Abstract
Long noncoding RNAs (lncRNAs) are emerging as major regulators of a variety of cell signaling processes. Many lncRNAs are expressed in immune cells and appear to play critical roles in the regulation of immune response. Here, we have investigated the potential role of a well-known lncRNA, HOTAIR, in inflammatory and immune response. Our studies demonstrate that HOTAIR expression is induced in immune cells (macrophages) upon treatment with lipopolysaccharide (LPS). Knockdown of HOTAIR reduces NF-κB-mediated inflammatory gene and cytokine expression in macrophages. Inhibition of NF-κB resulted in down-regulation of LPS-induced expression of HOTAIR as well as IL-6 and iNOS expression. We further demonstrated that HOTAIR regulates activation of NF-κB and its target genes (IL-6 and iNOS) expression via facilitating the degradation of IκBα. HOTAIR knockdown reduces the expression of NF-κB target gene expression via inhibiting the recruitment of NF-κB and associated cofactors at the target gene promoters. Taken together, our findings suggest that HOTAIR is a critical player in NF-κB activation in macrophages suggesting its potential functions in inflammatory and immune response.
Introduction
The mammalian immune system orchestrates innate and adaptive immune responses that are complex biochemical processes regulated by various protein and lipid mediators such as pattern recognition receptors, cytokines, chemokines, hormones, growth factors, and prostaglandins, etc1–4. Toll-like receptors (TLRs) play central roles in the innate reaction to bacterial infection and in immune responses against pathogens5–8. In particular, Toll-like receptor 4 (TLR4) is activated by endotoxin lipopolysaccharide (LPS) present in the outer membrane of Gram-negative bacteria9–13. TLR4 activation triggers a series of downstream signaling cascades including NF-κB activation14–19 and induction of cytokines, chemokines, and pro-inflammatory genes6,20–24. Increasing evidences suggest that noncoding RNAs (ncRNAs) are integral components of variety of cellular and physiological signaling processes including in immune response25–29. Notably ncRNAs are recently discovered classes of transcripts, which are coded by the genome, but remains mostly untranslated into proteins30–34. Noncoding RNAs, which are longer than 200 nucleotides, are classified as long-noncoding RNAs (lncRNAs)35–37. It is estimated that human genome encodes more than 28000 lncRNAs, though their functions and detailed characterization are yet to be revealed25,36,38. Examples of lncRNAs implicated in immune response include lincRNA-Cox2, linc1992/THRIL, nc-1L7R, NeST RNA, NEAT1 and others38–41. LincRNA-Cox2 regulates IL-6 expression, linc1992/THRIL regulates expression of IL-8, CXCL10, CCL1, and CSF139, nc-1L7R knockdown diminishes IL-6 and IL-8 mRNA levels40, NeST RNA induces secretion of IFNγ in CD8+ T cells41, NEAT1 causes transcriptional activation of IL-839 and so on. Here we explored potential roles of a well-known lncRNA called HOTAIR (HOX transcript antisense intergenic RNA) in cytokine regulation and immune response. HOTAIR is a 2.2 kb long lncRNA, an antisense transcript and is associated with gene repression42–44. HOTAIR interacts with gene silencing and chromatin modifying factors such as polycomb repressive complex 2 (PRC2)25 and lysine specific demethylase 1 (LSD1, a histone H3K4-demethylase) complexes42,45. EZH2 (Enhancer of zeste homolog 2) is a H3K27-methylase which is an interacting component of PRC246–48. H3K27-methylation by EZH2 and H3K4-demethylation by LSD1 are crucial to gene silencing42,49,50. HOTAIR facilitates recruitment of PRC2 and LSD1 multi-protein complexes at the target genes promoters, which induce H3K27-methylation and H3K4-demethylaytion respectively and contribute to gene silencing45,51. HOTAIR upregulation is associated with a variety of cancers37,52. Beyond gene repression, HOTAIR is also found to be involved in protein degradation associated with neuronal function and diseases53,54. Previous studies from our laboratory showed that HOTAIR is required for the viability of breast cancer cells and its expression is regulated by estradiol and hypoxia55–57. Here, we demonstrate that HOTAIR is a critical player in LPS-induced cytokine expression, immune, and inflammatory response in macrophages.
Materials and Methods
Cell culture and treatment with Lipopolysaccharide (LPS)
Mouse macrophage RAW264.7 cells were obtained from American Type Cell Culture Collection, and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated FBS (Fetal bovine serum), 2 mM L-glutamine, 100 units/mL penicillin and 0.1 mg/mL streptomycin in a humidified incubator with 5% CO2 and 95% air at 37 °C58. 2 × 106 cells were seeded in 60 mm cell culture plates and after overnight culture cells were treated with ultrapure E.coli-derived LPS (Invivogen) at 1.0 μg/mL concentration for different time periods58. Notably, this concentration of LPS has been widely used in various other laboratories to induce immune and pro-inflammatory response in macrophages58,59. Cells were harvested for the preparation of RNA and protein analysis.
RNA extraction, cDNA synthesis and real time PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. Briefly RAW264.7 cells treated with various conditions were harvested using TRIzol, mixed with chloroform and centrifuged at 12000 rpm for 15 min. The aqueous layer was mixed with equal volume of 100% ethanol and centrifuged at 12000 rpm for 10 min. The pellet was washed with 70% ethanol, and RNA was finally dissolved in 30–50 μL of RNase-free water (Sigma) and quantified using a Nanodrop spectrophotometer58. 1 μg of the isolated RNA was reverse transcribed into cDNA using iScript RT-supermix (Bio-Rad). For semi quantitative PCR the cDNA was PCR-amplified by using Taq DNA Polymerase (Genscript) as described earlier60 and primers are listed in Table 1. Real-time PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad), with gene specific PCR primers as listed in Table 1, in CFX96 real-time detection system (Bio-Rad). Each experiment was repeated three times with three parallel replicates each time. Expression data were normalized to GAPDH and expressed as 2−ΔCt 61,62.
Table 1.
Sequences of primers and antisense oligonucleotides.
| Primers | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| PCR Primers | ||
| HOTAIR | TCCAGATGGAAGGAACTCCAGACA | ATAGATGTGCGTGGTCAGATCGCT |
| IL-6 | CAAGAAAGACAAAGCCAGAGTC | GAAATTGGGGTAGGAAGGAC |
| iNOS | TGTGACACACAGCGCTACAACA | GAAACTATGGAGCACAGCCACAT |
| GAPDH | ACCCAGAAGACTGTGGATGG | CACATTGGGGGTAGGAACAC |
| FgB | CTATGGCTGCTGCTGCTATTG | GGCTCTTCCTTTCTCCTGTCAAC |
| Mt1 | AATGTGCCCAGGGCTGTGT | GCTGGGTTGGTCCGATACTATT |
| TNFα | TCCCAGGTTCTCTTCAAGGGA | GGTGAGGAGCACGTAGTCGG |
| MIP-1B | CCTGACCAAAAGAGGCAGAC | GAGGAGGCCTCTCCTGAAGT |
| ChIP PCR Primers | ||
| IL-6 promotera | GCACACTTTCCCCTTCCTAGTT | AGACTCATGGGAAAATCCCACATT |
| iNOS-promotera | GTGTACCTCAGACAAGGGCA | AAGCATTCACACATGGCATGGA |
| Antisense Oligonucleotides | ||
| HOTAIR Antisense | 5′-C*C*T*T*C*C*T*T*C*C*G*C*T*C*T*T*A*C*T*C*T-3′ | |
| Scramble Antisense | 5′-C*C*T*C*T*T*C*T*G*T*C*T*C*T*T*C*C*C*G*C*T-3′ | |
aChIP PCR primers are flanked around the NF-κB binding site.
*All phosphorothioate linkages instead of regular phosphodiester bonds.
Western blot analyses
The cells were washed in ice-cold PBS and then lysed in cell lysis buffer comprising 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 1% Igepal CA-630, 0.5% Na-deoxycholate, 2 mM Na3VO4, and complete protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche)63. The resulting cell lysates were centrifuged for 10 min at 13,000 rpm at 4 °C, and the protein concentrations in the supernatants were determined using a BCA protein assay kit (Pierce). 20 μg proteins were loaded onto 10% SDS-PAGE gels, followed by electro-transfer onto PVDF-membrane (Immobilon-P, Millipore). The membranes were blocked in 1× TBST (0.1% Tween-20, 20 mM Tris–Cl (pH 8.0), and 150 mM NaCl) containing 5% skimmed milk and then incubated with the primary antibodies against IκBα (1:1000 dilution, 4814S, Cell Signaling), Phospho-IκBα (1:1000 dilution, 2859S, Cell Signaling), Phospho-p65 (NF-κB subunit, 1:1000 dilution, 3033S, Cell Signaling), IL-6 (1:1000 dilution, 12912S, Cell Signaling), iNOS (1:1000 dilution, 13120S, Cell Signaling), and β-actin (1:1000 dilution, A2066, Sigma) overnight at 4 °C. Membranes were washed 3 times (1xTBST), incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature and then washed 3 times (1 × TBST). Finally, immunoreactive proteins were detected using ECL-super signal west femto substrate reagent (Thermo Scientific)63. The amount has been quantified by ImageLab5.2.1software.
Chromatin Immunoprecipitation (ChIP) assay
The ChIP assay was performed as described earlier55,60. Briefly, the cells were cross-linked with 1% formaldehyde for 10 min at 37 °C, washed twice in ice-cold PBS and harvested using SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris. HCl, pH 8.1) supplemented with complete protease inhibitor (Roche). Cells were subjected to sonication to shear the chromatin (∼200–300 bp range). The fragmented chromatin was pre-cleared with protein G agarose beads (16-266, EMD Millipore) and subjected to immunoprecipitation using antibodies specific to CBP (A22, Santa Cruz Biotechnology, Sc369), p300 (N15, Santa Cruz Biotechnology, Sc584), Phospho-p65 (3033, Cell Signaling), histone H3K4-trimethyl (07-473, EMD-Millipore), histone acetylation (06-599, EMD-Millipore), RNA Pol II (8WG16, Abcam), and β-actin (A2066, Sigma). Immunoprecipitated chromatin was washed, de-crosslinked and deproteinized at 65 °C in presence of 5 M NaCl followed by incubation with proteinase K (Sigma) at 45 °C for 1 h. Purified ChIP DNA fragments were analyzed by semi-quantitative PCR and real-time PCR using primers spanning NF-κB binding sites present in the IL-6 and iNOS promoters (Table 1)64–66.
Antisense-mediated knockdown of HOTAIR
For the antisense transfection, RAW 264.7 cells were grown up to 60% confluency in 60 mm culture plates and transfected with HOTAIR-antisense (HOTAIR-AS) and scramble-AS (no homology to HOTAIR)55,56 independently (Table 1) using GenMute transfection reagent (SL100568, SignaGen Laboratories) according to the manufacturer’s protocol. Prior to transfection, a cocktail of transfection reagent and antisense oligonucleotides was made as follows. Initially, 12 μL (12 μg) of GenMute reagent was mixed with 300 μL DMEM (without FBS and antibiotics) in an eppendorf tube. In a separate eppendorf, antisense oligonucleotide was mixed with 100 μL DMEM (without supplements), then mixed with diluted GenMute reagents and allowed to stand for 30 min in dark. In the meantime, cells were washed twice with supplement-free DMEM, and then 1.7 mL of supplement-free DMEM was added. Finally, antisense transfection reagents cocktail was applied, mixed gently and incubated for 48 h. Cells were then stimulated with LPS (1 μg/mL) for specified time period and then harvested for RNA/protein extraction or for ChIP assays.
SiRNA-mediated knockdown of HOTAIR
For the siRNA transfection, RAW 264.7 cells were grown up to 60% confluency in 60 mm culture plates and transfected with HOTAIR-siRNA, a pool of 4 different siRNA constructs (SI05685183, SI05685190, SI05685197, and SI05685204 Qiagen)67 and scramble siRNA (1027310 Qiagen) independently using GenMute transfection reagent (SL100568, SignaGen Laboratories) according to the manufacturer’s protocol. Prior to transfection, a cocktail of transfection reagent and antisense oligonucleotide was made as follows. Initially, 12 μL (12 μg) of GenMute reagent was mixed with 300 μL DMEM (without FBS and antibiotics) in an eppendorf tube. In a separate eppendorf, siRNA was mixed with 100 μL DMEM (without supplements). Then the diluted siRNA solution was mixed with diluted GenMute reagents and allowed to stand for 30 min in the dark. Cells were washed twice with supplement-free DMEM, 1.7 mL of supplement-free DMEM was added and finally, siRNA transfection reagents cocktail was applied to the cell plates, mixed gently and incubated for 48 h. Cells were then stimulated with LPS (1 μg/mL) for specified time period and harvested for RNA/protein extraction.
NF-κB inhibition assay
RAW264.7 macrophages (2 × 106) were seeded in 60 mm cell culture plates. After overnight incubation cells were initially treated with IKKβ inhibitor (25 μM, SC-514, Sigma)68 for 1 h to inhibit NF-kB signaling pathway and then cells were treated with LPS (1 μg/mL) and incubated for additional period of time 4 h. Cells were harvested, total RNA was isolated using TRIzol reagent, reverse transcribed to cDNA and analyzed by qPCR for the expression of HOTAIR, IL-6 and iNOS. GAPDH was used as control. Protein was also extracted (SC-514 1 h and additional1 h LPS treatment) after cell harvesting for Western blot.
Proteasomal inhibition assay
RAW264.7 macrophages (2 × 106) were seeded in 60 mm cell culture plates. After overnight incubation cells were initially treated with or without HOTAIR-AS for 48 h. The cells were then treated with MG132 (Sigma) (10 μM) for 2 h followed by LPS (1 μg/mL) and incubated for additional 1 h and followed the procedure for immunofluorescence microscopy analysis69.
Enzyme linked immunosorbent assay (ELISA)
HOTAIR was silenced in RAW264.7 macrophages by using HOTAIR-AS and HOTAIR-siRNA, a pool of 4 different siRNA constructs (SI05685183, SI05685190, SI05685197, and SI05685204 Qiagen) by the use of GenMute siRNA transfection reagent (SL100568, SignaGen Laboratories) according to the manufacturer’s protocol. 48 h following transfection RAW264.7 macrophages were stimulated with LPS for 12 h or kept untreated. Culture media were collected and amount of IL-6 secreted in culture media were measured using ELISA kits (DY406-05 R&D Systems) according to manufacturer’s instruction58.
Immunofluorescence microscopy analysis
For immunofluorescence staining of macrophages, cells were seeded on cover slips and fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature, washed with 1X PBS (3 times for 5 min each) and blocked with 1X PBS containing 5% goat normal serum and 0.3% Triton-X100 for 1 h. The cells were then incubated with primary antibodies (rabbit anti-P-p65, 1:200, 3033, CST; and mouse anti-IκBα (L35A5, 1:400, 4814, CST) overnight at 4 °C. After that the cells were washed 3 times with PBS followed by incubation with anti-rabbit Alexa Fluor 564 (Invitrogen) and anti-mouse Alexa Fluor 488 (Invitrogen), conjugated secondary antibodies for 1 h at RT. Finally the cells were washed 3 times with PBS and mounted with mounting media containing DAPI. Images were taken by fluorescence microscope (Nikon ECLIPSE TE2000-U). The amount has been quantified by the National Institutes of Health (NIH) ImageJ software.
Mice
Wild-type (C57BL6/J) mice were purchased from Jackson Laboratory. All mice are maintained in a specific pathogen free (SPF) facility at UT Southwestern Medical center. All studies were approved by the Institutional Animal Care and Use Committee (IACUC) and were conducted in accordance with the IACUC guidelines and the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Isolation and culture of primary macrophage (bone marrow derived macrophage, BMDM)
BMDM was isolated from mice bones and cultured as described earlier58,70. For the isolation of BMDMs, femur and tibia were collected from mouse legs. Using 25G needle bone marrows were flushed out with Iscove’s Modified Dulbecco’s Medium (IMDM), (12440061, Life technologies) and processed for single cell suspension by passing through 22G needle two times. The suspension was centrifuged at 1000 RPM for 5 min. The pellet was re-suspended with BMDM culture media (L-cell-conditioned IMDM medium supplemented with 15% L929 supernatant, 10% FBS, 1% nonessential amino acid, and 1% penicillin-streptomycin) followed by seeding in three 150 mm culture dishes and cultured for 6 days to differentiate into macrophages, while at day 3, 10 ml fresh BMDM culture media was added into each plate. After day 6, the culture plate was washed with ice cold PBS and cells were gently scraped with ice cold PBS. The BMDM was centrifuged at 1000 rpm for 5 min and re-suspended into BMDM media. The BMDM was counted and seeded in 6-well (2.5 × 106/well) cell culture plates. After overnight incubation the BMDM was treated with LPS or HOTAIR antisense oligonucleotides and processed for further experiments.
Statistical Analysis
Each experiment was done in two or three replicates, and then cells were pooled (and treated as one sample), subjected to RNA extraction, RT-PCR, and ChIP analysis, and each experiment was repeated at least three times (n = 3). The real-time PCR analysis of such samples were done in three replicate reactions and repeated in all three independent experiments (n = 3). Data are presented as means ± SD (as stated in the figure legends). Statistical significance was determined by unpaired Student’s t test (GraphPad Prism 6), and P ≤ 0.05 was considered statistically significant.
Results
LncRNA HOTAIR expression is induced by Lipopolysaccharide in macrophages
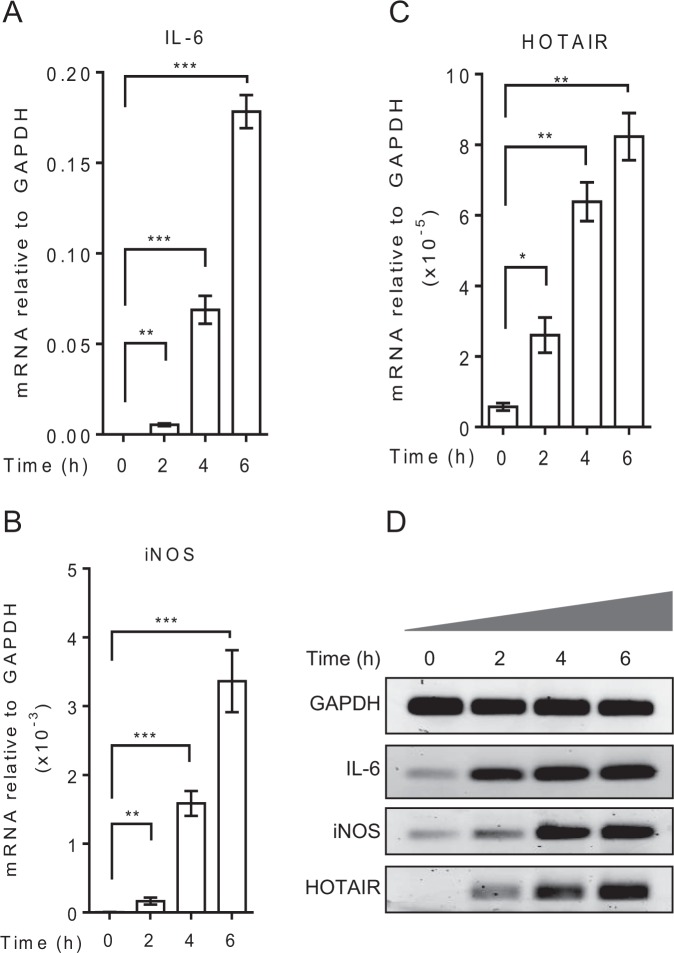
To investigate the roles of lncRNA in immune response, initially we explored potential involvement of lncRNA HOTAIR. As HOTAIR is well-known as a repressor49,50,71,72, initially we hypothesized that HOTAIR may be involved in repression of cytokine and inflammatory gene expression. To test this hypothesis, we treated macrophage cells (RAW264.7) with LPS and analyzed its impacts on the expression of HOTAIR along with well-known cytokines (e.g. interleukin-6, IL-6) and pro-inflammatory genes (e.g. inducible nitric oxide synthase, iNOS)73,74. RNA from the LPS-treated macrophages were analyzed by RT-qPCR. As expected, LPS-treatment induced the expression of IL-6 and iNOS in time-dependent manner (Fig. 1A,B). The expression of IL-6 was induced by 522, 6722 and 17321 folds at 2, 4 and 6 h post LPS-treatment, respectively (Figs 1A, S1A shows the fold change). Similarly, the expression of iNOS was also induced by 31, 292 and 621 folds at 2, 4 and 6 h respectively upon treatment with LPS (Figs 1B and S1A). Notably, along with IL-6 and iNOS, we also examined the LPS-induced expression of additional cytokines and pro-inflammatory genes, such as tumor necrosis factor α (TNFα), macrophage inflammatory protein-1β (MIP-1β), beta-fibrinogen (FgB) and metallothionein1 (Mt1) (Fig. S1B)75,76. TNFα and MIP-1β were significantly induced by LPS-treatment, while LPS-induced expressions of FgB and Mt1 were relatively low (Fig. S1B). Interestingly, along with IL-6, iNOS and other cytokines and inflammatory genes, lncRNA HOTAIR expression was also induced upon treatment with LPS (5, 11 and 14 folds at 2, 4 and 6 h, respectively) (Figs 1C and S1A). Notably, LPS-dependent induction of IL-6 and iNOS is much higher compared to HOTAIR expression, though the LPS-induced HOTAIR expression is significant. The cDNA was also analyzed by semi-quantitative PCR and products were analyzed on agarose gel showing the LPS-dependent induction of IL-6, iNOS and HOTAIR (Figs 1D and S1C). Overall, these observations demonstrated that along with IL-6, iNOS, lncRNA HOTAIR expression is induced upon stimulation with LPS and it may be associated with inflammatory and immune response in macrophages.
Figure 1.
LPS induces HOTAIR expression in macrophages. RAW264.7 cells were treated with LPS (1 μg/mL) for varying period of time, total RNA was isolated, reverse transcribed to cDNA and analyzed by qPCR for expression of IL-6 (A) iNOS (B) and HOTAIR (C). cDNA was also analyzed by semi-quantitative PCR and agarose gel (D). Each experiment was repeated at least with three parallel replicates. GAPDH was used as loading control. Data represent mean ± SD (n = 3); *p < 0.05, **p < 0.001, ***p < 0.0001.
Inhibition of NF-κB downregulates LPS-induced HOTAIR expression
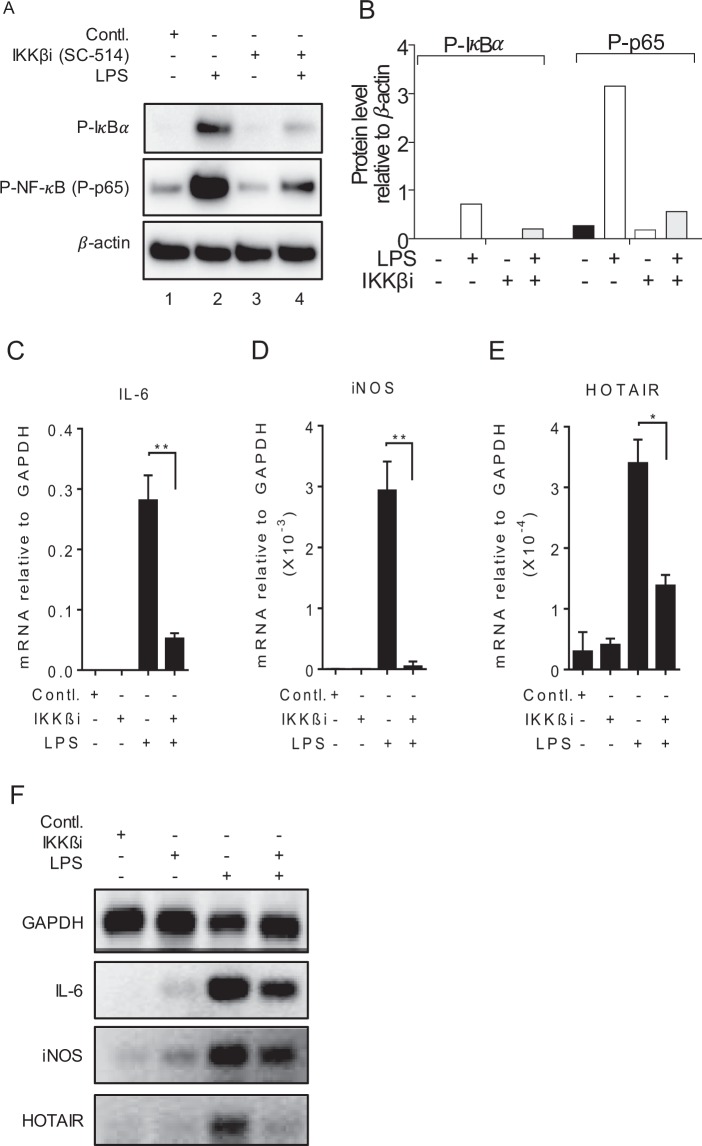
The activation of transcription factor NF-κB is well-known to be associated with LPS-induced cytokine expression (including IL-6 and iNOS) and inflammatory response77,78. To investigate if NF-κB activation is associated with LPS-induced HOTAIR expression, we treated macrophages with an inhibitor of IKKβ (SC-514)79 and analyzed its impacts on LPS-induced HOTAIR, IL-6 and iNOS expressions. Notably, IKKβ is a kinase which phosphorylates IκBα allowing its poly-ubiquitination, proteasomal degradation, and hence NF-κB activation6,77,80,81. Thus, inhibition of IKKβ (by SC-514) results in deactivation of NF-κB6,77. Briefly, RAW264.7 cells were treated with IKKβ-inhibitor SC-514 (25 µM, 1 h) and stimulated with LPS. The concentration of the inhibitor (SC-514) used is chosen based on previous literature68. Proteins from the control and SC-514 treated cells were analyzed by Western blot to detect the levels of phospho-IκBα and phospho-p6580,81. As expected SC-514 treatment resulted in a decrease in phospho-IκBα and phospho-p65 (NF-κB submit) levels, indicating the effective inhibition of IKKβ kinase activity and consequent decrease in NF-κB activation (Fig. 2A, quantifications in panel 2B, Supplementary Fig. S2A). The RNA from the control and SC-514 treated cells were also then analyzed by RT-qPCR (Fig. 2C–E) and semi-quantitative PCR (Figs 2F and S2B) for the expression of HOTAIR, IL-6 and iNOS. Interestingly, treatment with IKKβ inhibitor down-regulated LPS-induced expression of NF-κB target genes IL-6 (5.4 folds) and iNOS (15.3 folds) along with a decrease in HOTAIR (2.4 folds) expression (Fig. 2C–F). The treatment of SC-154 alone in the absence of LPS has no significant impact on IL-6, iNOS and HOTAIR expression (Fig. 2C–F). These observations suggest that similar to IL-6 and iNOS, HOTAIR expression is also regulated by NF-κB upon LPS-stimulation.
Figure 2.
Inhibition of NF-κB downregulates LPS-induced HOTAIR expression in macrophages. RAW264.7 cells were initially treated with IKKβ-inhibitor SC-514 (for 1 h) and then treated with LPS for additional 1 h for protein and 4 h for RNA. Proteins were analyzed by Western blotting using antibody against phospho-IκBα, phospho-p65 (NF-κB subunit) and β-actin (loading control) (panel A, quantifications using ImageLab5.2.1software is shown in panel B). RNA was isolated and expressions of IL-6 (C) iNOS (D) and HOTAIR (E) were measured by RT-qPCR. cDNA was also analyzed by semi-quantitative PCR and agarose gel (F). GAPDH was used as control for PCR experiments. Data represent mean ± SD (n = 3); *p < 0.05, **p < 0.001.
HOTAIR knockdown abolishes LPS-induced IL-6 and iNOS expression
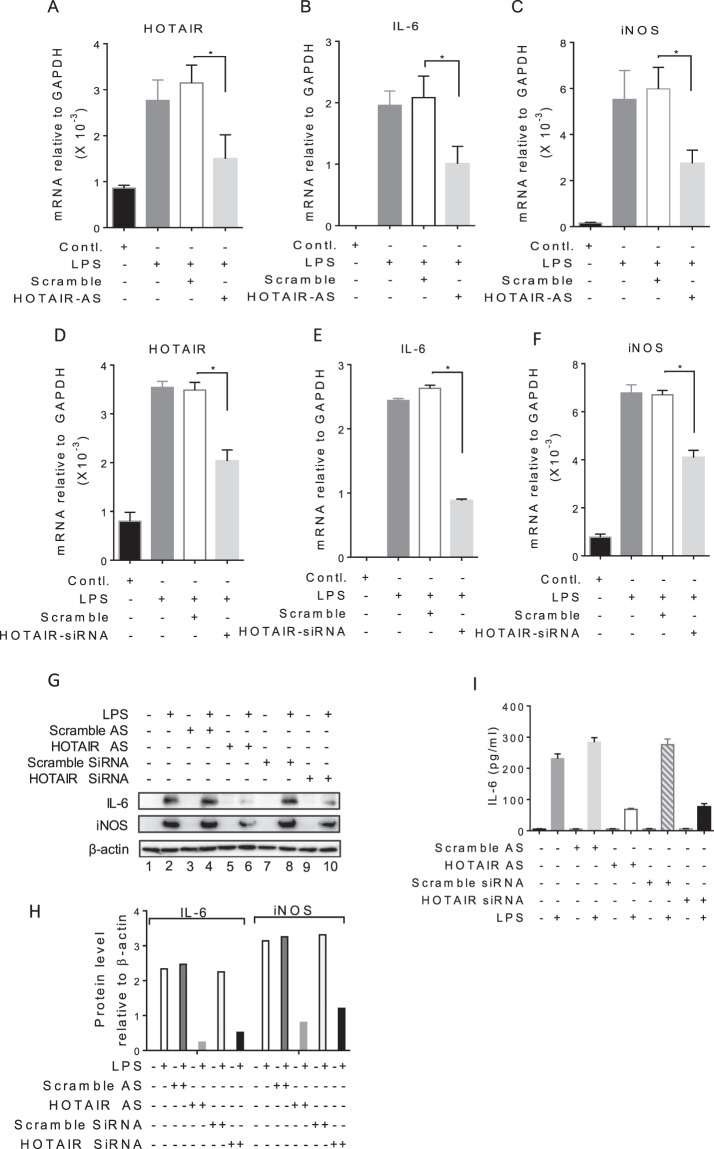
To investigate any potential contribution of HOTAIR in cytokine regulation and inflammatory response, we knocked down HOTAIR in macrophages and analyzed its impacts on LPS-induced expression of IL-6 and iNOS. In brief, RAW264.7 cells were transfected with HOTAIR-antisense (HOTAIR-AS) and scramble-antisense (control) for 48 h, then treated with LPS (1 µg/mL, 4 h) and RNA was analyzed by RT-qPCR. As expected, LPS treatment induced the expression of HOTAIR as well as IL-6 and iNOS (Fig. 3A–C). The application of HOTAIR-AS knocked down the levels of LPS-induced HOTAIR (Fig. 3A). Scramble-AS did not downregulate HOTAIR level (Fig. 3A). Interestingly, the knockdown of HOTAIR resulted in down-regulation of the LPS-induced expression levels of IL-6 and iNOS significantly (Fig. 3B,C). To further confirm the antisense-specificity of HOTAIR, we knocked down HOTAIR using HOTAIR-specific siRNA (a pool of 4 different siRNAs, targeting a different region 450–800 bp, 800–1050 bp, 1050–1550 bp and 1550–1900 bp of HOTAIR)67 and then analyzed it impacts in LPS-induced expression of IL-6 and iNOS. Interestingly, HOTAIR-siRNA knocked down HOTAIR and also down-regulated LPS-induced IL-6 and iNOS expression levels (Fig. 3D–F). To understand further the roles of HOTAIR in immune and inflammatory signaling, we analyzed the expression of additional cytokine and inflammatory genes such as TNFα, MIP-1β and others, in the absence and presence of LPS and under HOTAIR knockdown (antisense or siRNA-treatments) conditions. Interestingly, HOTAIR-knockdown (using antisense or siRNA) also down-regulated the LPS-induced expression of TNFα, MIP-1β and others, indicating further the importance of HOTAIR in immune and inflammatory response (Fig. S3A,B).
Figure 3.
Knockdown of HOTAIR reduces LPS-induced IL-6 and iNOS expressions in macrophages. (A–C) Antisense-mediated knockdown of HOTAIR followed by treatment with LPS. RAW264.7 cells were transfected with HOTAIR-AS and scramble-AS, then treated with LPS for 4 h. RNA was analyzed by RT-qPCR for the expression of HOTAIR, IL-6, and iNOS. (D–F) siRNA-mediated knockdown of HOTAIR followed by treatment with LPS. RAW264.7 cells were transfected with HOTAIR-siRNA and scramble, RNA was analyzed RT-qPCR for the expression of HOTAIR, IL-6, and iNOS. GAPDH was used as control. Data represent mean ± SD (n = 3); *p < 0.05, **p < 0.001, ***p < 0.0001. (G,H) Proteins from HOTAIR-antisense or HOTAIR-siRNA treatment followed by 6 h LPS-treated RAW264.7 cells were analyzed by Western blotting using antibodies against IL-6, iNOS and β-actin (loading control) (G). The changes in amounts of IκBα and NF-κB have been quantified by ImageLab5.2.1 software (H). (I) ELISA for IL-6 expression. HOTAIR was silenced in RAW264.7 macrophages by using HOTAIR-antisense and HOTAIR-siRNA separately, treated with LPS (12 h). Culture media were collected and amount of IL-6 secreted in culture media were measured using ELISA (n = 4).
In addition to mRNA levels, we also analyzed the protein levels of IL-6 and iNOS in the absence and presence of LPS and under HOTAIR-conditions, using Western blots. As expected, IL-6 and iNOS were increased upon treatment with LPS (compare lanes 1 and 2) and their levels were decreased upon treatment HOTAIR-AS (compare lane 2 and 6) or HOTAIR-siRNA (compare lanes 2 and 10) (Fig. 3G, quantification in H and Supplementary Fig. S3C). Scramble antisense or siRNA has no significant impacts on the LPS-induced expression of IL-6 and iNOS. Furthermore, we also measured the secretion of IL-6 in culture media using ELISA and found that secreted IL-6 level was increased upon treatment with LPS and that were decreased upon treatment HOTAIR-AS or HOTAIR-siRNA (Fig. 3I). Taken together, our analysis demonstrated HOTAIR is required for LPS-induced expression of cytokines and inflammatory response genes in macrophages.
HOTAIR knockdown results in inactivation of NF-κB
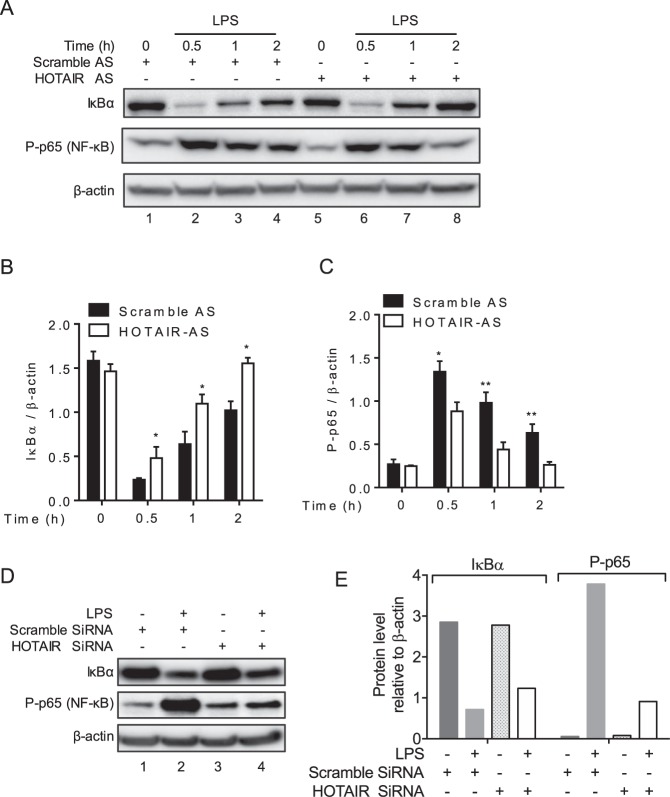
As HOTAIR is required for LPS-induced expression of IL-6 and iNOS, we investigated its potential mechanism of action in this process. Notably, IL-6 and iNOS are regulated via activation of transcription factor NF-κB82–84 and therefore, we explored if HOTAIR may be involved in NF-κB activation. We knocked down HOTAIR (using HOTAIR-antisense) in RAW264.7 macrophages and treated with LPS (varying time) and then analyzed the protein levels of IκBα and NF-κB (phospho-p65 subunit) (Figs 4A–C and S4A). Beta-actin was used a loading control. As expected, IκBα level was significantly decreased upon treatment with LPS (compare lane 1 with lanes 2–4, Fig. 4A, quantification of IκBα levels relative to β-actin in Fig. 4B). The degradation of IκBα was highest at 0.5 h of LPS-treatment and it was slightly increased over time (Fig. 4A,B). As expected the level of phospho-p65 (NF-κB subunit) was also increased (NF-κB activation) upon LPS-treatment in comparison to the control (compare lane 1 with lanes 2–4, Fig. 4A and quantification in 4C). The decrease in IκBα and concomitant increase in phospho-p65 upon LPS treatment suggest that LPS-stimulation has induced proteasomal degradation of IκBα resulting in activation of NF-κB. Interestingly, upon treatment with HOTAIR-AS (HOTAIR-knockdown), the level of LPS-induced IκBα degradation was decreased in comparison to LPS alone (compare lanes 2 with 6, 3 with 7, 4 with 8, Fig. 4A,B). HOTAIR-knockdown also reduced phospho-p65 levels compared LPS-alone (compare lanes 2 and 6; 3 and 7; 4 and 8; Fig. 4A,C). To cross verify the impact of HOTAIR-knockdown on deactivation of NF-κB, we also examined the IκBα and NF-κB (phospho-p65 subunit) levels under HOTAIR-siRNA treatment followed by LPS-stimulation (Figs 4D,E and S4B). Interestingly, siRNA-mediated knockdown of HOTAIR resulted in decrease in LPS-induced IκBα degradation and reduced phospho-p65 levels (compare lanes 2 with 4 in Fig. 4D, quantifications in 4E). Scramble-siRNA has no significant impacts on LPS-induced level of IκBα degradation and phospho-p65 (Fig. 4D,E). These observations demonstrate that HOTAIR is required for LPS-induced proteasomal degradation of IκBα and NF-κB activation.
Figure 4.
HOTAIR promotes IκBα degradation thereby activates NF-κB. (A–C) Antisense-mediated knockdown of HOTAIR followed by LPS-treatment. RAW264.7 macrophages were transfected with HOTAIR- or scramble antisense for 48 h, treated with LPS for different time periods (0.5 h, 1 h and 2 h) and protein was isolated. The protein was resolved on SDS-PAGE and immunoblotted with antibody against IκBα, phospho p65 (NF-κB subunit) and β-actin (loading control). The changes in amounts of IκBα and NF-κB have been quantified by ImageLab5.2.1 software (B,C). (D,E) siRNA-mediated knockdown of HOTAIR followed by LPS-treatment. Proteins from HOTAIR- or scramble siRNA (48 h-treatment) followed by LPS (1 h) -treated RAW264.7 were analyzed by Western blot using antibody against IκBα, phospho p65 and β-actin. Quantifications (ImageLab5.2.1software) are shown in panel E. Data represent mean ± SD; *p < 0.05, **p < 0.001.
HOTAIR promotes IκBα degradation and nuclear translocation of NF-κB
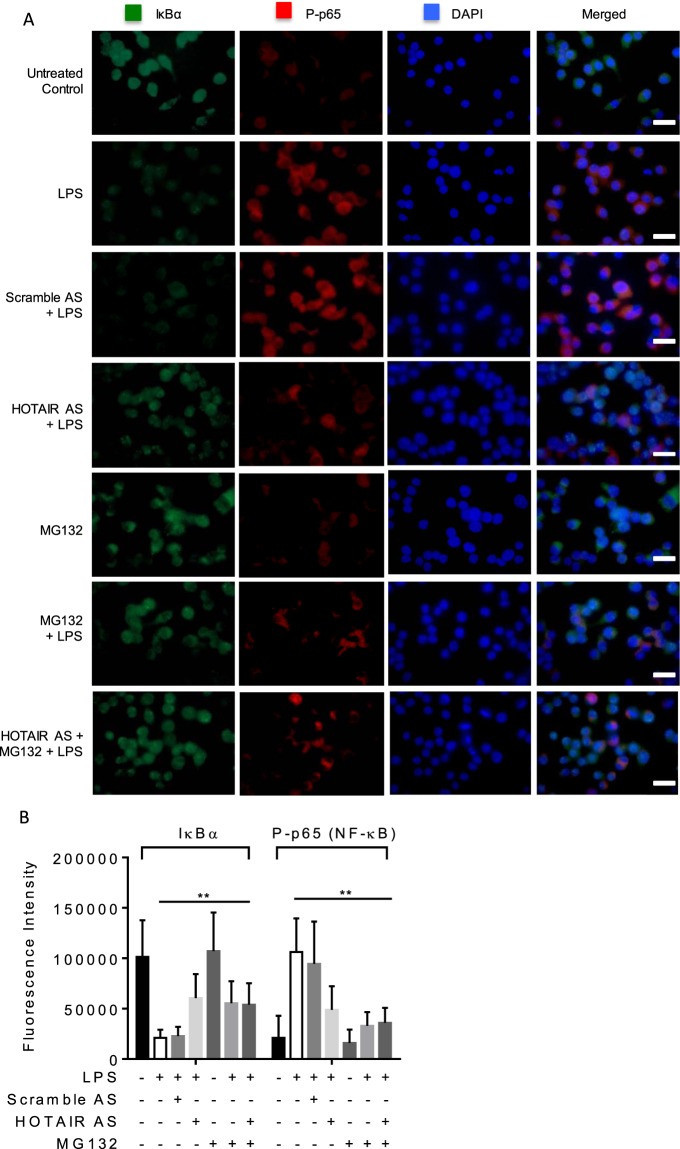
To further understand the function of HOTAIR in NF-κB activation, we measured the co-expression of IκBα and phospho-p65 (NF-κB) in macrophages by using immunofluorescence assay. We knocked down HOTAIR in RAW264.7 cells by using HOTAIR-AS and scramble-AS (control) and then treated LPS. Control and antisense-treated cells were subjected to immunostaining with IκBα and phospho-p65 (NF-κB) antibodies. Cell nucleus was stained with DNA binding dye DAPI. We observed that IκBα protein levels were higher while phospho NF-κB protein levels were lower in unstimulated (in the absence of LPS) control cells (Fig. 5A, top panel, quantification of the immunofluorescence staining images are shown in Fig. 5B). However, when stimulated with LPS we found a decrease in IκBα levels and increase in phospho NF-κB in macrophages compared to LPS-untreated control cells (compare top two panels, Fig. 5A). Interestingly, upon HOTAIR-knockdown (HOTAIR-AS and LPS treatments), IκBα level was increased in comparison to LPS-treatment alone (compare panels 4 and 2, Fig. 5A,B). The level of phospho-p65 protein (NF-κB) was decreased concomitantly upon knockdown of HOTAIR (HOTAIR-AS and LPS treatment) relative to LPS alone (compare panels 4 and 2, Fig. 5A,B). Scramble antisense has no significant impact on LPS-induced expression of IκBα and phospho-p65 levels (Fig. 5A,B). These results further indicate that HOTAIR is required for LPS-induced proteasomal degradation of IκBα and activation of NF-κB in immune cells. Notably, to further understand the potential involvement of HOTAIR in proteasomal degradation of IκBα, we applied a well-known proteasomal inhibitor MG132 followed by treatment with LPS in the presence and absence of HOTAIR knockdowns. Interestingly, application of MG132 (followed by treatment with LPS) also inhibited the level of LPS-induced degradation of IκBα and lowered phospho-p65 level (compare panels 6 and 2, Fig. 5A,B). The effects of independent treatment of MG132 or HOTAIR-AS on inhibition of LPS-induced degradation of IκBα and phospho-p65 level are comparable (compare panels 6 and 4, Fig. 5A,B). The application of MG132, in combination with HOTAIR-AS followed by LPS-treatment, showed no further significant impact in comparison to MG132 or HOTAIR-AS treatments alone (compare panels 7 with 6 and 4, respectively, Fig. 5A,B). Overall, these studies demonstrate that HOTAIR plays potential roles in the regulation of proteasomal degradation of IκBα and subsequent activation of NF-κB in response to LPS in immune cells.
Figure 5.
HOTAIR promotes IκBα degradation and nuclear translocation of NF-κB. HOTAIR was silenced in RAW 264.7 cells by using HOTAIR antisense and scramble (control) for 48 h. Additionally, cells were also treated with proteasomal inhibitor MG132 (2 h) alone or in combination with HOTAIR-knockdown and then treated with LPS (1 h). The cells were then fixed with paraformaldehyde and immunostained with antibody against IκBα and phospho NF-κB (P-p65), and counterstained with DAPI. Images were taken by fluorescence microscope (Nikon ECLIPSE TE2000-U) (A) and fluorescence intensity showing the expressions of IκBα and phospho-p65 was quantified and plotted by ImageJ software (B). Data represent mean ± SD; *p < 0.05, **p < 0.001.
HOTAIR knockdown abolishes the recruitment of NF-κB at IL-6 and iNOS promoters
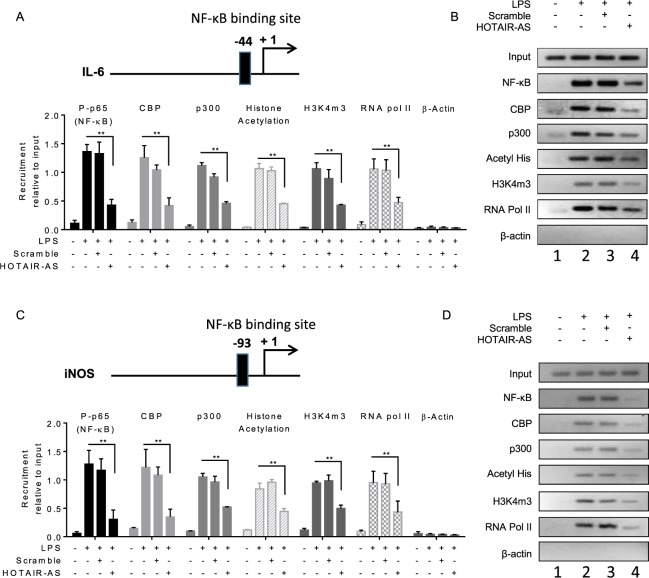
The pro-inflammatory cytokine and inflammatory response genes such as IL-6 and iNOS are regulated via transcription factor NF-κB85,86. The promoters of IL-6 and iNOS contain NF-κB binding sites (NF-κB response elements) (Figs 6A,C and S5A,B). As HOTAIR is required for NF-κB activation and expression of NF-κB regulated genes IL-6 and iNOS expression, we analyzed HOTAIR-dependent recruitment of NF-κB at the NF-κB binding sites present in IL-6 and iNOS promoters as a function of LPS-treatment, using chromatin immunoprecipitation (ChIP) assay. Briefly, control and HOTAIR-knocked down RAW264.7 treated with LPS were subjected ChIP using phosphorylated p65 (NF-κB subunit) and β-actin (control) and ChIP DNA fragments were PCR-amplified using primers specific to NF-κB binding sites present in IL-6 and iNOS promoters60,87. Interestingly, these analyses demonstrated that phospho-p65 (NF-κB) levels were enriched at the IL-6 and iNOS promoters (NF-κB response element regions) upon treatment with LPS and these LPS-induced NF-κB binding was reduced upon HOTAIR-knockdown in both IL-6 and iNOS promoters (see p-65 ChIP qPCR data in Fig. 6A,C and also compare lanes 2 and 4, Fig. 6B,D for regular PCR analysis). Notably, along with NF-κB, there are other coactivators which are associated with LPS-induced IL-6 and iNOS expression. For example, histone acetyltransferase CBP/p300 is known to interact with NF-κB to regulate NF-κB target genes88–90. Our ChIP analysis demonstrates that similar to NF-κB, the levels of CBP/p300 were also enriched at IL-6 and iNOS promoters and these LPS-induced enrichments were alleviated upon knockdown of HOTAIR (Fig. 6A,B for qPCR analysis and C-D for regular PCR analysis, compare lanes 2 and 4). Scramble antisense treatment has no significant impact on the LPS-induced enrichment of NF-κB and CBP/p300 at the IL-6 and iNOS promoters. As a control, we measured β-actin (antibody control), but no enrichment was observed irrespective of the LPS or HOTAIR-AS treatment (Figs 6A–D, S5A,B). These observations suggest that HOTAIR plays key roles in LPS-induced NF-κB activation and hence its enrichment at the promoters of NF-κB target genes such as IL-6 and iNOS to regulate their expression.
Figure 6.
Knockdown of HOTAIR reduces the recruitment of transcription factors and coactivators at NF-κB binding sites on IL-6 and iNOS promoters. RAW264.7 macrophage cells were transfected with HOTAIR or scramble-antisense, then treated with LPS (1.5 h). Cells were then fixed with formaldehyde and subjected to ChIP assay using antibodies specific to phospho-p65, CBP, p300, histone acetylation, H3K4m3, RNA pol II and β-actin (control). The immunoprecipitated DNA fragments were analyzed by qPCR (panels A shows the ChIP analysis for IL-6, and C for iNOS) and semi-quantative PCR (panels B and D) using primers specific to the NF-κB binding regions on IL-6 and iNOS promoters. The location of NF-κB binding sites at the IL-6 and iNOS promoters are shown at the top of panels A and C respectively. Each experiment was repeated at least thrice (n = 3). Data represent mean ± SD; *p < 0.05, **p < 0.001.
Histone H3K4-trimethylation and histone acetylation are also well known marks for gene activation60,87,91–93. ChIP analysis demonstrated that levels of H3K4-trimethylation and histone acetylation as well as the level of RNA polymerase II (RNA pol II) were enriched at IL-6 and iNOS promoters in the presence of LPS and this was decreased upon HOTAIR knockdown (Fig. 6A–D). These observations suggested that HOTAIR is required for promoter activation (H3K4-methylation and histone acetylation) of IL-6 and iNOS and this is mediated via activation of NF-κB followed by recruitment of NF-κB and its coregulators including histone methyl-transferases and histone acetyl-transferases at the target gene promoters. Taken together our ChIP analysis demonstrated that LPS-induced expression of IL-6 and iNOS are regulated via transcription factors NF-κB, CBP/p300 and other coactivators and this is regulated by HOTAIR via regulation of NF-κB activation.
HOTAIR expression is induced by LPS in primary macrophages and is required for LPS-induced cytokine expression
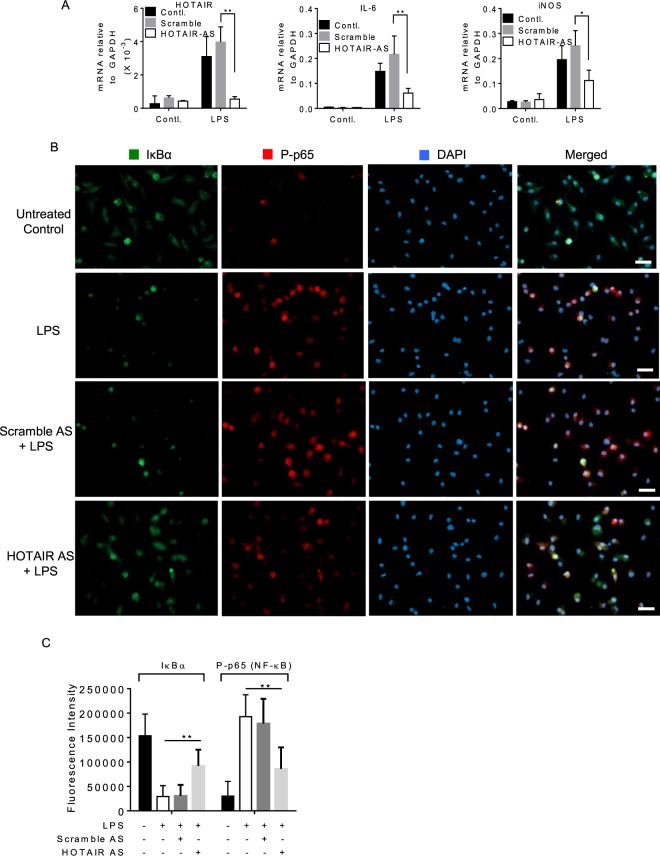
We investigated further the importance of HOTAIR in cytokine expression and immune response in primary macrophages, bone marrow derived macrophages. Briefly, BMDM cells were treated with LPS (1 µg/mL, for 4 h) in the presence and absence of HOTAIR and scramble (control) antisense, RNA was analyzed by RT-qPCR. Interestingly, the levels of IL-6 and iNOS as well as HOTAIR expression were induced upon treatment with LPS in BMDM (compare 1st and 4th bar graphs in each panel, Fig. 7A). Application of HOTAIR-AS resulted in significant knockdown of LPS-induced HOTAIR expression level (compare 4th or 5th with 6th bar graphs, left panel, Fig. 7A). Importantly, the levels of LPS- induced expression of IL-6 and iNOS were decreased upon HOTAIR knockdown (compare 4th or 5th with 6th bar graphs for IL-6 and iNOS panels, Fig. 7A). HOTAIR-AS has no significant impact on IL-6 and iNOS expression in the absence of LPS and also scramble antisense has no significant impacts on LPS-induced IL-6 and iNOS expression (Fig. 7A). These observations demonstrated that HOTAIR expression is induced upon LPS-stimulation in primary macrophages and it is required for the LPS-induced expression of IL-6 and iNOS.
Figure 7.
HOTAIR expression is induced by LPS in primary macrophages (Bone marrow derived macrophages, BMDM). (A) BMDM cells were treated by HOTAIR-AS and scramble-AS followed by LPS treatment and RNA was extracted. The expression of HOTAIR, IL-6, and iNOS was measured by real time PCR. (B,C) BMDM cells were treated by HOTAIR-AS and scramble-AS, treated with LPS and were immunostained with antibodies against IκBα and phospho NF-κB (P-p65), and counterstained with DAPI to visualize the nucleus. Images were taken by fluorescence microscope (Nikon ECLIPSE TE2000-U) (B) and fluorescence intensity showing the expressions of IκBα and phospho NF-κB (P-p65) was quantified by ImageJ software and plotted in panel C. Each experiment was repeated at least thrice (n = 3). Data represent mean ± SD; *p < 0.05, **p < 0.001.
HOTAIR-knocked down and LPS-treated BMDM cells were also analyzed by immunostaining to understand the function of HOTAIR in NF-κB activation using similar experiments outlined in Fig. 5. Interestingly, we found that IκBα protein levels were higher in the control BMDM (no LPS) and this was decreased upon treatment with LPS (Fig. 7B). The level of NF-κB (phospo-p65) protein was increased upon treatment with LPS (Fig. 7B). Interestingly, we also found that upon HOTAIR-knockdown, the level of LPS-induced decrease in IκBα protein level was rescued significantly while LPS-induced phospho-p65 NF-κB protein level was decreased. Scramble antisense has no significant impacts on LPS-induced expression of IκBα and NF-κB (phospho-p65) protein levels. Quantification of immunofluorescence images showing the expression of IκBα and NF-κB (phospho-p65) protein levels under different treatments are shown in Fig. 7C. These observations based on primary macrophage analysis further demonstrate that HOTAIR plays a critical role in LPS-induced NF-κB activation and immune response.
Discussion
The human genome contains about 3 billion base pairs of which only 1.5% codes for proteins94–96. The ENCODE project has suggested that more than 80% of the genome is functionally active and codes for ncRNA and regulatory sequences96,97. NcRNAs, even though are not translated into proteins, appear to play critical roles in a variety of cellular and physiological processes including gene regulation, cells signaling, differentiation and development37,98. NcRNAs are misregulated in human diseases37,99. Increasing numbers of studies indicate that lncRNAs are associated with immune signaling and inflammatory response100. LncRNAs are expressed in immune cells including monocytes, macrophages, dendritic cells, neutrophils, T cells and B cells101. For example, lncRNA lincR-Ccr2–5′AS is associated with CD4+ T cell differentiation98. LncRNAs are linked to pathogen-response pathways such as lincRNA-Cox2 expression is elevated upon activation of the toll-like receptors in bone-marrow-derived dendritic cells and macrophages31,39,98. LincRNA-Cox2 is required for the induction of other immune-related genes, such as IL-6, Tlr1, and IL-23a in bone marrow-derived macrophages by Pam3CSK4 treatment39,102. NRON (non-protein coding RNA, repressor of NFAT) acts as a transcription regulator for immune regulation by inhibiting nucleocytoplasmic shuttling of NFAT (Nuclear Factor of Activated T cells)103. Lethe acts as a decoy lncRNA and is a negative feedback inhibitor of NF-kB signaling in inflammation by being increased with proinflammatory cytokines such as TNFα and IL-1β31,104. THRIL (TNFα and heterogeneous nuclear ribonucleoprotein L related immunoregulatory lincRNA) plays immunoregulatory roles by binding with heterogeneous nuclear ribonucleoprotein L (hnRNPL) to induce the expression of TNFα, IL-6, IL-8, CXCL10, CCL1 and CSF139,105,106. Here, we investigated the importance of lncRNA HOTAIR in immune and inflammatory response in macrophages.
Our studies demonstrate that HOTAIR expression is induced in macrophage cells in response to LPS treatment. LPS, present on the Gram-negative bacteria cell wall, is one of the most potent pathogen-associated molecular patterns (PAMPs) known and responsible for the inflammatory response observed during endotoxic shock83,107–109. LPS stimulation induces variety of cytokines, chemokines and inflammatory genes such as ILs, TNFs, interferons (IFNs), iNOS etc3,85,110–113. In our studies, we observed that LPS-treatment (macrophage RAW264.7) induced the expression of various cytokines and inflammatory genes including IL-6, iNOS, TNFα, MIP-1B and others along with HOTAIR. Interestingly, we also observed that HOTAIR is required for the LPS-induced cytokines and inflammatory genes expression. Antisense or siRNA-mediated knockdown of HOTAIR abolished the LPS-induced activation of IL-6, iNOS, TNFα, and MIP-1B. HOTAIR-knockdown down regulated the expression of IL-6 and iNOS, both at mRNA and protein levels. Independent knockdowns of HOTAIR with antisense or siRNAs targeting different regions of HOTAIR confirmed the HOTAIR-target specificity. These observations demonstrate that HOTAIR is a critical player in cytokine expression and inflammatory response in macrophages upon LPS-stimulation.
It is well known that LPS-induces activation of TLRs which activates down-stream signaling cascades and this ultimately results in degradation of IκBα and activation of NF-κB; the NF-κB activation triggers its target gene expression and induces immune and inflammatory response114–118. To investigate if LPS-induced HOTAIR expression is regulated via NF-κB activation and if HOTAIR is required for LPS-induced NF-κB activation and its target gene expression, we blocked NF-κB activation via application of IKKβ-inhibitor (SC-514). We observed that upon treatment with IKKβ-inhibitor, the levels of phospho-IκBα as well as phospho-p65 were decreased. LPS-induced HOTAIR expression is also reduced upon IKKβ-inhibition. Notably, the kinase IKKβ phosphorylates IκBα at Ser 32/36, which triggers its polyubiquitination and proteasomal degradation, resulting in release and activation of NF-κB6,77. Activation of NF-κB is accompanied by phosphorylation of its subunit p65 (at Ser 536)80,81. Thus, the decrease in phospho-IκBα as well as phospho-p65 level indicated effective inhibition of NF-κB by SC-514. The decrease in HOTAIR expression upon IKKβ-inhibition indicated potential roles of NF-κB in LPS-induced HOTAIR expression. Furthermore, our biochemical and immunofluorescence studies demonstrated that HOTAIR is required for LPS-induced degradation of IκBα and activation of NF-κB and its nuclear translocation. HOTAIR-knockdown inhibited the LPS-induced degradation of IκBα and decrease on phosphorylation of p-65 (NF-κB), similar to the effects observed upon independent application of a proteasomal inhibitor MG132. This observation indicated that HOTAIR may be important for facilitating LPS-induced degradation of IκBα and activation of NF-κB. ChIP analysis demonstrated that HOTAIR is required for the recruitment of NF-κB and its coregulators at NF-κB target genes promoters regulating their expression. Experiments using primary macrophages isolated mouse bones further support that HOTAIR is induced by LPS and its expression is required for LPS-mediated NF-κB activation and hence IL-6 and iNOS expression. Thus, based on our biochemical studies in RAW264.7 macrophage cell lines and primary cells (BMDM) we demonstrated that HOTAIR plays a central role in NF-κB activation and pro-inflammatory response in immune cells upon stimulation with LPS.
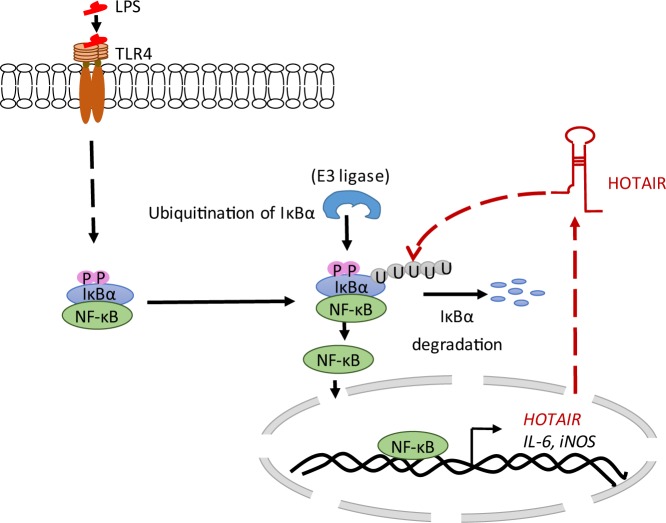
Notably, lncRNA HOTAIR is well known for its functions as repressor via interaction with chromatin modifying enzymes such as histone methylase EZH2 containing complex PRC2 and H3K4-demethylase LSD145,119–121. HOTAIR facilitates the recruitment of PRC2 and LSD1 at the target genes and induces gene silencing37,122,123. HOTAIR is also implicated in DNA damage response124, proteasomal degradation via assembling E3-ubiquitin ligases as associated neuronal siganling53,125 HOTAIR induces ubiquitin-mediated proteolysis via interaction with E3 ubiquitin ligases Dzip3 and Mex3b, along with their respective ubiquitination substrates Ataxin-1 and Snurportin-153. This leads to the degradation of Ataxin-1 and Snurportin-137,53,126. A recent study also demonstrated that HOTAIR is involved in NF-κB activation and DNA damage response in ovarian cancer cells124. HOTAIR is overexpressed in variety of cancers127. Previous studies from our lab show that HOTAIR is required for the viability of breast cancer cells and its expression is transcriptionally regulated by estradiol via coordination of estrogen receptors (ERs) and ER-coregulators such as MLL (mixed lineage leukemia)-family of histone methyltransferases and CBP/p300 in breast cancer cells55,56,60,128. Here, we have demonstrated that HOTAIR expression is induced in immune cells and is required for pathogen-induced activation of cytokine expression and pro-inflammatory response. HOTAIR plays critical roles in IκBα degradation, which results in NF-κB activation followed by its target gene (cytokine, chemokines, and pro-inflammatory genes) expression. A model, showing the potential roles of HOTAIR in regulation of IκBα degradation and hence NF-κB activation, its translocation into the nucleus and binding to the target gene promoters inducing their expressions, is shown in Fig. 8. The detailed mechanism by which HOTAIR regulates IκBα degradation remains elusive. Overall, our observations demonstrate for the first time that HOTAIR plays a central role in inflammation and immune signaling in immune cells and this may shed light into the novel immune signaling pathway that may aid development of novel therapeutics.
Figure 8.
Proposed model for LPS-mediated HOTAIR induction in immune cells. When TLR4 senses LPS, NF-κB is activated that induces IL-6, iNOS and HOTAIR expression. In turn, HOTAIR facilitates IκBα degradation and enhances NF-κB activation, nuclear translocation and binding to NF-κB regulated genes (pro-inflammatory genes such as IL-6, iNOS) inducing their expressions.
Electronic supplementary material
Acknowledgements
We thank all the Mandal lab members for helpful discussions. Research in Mandal laboratory is supported by grants from National Institute of Health (1R15 ES019129-01 and 1R15HL142032). M.H.Z. is supported by Cancer Prevention and Research Institute of Texas (CPRIT) Individual Investigator Award (RP160169).
Author Contributions
M.O. did all the experiments, was involved in experimental design and data analysis and prepared the manuscript in coordination with S.S.M. S.M.N.U. isolated the primary macrophage (BMDM) and contributed in data analysis. P.D. helped in designing primers and promoter analysis. N.S. assisted M.O. in cell culture and LPS treatment experiments. H.Z. supervised the animal experiments and isolation of BMDM from mouse bone marrow. S.S.M. conceived the idea and supervised the overall project including experimental, data analysis and writing the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33722-2.
References
- 1.Cekici A, Kantarci A, Hasturk H, Van Dyke TE. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000. 2014;64:57–80. doi: 10.1111/prd.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014;5:491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia E, Aguilar-Cevallos J, Silva-Garcia R, Ibarra A. Cytokine and Growth Factor Activation In Vivo and In Vitro after Spinal Cord Injury. Mediators Inflamm. 2016;2016:9476020. doi: 10.1155/2016/9476020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasselon T, Detmers PA. Toll receptors: a central element in innate immune responses. Infect Immun. 2002;70:1033–1041. doi: 10.1128/IAI.70.3.1033-1041.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Ferrero MC, et al. Key role of Toll-like receptor 2 in the inflammatory response and major histocompatibility complex class ii downregulation in Brucella abortus-infected alveolar macrophages. Infect Immun. 2014;82:626–639. doi: 10.1128/IAI.01237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baral P, Batra S, Zemans RL, Downey GP, Jeyaseelan S. Divergent functions of Toll-like receptors during bacterial lung infections. Am J Respir Crit Care Med. 2014;190:722–732. doi: 10.1164/rccm.201406-1101PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosmann M, Ward PA. Modulation of inflammation by interleukin-27. J Leukoc Biol. 2013;94:1159–1165. doi: 10.1189/jlb.0213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LJ, et al. The protective effect of lidocaine on lipopolysaccharide-induced acute lung injury in rats through NF-kappaB and p38 MAPK signaling pathway and excessive inflammatory responses. Eur Rev Med Pharmacol Sci. 2018;22:2099–2108. doi: 10.26355/eurrev_201804_14743. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, et al. Cirsitakaoside isolated from Premna szemaoensis reduces LPS-induced inflammatory responses in vitro and in vivo. Int Immunopharmacol. 2018;59:384–390. doi: 10.1016/j.intimp.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Mogensen T. H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clinical Microbiology Reviews. 2009;22(2):240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oviedo-Boyso J, Bravo-Patino A, Baizabal-Aguirre VM. Collaborative action of Toll-like and NOD-like receptors as modulators of the inflammatory response to pathogenic bacteria. Mediators Inflamm. 2014;2014:432785. doi: 10.1155/2014/432785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steimle A, Autenrieth IB, Frick JS. Structure and function: Lipid A modifications in commensals and pathogens. Int J Med Microbiol. 2016;306:290–301. doi: 10.1016/j.ijmm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Wang YY, et al. GABA regulates the proliferation and apoptosis of MAC-T cells through the LPS-induced TLR4 signaling pathway. Res Vet Sci. 2018;118:395–402. doi: 10.1016/j.rvsc.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, et al. Lipopolysaccharide activated TLR4/NF-kappaB signaling pathway of fibroblasts from uterine fibroids. Int J Clin Exp Pathol. 2015;8:10014–10025. [PMC free article] [PubMed] [Google Scholar]
- 17.Guijarro-Munoz I, Compte M, Alvarez-Cienfuegos A, Alvarez-Vallina L, Sanz L. Lipopolysaccharide activates Toll-like receptor 4 (TLR4)-mediated NF-kappaB signaling pathway and proinflammatory response in human pericytes. J Biol Chem. 2014;289:2457–2468. doi: 10.1074/jbc.M113.521161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 19.Kuzmich Nikolay, Sivak Konstantin, Chubarev Vladimir, Porozov Yuri, Savateeva-Lyubimova Tatiana, Peri Francesco. TLR4 Signaling Pathway Modulators as Potential Therapeutics in Inflammation and Sepsis. Vaccines. 2017;5(4):34. doi: 10.3390/vaccines5040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. 2007;39:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- 21.Kong W, et al. GTS-21 Protected Against LPS-Induced Sepsis Myocardial Injury in Mice Through alpha7nAChR. Inflammation. 2018;41:1073–1083. doi: 10.1007/s10753-018-0759-x. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki N, et al. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416:750–756. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 23.Cui H, Banerjee S, Guo S, Xie N, Liu G. IFN Regulatory Factor 2 Inhibits Expression of Glycolytic Genes and Lipopolysaccharide-Induced Proinflammatory Responses in Macrophages. J Immunol. 2018;200:3218–3230. doi: 10.4049/jimmunol.1701571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schappe MS, et al. Chanzyme TRPM7 Mediates the Ca(2+) Influx Essential for Lipopolysaccharide-Induced Toll-Like Receptor 4 Endocytosis and Macrophage Activation. Immunity. 2018;48:59–74 e55. doi: 10.1016/j.immuni.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 27.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- 28.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scaria V, Pasha A. Long Non-Coding RNAs in Infection Biology. Front Genet. 2012;3:308. doi: 10.3389/fgene.2012.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng H, Tan XD. Functional diversity of long non-coding RNAs in immune regulation. Genes Dis. 2016;3:72–81. doi: 10.1016/j.gendis.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R, et al. A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget. 2016;7:72546–72558. doi: 10.18632/oncotarget.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. RNA-Seq analysis of non-small cell lung cancer in female never-smokers reveals candidate cancer-associated long non-coding RNAs. Pathol Res Pract. 2016;212:549–554. doi: 10.1016/j.prp.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, et al. Sialidase Deficiency in Porphyromonas gingivalis Increases IL-12 Secretion in Stimulated Macrophages Through Regulation of CR3, IncRNA GAS5 and miR-21. Front Cell Infect Microbiol. 2018;8:100. doi: 10.3389/fcimb.2018.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan X, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Cui H, et al. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. 2014;44:2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez JA, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsayed, E. T., Salem, P. E., Darwish, A. M. & Fayed, H. M. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int J Biol Markers 1724600818760244, 10.1177/1724600818760244 (2018). [DOI] [PubMed]
- 44.Xiao Z, et al. LncRNA HOTAIR is a Prognostic Biomarker for the Proliferation and Chemoresistance of Colorectal Cancer via MiR-203a-3p-Mediated Wnt/ss-Catenin Signaling Pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 45.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jene-Sanz A, et al. Expression of polycomb targets predicts breast cancer prognosis. Mol Cell Biol. 2013;33:3951–3961. doi: 10.1128/MCB.00426-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 48.Michalak EM, et al. Polycomb group gene Ezh2 regulates mammary gland morphogenesis and maintains the luminal progenitor pool. Stem Cells. 2013;31:1910–1920. doi: 10.1002/stem.1437. [DOI] [PubMed] [Google Scholar]
- 49.Kogo R, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 50.Milhem, M. M. et al. Correlation of MTDH/AEG-1 and HOTAIR Expression with Metastasis and Response to Treatment in Sarcoma Patients. J Cancer Sci TherS5 (2011). [PMC free article] [PubMed]
- 51.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932–1956. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 53.Yoon JH, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, et al. PLK1 and HOTAIR Accelerate Proteasomal Degradation of SUZ12 and ZNF198 during Hepatitis B Virus-Induced Liver Carcinogenesis. Cancer Res. 2015;75:2363–2374. doi: 10.1158/0008-5472.CAN-14-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhan A, et al. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhan A, et al. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhan A, et al. Histone methyltransferase EZH2 is transcriptionally induced by estradiol as well as estrogenic endocrine disruptors bisphenol-A and diethylstilbestrol. J Mol Biol. 2014;426:3426–3441. doi: 10.1016/j.jmb.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 58.Udden SMN, et al. NOD2 Suppresses Colorectal Tumorigenesis via Downregulation of the TLR Pathways. Cell Rep. 2017;19:2756–2770. doi: 10.1016/j.celrep.2017.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu MX, et al. LPS at low concentration promotes the fracture healing through regulating the autophagy of osteoblasts via NF-kappaB signal pathway. Eur Rev Med Pharmacol Sci. 2018;22:1569–1579. doi: 10.26355/eurrev_201803_14561. [DOI] [PubMed] [Google Scholar]
- 60.Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2013;32:3359–3370. doi: 10.1038/onc.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugden K, Tichopad A, Khan N, Craig IW, D’Souza UM. Genes within the serotonergic system are differentially expressed in human brain. BMC Neurosci. 2009;10:50. doi: 10.1186/1471-2202-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 63.Udden SM, Morita-Fujimura Y, Satake M, Ikawa S. c-ABL tyrosine kinase modulates p53-dependent p21 induction and ensuing cell fate decision in response to DNA damage. Cell Signal. 2014;26:444–452. doi: 10.1016/j.cellsig.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Deb P, et al. Endocrine disrupting chemical, bisphenol-A, induces breast cancer associated gene HOXB9 expression in vitro and in vivo. Gene. 2016;590:234–243. doi: 10.1016/j.gene.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shrestha B, et al. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. FEBS J. 2012;279:3715–3726. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ansari KI, Kasiri S, Mishra BP, Mandal SS. Mixed lineage leukaemia-4 regulates cell-cycle progression and cell viability and its depletion suppresses growth of xenografted tumour in vivo. Br J Cancer. 2012;107:315–324. doi: 10.1038/bjc.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C, et al. KH-type splicing regulatory protein mediate inflammatory response in gastric epithelial cells induced by lipopolysaccharide. Cell Biol Int. 2017;41:871–878. doi: 10.1002/cbin.10804. [DOI] [PubMed] [Google Scholar]
- 69.Ortiz-Lazareno PC, et al. MG132 proteasome inhibitor modulates proinflammatory cytokines production and expression of their receptors in U937 cells: involvement of nuclear factor-kappaB and activator protein-1. Immunology. 2008;124:534–541. doi: 10.1111/j.1365-2567.2008.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit14 11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battistelli C, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soromou LW, et al. Regulation of inflammatory cytokines in lipopolysaccharide-stimulated RAW 264.7 murine macrophage by 7-O-methyl-naringenin. Molecules. 2012;17:3574–3585. doi: 10.3390/molecules17033574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aldridge C, Razzak A, Babcock TA, Helton WS, Espat NJ. Lipopolysaccharide-stimulated RAW 264.7 macrophage inducible nitric oxide synthase and nitric oxide production is decreased by an omega-3 fatty acid lipid emulsion. J Surg Res. 2008;149:296–302. doi: 10.1016/j.jss.2007.12.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Denlinger LC, et al. Regulation of inducible nitric oxide synthase expression by macrophage purinoreceptors and calcium. J Biol Chem. 1996;271:337–342. doi: 10.1074/jbc.271.1.337. [DOI] [PubMed] [Google Scholar]
- 75.Everhardt Queen A, Moerdyk-Schauwecker M, McKee LM, Leamy LJ, Huet YM. Differential Expression of Inflammatory Cytokines and Stress Genes in Male and Female Mice in Response to a Lipopolysaccharide Challenge. PLoS One. 2016;11:e0152289. doi: 10.1371/journal.pone.0152289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lampiasi N, Montana G. The molecular events behind ferulic acid mediated modulation of IL-6 expression in LPS-activated Raw 264.7 cells. Immunobiology. 2016;221:486–493. doi: 10.1016/j.imbio.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 77.Palsson-McDermott EM, O’Neill LA. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology. 2004;113:153–162. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 79.Kishore N, et al. A selective IKK-2 inhibitor blocks NF-kappa B-dependent gene expression in interleukin-1 beta-stimulated synovial fibroblasts. J Biol Chem. 2003;278:32861–32871. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- 80.Jeong SJ, Pise-Masison CA, Radonovich MF, Park HU, Brady JN. A novel NF-kappaB pathway involving IKKbeta and p65/RelA Ser-536 phosphorylation results in p53 Inhibition in the absence of NF-kappaB transcriptional activity. J Biol Chem. 2005;280:10326–10332. doi: 10.1074/jbc.M412643200. [DOI] [PubMed] [Google Scholar]
- 81.Hochrainer K, Racchumi G, Anrather J. Site-specific phosphorylation of the p65 protein subunit mediates selective gene expression by differential NF-kappaB and RNA polymerase II promoter recruitment. J Biol Chem. 2013;288:285–293. doi: 10.1074/jbc.M112.385625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 83.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 84.Lubberts E. IL-17/Th17 targeting: on the road to prevent chronic destructive arthritis? Cytokine. 2008;41:84–91. doi: 10.1016/j.cyto.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 85.Wojdasiewicz P, Poniatowski LA, Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm. 2014;2014:561459. doi: 10.1155/2014/561459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ansari KI, et al. MLL histone methylases regulate expression of HDLR-SR-B1 in presence of estrogen and control plasma cholesterol in vivo. Mol Endocrinol. 2013;27:92–105. doi: 10.1210/me.2012-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van den Berghe N, Cool RH, Wittinghofer A. Discriminatory residues in Ras and Rap for guanine nucleotide exchange factor recognition. J Biol Chem. 1999;274:11078–11085. doi: 10.1074/jbc.274.16.11078. [DOI] [PubMed] [Google Scholar]
- 89.Shenkar R, Yum HK, Arcaroli J, Kupfner J, Abraham E. Interactions between CBP, NF-kappaB, and CREB in the lungs after hemorrhage and endotoxemia. Am J Physiol Lung Cell Mol Physiol. 2001;281:L418–426. doi: 10.1152/ajplung.2001.281.2.L418. [DOI] [PubMed] [Google Scholar]
- 90.Mukherjee SP, et al. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF-kappaB-driven transcription. PLoS Biol. 2013;11:e1001647. doi: 10.1371/journal.pbio.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 92.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bayarsaihan D. Epigenetic mechanisms in inflammation. J Dent Res. 2011;90:9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 95.Mattick J. S. The Functional Genomics of Noncoding RNA. Science. 2005;309(5740):1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 96.Mattick JS, Makunin IV. Non-coding RNA. Human Molecular Genetics. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 97.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 100.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Willingham A. T. A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 104.Rapicavoli NA, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Z, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Imamura K, Akimitsu N. Long Non-Coding RNAs Involved in Immune Responses. Front Immunol. 2014;5:573. doi: 10.3389/fimmu.2014.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leone S, et al. Molecular structure of endotoxins from Gram-negative marine bacteria: an update. Mar Drugs. 2007;5:85–112. doi: 10.3390/md503085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Munford RS. Sensing gram-negative bacterial lipopolysaccharides: a human disease determinant? Infect Immun. 2008;76:454–465. doi: 10.1128/IAI.00939-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schulte W, Bernhagen J, Bucala R. Cytokines in sepsis: potent immunoregulators and potential therapeutic targets–an updated view. Mediators Inflamm. 2013;2013:165974. doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scott MG, Rosenberger CM, Gold MR, Finlay BB, Hancock RE. An alpha-helical cationic antimicrobial peptide selectively modulates macrophage responses to lipopolysaccharide and directly alters macrophage gene expression. J Immunol. 2000;165:3358–3365. doi: 10.4049/jimmunol.165.6.3358. [DOI] [PubMed] [Google Scholar]
- 111.Ivanenkov YA, Balakin KV, Tkachenko SE. New approaches to the treatment of inflammatory disease: focus on small-molecule inhibitors of signal transduction pathways. Drugs R D. 2008;9:397–434. doi: 10.2165/0126839-200809060-00005. [DOI] [PubMed] [Google Scholar]
- 112.Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol. 2007;8:1. doi: 10.1186/1471-2172-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 114.Phelps CB, Sengchanthalangsy LL, Huxford T, Ghosh G. Mechanism of I kappa B alpha binding to NF-kappa B dimers. J Biol Chem. 2000;275:29840–29846. doi: 10.1074/jbc.M004899200. [DOI] [PubMed] [Google Scholar]
- 115.Phelps CB, Sengchanthalangsy LL, Malek S, Ghosh G. Mechanism of kappa B DNA binding by Rel/NF-kappa B dimers. J Biol Chem. 2000;275:24392–24399. doi: 10.1074/jbc.M003784200. [DOI] [PubMed] [Google Scholar]
- 116.Mann PB, Kennett MJ, Harvill ET. Toll-like receptor 4 is critical to innate host defense in a murine model of bordetellosis. J Infect Dis. 2004;189:833–836. doi: 10.1086/381898. [DOI] [PubMed] [Google Scholar]
- 117.Hallman M, Ramet M, Ezekowitz RA. Toll-like receptors as sensors of pathogens. Pediatr Res. 2001;50:315–321. doi: 10.1203/00006450-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 118.Harju K, Glumoff V, Hallman M. Ontogeny of Toll-like receptors Tlr2 and Tlr4 in mice. Pediatr Res. 2001;49:81–83. doi: 10.1203/00006450-200101000-00018. [DOI] [PubMed] [Google Scholar]
- 119.Pasini D, et al. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Selective targeting of histone methylation. Cell Cycle. 2011;10:413–424. doi: 10.4161/cc.10.3.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ansari KI, Shrestha B, Hussain I, Kasiri S, Mandal SS. Histone methylases MLL1 and MLL3 coordinate with estrogen receptors in estrogen-mediated HOXB9 expression. Biochemistry. 2011;50:3517–3527. doi: 10.1021/bi102037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol. 2015;16:499–513. doi: 10.1038/nrm4029. [DOI] [PubMed] [Google Scholar]
- 123.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ozes AR, et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Carlisle Casey, Prill Kendal, Pilgrim Dave. Chaperones and the Proteasome System: Regulating the Construction and Demolition of Striated Muscle. International Journal of Molecular Sciences. 2017;19(1):32. doi: 10.3390/ijms19010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5. [DOI] [PubMed] [Google Scholar]
- 127.Huang L, et al. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet. 2014;290:717–723. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 128.Hussain I, et al. Bisphenol-A induces expression of HOXC6, an estrogen-regulated homeobox-containing gene associated with breast cancer. Biochim Biophys Acta. 2015;1849:697–708. doi: 10.1016/j.bbagrm.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.