoides (Nevo et al. 2012). All these resurrection studies certified how fast annual plant populations can change over relatively short periods of time as a result of various environmental pressures.
We applied a resurrection approach on A. thaliana by sampling four Spanish natural populations twice over a decade. Aerial orthophotographs indicated that populations did not experience major disturbances over the study period [seeSupporting Information—Fig. S1]. Based on that, we assume that temporal variation in phenotypic and genetic attributes chiefly responded to the variation in the climatic conditions, although we ignore how other biotic factors, such as herbivores or pathogens, might have affected these populations. Based on the comparison between annual weather records over the study period and historical records, we detected a trend for warming in all populations over the study period when compared to their historical temperature records (Fig. 2). Only CDC exhibited more years with average annual minimum temperature slightly below the historical record for that population. Overall, this trend is in agreement with the warming patterns predicted to occur in the region throughout the 21st century (Gómez-Navarro et al. 2010). In contrast, total annual precipitation exhibited a different pattern, with two populations above and other two below their respective historical records over the study period. This result also supports the accepted view that regional-scale climate change projections for precipitation still remain largely uncertain (Christensen et al. 2008; Schaller et al. 2011; Mahlstein et al. 2012; Tsanis et al. 2013), which is a problem to figure out the actual climatic scenarios for the near future. Hence, the four A. thaliana populations represented two distinct climatic scenarios: intense warming with increased precipitation, i.e. AGU and MAR, and moderate warming with decreased precipitation, i.e. CDC and SAN.
Our field experiments, designed to quantify differentiation in flowering time in A. thaliana over time, indicated that mean flowering time of A. thaliana populations did not differ substantially between samplings. However, the interaction between sampling and population was significant for the first two experiments in mid and late autumn including all populations. In CDC and MAR, A. thaliana advanced flowering with time, whereas in SAN the species exhibited the opposite behaviour, particularly in the mid autumn experiment where individuals from the second sampling flowered on average 6.5 days later than those from the first sampling (Table 1). A recent resurrection approach on A. thaliana encompassing 8 years also found a mean delay of 6.1 days in bolting time in a French population that experienced a significant warming of more than 1 °C over the last 30 years (Frachon et al. 2017). Although it is difficult to provide an explanation for these patterns, such a great difference in flowering time in SAN between the two samplings was only evident in the mid autumn experiment, when individuals experienced the longest vegetative phase prior to reproduction. This result stresses the important effect that the environment experienced by individuals during their development may have on key life-history traits (Donohue 2005, 2009, 2014), as well as the enormous complexity of the genotype × environment interaction in the expression of flowering time in A. thaliana (Stratton 1998; Méndez-Vigo et al. 2013, 2016; El-Soda et al. 2014; Sasaki et al. 2015; Ågren et al. 2017).
Beyond the trends observed for mean flowering time, broad sense heritability (h2) values for flowering time did show a more meaningful pattern of variation between individuals from the two samplings. In particular, there was a trend for reduction in h2 values for flowering time in populations from the second sampling (Table 2). This result indicated that the amount of among-individual variance in flowering time shrank over the study period. We hypothesize that environmental variation over the study period might have imposed stabilizing selection on flowering time, reducing therefore among-individual variation in this trait, although the exact mechanisms underlying stabilizing selection on flowering time cannot be determined from these data. When looking at the two extreme populations in terms of mild and sharp reduction in h2 values for flowering time, MAR and SAN, respectively, we speculate that the scenario of moderate warming and decreased precipitation in SAN would have stronger effect on h2 values for flowering time than intense warming and increased precipitation in MAR. Hence, it is plausible to consider drought stress as a major force in determining plant response and population performance in A. thaliana, as shown by various studies quantifying the effects of drought on a suite of A. thaliana traits dealing with life history, physiology, resource allocation and resistance to herbivores (McKay et al. 2003; Rizhsky et al. 2004; Riboni et al. 2013; Wolfe and Tonsor 2014; El-Soda et al. 2014; Zhang et al. 2015; Davila Olivas 2017a, b; Exposito-Alonso et al. 2018b). However, further work will be needed in order to test this hypothesis by increasing the number of populations resampled in two points in time and differing in the degree of warming and precipitation intensity.
From a genetic viewpoint, A. thaliana populations also showed distinctive genetic changes over the study period. As far as neutral microsatellites are concerned, AGU and MAR increased their genetic diversity between samplings, CDC decreased it and SAN barely maintained the same value (Table 3). It is worth noting that the two populations with intense warming and increased precipitation, i.e. AGU and MAR, where also those that showed increased genetic diversity over time. This result highlights the important role that precipitation patterns may have for the genetic make-up of A. thaliana populations affected by different warming intensities. Although this conclusion seems plausible, we must assume that these changes probably reflect natural demographic fluctuations affecting allele frequencies over time. This assumption is supported by the effective population size (Ne) estimates indicating that the effects of genetic drift on these populations were rather similar, except for AGU that showed the lowest Ne value.
This demographic scenario would also be applicable to the temporal patterns of variation in silent nucleotide diversity of flowering time and seed dormancy genes, which were rather erratic and gene dependent, probably as a result of the lower number of polymorphisms found in these genes (Table 4). Despite the fact that we also detected non-synonymous substitutions, all of them, except a FRI truncation only detected in AGU in the first sampling, were not associated to phenotypic changes in flowering time (Méndez-Vigo et al. 2011, 2013) or seed dormancy (Kronholm et al. 2012). This supports the assumption that changes detected in these flowering time and seed dormancy genes are probably determined by natural demographic fluctuations. Although these genes underlie major quantitative trait loci for flowering time and seed dormancy, it is clear that understanding the spatio-temporal variation in the genetic basis of these polygenic key life-history traits in natural populations is still an outstanding question in plant biology.
In contrast, genetic differentiation and genetic structure analyses did provide meaningful results when comparing the same populations between the two samplings. For example, genetic differentiation between samplings remarkably differed from a low of 20 % to a high of 34 % in just up to a decade. This is in agreement with other resurrection studies in annuals reporting substantial genetic changes in plant populations over decades (Vigouroux et al. 2011; Nevo et al. 2012; Van Dijk and Hautekèete 2014; Kuester et al. 2016). Based on this result, it can be concluded that natural A. thaliana populations exhibited a quite dynamic demographic behaviour, which is known to be characterized by dramatic year-to-year variation in plant abundance (Picó 2012) and a relatively short seed permanency in the soil seed bank (2–4 years; Lundemo et al. 2009; Montesinos et al. 2009; Falahati-Anbaran et al. 2014; Postma et al. 2016), features that easily lead to rapid temporal genetic differentiation. Besides, A. thaliana is known to exhibit low inter-population migration rates and limited dispersal distances (Lundemo et al. 2009; Bomblies et al. 2010; Falahati-Anbaran et al. 2014), which minimizes the effect of gene flow on genetic differentiation over A. thaliana generations. Thus, at this particular local scale and for non-urban populations, it is well accepted that genetic variation in A. thaliana is mostly accounted for by novel mutations, outcrossing and recombination, enhanced by the species’ low linkage disequilibrium, that altogether have the potential to generate genetic novelty in A. thaliana in a relatively short period of time (Bomblies et al. 2010; Gomaa et al. 2011).
In this resurrection study, we have been able to quantify the actual impact of all the processes mentioned above on natural A. thaliana populations over time. The best picture of the genetic temporal change in A. thaliana was given by the genetic structure between samplings based on non-redundant multilocus genotypes. SAN and MAR were the populations with the lowest and the highest genetic divergence over the study period, respectively. Interestingly, the two most different populations, in terms of warming, precipitation patterns and broad sense heritability values for flowering time, were also the ones with the most different genetic structuring over time. In conclusion, we hypothesize that dramatically drier conditions, such as those experienced in SAN over the study period, would have the potential to homogenize A. thaliana populations by reducing among-individual variation in key quantitative traits as well as by constraining genetic differentiation over time. It must be noted that this trend does not come into conflict with maintaining demographically viable populations with high levels of microsatellite and gene genetic diversity, as occurred in SAN and MAR. Hence, these results might illustrate the ability of Iberian A. thaliana populations to cope with harsher environmental conditions imposed by climate change (see experimental evidence in Exposito-Alonso et al. 2018a).
It remains to be seen whether the significant and dynamic phenotypic and genetic changes detected in A. thaliana populations in just a decade affect the long-term population persistence under a scenario of increasing warming with increased or decreased precipitation. However, we want to stress the importance of quantifying the pace and intensity of temporal change in plant populations by resampling populations periodically and conducting resurrection experiments. In the case of A. thaliana, we need to scale up the study presented here to a larger number of A. thaliana populations across different environments in the Iberian Peninsula from which we possess seed collected and preserved since early 2000s. Recently, we have shown that warm and cool Iberian environments might be exerting very different selective pressures on both flowering time and seed dormancy, but also on the correlation between the two traits in A. thaliana (Vidigal et al. 2016; Marcer et al. 2018). Thus, resurrection experiments dealing with warm and cool Iberian populations would provide a proof of concept of the microevolutionary implications of climate change in annual and short-lived plants.
Data
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.84vc24v.
Sources of Funding
F.X.P., A.M. and R.G. were funded by grants CGL2012-33220/BOS and CGL2016-77720-P (AEI/FEDER, UE). A.M. acknowledges the Agency for Management of University and Research Grants of the Generalitat de Catalunya (2014-SGR-913).
Contributions by the Authors
F.X.P. and C.A.-B. planned and designed the research. R.G., B.M.-V., A.M., C.A.-B. and F.X.P. conducted field work, performed experiments and/or analysed data. F.X.P. wrote the first draft of the manuscript and all authors contributed to the final version of it.
Conflict of Interest
None declared.
Supplementary Material
Acknowledgements
We thank M. Ramiro and D. Ragel for laboratory and field assistance. We are grateful to AEMET for providing meteorological data. We also thank the administration of the Sierra de Grazalema Natural Park for permission to work at the El Castillejo Botanical Garden. The associate editor (H. Huber), A. Castilla and two reviewers provided helpful comments on an earlier version of this manuscript.
Supporting Information
The following additional information is available in the online version of this article—
Figure S1. Aerial orthophotographs of Arabidopsis thaliana populations. Orthophotographs were chosen from those available to the closer years for the first and second samplings. The circular area (500 m radius) around the GPS coordinate is indicated, which was previously used to describe the ecological characteristics of populations.
Literature Cited
- Ågren J, Oakley CG, Lundemo S, Schemske DW.. 2017. Adaptive divergence in flowering time among natural populations of Arabidopsis thaliana: estimates of selection and QTL mapping. Evolution 71:550–564. [DOI] [PubMed] [Google Scholar]
- Ågren J, Schemske DW.. 2012. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. The New Phytologist 194:1112–1122. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M.. 2003. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164:711–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Panetta AM, Mitchell-Olds T.. 2012. Evolutionary and ecological responses to anthropogenic climate change: update on anthropogenic climate change. Plant Physiology 160:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont LJ, Hughes L, Pitman AJ.. 2008. Why is the choice of future climate scenarios for species distribution modelling important?Ecology Letters 11:1135–1146. [DOI] [PubMed] [Google Scholar]
- Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F.. 2012. Impacts of climate change on the future of biodiversity. Ecology Letters 15:365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennington CC, McGraw JB, Vavrek MC.. 1991. Ecological genetic variation in seed banks. II. Phenotypic and genetic differences between young and old subpopulations of Luzula parviflora. Journal of Ecology 79:627–643. [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, Blankestijn-de Vries H, Coltrane C, Keizer P, El-Lithy M, Alonso-Blanco C, de Andrés MT, Reymond M, van Eeuwijk F, Smeekens S, Koornneef M.. 2010. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proceedings of the National Academy of Sciences 107:4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M.. 2006. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences 103:17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodbyl Roels SA, Kelly JK.. 2011. Rapid evolution caused by pollinator loss in Mimulus guttatus. Evolution 65:2541–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, Fitz J, Weigel D.. 2010. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genetics 6:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos-Segura C, Fornoni J, Núñez-Farfán J.. 2014. Evolutionary changes in plant tolerance against herbivory through a resurrection experiment. Journal of Evolutionary Biology 27:488–496. [DOI] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Dittmar E, Kramer EM, de Casas RR, Donohue K.. 2013. Pleiotropy in the wild: the dormancy gene DOG1 exerts cascading control on life cycles. Evolution 67:883–893. [DOI] [PubMed] [Google Scholar]
- Christensen JH, Boberg F, Christensen OB, Lucas-Picher P.. 2008. On the need for bias correction of regional climate change projections of temperature and precipitation. Geophysical Research Letters 35:L20709. [Google Scholar]
- Davila Olivas NH, Frago E, Thoen MPM, Kloth KJ, Becker FFM, van Loon JJA, Gort G, Keurentjes JJB, van Heerwaarden J, Dicke M.. 2017. a Natural variation in life history strategy of Arabidopsis thaliana determines stress responses to drought and insects of different feeding guilds. Molecular Ecology 26:2959–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila Olivas NH, Kruijer W, Gort G, Wijnen CL, van Loon JJ, Dicke M.. 2017. b Genome-wide association analysis reveals distinct genetic architectures for single and combined stress responses in Arabidopsis thaliana. The New Phytologist 213:838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MB, Shaw RG, Etterson JR.. 2005. Evolutionary responses to changing climate. Ecology 86:1704–1714. [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR.. 2014. NeEstimator V2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14:209–214. [DOI] [PubMed] [Google Scholar]
- Donohue K. 2005. Niche construction through phenological plasticity: life history dynamics and ecological consequences. The New Phytologist 166:83–92. [DOI] [PubMed] [Google Scholar]
- Donohue K. 2009. Completing the cycle: maternal effects as the missing link in plant life histories. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 364:1059–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K. 2014. Why ontogeny matters during adaptation: developmental niche construction and pleiotropy across the life cycle in Arabidopsis thaliana. Evolution 68:32–47. [DOI] [PubMed] [Google Scholar]
- El-Soda M, Malosetti M, Zwaan BJ, Koornneef M, Aarts MG.. 2014. Genotype × environment interaction QTL mapping in plants: lessons from Arabidopsis. Trends in Plant Science 19:390–398. [DOI] [PubMed] [Google Scholar]
- Etterson JR, Franks SJ, Mazer SJ, Shaw RG, Gorden NL, Schneider HE, Weber JJ, Winkler KJ, Weis AE.. 2016. Project baseline: an unprecedented resource to study plant evolution across space and time. American Journal of Botany 103:164–173. [DOI] [PubMed] [Google Scholar]
- Exposito-Alonso M, Brennan AC, Alonso-Blanco C, Picó FX.. 2018. a Spatio-temporal variation in fitness responses to contrasting environments in Arabidopsis thaliana. Evolution 72:1570–1586. [DOI] [PubMed] [Google Scholar]
- Exposito-Alonso M, Vasseur F, Ding W, Wang G, Burbano HA, Weigel D.. 2018. b Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nature Ecology & Evolution 2:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati-Anbaran M, Lundemo S, Stenøien HK.. 2014. Seed dispersal in time can counteract the effect of gene flow between natural populations of Arabidopsis thaliana. The New Phytologist 202:1043–1054. [DOI] [PubMed] [Google Scholar]
- Frachon L, Libourel C, Villoutreix R, Carrère S, Glorieux C, Huard-Chauveau C, Navascués M, Gay L, Vitalis R, Baron E, Amsellem L, Bouchez O, Vidal M, Le Corre V, Roby D, Bergelson J, Roux F.. 2017. Intermediate degrees of synergistic pleiotropy drive adaptive evolution in ecological time. Nature Ecology & Evolution 1:1551–1561. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Avise JC, Bradshaw WE, Conner JK, Etterson JR, Mazer SJ, Shaw RG, Weis AE.. 2008. The resurrection initiative: storing ancestral genotypes to capture evolution in action. Bioscience 58:870–873. [Google Scholar]
- Franks SJ, Hamann E, Weis AE.. 2018. Using the resurrection approach to understand contemporary evolution in changing environments. Evolutionary Applications 11:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Hoffmann AA.. 2012. Genetics of climate change adaptation. Annual Review of Genetics 46:185–208. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Kane NC, O’Hara NB, Tittes S, Rest JS.. 2016. Rapid genome-wide evolution in Brassica rapa populations following drought revealed by sequencing of ancestral and descendant gene pools. Molecular Ecology 25:3622–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE.. 2007. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proceedings of the National Academy of Sciences 104:1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Weber JJ, Aitken SN.. 2014. Evolutionary and plastic responses to climate change in terrestrial plant populations. Evolutionary Applications 7:123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Weis AE.. 2008. A change in climate causes rapid evolution of multiple life-history traits and their interactions in an annual plant. Journal of Evolutionary Biology 21:1321–1334. [DOI] [PubMed] [Google Scholar]
- Fukano Y, Tanaka K, Yahara T.. 2013. Directional selection for early flowering is imposed by a re-associated herbivore - but no evidence of directional evolution. Basic and Applied Ecology 14:387–395. [Google Scholar]
- Gilbert GS, Parker IM.. 2010. Rapid evolution in a plant-pathogen interaction and the consequences for introduced host species. Evolutionary Applications 3:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa NH, Montesinos-Navarro A, Alonso-Blanco C, Picó FX.. 2011. Temporal variation in genetic diversity and effective population size of Mediterranean and subalpine Arabidopsis thaliana populations. Molecular Ecology 20:3540–3554. [DOI] [PubMed] [Google Scholar]
- Gómez-Navarro JJ, Montávez JP, Jimenez-Guerrero P, Jerez S, García-Valero JA, González-Rouco JF.. 2010. Warming patterns in regional climate change projections over the Iberian Peninsula. Meteorologische Zeitschrift 19:275–285. [Google Scholar]
- Gomulkiewicz R, Shaw RG.. 2013. Evolutionary rescue beyond the models. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 368:20120093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM.. 2003. Regulation of flowering time by histone acetylation in Arabidopsis. Science 302:1751–1754. [DOI] [PubMed] [Google Scholar]
- Holt RD. 1990. The microevolutionary consequences of climate change. Trends in Ecology & Evolution 5:311–315. [DOI] [PubMed] [Google Scholar]
- Horgan-Kobelski T, Matesanz S, Sultan SE.. 2016. Limits to future adaptation in the invasive plant Polygonum cespitosum: expression of functional and fitness traits at elevated CO2. The Journal of Heredity 107:42–50. [DOI] [PubMed] [Google Scholar]
- Karell P, Ahola K, Karstinen T, Valkama J, Brommer JE.. 2011. Climate change drives microevolution in a wild bird. Nature Communications 2:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM.. 2009. Vernalization: winter and the timing of flowering in plants. Annual Review of Cell and Developmental Biology 25:277–299. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sung S.. 2014. Genetic and epigenetic mechanisms underlying vernalization. The Arabidopsis Book 12:e0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W.. 1998. Genetic control of flowering time in Arabidopsis. Annual Review of Plant Physiology and Plant Molecular Biology 49:345–370. [DOI] [PubMed] [Google Scholar]
- Kopp M, Matuszewski S.. 2014. Rapid evolution of quantitative traits: theoretical perspectives. Evolutionary Applications 7:169–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves TM, Schmid KJ, Caicedo AL, Mays C, Stinchcombe JR, Purugganan MD, Schmitt J.. 2007. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. The American Naturalist 169:E141–E157. [DOI] [PubMed] [Google Scholar]
- Kronholm I, Picó FX, Alonso-Blanco C, Goudet J, de Meaux J.. 2012. Genetic basis of adaptation in Arabidopsis thaliana: local adaptation at the seed dormancy QTL DOG1. Evolution 66:2287–2302. [DOI] [PubMed] [Google Scholar]
- Kuester A, Wilson A, Chang SM, Baucom RS.. 2016. A resurrection experiment finds evidence of both reduced genetic diversity and potential adaptive evolution in the agricultural weed Ipomoea purpurea. Molecular Ecology 25:4508–4520. [DOI] [PubMed] [Google Scholar]
- Le Corre V. 2005. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Molecular Ecology 14:4181–4192. [DOI] [PubMed] [Google Scholar]
- Le Rouzic A, Carlborg O.. 2008. Evolutionary potential of hidden genetic variation. Trends in Ecology & Evolution 23:33–37. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J.. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Lloret F, Peñuelas J, Estiarte M.. 2004. Experimental evidence of reduced diversity of seedlings due to climate modification in a Mediterranean-type community. Global Change Biology 10:248–258. [Google Scholar]
- Lundemo S, Falahati-Anbaran M, Stenøien HK.. 2009. Seed banks cause elevated generation times and effective population sizes of Arabidopsis thaliana in northern Europe. Molecular Ecology 18:2798–2811. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B.. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Mahlstein I, Portmann RW, Daniel JS, Solomon S, Knutti R.. 2012. Perceptible changes in regional precipitation in a future climate. Geophysical Research Letters 39:L05701. [Google Scholar]
- Manzano-Piedras E, Marcer A, Alonso-Blanco C, Picó FX.. 2014. Deciphering the adjustment between environment and life history in annuals: lessons from a geographically-explicit approach in Arabidopsis thaliana. PLoS One 9:e87836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcer A, Méndez-Vigo B, Alonso-Blanco C, Picó FX.. 2016. Tackling intraspecific genetic structure in distribution models better reflects species geographical range. Ecology and Evolution 6:2084–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcer A, Vidigal DS, James PMA, Fortin MJ, Méndez-Vigo B, Hilhorst HWM, Bentsink L, Alonso-Blanco C, Picó FX.. 2018. Temperature fine-tunes Mediterranean Arabidopsis thaliana life-cycle phenology geographically. Plant Biology 20:148–156. [DOI] [PubMed] [Google Scholar]
- Maron JL, Vilà M, Bommarco R, Elmendorf S, Beardsley P.. 2004. Rapid evolution of an invasive plant. Ecological Monographs 74:261–280. [Google Scholar]
- McGraw JB, Vavrek M, Bennington CC.. 1991. Ecological genetic variation in seed banks. I. Establishment of a time transect. Journal of Ecology 79:617–625. [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T.. 2003. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology 12:1137–1151. [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Gomaa NH, Alonso-Blanco C, Picó FX.. 2013. Among- and within-population variation in flowering time of Iberian Arabidopsis thaliana estimated in field and glasshouse conditions. The New Phytologist 197:1332–1343. [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C.. 2011. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiology 157:1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vigo B, Savic M, Ausín I, Ramiro M, Martín B, Picó FX, Alonso-Blanco C.. 2016. Environmental and genetic interactions reveal FLOWERING LOCUS C as a modulator of the natural variation for the plasticity of flowering in Arabidopsis. Plant, Cell & Environment 39:282–294. [DOI] [PubMed] [Google Scholar]
- Menzel A, Sparks TH, Estrella N, Koch E, Aasa A, Ahas R, Alm-Kübler K, Bissolli P, Braslavská O, Briede A, Chmielewski FM, Crepinsek Z, Curnel Y, Dahl A, Defila C, Donnelly A, Filella Y, Jatcza K, Måge F, Mestre A, Nordli Ø, Peñuelas J, Pirinen P, Remisová V, Scheifinger H, Striz M, Susnik A, Van Vliet AJH, Wielgolaski FE, Zach S, Zust A.. 2006. European phenological response to climate change matches the warming pattern. Global Change Biology 12:1969–1976. [Google Scholar]
- Messer PW, Ellner SP, Hairston NG Jr.. 2016. Can population genetics adapt to rapid evolution?Trends in Genetics 32:408–418. [DOI] [PubMed] [Google Scholar]
- Montesinos A, Tonsor SJ, Alonso-Blanco C, Picó FX.. 2009. Demographic and genetic patterns of variation among populations of Arabidopsis thaliana from contrasting native environments. PLoS One 4:e7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Tajima F.. 1981. Genetic drift and estimation of effective population size. Genetics 98:625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E, Fu YB, Pavlicek T, Khalifa S, Tavasi M, Beiles A.. 2012. Evolution of wild cereals during 28 years of global warming in Israel. Proceedings of the National Academy of Sciences 109:3412–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW.. 1997. GeneDoc: analysis and visualization of genetic variation. EMBnet.news 4:1–4. [Google Scholar]
- O’Hara NB, Rest JS, Franks SJ.. 2016. Increased susceptibility to fungal disease accompanies adaptation to drought in Brassica rapa. Evolution 70:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C. 2007. Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Global Change Biology 13:1860–1872. [Google Scholar]
- Parmesan C, Hanley ME.. 2015. Plants and climate change: complexities and surprises. Annals of Botany 116:849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE.. 2012. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RG, Dawson TP.. 2003. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful?Global Ecology and Biogeography 12:361–371. [Google Scholar]
- Peñuelas J, Filella I.. 2001. Phenology. Responses to a warming world. Science 294:793–795. [DOI] [PubMed] [Google Scholar]
- Peñuelas J, Sardans J, Estiarte M, Ogaya R, Carnicer J, Coll M, Barbeta A, Rivas-Ubach A, Llusià J, Garbulsky M, Filella I, Jump AS.. 2013. Evidence of current impact of climate change on life: a walk from genes to the biosphere. Global Change Biology 19:2303–2338. [DOI] [PubMed] [Google Scholar]
- Picó FX. 2012. Demographic fate of Arabidopsis thaliana cohorts of autumn- and spring-germinated plants along an altitudinal gradient. Journal of Ecology 100:1009–1018. [Google Scholar]
- Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C.. 2008. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian Peninsula. Genetics 180:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma FM, Lundemo S, Ågren J.. 2016. Seed dormancy cycling and mortality differ between two locally adapted populations of Arabidopsis thaliana. Annals of Botany 117:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoné B, Vitalis R, Goldringer I, Bonnin I.. 2010. Evolution of flowering time in experimental wheat populations: a comprehensive approach to detect genetic signatures of natural selection. Evolution 64:2110–2125. [DOI] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L.. 2013. GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiology 162:1706–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R.. 2004. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134:1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Zhang P, Atwell S, Meng D, Nordborg M.. 2015. “Missing” G x E variation controls flowering time in Arabidopsis thaliana. PLoS Genetics 11:e1005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller N, Mahlstein I, Cermak J, Knutti R.. 2011. Analyzing precipitation projections: a comparison of different approaches to climate model evaluation. Journal of Geophysical Research 116:D10118. [Google Scholar]
- Shaw RG, Etterson JR.. 2012. Rapid climate change and the rate of adaptation: insight from experimental quantitative genetics. The New Phytologist 195:752–765. [DOI] [PubMed] [Google Scholar]
- Stratton DA. 1998. Reaction norm functions and QTL-environment interactions for flowering time in Arabidopsis thaliana. Heredity 81:144–155. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Horgan-Kobelski T, Nichols LM, Riggs CE, Waples RK.. 2013. A resurrection study reveals rapid adaptive evolution within populations of an invasive plant. Evolutionary Applications 6:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann M, Imbert E, Engstrand RC, Cheptou PO.. 2015. Contemporary evolution of plant reproductive strategies under global change is revealed by stored seeds. Journal of Evolutionary Biology 28:766–778. [DOI] [PubMed] [Google Scholar]
- Tsanis IK, Grillakis MG, Koutroulis AG, Jacob D.. 2013. Reducing uncertainty on global precipitation projections. Journal of Earth Science & Climate Change 5:178. [Google Scholar]
- Van Dijk H, Hautekèete NC.. 2014. Evidence of genetic change in the flowering phenology of sea beets along a latitudinal cline within two decades. Journal of Evolutionary Biology 27:1572–1581. [DOI] [PubMed] [Google Scholar]
- Vidigal DS, Marques AC, Willems LA, Buijs G, Méndez-Vigo B, Hilhorst HW, Bentsink L, Picó FX, Alonso-Blanco C.. 2016. Altitudinal and climatic associations of seed dormancy and flowering traits evidence adaptation of annual life cycle timing in Arabidopsis thaliana. Plant, Cell & Environment 39:1737–1748. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Mariac C, De Mita S, Pham JL, Gérard B, Kapran I, Sagnard F, Deu M, Chantereau J, Ali A, Ndjeunga J, Luong V, Thuillet AC, Saïdou AA, Bezançon G.. 2011. Selection for earlier flowering crop associated with climatic variations in the Sahel. PLoS One 6:e19563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MD, Wahren CH, Hollister RD, Henry GHR, Ahlquist LE, Alatalo JM, Bret-Harte MS, Calef MP, Callaghan TV, Carroll AB, Epstein HE, Jónsdóttir IS, Klein JA, Magnusson B, Molau U, Oberbauer SF, Rewa SP, Robinson CH, Shaver GR, Suding KN, Thompson CC, Tolvanen A, Totland Ø, Turner PL, Tweedie CE, Webber PJ, Wookey PA.. 2006. Plant community responses to experimental warming across the tundra biome. Proceedings of the National Academy of Sciences 103:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. 2005. Estimation of effective population sizes from data on genetic markers. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 360:1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. 1989. A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C, Ungerer MC, Dorn LA, Kane NC, Toyonaga Y, Halldorsdottir SS, Mackay TF, Purugganan MD, Schmitt J.. 2002. Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162:1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welt RS, Litt A, Franks SJ.. 2015. Analysis of population genetic structure and gene flow in an annual plant before and after a rapid evolutionary response to drought. AoB Plants 7:plv026; doi: 10.1093/aobpla/plv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MD, Tonsor SJ.. 2014. Adaptation to spring heat and drought in northeastern Spanish Arabidopsis thaliana. The New Phytologist 201:323–334. [DOI] [PubMed] [Google Scholar]
- Wolkovich EM, Cook BI, Allen JM, Crimmins TM, Betancourt JL, Travers SE, Pau S, Regetz J, Davies TJ, Kraft NJ, Ault TR, Bolmgren K, Mazer SJ, McCabe GJ, McGill BJ, Parmesan C, Salamin N, Schwartz MD, Cleland EE.. 2012. Warming experiments underpredict plant phenological responses to climate change. Nature 485:494–497. [DOI] [PubMed] [Google Scholar]
- Zhang N, Belsterling B, Raszewski J, Tonsor SJ.. 2015. Natural populations of Arabidopsis thaliana differ in seedling responses to high-temperature stress. AoB Plants 7:plv101; doi: 10.1093/aobpla/plv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.