Abstract
Background
Basal cell carcinoma (BCC) is a frequent malignant tumor of skin cancers with high morbidity. The objective of this study was to identify critical genes and pathways related to the carcinogenesis of BCC and gain more insights into the underlying molecular mechanisms of BCC.
Materials and methods
The gene expression profiles of GSE7553 and GSE103439 were downloaded from the Gene Expression Omnibus database with 19 tumors and 6 normal skin tissues. Differentially expressed genes (DEGs) were screened between BCC samples and normal tissues, followed by gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis. Subsequently, protein–protein interaction (PPI) network was constructed for these DEGs, and module analysis was performed.
Results
A total of 313 DEGs were obtained. Among them, 222 genes were upregulated and 91 genes were downregulated. Enrichment analysis indicated that the upregulated genes were significantly enriched in cell cycle and mitosis, while the downregulated genes were mainly associated with unsaturated fatty acid metabolic process and cell differentiation. In addition, TOP2A, CDK1, and CCNB1 were identified as the top three hub genes ranked by degrees in the PPI network. Meanwhile, three subnetworks were derived, which indicated that these DEGs were significantly enriched in pathways, including “cell cycle”, “extracellular matrix–receptor interaction”, “basal cell carcinoma”, and “hedgehog signaling pathway”.
Conclusions
The novel critical DEGs and pathways identified in this study may serve pivotal roles in the carcinogenesis of BCC and indicate more molecular targets for the treatment of BCC.
Keywords: basal cell carcinoma, differentially expressed genes, enrichment analysis, bioinformatics analysis
Introduction
Cutaneous basal cell carcinoma (BCC) is recognized as a common subtype of nonmelanoma skin malignancies with high morbidity, which accounts for ~80% of newly diagnosed nonmelanoma skin carcinomas.1 In the last decade, there has been a substantial increase in the incidence of BCC.2 Due to the characteristics of slow-growing and locally aggressive, metastasis rarely occurred in patients with BCC, which resulted in a relatively good prognosis. As we all know, long-term exposure to sunlight, especially ultraviolet light, is considered as the main risk factor of skin cancers.3 However, the underlying molecular mechanisms for the development of BCC has not been completely illuminated. Meanwhile, the treatments of BCC are limited and drug resistance is ubiquitous in advanced or metastatic BCC patients. Therefore, an urgent need exists for further exploring the potential mechanisms of BCC and finding more effective molecular targets for the treatment of BCC.
To date, several signaling pathways and molecules have been demonstrated to be involved in the tumorigenesis and progression of BCC at the molecular level, such as the hedgehog signaling pathway.4 Genes included in this pathway, such as the hedgehog receptors patched (PTCH1) or smoothened (SMO), have been extensively studied.5,6 Mutations in these genes may cause constitutive hedgehog pathway activation, which promote the development of BCC. Recently, two new hedgehog pathway inhibitors, Vismodegib and Sonidegib, have been approved by the Food and Drug Administration for the targeted treatment of BCC.7,8 However, the response rate of advanced or metastatic BCC is not promising and the secondary drug resistance may also occur.
With the development of high-throughput technology, more and more new potential targets have been uncovered in BCC. In addition to canonical hedgehog pathway components, the transcription factor serum response factor was identified as a noncanonical hedgehog activator by multidimensional genomics analysis, which leads to the amplification of the hedgehog transcription factor glioma-associated oncogene family zinc finger-1 (GLI1).9 At the DNA level, Bonilla et al performed a genomic analysis of 293 BCC samples and revealed that mutations in other cancer-related genes also drove the initiation of BCC, including MYCN, PTPN14, and LATS1.10 Thus, much more molecular targets remain to be elucidated.
Bioinformatics analysis of gene expression profiles or other high-throughput data are now playing a critical role in investigating the mechanisms of human disease, particularly in tumors. Accordingly, in the present study, we first time integratively reanalyzed the gene expression profiles of 19 BCC and 6 normal tissues deposited in two datasets by differentially expressed genes (DEGs) screening and functional and pathway enrichment analysis. By protein–protein interaction (PPI) network analysis, we identified top three hub genes (TOP2A, CDK1, and CCNB1). Finally, module analysis revealed that several critical pathways were mainly associated with the carcinogenesis of BCC, which might be used as molecular targets for the treatment of BCC.
Materials and methods
Microarray data
Two datasets (GSE7553 and GSE103439) were respectively retrieved from Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), including 19 BCC and 6 normal tissues (Table 1).11 These gene expression profiles were generated by GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) containing 54,675 probes. The latest annotation file of GPL570 platform was downloaded from Affymetrix official website (http://www.affymetrix.com/), in which 54,675 probes now mapped to 21,297 genes.
Table 1.
The basal information of two datasets in this study
Abbreviations: BCC, basal cell carcinoma; GEO, Gene Expression Omnibus; NS, normal skin.
Data preprocessing and DEGs screening
The raw data files (.CEL files) of these 25 samples were processed by the R package “affy”.12 Background adjustment and normalization were performed using the Robust Multichip Average algorithm. Once multiple probes mapped to the same gene, the average value was finally selected to represent the gene expression value. DEGs were screened between BCC and normal tissues by the “limma” package in R.13 Then, hierarchical clustering analysis was applied to the DEGs by the “pheatmap” package in R based on the Euclidean distance. The criteria of DEGs was set as |log2fold change|>1 and false discovery rate (FDR) <0.05.
Functional and pathway enrichment analysis
Gene ontology (GO) analysis defines the functions of gene products covering three domains, including biological process, molecular function, and cellular component.14,15 The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database is widely used to map large-scale datasets to pathway maps for higher-order functional information.16 The Database for Annotation, Visualization and Integrated Discovery (DAVID version 6.8, http://david.abcc.ncifcrf.gov/) consists of an integrated biological knowledgebase and analytic tools, which can systematically extract biological meaning from large gene/protein lists.17 With the online DAVID tool, we performed functional and pathway enrichment analysis for these DEGs. P-value <0.05 was considered as significant.
Construction of PPI network and module analysis
Given the large number of DEGs, the “STRINGdb” package in R was used to investigate the potential interactions that existed in these DEGs.18 Briefly, 313 DEGs were mapped to their corresponding proteins in the Search Tool for the Retrieval of Interacting Genes/Proteins database. Only interactions with a combined score of >0.4 were imported into Cytoscape software to visualize the PPI network.19 Each node in the network represents one protein, and the degree of each protein was termed as the number of its interactions. Then, the Molecular Complex Detection (MCODE) plug-in was used to analyze the PPI network to identify significant modules.20 In addition, the functional and pathway enrichment analysis of genes in the subnetworks were performed. P-value <0.05 was set as the threshold.
Results
Identification of DEGs
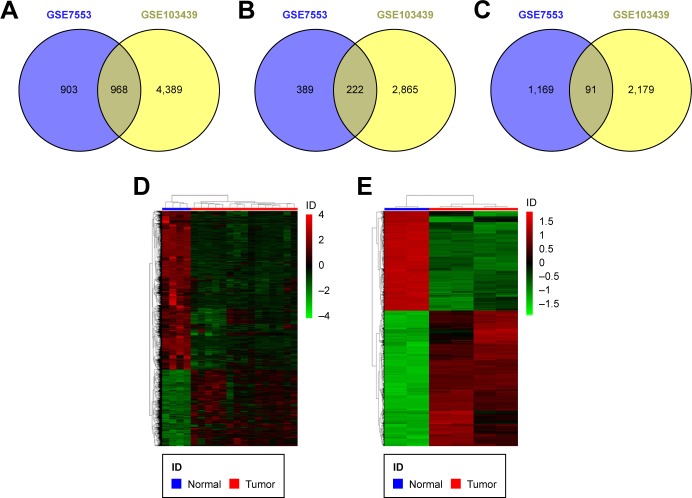
We screened DEGs in the two datasets (GSE7553 and GSE103439). Compared with normal skin tissues, 1,871 DEGs and 5,357 DEGs were obtained, respectively (Figure 1A). Finally, a total of 313 aberrantly expressed genes (222 upregulated genes and 91 downregulated genes) were identified by integrated analysis (Figure 1B and C). Strikingly, the number of upregulated genes were largely more than down-regulated genes (Table S1). The heatmap of hierarchical clustering analysis showed that these DEGs could clearly distinguish BCC samples from the normal skin samples (Figure 1D and E).
Figure 1.
DEGs in the two datasets.
Notes: (A) Common DEGs between GSE7553 and GSE103439. (B) Common upregulated DEGs between GSE7553 and GSE103439. (C) Common downregulated DEGs between GSE7553 and GSE103439. (D, E) Hierarchical clustering analysis of the DEGs in GSE7553 and GSE103439, respectively. Red and green indicate higher expression and lower expression, respectively.
Abbreviation: DEGs, differentially expressed genes.
GO and KEGG pathway enrichment analysis
To further investigate the potential functions of these 313 DEGs, GO and KEGG pathways enrichment analysis was performed by the online DAVID tool. The results of GO analysis indicated that upregulated genes enriched in biological process were mainly involved in cell cycle and mitosis, such as the cell division (P=4.39×10−11) and the mitotic nuclear division (P=5.90×10−8) (Table 2). Meanwhile, downregulated genes were significantly enriched in unsaturated fatty acid metabolic process (P=2.10×10−3) and cell differentiation (P=6.76×10−3) (Table 3). With regard to pathway enrichment analysis, the most significant pathway of upregulated genes was cell cycle (P=4.75×10−9) containing 13 genes. Interestingly, another five genes (LEF1, PTCH1, GLI2, FZD7, and GLI1) were enriched in the pathway named “basal cell carcinoma” (P=2.60×10−3) (Table 2), while downregulated genes were most significantly involved in the biosynthesis and metabolism of unsaturated fatty acids (P=5.26×10−3) (Table 3).
Table 2.
The top 10 GO terms and KEGG pathways of upregulated genes
| Term | Count | P-value | Genes |
|---|---|---|---|
| GO:0051301:cell division | 24 | 4.39E-11 | KIF14, CDK1, KIF11, NEK2, NUF2, KIF18B, NDC80, BIRC5, CDC20, CDC25C, MCM5, CCNE2, CCNB1, SPC25, MAD2L1, CCNB2, HMCN1, SGO2, SPAG5, NCAPG, NCAPG2, ZWINT, CENPW, BUB1B |
| GO:0007067:mitotic nuclear division | 17 | 5.90E-08 | CDK1, KIF11, NEK2, KIF15, NUF2, BIRC5, NDC80, CDC20, PBK, CEP55, CDC25C, SPC25, CCNB2, NCAPG2, BUB1B, CENPW, ASPM |
| GO:0000070:mitotic sister chromatid segregation | 7 | 4.24E-07 | MAD2L1, NEK2, SPAG5, ZWINT, NUSAP1, KIF18B, NDC80 |
| GO:0007062:sister chromatid cohesion | 11 | 4.75E-07 | SPC25, MAD2L1, SGO2, ZWINT, KIF18A, NUF2, BUB1B, NDC80, BIRC5, CDC20, CENPK |
| GO:0007052:mitotic spindle organization | 7 | 1.35E-06 | CCNB1, SPC25, KIF11, PCNT, TTK, NDC80, STMN1 |
| GO:0007019:microtubule depolymerization | 5 | 4.11E-06 | KIF14, STMN3, KIF18A, KIF18B, STMN1 |
| GO:0045893:positive regulation of transcription, DNA templated | 21 | 4.56E-06 | SOX11, PAX6, TGFB3, ATAD2, LEF1, TBX1, CREB5, SOX9, GLI2, MDK, FZD7, GLI1, MYCN, SMARCD3, LHX2, ZNF711, TFAP2B, CAND2, RFX3, PTCH1, SOX18 |
| GO:0030574:collagen catabolic process | 8 | 1.18E-05 | MMP10, COL6A3, COL6A2, COL6A1, ADAMTS3, COL11A1, COL5A2, MMP12 |
| GO:0007059:chromosome segregation | 8 | 1.77E-05 | SPC25, KIF11, NEK2, SPAG5, NUF2, CENPW, NDC80, TOP2A |
| GO:0006260:DNA replication | 11 | 1.92E-05 | CDK1, GINS2, POLE2, DTL, RRM2, BRIP1, CDC25C, MCM5, FEN1, MCM6, NFIB |
| hsa04110:cell cycle | 13 | 4.75E-09 | CCNE2, CCNB1, CDK1, MAD2L1, CCNB2, GADD45G, TGFB3, TTK, BUB1B, CDC20, CDC25C, MCM5, MCM6 |
| hsa04115:p53 signaling pathway | 6 | 6.65E-04 | CCNB1, CCNE2, CDK1, CCNB2, RRM2, GADD45G |
| hsa04974:protein digestion and absorption | 6 | 0.002273 | COL6A3, COL6A2, COL6A1, COL11A1, COL5A2, DPP4 |
| hsa05217:basal cell carcinoma | 5 | 0.002604 | LEF1, PTCH1, GLI2, FZD7, GLI1 |
| hsa03030:DNA replication | 4 | 0.006291 | POLE2, MCM5, FEN1, MCM6 |
| hsa04512:ECM-receptor interaction | 5 | 0.013198 | COL6A3, COL6A2, COL6A1, COL11A1, COL5A2 |
| hsa04914:progesterone-mediated oocyte maturation | 5 | 0.013198 | CCNB1, CDK1, MAD2L1, CCNB2, CDC25C |
| hsa05200:pathways in cancer | 10 | 0.021829 | CCNE2, TGFB3, RUNX1T1, LEF1, BIRC5, PTCH1, GLI2, FZD7, GNG7, GLI1 |
| hsa04114:oocyte meiosis | 5 | 0.027764 | CCNE2, CDK1, MAD2L1, CDC20, CDC25C |
| hsa04340:hedgehog signaling pathway | 3 | 0.032592 | PTCH1, GLI2, GLI1 |
Abbreviations: ECM, extracellular matrix; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 3.
The top 10 GO terms and KEGG pathways of downregulated genes
| Term | Count | P-value | Genes |
|---|---|---|---|
| GO:0048704:embryonic skeletal system morphogenesis | 4 | 7.52E-04 | HOXB2, HOXB7, HOXA5, HOXA6 |
| GO:0036109:alpha-linolenic acid metabolic process | 3 | 0.001566736 | ELOVL5, FADS1, FADS2 |
| GO:0006636:unsaturated fatty acid biosynthetic process | 3 | 0.002096572 | ELOVL5, FADS1, FADS2 |
| GO:0043651:linoleic acid metabolic process | 3 | 0.002699483 | ELOVL5, FADS1, FADS2 |
| GO:0001558:regulation of cell growth | 4 | 0.005913176 | MELTF, BCAR1, NANOS1, CYR61 |
| GO:0009952:anterior/posterior pattern specification | 4 | 0.005913176 | HOXB2, HOXB7, HOXA5, HOXA6 |
| GO:0007267:cell–cell signaling | 6 | 0.006210487 | BMP2, ADRB2, FADS1, AREG, GDF15, CYR61 |
| GO:0055007:cardiac muscle cell differentiation | 3 | 0.006763636 | BMP2, SIK1, NRG1 |
| GO:0060325:face morphogenesis | 3 | 0.008308263 | DKK1, TIPARP, RRAS |
| GO:2000726:negative regulation of cardiac muscle cell differentiation | 2 | 0.013694378 | BMP2, DKK1 |
| hsa01040:biosynthesis of unsaturated fatty acids | 3 | 0.005255803 | ELOVL5, FADS1, FADS2 |
| hsa01212:fatty acid metabolism | 3 | 0.02175659 | ELOVL5, FADS1, FADS2 |
| hsa05230:central carbon metabolism in cancer | 3 | 0.037090419 | SLC1A5, HKDC1, MYC |
Abbreviations: GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
PPI network analysis and module analysis
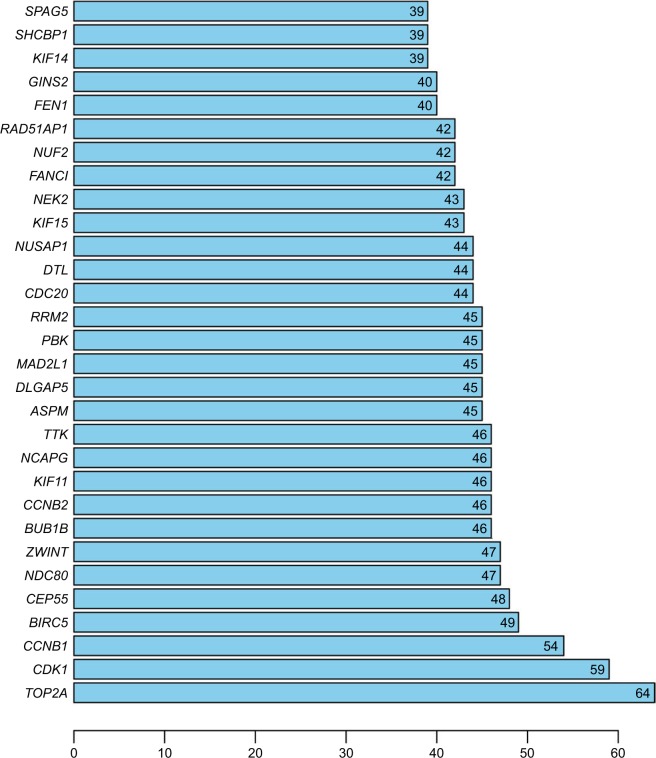
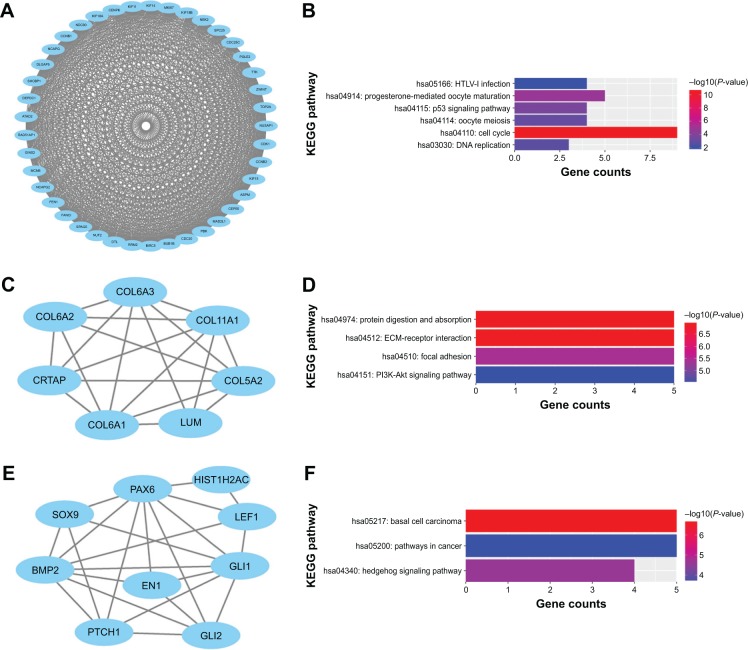
After data of interactions imported into Cytoscape software, the PPI network with 202 nodes and 1,245 edges was constructed. Based on this network, TOP2A (degree =64), CDK1 (degree =59), and CCNB1 (degree =54) were screened as the top three hub genes due to the higher degrees (Figure 2). Subsequently, we performed module analysis of the whole network by the MCODE plug-in. Three modules were identified and created as subnetworks. In addition, pathway enrichment analysis of genes included in each subnetwork was performed, which revealed that DEGs in modules 1–3 were mainly associated with “cell cycle”, “extracellular matrix (ECM)-receptor interaction”, “basal cell carcinoma”, and “hedgehog signaling pathway” (Figure 3).
Figure 2.
Histogram of degrees of the top 30 genes in the protein–protein interaction network.
Note: The number displayed on each column is the degree of each gene.
Figure 3.
Three subnetworks obtained from the whole protein–protein interaction network.
Notes: (A, B) Module 1 and the pathway enrichment analysis of genes in module 1. (C, D) Module 2 and the pathway enrichment analysis of genes in module 2. (E, F) Module 3 and the pathway enrichment analysis of genes in module 3. Vertical axis represents GO or pathway terms. P-values are displayed by gradient colors.
Abbreviations: ECM, extracellular matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
BCC, with low malignancy, is the most common skin cancer worldwide. Although rarely metastasize, BCC can cause substantial local tissue damage along with disfigurement and involve other adjacent areas of soft tissue, cartilage, and bone.7 Currently, the targeted treatments of BCC implicated in clinical practice mainly focus on the hedgehog signaling pathway.21 However, the issue of drug resistance and poor response rate cannot be ignored. In order to explore more potential therapeutic targets, the gene expression profiles of BCC need to be comprehensively studied. In our present study, a bioinformatics approach was conducted to reanalyze the gene expression profiles of 19 BCC and 6 normal skin tissues. A total of 313 DEGs were identified with 222 upregulated genes and 91 downregulated genes. Functional and pathway enrichment analysis indicated that these DEGs were significantly associated with mitosis, cell cycle, and unsaturated fatty acid metabolic process. By PPI network and module analysis, three critical genes and four pathways were finally identified, which may play a key role in the carcinogenesis of BCC.
With regard to functional and pathway enrichment analysis, upregulated DEGs were mainly involved in the process of mitosis and cell cycle. Deregulation of cell cycle is a common feature in the initiation and progression of various cancers, which is often mediated by alterations in cyclin and cyclin-dependent kinase (CDK) activity.22 CDK1, as a mitotic CDK, is sufficient to drive the mammalian cell cycle without other interphase CDKs.23 Accumulating evidences indicated that dysregulation of CDK1 activity was participated in a variety of tumors, including lung cancer,24 prostate cancer,25 and colorectal cancer.26 Schmit et al also discovered that increased level of CDK1 and CCNB1 presented in nonmelanoma skin cancer cells (BCC and squamous cell carcinoma) compared with normal human epidermal keratinocytes growth.27 Moreover, patched1, the BCC-related protein, was found to be interacted with cyclin B1 to regulate cell-cycle progression in BCC.28,29 Recently, targeting cyclin-dependent kinases has become a promising approach in cancer therapy. AZD5438, as a highly specific inhibitor of CDK1, 2, and 9, was discovered to enhance the radiosensitivity of non-small-cell lung cancer.30 In the present study, our results revealed that CDK1 was significantly upregulated in BCC samples and enriched in many cell cycle-related GO terms, which indicated the potential to be a therapeutic target in BCC.
Topoisomerases have been considered as important therapeutic targets for human malignancies. TOP2A, the major isoform of topoisomerase II, is capable of resolving catenanes and supercoils during DNA metabolic processes and plays a critical role in condensation and segregation of chromosomes at mitosis. Accumulating studies highlighted that higher TOP2A expression level was correlated to advanced tumor stage and poor patients’ survival in human cancers. At the protein level, increased expression of topoisomerase IIα was demonstrated to be associated with elevated cell replication in BCC compared with squamous cell carcinoma.31 In our study, TOP2A was screened as the most significant gene with the highest degree and was up-regulated in BCC. Elevated expression of TOP2A was implicated in cell cycle, and targeting TOP2A was also considered as an important therapy for human cancers.32 Thus, TOP2A could be a critical target in BCC.
COL6A1, COL6A2, COL6A3, COL5A2, and COL11A1 are members of the collagen family, and these five genes are enriched in the pathway of “ECM–receptor interaction”, which leads to a direct or indirect control of cellular activities such as adhesion, migration, differentiation, proliferation, and apoptosis. Accumulating evidence indicated that the “ECM–receptor interaction” pathway served as a critical role in the carcinogenesis and metastasis of human cancers, such as prostate cancer,33 breast cancer,34 and colorectal cancer.35 In this study, we also screened “ECM–receptor interaction” as an important pathway by module analysis, which indicated the potential role in the pathogenesis of BCC.
Hedgehog signaling pathway, a highly conserved evolutionary pathway of signal transmission from the cell membrane to the nucleus, has been revealed to be associated with the development of cancers, especially in BCC.5 The main downstream genes of hedgehog signaling pathway include PTCH1, GLI1, and GLI2. In the module 3 analysis, these three genes were significantly enriched in “basal cell carcinoma”, “hedgehog signaling pathway”, and “pathways in cancer”. Currently, targeting the hedgehog signaling pathway has been an important strategy for cancer therapy, which has achieved a promising success in BCC.21 However, the targeted genes were restricted to two genes (PTCH1 and SMO). Therefore, the other critical genes in this pathway are expected to be studied.
Of note, several limitations also existed in our work. First, the inclusive criteria for BCC patients and normal controls was not available due to lack of data from the public database. Second, the same as most previous studies, two relatively small patient cohorts were performed in this study. Third, there was a lack of validation in biological experiments or another dataset, which might increase the FDR in our results.
In conclusion, we performed a comprehensive bioinformatics analysis of DEGs obtained from 19 BCC and 6 normal skin tissues. Three hub genes and four pathways were finally identified, which might play a critical role in BCC. Our results further revealed the potential molecular mechanisms during the initiation of BCC and laid the foundation of exploring effective molecular targets for the treatment of BCC. However, future biology experiments are required to confirm these findings.
Supplementary materials
Table S1.
Differentially expressed genes between basal cell carcinoma and normal skin tissues
| Upregulated genes |
| ADAMTS3 |
| LHX2 |
| CHGA |
| LGR5 |
| SOX11 |
| S100A9 |
| PTCH1 |
| FBN3 |
| FAT3 |
| TMSB15A |
| KCNE1 |
| MYCN |
| CRNN |
| COL11A1 |
| MMP10 |
| BNC2 |
| GAS2 |
| TOX2 |
| SPON2 |
| TFAP2B |
| GLI2 |
| HEPH |
| LMO3 |
| ADAMTS17 |
| VASH2 |
| LINGO1 |
| DIO2 |
| CHST2 |
| PCDHB2 |
| PCDH8 |
| NPNT |
| SOX18 |
| PITX2 |
| UHRF1 |
| TBX1 |
| CREB5 |
| ABI3BP |
| LINC00865 |
| EDIL3 |
| GPC4 |
| SHCBP1 |
| SLC6A1 |
| SERPINB4 |
| GJB6 |
| APCDD1L |
| SOSTDC1 |
| LRRN1 |
| VCAN |
| BGN |
| FZD7 |
| SFRP5 |
| TNRC6C |
| MARCH1 |
| MUM1L1 |
| ZNF711 |
| SHOX2 |
| LOC101929122 |
| F2RL2 |
| DTL |
| TSPYL5 |
| CASC15 |
| PELI2 |
| NRTN |
| GLI1 |
| SETBP1 |
| FNDC1 |
| MEGF6 |
| RAD51AP1 |
| PAPPA |
| SOBP |
| HUNK |
| NINL |
| UCP2 |
| HIST1H4C |
| ADGRL3 |
| CHRDL2 |
| NAP1L3 |
| NTRK3 |
| TOP2A |
| SOX9 |
| TSPAN18 |
| H2BFXP |
| DLGAP5 |
| MAD2L1 |
| S100A8 |
| PLEKHG4B |
| NUF2 |
| GMPR |
| NDC80 |
| LRIG3 |
| SLC7A2 |
| CENPK |
| KRT85 |
| ALDH1A3 |
| MMP12 |
| BIRC5 |
| PCNT |
| KALRN |
| KCNS3 |
| SDC2 |
| CYFIP2 |
| KIF11 |
| COL5A2 |
| CNTN4 |
| GBP6 |
| BACH2 |
| HS3ST3A1 |
| LEF1 |
| SGO2 |
| GINS1 |
| CDH11 |
| TM4SF1 |
| KIF14 |
| MARCKSL1 |
| STMN1 |
| LOC440173 |
| PCDHB10 |
| NEK2 |
| APOBEC3A |
| SMARCD3 |
| IFI27 |
| SH3GL3 |
| SLC6A6 |
| NUDT10 |
| MDK |
| CMPK2 |
| APELA |
| SHANK2 |
| GADD45G |
| RUNX1T1 |
| TTK |
| CCNB1 |
| TET1 |
| LTBP1 |
| PBK |
| KRT13 |
| CDC20 |
| ABCC12 |
| DENND2A |
| BEND5 |
| ASPM |
| NUSAP1 |
| RRM2 |
| CENPW |
| DMD |
| BRIP1 |
| SLAIN1 |
| SYNPO |
| PTPRN2 |
| NRXN3 |
| STMN3 |
| MXRA5 |
| FANCD2 |
| DCHS1 |
| MCM6 |
| CDC25C |
| CDH22 |
| COL6A2 |
| SORCS2 |
| DPP4 |
| TIGD1 |
| SPAG5 |
| MTFR2 |
| KIF15 |
| KIF18A |
| RFX3 |
| PRIMA1 |
| CCNB2 |
| STON1 |
| CDC42EP4 |
| MCM5 |
| ZWINT |
| SEMA6A |
| ZNF367 |
| CEP55 |
| HMCN1 |
| TMTC1 |
| BUB1B |
| PABPC4L |
| FANCI |
| ZNF566 |
| CAND2 |
| POLE2 |
| CDK1 |
| STC1 |
| ALMS1 |
| PLCE1 |
| NFIB |
| PCSK5 |
| PLPPR1 |
| COL6A1 |
| NCAPG |
| BHLHE41 |
| CCNE2 |
| PAX6 |
| MIR3682 |
| PRRT2 |
| GINS2 |
| NCAPG2 |
| CFAP44 |
| COL6A3 |
| FEN1 |
| TNFSF10 |
| PLEKHO1 |
| TNS3 |
| KIF18B |
| ATAD2 |
| TMEM173 |
| ZNF853 |
| SLCO2A1 |
| TBC1D1 |
| GNG7 |
| DEPDC1 |
| SPC25 |
| OSBPL7 |
| TAGLN |
| NTN1 |
| TGFB3 |
| BICC1 |
| IFI44 |
| MKI67 |
| LUM |
| Downregulated genes |
| CRTAP |
| ID3 |
| CNTN1 |
| NRG1 |
| SLC22A15 |
| TIPARP |
| MMP28 |
| IDH1-AS1 |
| KLHDC8B |
| XG |
| SMIM21 |
| HOXB2 |
| NEFL |
| CRELD1 |
| HKDC1 |
| C11orf70 |
| MYADM |
| LPAR3 |
| IL17RC |
| FAM110A |
| BMP2 |
| CBS |
| NUDT16P1 |
| PHLDB2 |
| IMPACT |
| HOXB7 |
| RTN4RL1 |
| SLC1A5 |
| RRAS |
| SNORA4 |
| GDF15 |
| C8orf88 |
| CDKN2AIPNL |
| ESPN |
| ADGRF4 |
| KLF6 |
| PTPN20 |
| BCAR1 |
| PRSS21 |
| MOCOS |
| PLPP2 |
| LURAP1L |
| MINDY2 |
| MAFF |
| ERRFI1 |
| DLK2 |
| CTH |
| GNAI1 |
| HOXA6 |
| CORO2A |
| C1orf21 |
| MST1R |
| EVPLL |
| PYROXD2 |
| CYR61 |
| SYBU |
| ELOVL5 |
| HOOK2 |
| SERPINB2 |
| FAS |
| AREG |
| FAM89A |
| HIST1H2BD |
| IL13RA2 |
| QPRT |
| AP1S1 |
| BTBD16 |
| ACP5 |
| MFSD2A |
| ANTXR2 |
| RAB5C |
| NANOS1 |
| CCPG1 |
| C11orf63 |
| FADS1 |
| HIST1H2AC |
| EML2 |
| TSC22D3 |
| DKK1 |
| FKBP5 |
| MYC |
| ATF3 |
| HOXA5 |
| EN1 |
| SIK1 |
| ECM2 |
| CHMP4C |
| RNF128 |
| FADS2 |
| MELTF |
| ADRB2 |
Acknowledgments
This project was supported by grants from the National Nature Science Foundation of China (No 81472027) to S-KW; Key Project of Science and Technology Development of Nanjing Medicine (ZDX16001); and innovation team of Jiangsu provincial health-strengthening engineering by science and education (CXTDB2017008) to S-KW.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Eisemann N, Waldmann A, Geller AC, et al. Non-melanoma skin cancer incidence and impact of skin cancer screening on incidence. J Invest Dermatol. 2014;134(1):43–50. doi: 10.1038/jid.2013.304. [DOI] [PubMed] [Google Scholar]
- 2.Devine C, Srinivasan B, Sayan A, Ilankovan V. Epidemiology of basal cell carcinoma: a 10-year comparative study. Br J Oral Maxillofac Surg. 2018;56(2):101–106. doi: 10.1016/j.bjoms.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 3.Narayanan DL, Saladi RN, Fox JL. Ultraviolet radiation and skin cancer. Int J Dermatol. 2010;49(9):978–986. doi: 10.1111/j.1365-4632.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- 4.Lesiak A, Sobolewska-Sztychny D, Majak P, et al. Relation between sonic hedgehog pathway gene polymorphisms and basal cell carcinoma development in the Polish population. Arch Dermatol Res. 2016;308(1):39–47. doi: 10.1007/s00403-015-1612-9. [DOI] [PubMed] [Google Scholar]
- 5.Skoda AM, Simovic D, Karin V, et al. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci. 2018;18(1):8–20. doi: 10.17305/bjbms.2018.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durmaz CD, Evans G, Smith MJ, et al. A novel PTCH1 frameshift mutation leading to nevoid basal cell carcinoma syndrome. Cytogenet Genome Res. 2018;154(2):57–61. doi: 10.1159/000487747. [DOI] [PubMed] [Google Scholar]
- 7.Eberl M, Mangelberger D, Swanson JB, et al. Tumor architecture and notch signaling modulate drug response in basal cell carcinoma. Cancer Cell. 2018;33(2):229–243. doi: 10.1016/j.ccell.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migden MR, Guminski A, Gutzmer R, et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol. 2015;16(6):716–728. doi: 10.1016/S1470-2045(15)70100-2. [DOI] [PubMed] [Google Scholar]
- 9.Whitson RJ, Lee A, Urman NM, et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat Med. 2018;24(3):271–281. doi: 10.1038/nm.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonilla X, Parmentier L, King B, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 11.Riker AI, Enkemann SA, Fodstad O, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy – analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium Gene Ontology Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein– protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xin M, Ji X, de La Cruz LK, Thareja S, Wang B. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med Res Rev. 2018;38(3):870–913. doi: 10.1002/med.21482. [DOI] [PubMed] [Google Scholar]
- 22.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 23.Santamaría D, Barrière C, Cerqueira A, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448(7155):811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Elkahloun AG, Robertson M, et al. Loss of cytoplasmic CDK1 predicts poor survival in human lung cancer and confers chemotherapeutic resistance. PLoS One. 2011;6(8):e23849. doi: 10.1371/journal.pone.0023849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willder JM, Heng SJ, Mccall P, et al. Androgen receptor phosphorylation at serine 515 by Cdk1 predicts biochemical relapse in prostate cancer patients. Br J Cancer. 2013;108(1):139–148. doi: 10.1038/bjc.2012.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung WW, Lin YM, Wu PR, et al. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer. 2014;14:951. doi: 10.1186/1471-2407-14-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmit TL, Zhong W, Nihal M, Ahmad N. Polo-like kinase 1 (Plk1) in non-melanoma skin cancers. Cell Cycle. 2009;8(17):2697–2702. doi: 10.4161/cc.8.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes EA, Kong M, Ollendorff V, Donoghue DJ. Patched1 interacts with cyclin B1 to regulate cell cycle progression. Embo J. 2001;20(9):2214–2223. doi: 10.1093/emboj/20.9.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66(4):2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 30.Raghavan P, Tumati V, Yu L, et al. AZD5438, an inhibitor of Cdk1, 2, and 9, enhances the radiosensitivity of non-small cell lung carcinoma cells. Int J Radiat Oncol Biol Phys. 2012;84(4):e507–e514. doi: 10.1016/j.ijrobp.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stelkovics E, Kiszner G, Meggyeshazi N, et al. Selective in situ protein expression profiles correlate with distinct phenotypes of basal cell carcinoma and squamous cell carcinoma of the skin. Histol Histopathol. 2013;28(7):941–954. doi: 10.14670/HH-28.941. [DOI] [PubMed] [Google Scholar]
- 32.Delgado JL, Hsieh CM, Chan NL, Hiasa H. Topoisomerases as anticancer targets. Biochem J. 2018;475(2):373–398. doi: 10.1042/BCJ20160583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Guo W, Xu H, et al. An extensive study of the mechanism of prostate cancer metastasis. Neoplasma. 2018;65(2):253–261. doi: 10.4149/neo_2018_161217N648. [DOI] [PubMed] [Google Scholar]
- 34.Fang E, Zhang X. Identification of breast cancer hub genes and analysis of prognostic values using integrated bioinformatics analysis. Cancer Biomark. 2017;21(1):373–381. doi: 10.3233/CBM-170550. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, Wang Y, Zhang L, et al. Exploration of the mechanism of colorectal cancer metastasis using microarray analysis. Oncol Lett. 2017;14(6):6671–6677. doi: 10.3892/ol.2017.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Differentially expressed genes between basal cell carcinoma and normal skin tissues
| Upregulated genes |
| ADAMTS3 |
| LHX2 |
| CHGA |
| LGR5 |
| SOX11 |
| S100A9 |
| PTCH1 |
| FBN3 |
| FAT3 |
| TMSB15A |
| KCNE1 |
| MYCN |
| CRNN |
| COL11A1 |
| MMP10 |
| BNC2 |
| GAS2 |
| TOX2 |
| SPON2 |
| TFAP2B |
| GLI2 |
| HEPH |
| LMO3 |
| ADAMTS17 |
| VASH2 |
| LINGO1 |
| DIO2 |
| CHST2 |
| PCDHB2 |
| PCDH8 |
| NPNT |
| SOX18 |
| PITX2 |
| UHRF1 |
| TBX1 |
| CREB5 |
| ABI3BP |
| LINC00865 |
| EDIL3 |
| GPC4 |
| SHCBP1 |
| SLC6A1 |
| SERPINB4 |
| GJB6 |
| APCDD1L |
| SOSTDC1 |
| LRRN1 |
| VCAN |
| BGN |
| FZD7 |
| SFRP5 |
| TNRC6C |
| MARCH1 |
| MUM1L1 |
| ZNF711 |
| SHOX2 |
| LOC101929122 |
| F2RL2 |
| DTL |
| TSPYL5 |
| CASC15 |
| PELI2 |
| NRTN |
| GLI1 |
| SETBP1 |
| FNDC1 |
| MEGF6 |
| RAD51AP1 |
| PAPPA |
| SOBP |
| HUNK |
| NINL |
| UCP2 |
| HIST1H4C |
| ADGRL3 |
| CHRDL2 |
| NAP1L3 |
| NTRK3 |
| TOP2A |
| SOX9 |
| TSPAN18 |
| H2BFXP |
| DLGAP5 |
| MAD2L1 |
| S100A8 |
| PLEKHG4B |
| NUF2 |
| GMPR |
| NDC80 |
| LRIG3 |
| SLC7A2 |
| CENPK |
| KRT85 |
| ALDH1A3 |
| MMP12 |
| BIRC5 |
| PCNT |
| KALRN |
| KCNS3 |
| SDC2 |
| CYFIP2 |
| KIF11 |
| COL5A2 |
| CNTN4 |
| GBP6 |
| BACH2 |
| HS3ST3A1 |
| LEF1 |
| SGO2 |
| GINS1 |
| CDH11 |
| TM4SF1 |
| KIF14 |
| MARCKSL1 |
| STMN1 |
| LOC440173 |
| PCDHB10 |
| NEK2 |
| APOBEC3A |
| SMARCD3 |
| IFI27 |
| SH3GL3 |
| SLC6A6 |
| NUDT10 |
| MDK |
| CMPK2 |
| APELA |
| SHANK2 |
| GADD45G |
| RUNX1T1 |
| TTK |
| CCNB1 |
| TET1 |
| LTBP1 |
| PBK |
| KRT13 |
| CDC20 |
| ABCC12 |
| DENND2A |
| BEND5 |
| ASPM |
| NUSAP1 |
| RRM2 |
| CENPW |
| DMD |
| BRIP1 |
| SLAIN1 |
| SYNPO |
| PTPRN2 |
| NRXN3 |
| STMN3 |
| MXRA5 |
| FANCD2 |
| DCHS1 |
| MCM6 |
| CDC25C |
| CDH22 |
| COL6A2 |
| SORCS2 |
| DPP4 |
| TIGD1 |
| SPAG5 |
| MTFR2 |
| KIF15 |
| KIF18A |
| RFX3 |
| PRIMA1 |
| CCNB2 |
| STON1 |
| CDC42EP4 |
| MCM5 |
| ZWINT |
| SEMA6A |
| ZNF367 |
| CEP55 |
| HMCN1 |
| TMTC1 |
| BUB1B |
| PABPC4L |
| FANCI |
| ZNF566 |
| CAND2 |
| POLE2 |
| CDK1 |
| STC1 |
| ALMS1 |
| PLCE1 |
| NFIB |
| PCSK5 |
| PLPPR1 |
| COL6A1 |
| NCAPG |
| BHLHE41 |
| CCNE2 |
| PAX6 |
| MIR3682 |
| PRRT2 |
| GINS2 |
| NCAPG2 |
| CFAP44 |
| COL6A3 |
| FEN1 |
| TNFSF10 |
| PLEKHO1 |
| TNS3 |
| KIF18B |
| ATAD2 |
| TMEM173 |
| ZNF853 |
| SLCO2A1 |
| TBC1D1 |
| GNG7 |
| DEPDC1 |
| SPC25 |
| OSBPL7 |
| TAGLN |
| NTN1 |
| TGFB3 |
| BICC1 |
| IFI44 |
| MKI67 |
| LUM |
| Downregulated genes |
| CRTAP |
| ID3 |
| CNTN1 |
| NRG1 |
| SLC22A15 |
| TIPARP |
| MMP28 |
| IDH1-AS1 |
| KLHDC8B |
| XG |
| SMIM21 |
| HOXB2 |
| NEFL |
| CRELD1 |
| HKDC1 |
| C11orf70 |
| MYADM |
| LPAR3 |
| IL17RC |
| FAM110A |
| BMP2 |
| CBS |
| NUDT16P1 |
| PHLDB2 |
| IMPACT |
| HOXB7 |
| RTN4RL1 |
| SLC1A5 |
| RRAS |
| SNORA4 |
| GDF15 |
| C8orf88 |
| CDKN2AIPNL |
| ESPN |
| ADGRF4 |
| KLF6 |
| PTPN20 |
| BCAR1 |
| PRSS21 |
| MOCOS |
| PLPP2 |
| LURAP1L |
| MINDY2 |
| MAFF |
| ERRFI1 |
| DLK2 |
| CTH |
| GNAI1 |
| HOXA6 |
| CORO2A |
| C1orf21 |
| MST1R |
| EVPLL |
| PYROXD2 |
| CYR61 |
| SYBU |
| ELOVL5 |
| HOOK2 |
| SERPINB2 |
| FAS |
| AREG |
| FAM89A |
| HIST1H2BD |
| IL13RA2 |
| QPRT |
| AP1S1 |
| BTBD16 |
| ACP5 |
| MFSD2A |
| ANTXR2 |
| RAB5C |
| NANOS1 |
| CCPG1 |
| C11orf63 |
| FADS1 |
| HIST1H2AC |
| EML2 |
| TSC22D3 |
| DKK1 |
| FKBP5 |
| MYC |
| ATF3 |
| HOXA5 |
| EN1 |
| SIK1 |
| ECM2 |
| CHMP4C |
| RNF128 |
| FADS2 |
| MELTF |
| ADRB2 |